Abstract
Melanoma cells release different types of extracellular vesicles (EVs) into the extracellular milieu that are involved with communication and signaling in the tumor microenvironment. Subsets of EVs include exosomes, microvesicles, and apoptotic bodies that carry protein and genetic (RNA) cargos. To define the contribution of the RNA cargo of melanoma cell derived EVs we performed small RNA sequencing to identify different small RNAs in the EV subsets. Using validated centrifugation protocols, we separated these EV subsets released by the melanoma cell line MML-1, and performed RNA sequencing with the Ion Torrent platform. Various, but different, non-coding RNAs were detected in the EV subsets, including microRNA, mitochondrial associated tRNA, small nucleolar RNA, small nuclear RNA, Ro associated Y-RNA, vault RNA and Y-RNA. We identified in total 1041 miRNAs in cells and EV subsets. Hierarchical clustering showed enrichment of specific miRNAs in exosomes, including hsa-miR-214-3p, hsa-miR-199a-3p and hsa-miR-155-5p, all being associated with melanoma progression. Comparison of exosomal miRNAs with miRNAs in clinical melanoma samples indicate that multiple miRNAs in exosomes also are expressed specifically in melanoma tissues, but not in benign naevi. This study shows for the first time the presence of distinct small RNAs in subsets of EVs released by melanoma cells, with significant similarities to clinical melanoma tissue, and provides unique insights into the contribution of EV associated extracellular RNA in cancer.
Keywords: cancer, extracellular RNA, malignant melanoma, membrane vesicles, non-coding RNA, next-generation sequencing
Introduction
Melanoma is one of the most fatal forms of skin cancers, especially in young adults,1 and often involves disturbance of the MAPK signaling pathway.2,3 Tumor cells, including melanoma cells, are known to secrete several subsets of extracellular vesicles (EVs), including apoptotic bodies, microvesicles and exosomes.4-10 Further, EVs have been implicated in tumor microenvironment interactions, including angiogenesis, tumor cell proliferation and epithelial mesenchymal transition,11–13 and malignant cells release different EV RNA molecules compared to normal cells.14 Hence, describing the detailed molecular and genetic content of melanoma EVs is important to understand the complexity of cell-to-cell interaction in melanoma, including disease progression.15,16
Multiple small non-coding RNAs, including microRNA, small interfering RNA, small nucleolar RNA and long non coding RNA have the capacity to control mRNA targets. Currently, miRNAs are the most extensively studied, with more than 2000 miRNA species discovered so far.17-21 Importantly, many small RNAs are released in the extracellular environment as EV cargo, which has been specifically documented for the exosomes.22-27 Importantly, many cellular RNAs, including miRNAs are detected in the extracellular space, especially in extracellular vesicles.28,29 Currently, extracellular RNA communication is also considered to be putatively important in animals, with important implications for biology, disease and medicine.28
Recently, our group has demonstrated the presence of distinct RNA profiles in separate types of EVs released by different cell lines, including apoptotic bodies, microvesicles and exosomes.6 We therefore hypothesized that different types of EVs might contain distinct group of small RNA species. We use small RNA sequencing (Ion Torrent platform) to define the RNA species present in apoptotic bodies, microvesicles and exosomes isolated from the melanoma cell line MML-1. Further, we also show that the miRNA cargo from the MML-1 cells may be clinically relevant, by comparing the exosomal miRNA sequences with public clinical miRNA datasets from melanoma tissue samples. Taken together, these data provide the first detailed analysis of small RNA species contained in the several distinct subclasses of EVs derived from the single cell line, and provide a workflow for performing similar analyses in other cancer types and disease models.
Results
RNA profiles in subsets of EVs
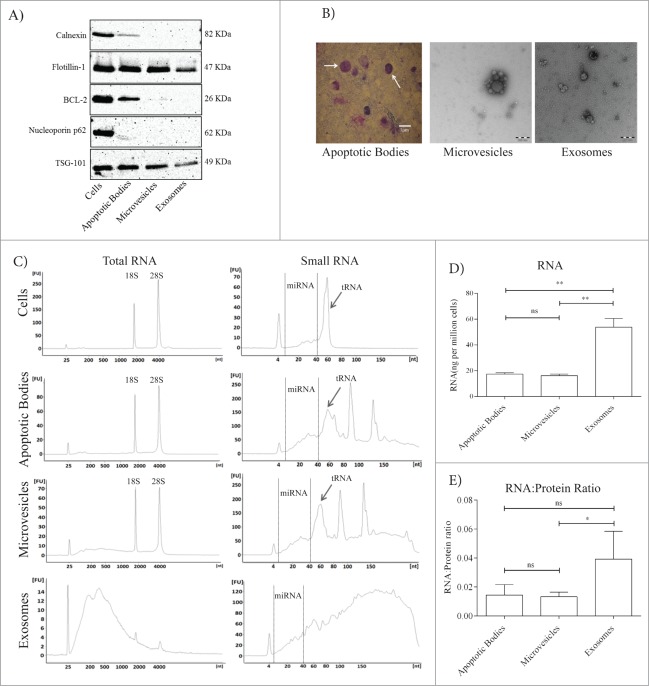
Using the MML-1 cell model of melanoma, EV subtypes were isolated using a previously published and validated sequential centrifugation protocol.6 We first characterized the EVs by using Western blot to detect several markers (Fig. 1A). Calnexin, which is an endoplasmic reticulum marker was detected in apoptotic bodies, but not in microvesicles or exosomes. Flotillin-1 and TSG 101, known to be present in exosomes,30,31 were detected in exosomes but also in apoptotic bodies and microvesicles. The mitochondrial marker Bcl-2 was specifically enriched in apoptotic bodies, compared to microvesicles, but was not detected in exosomes. Also, the nuclear envelope marker Nucleoporin p62 was absent in microvesicles and exosomes but slightly expressed in apoptotic bodies (Fig. 1A), which is similar to previously published data.22 These results show that our protocol for specifically exosome isolation results in no contamination of nuclear, mitochondrial, or endoplasmic reticulum membranes components, whereas the other two EV subsets seem to contain such markers. The presence of different types of vesicles in the different EV isolates was also confirmed by electron microscopy (Fig. 1B). Apoptotic body preparations were not suitable for electron microscopy, but were visualized in Cytospins using Giemsa stain. Some of the observed apoptotic bodies did not contain any nuclear dense components, as indicated with the uniform pink cytoplasmic vesicles, while some vesicles do contain a nuclear dense component seen as purple areas (indicated by arrow marks, Fig. 1B first panel). These results are coherent with the results of Western blot, where apoptotic bodies were positive for Nucleoporin p62 (Fig. 1A). The size of the microvesicles were also relatively larger than size of the exosomes as depicted by the electron microscopy, again confirming that the differential centrifugation do separate vesicles based on their sizes.
Figure 1.
Characterization of extracellular vesicles including RNA profiles. Protein and RNA was extracted from MML-1 cells as well as apoptotic bodies, microvesicles and exosomes released by these cells. (A) Immunoblotting of the endoplasmic reticulum protein Calnexin, the mitochondrial protein Bcl-2, the nucleus protein nucleoporin p62 and the exosomal protein TSG101 and Flotillin-1 in isolates of EVs from MML-1 cells and cellular extract. All the antibodies were used in 1:1000 dilutions. (B) Characterization of EVs by Giemsa staining and transmission electron microscopy (TEM). Cytospin preparations were used for apoptotic bodies (50μg) and was stained with Giemsa. Scale bar 10μm. Ten microgram was used for the TEM for microvesicles and exosomes. Scale bar 200nm. (C) Bioanalyzer analysis of RNA isolated from cells, apoptotic bodies, microvesicles and exosomes on total RNA NanoChip and Small RNA chip. The rRNA peaks are indicated as 18S and 28S subunits in cells, apoptotic bodies and microvesicles, whereas exosomes shows reduced presence of rRNA. Cells and EVs all exhibited the miRNA region from 4-40nt whereas tRNA was enriched in cells, apoptotic bodies and microvesicles but absent in exosomes. These images are representative of 2 replicates, and the second replicate display a similar RNA profile. (D) The RNA yield is represented as ng (RNA)/million cells (n = 3). Data are presented as mean ± SEM. (E) Protein to RNA ratio in EVs is calculated by measuring the total amount of protein in micrograms (μg) and the total amount of RNA in micrograms (μg) per million cells. Data are presented as mean ± SEM. **P < 0.01, ns; non-significant.
To determine whether EVs from MML-1 cells also contain distinct profiles of RNAs, RNAs were isolated from apoptotic bodies, microvesicles and exosomes. The Bioanalyzer RNA profiles from different EV subpopulations are shown in Figure 1C, which documents the presence of distinct rRNA peaks (18S and 28S) with relatively moderate levels of small RNA in apoptotic bodies and microvesicles. By contrast, exosomes contained less rRNA and relatively more small RNA species. The EV populations from the melanoma cell lines SK-MEL-28 and A375 showed similar RNA profiles as the MML-1 cell line (Fig. S1). The RNA profiles were not affected by RNAse treatment, as shown in Supplemental Figure 2A, which indicates that the RNA is protected in the vesicles. Although we observed the depletion of rRNA in the microvesicles fraction, it was not possible to determine whether the rRNA is inside or outside of the vesicles in this sample. During sample preparation, rRNA was depleted as it covers a major population for the small RNA sequencing.
Analysis of small RNA, using a small RNA chip for the Bioanalyzer showed typical peaks representing the presence of various RNA species including miRNA, tRNA, and other RNAs (Fig. 1C). Furthermore, the total RNA yield obtained from exosomes was significantly greater than that from apoptotic bodies and microvesicles (Fig. 1D). Even though the RNA content per particle shows no significant difference between the exosomes and either microvesicles or apoptotic bodies, we did observe a significant difference between apoptotic bodies and microvesicles, suggesting that RNA cargo per vesicle is not always the same and can change during each isolation (Fig. S2B). Importantly, there may be aggregation of the vesicles/particles during the analysis, which can affect the numbers retrieved by the nanotracking analysis (NTA), and thus downstream analysis of the RNA load per particle. In addition, the RNA to protein ratio showed that exosomes contains significantly higher quantities of RNA to protein as compared to microvesicles whereas no statistically significant differences were observed between apoptotic bodies and exosomes (Fig. 1E). Collectively, these data argue that exosomes potentially contain relatively more RNA than apoptotic bodies and microvesicles.
Differential expression of non-coding RNA
To determine whether the distinct profiles found in each EV subtype represent different RNA transcripts, we performed small RNA sequencing on MML-1 cells and the three EV subtypes isolated from that cells. This approach allow us to determine whether differences in the expression of RNA can be observed in the parent cell and the EV subset, and can also explain whether the subsets are distinct in relation to their RNA cargo. Sequencing data underwent a quality check using FastQC prior to alignment to the human genome (HG19), which revealed that more than 80% of the small RNA reads were aligned to the reference genome (Table 1). Even though microvesicles and cells showed differences in the number of reads between the replicates, more than 90% of the reads were mapped to the human genome, and were normalized for further analysis. We firstly identified various miRNA species in all the samples by mapping to miRBase (release 20). In total, 1041 miRNAs were identified which is ∼40% of all mature miRNAs known to date (2578 miRNAs according to miRBase release 20). This confirms that miRNA are abundantly present in EVs.9,32
Table 1.
Total number of sequence reads from small RNA sequencing with number of alignments mapped to human genome (HG19) and number of miRNA identified in each sample replicate. The samples were tested for the quality control and then aligned to the human genome. Number of miRNAs identified in each sample replicate represent the number of all the total miRNA with reads more than or equal to 1
| Cells-1 | Cells-2 | ABs-1 | ABs-2 | MVs-1 | MVs-2 | EXO-1 | EXO-2 | |
|---|---|---|---|---|---|---|---|---|
| Total raw reads | 753,813 | 1,172,620 | 1,107,968 | 995,806 | 1,394,982 | 755,131 | 310,528 | 331,522 |
| Quality score (FastQC) | Passed | Passed | Passed | Passed | Passed | Passed | Passed | Passed |
| Number of alignments to HG19 | 677,377 | 1,092,573 | 974,814 | 927,889 | 1,312,876 | 682,905 | 268,095 | 260,315 |
| Percentage of alignments | 89.86 | 93.17 | 87.98 | 93.179 | 94.11 | 90.43 | 86.33 | 78.52 |
| Number of reads for miRNAs | 77,200 | 93,752 | 63,347 | 30,779 | 50,200 | 16,770 | 21,856 | 14,755 |
| Number of identified miRNAs | 544 | 570 | 532 | 433 | 474 | 360 | 411 | 385 |
| Number of miRNA with read=1 | 147 | 141 | 153 | 143 | 122 | 108 | 109 | 113 |
ABs – Apoptotic Bodies, MVs – Microvesicles, EXO – Exosomes
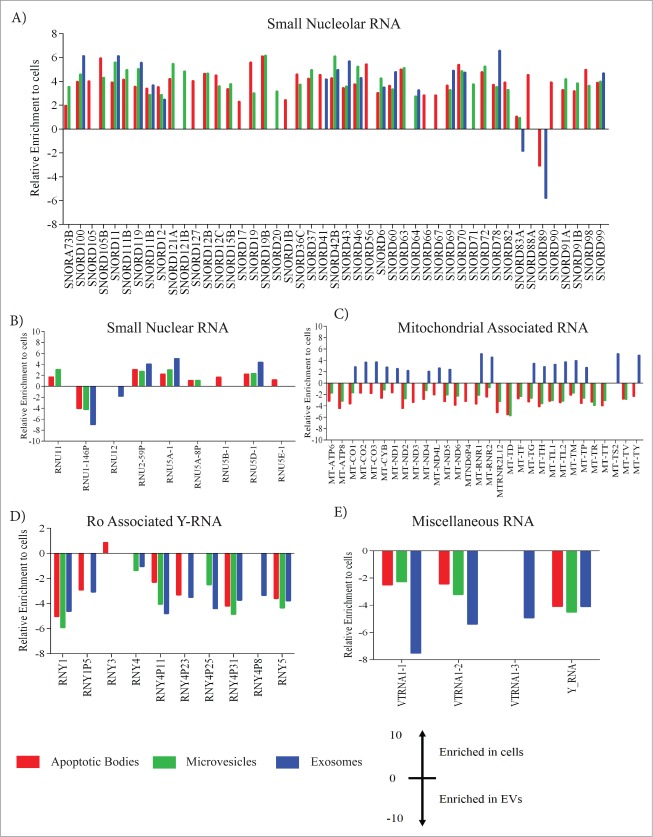
To annotate other small RNA species, the aligned reads were counted using HTSeq-count using Gencode (version 19) human annotation. The comparison of non-coding RNAs, between cells and EVs is shown in Figure 2. The data shows the enrichment of non-coding RNA, where the positive values represent a relative enrichment of noncoding RNA in cells compared to EVs and the negative values represent a relative enrichment in the EVs compared to cells. While comparing the snoRNA data in Figure 2A, we could see clearly that most of the snoRNA are enriched in cells; however exosomes specifically exhibited more than 2 fold enrichment of snord83A and snord89 specifically. SnoRNA usually guides chemical modification of tRNA, rRNA and most of small nuclear RNA. We also see more than a 3 fold enrichment of snord89 in apoptotic bodies which was clearly absent in microvesicles. Next we looked at the snRNAs which are mostly expressed in the nucleus of proliferating cells and observed that most of these RNA species are enriched in the cells, whereas RNU1-146P is more than 3.5 fold highly enriched in all EVs compared to cells (Fig. 2B). This small RNA is a pseudogene having a 165 bp transcript, but the exact function of this snRNA remains to be deciphered. The presence of snRNA in exosomes is interesting, as to date most of the RNA species in exosomes are considered to be of cytoplasmic origin.23 We also observed an enrichment of RNU12 snRNA in exosomes - which was absent in apoptotic bodies and microvesicles. The enrichment of mitochondrial associated RNA (mt-RNA) in apoptotic bodies and microvesicles suggest that mt-RNA is more prone to be loaded into apoptotic bodies and microvesicles than into the exosomes (Fig. 2C). In addition, we could also detect mitochondrial associated tRNA carrying each amino acid including MT-TD, MT-TF, MT-TH, MT-TL1 among others, as described in Figure 2C. These results are slightly in contrast from those of Vojtech, et. al., that described the presence of tRNA in seminal fluid exosomes in 2 different libraries,33 although both sources of the exosomes were different, and the vesicular isolation protocols used different centrifugations. Lastly, we could also observe enrichment of Ro associated Y-RNA and miscellaneous RNA such as vault RNA and other Y-RNAs in all vesicles (Fig. 2D, E). One of the isoform of vault RNA “vtRNA 1–3” is relatively more enriched in exosomes compared to microvesicles and apoptotic bodies. Our data of vtRNA and Y-RNA are in accordance with previous study on T-cell/dendritic cell exosomes, also describing enrichment of Y-RNA and vault RNA in exosomes.22,23 Together, these data show that the different subsets of EVs contain different types of small non-coding RNAs, which putatively could be used as a surrogate signature to distinguish the EV subsets.
Figure 2.
For figure legend, See page 815. Figure 2. (see previous page) Enrichment of non-coding RNA in EVs. Relative enrichment of small non-coding RNA (A) small nucleolar RNA (snoRNA) (B) small nuclear RNA (snRNA) (C) mitochondrial associated RNA (D) Ro associated Y-RNA and (E) Miscellaneous RNA, compared to cells with negative binomial distribution. The data are presented as significant enrichment of non-coding RNA that is analyzed by DESeq2. A negative value means the enrichment of all noncoding RNA in EVs in relation to cells and positive value means the enrichment of all noncoding RNA in cells. (A) Enrichment of 47 small nucleolar RNA across all EVs. Snord89 and snord83A was enriched in exosomes and apoptotic bodies shown in negative fold expression. All other snoRNA was expressed in cells. (B) Small nuclear RNA enrichment in cells and EVs. There is presence of pseudogene snRNA enriched in all EVs and absent in cells. (C) Enrichment of 30 mitochondrial associated tRNA in cells and sub populations of EVs. Apoptotic bodies and microvesicles are enriched in most of the mtRNA and exosomes does not have any expression. (D and E) Enrichment of Ro associated y-RNA and miscellaneous RNA in EVs and absent in cells. All the data are the average of the duplicates analyzed from DESeq2. The data represented is the significant enrichment having P < 0.05.
Relationship between miRNA in subsets of EVs and cells
To determine whether EVs and cells differ in their respective miRNA expression, we performed principal component analysis (PCA). The PCA plot shows that all the EV subtypes and cells display higher variance between the different sample types (Fig. 3A) than to their respective replicates. Indeed, the replicates for cells, apoptotic bodies and exosomes cluster relatively close, but slightly less so for microvesicles. This discrepancy may be explained by the difference in the read counts in the microvesicles samples. Overall, the PCA analysis visualizes distinct components among the different EV subsets, but also between all EV subsets and the cells.
Figure 3.
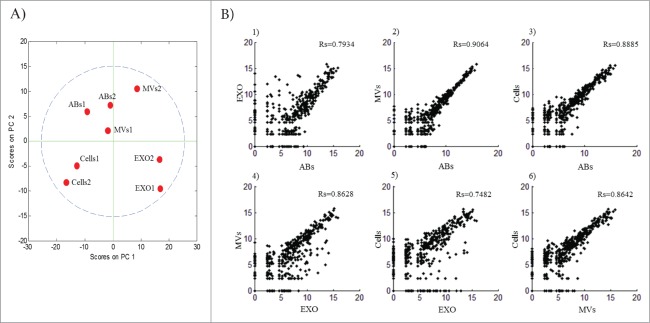
Relationship of miRNA between cells and subsets of extracellular vesicles. (A) Principal component analysis was performed on the 8 samples using MATLAB (4 sample types with 2 biological replicates), presenting each sample replicates lying in each individual component. (B) Scatter plots showing correlation between the average expressions of the 2 biological replicates. All the correlation curves were visualized and calculated using MATLAB. 1 - EXO vs ABs Rs = 0.7934; 2 – MVs vs ABs Rs = 0.9064; 3 – Cells vs ABs Rs = 0.8885; 4 – MVs vs EXO Rs = 0.8628; 5 – Cells vs EXO Rs = 0.7482; 6 – Cells vs MVs Rs = 0.8642. False Discovery Rate (FDR) < 1%. ABs – Apoptotic Bodies, MVs – Microvesicles, EXO – Exosomes.
Next, we compared the expression of all miRNAs identified by small RNA sequencing to determine whether there are any close relationships between sample types. Firstly, we investigated sample-to-sample variation by comparing the average correlation of duplicate samples of cells, apoptotic bodies, microvesicles and exosomes (Fig. 3B). Overall, we found a close relationship between apoptotic bodies and microvesicles (2; Rs = 0.91), apoptotic bodies and MML-1 cells (3; Rs = 0.89), microvesicles and exosomes (4; Rs = 0.86), and microvesicles and cells (6; Rs = 0.86). However, we also observed a somewhat less strong relationship between apoptotic bodies and exosomes (1; Rs = 0.79) and exosomes and cells (5; Rs=0.75) (Fig. 3B). (All the P-values are < 0.01). This data collectively suggest that the vesicles subsets at least to some degree are different but also is in positive correlation from the aspect of their miRNA cargo.
Identification of unique miRNAs in each EV subpopulation
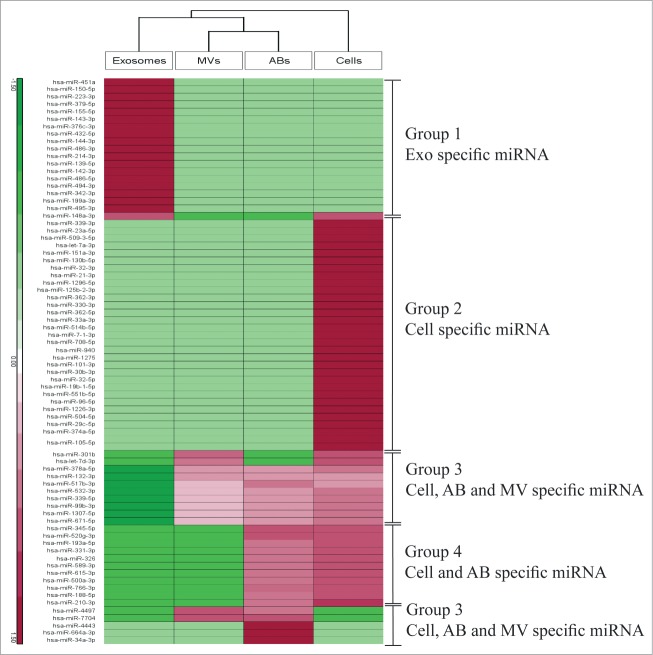
We also compared the overlap of miRNA species among the various EVs and the MML-1 cells. By using an integrative statistical hypothesis testing method that combines adjusted P-values from paired two-tailed t-tests and the median ratio test,34 we identified 252 miRNA in all samples having p-values less than 0.05, out of which 113 miRNAs were shared between all EVs and the cells (Fig. 4). Notably, exosomes contained 23 unique miRNAs that were absent in the other samples, indicating loading of specific miRNAs into exosomes. Overall, microvesicles did not show a distinct miRNA distribution pattern per say, but we did find 3 miRNAs were shared between cells and microvesicles, and 3 were detected in both apoptotic bodies and microvesicles. Interestingly, 3 unique miRNAs were found in apoptotic bodies and 26 were shared between cells, apoptotic bodies and microvesicles but were absent in exosomes. A list of miRNAs that are exclusively present in exosomes, apoptotic bodies and cells are represented in Supplemental Table 1. Taken together, these data suggest that the different isolates of vesicles represent distinct subsets of extracellular RNA populations.
Figure 4.
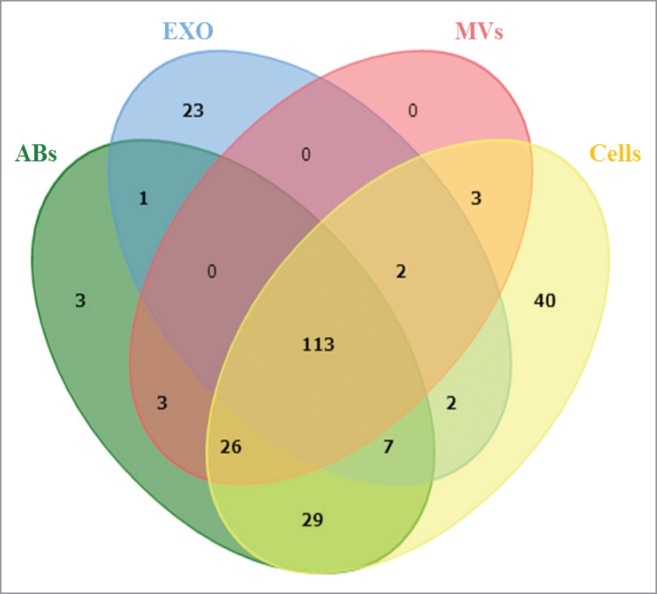
Distribution of miRNA among the different samples. Venn diagram analysis was used to determine shared and uniquely identified miRNA in the different samples. All the miRNA identified by combined P-values were used to analyze the sharing of miRNA between cells, apoptotic bodies, microvesicles and exosomes. ABs – Apoptotic Bodies, MVs – Microvesicles, EXO – Exosomes.
To illustrate putative mRNA targets for the unique miRNA expressed in exosomes, we used miRTarBase (release 4.5)35 which contains validated information based on real time PCR, western blot or reporter assays, thus increasing the confidence that any discovered interaction may be relevant in recipient cells. A list of possible exosomal miRNA-mRNA targets is summarized as Supplemental Table 2. The table shows that miR-155-5p has multiple targets compared with the other identified miRNAs, suggesting that this miRNA may regulate many genes that have multiple functions in melanoma and other disease models.
Cluster analysis of miRNA patterns in EV subsets
Four groups of miRNA cluster were identified using hierarchical clustering with log2 expression of miRNAs where P-values were less than 0.05 in any sample pairs. (Fig. 5). Importantly, group 1 contained miRNAs that were uniquely enriched in exosomes (Fig. 5). Several miRNAs in this cluster group such as hsa-miR-214-3p, hsa-miR-199a-3p and hsa-miR-155-5p have been associated with metastatic phenotype in melanoma.36-38 The detection of these miRNAs were validated in the exosome samples by inclusion of miR-486-5p which is found to be abundantly present in exosomes using SYBR green detection qPCR (Table 2). The validation data for miR-214-3p is not shown since the melting curve for this miRNA was below threshold and shows the presence of primer dimer formation and other fragmented product when resolved on 1.5% agarose gel (data not shown). Group 2 defines cell specific miRNAs and constitutes ∼40% of the identified miRNAs. Furthermore, group 3 constitutes cells, apoptotic bodies and microvesicles specific miRNAs, and group 4 constitutes cells and apoptotic bodies specific miRNAs. Microvesicles did not contain any unique miRNA cluster, but overlap with apoptotic bodies and cell miRNA. Furthermore, some miRNAs present in microvesicles did not show significant differences in expression as indicated by the white shade in the hierarchy. This analysis describes a clear discrepancy of miRNAs based on the clusters groups, arguing that each isolated EV-subpopulation indeed are unique entities. This may also imply different functional roles of the EV subsets in malignant disease. We also performed the pathway enrichment using KEGG pathway using the miRNA from group 1, focusing on gene targets involved in pathways of melanoma. The DAVID bioinformatics tool was used to associate the enriched miRNA from group 1 with at most −0.3 context+ scores.39 This analysis showed the specific miRNA targets such as growth factor (GF), receptor tyrosine kinase (RTK) and the transcription factor E2F which is mostly involved in cell cycle regulation (All 3 marked with red star in KEGG pathway, Fig. S4). This data shows that group 1 associated miRNA are involved in signaling pathways as well as in cell cycle regulation.
Figure 5.
Clustering of uniquely identified miRNA and their association with biological processes. Hierarchical clustering of identified miRNAs in cells, apoptotic bodies, microvesicles and exosomes. The clusters are assembled together in 4 groups. The miRNA are indicated on the right and samples are indicated below. Group 1 represent Exosomes specific miRNA, group 2 – cell specific miRNA, group 3 – cell, apoptotic bodies, microvesicles shared miRNA and group 4 – cell and apoptotic bodies shared miRNA.
Table 2.
Validation of miRNA that are enriched in exosomes. The miRNA present in exosomes were validated with qPCR across all extracellular vesicles
| miR-155-5p | miR-199a-3p | miR-486-5p | |
|---|---|---|---|
| Cells | 29.85 ± 1.11 | 27.70 ± 1.04 | 24.24 ± 0.89 |
| Apoptotic Bodies | 33.20 ± 1.29 | 31.91 ± 0.69 | 28.70 ± 0.43 |
| Microvesicles | 33.94 ± 0.27 | 31.81 ± 0.13 | 30.53 ± 0.86 |
| Exosomes | 33.72 ± 1.4 | 29.99 ± 0.8 | 28.93 ± 1.17 |
Differential expression of statistically significant miRNA in melanoma biopsies and tissues
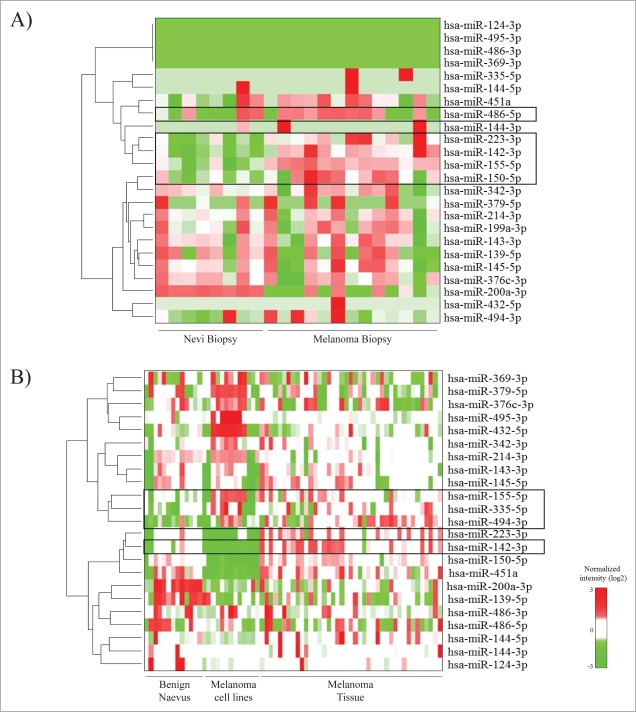
To further evaluate whether the unique miRNAs in the MML-1 derived exosomes are of clinical relevance, we compared them with 2 publically available microarray data sets (GSE34460 and GSE35579) in the GEO database. In the GSE34460 dataset,40 we found that hsa-miR-142-3p, hsa-miR-150-5p, hsa-miR-155-5p, hsa-miR-223-3p, and hsa-miR-486-5p are significantly differentially expressed in melanoma biopsies as compared to the benign naevi (Fig. 6A). In the GSE35579 data set, we additionally found that hsa-miR-335-5p and hsa-miR-494-3p to be significantly differentially expressed in melanoma tissues and melanoma cell lines as compared to benign naevi (Fig 6B). This finding demonstrates that miRNA we found to be significantly enriched in exosomes from MML-1 cells are associated with disease progression in patients.
Figure 6.
Comparative analysis of exosome specific miRNA and miRNA identified previously in cell lines and melanoma tissues and biopsies: Comparison of unique MML-1 exosomal microRNAs with databases of melanoma tissue and benign naevi microRNAs. (A) Heatmap showing the significant expression of hsa-miR-142-3p, hsa-miR-150, hsa-miR-155, hsa-miR-223, and hsa-miR-486-5p in melanoma biopsy as compared to the Nevi Biopsy. (B) Heatmap showing the significant expression of hsa-miR-142-3p, hsa-miR-335, hsa-miR-155, and hsa-miR-494 in melanoma tissue and melanoma cell lines as compared to benign naevus. This data was accessed from GEO database with accession number A) GSE34460 and B) GSE35579. An integrative statistical hypothesis testing method was applied for analyzing the P values of the statistically significant candidates. P < 0.05 was considered as statistically significant.
Discussion
The presence of RNAs, including miRNAs in exosomes, was published only 8 years ago.9 Specifically, exosomes contain significant amounts of miRNAs that can induce functional effects in a recipient cell.41 Most studies describing RNA species in exosomes and other extracellular vesicles have applied array-based methods,14 and only few have utilized deep sequencing to illustrate the RNA content in detail.22,23 Until today, no study has described the RNA content in different vesicle subsets secreted from a single cell type, using any methodology. This is thus the first study that utilizes RNA sequencing to profile small RNAs concomitantly in apoptotic bodies, microvesicles and exosomes released by any single cell type under any condition. The sequencing identified many types of non-coding RNA molecules in all subsets, including miRNAs, and can by itself distinguish the subsets of EVs.
Many research groups are describing different miRNAs primarily in exosomes, whereas other RNAs may also be incorporated into the different vesicle subsets, and could contribute to EV biology.22,23 Interestingly, we found enrichment of multiple types of non-coding RNAs in all EV subsets. Despite our focus on short RNA molecules up to 200 nucleotides, fragmented mRNA can also be detected in the vesicular RNA, and are captured by the cDNA library preparation (data not shown). Notably y-RNAs, a type of structural RNA present in ribonucleoprotein particles, were observed to be enriched in all EV subsets compared to the cells, which is in line with previously published RNA sequencing of exosomes alone.23 The collective evidence evolving from the few studies so far published, argue that exosomes released by most cells indeed carry significant amounts of ncRNAs and protein coding RNAs, which should be considered when studying effects on recipient cells. Interestingly, small RNA profiles showed tRNA peaks clearly present in cells, apoptotic bodies and microvesicles, but less so in exosomes. The proportion of tRNAs associated with mitochondria in cells and EVs was strikingly low based on the sequencing, suggesting that the RNA profiles alone are insufficient to describe the complexity of the transcriptome. This is at least partly in contrast to Vojtech et. al., that described a repertoire of tRNAs among other non-coding RNAs in exosomes.33 We cannot exclude that matured tRNAs in our study was degraded during sample processing, or that the tRNA content varies among vesicles from different cellular sources. Importantly, although our data was aligned with more than 80% to the HG19, we found the presence of a significant number of transcripts (>70%) that are un-annotated with known functions.
As shown earlier by the presence of small RNA in the studied EV subpopulations,6 we have here established a workflow for analyzing miRNA in these samples using small RNA sequencing data. This approach is unbiased, and can profile a large number of RNA species from the transcriptome, which is different from analyzing miRNAs using microarray systems with limited number of pre-known miRNA candidates. In this study we identified 1041 miRNAs altogether in the extracellular vesicle and cell samples studied, which are more than most array systems would detect.22–27,42,43 Further, comparison among expression of miRNAs is possible with deep sequencing approaches, if the absolute number of copies in the sample can be quantified.44 In this study, by using the Quantile normalization method for all samples, miRNA expression showed positive relationship between the samples, although many detected miRNA are unique in the different subsets of vesicles (Fig. 3; Fig. S3). Importantly, we here describe a drastic variance among the different vesicle types which allowed us to distinguish the miRNA functions in each sample using PCA, clarifying that the vesicle population studied are different from each other. The relationship between the samples using transformed data showed relatively strong relationships, but this varied to some degree among samples, with exosomes correlating less with cells and apoptotic bodies, which is in support of previous studies suggesting specific RNAs are loaded into exosomes.9 In a previous transcriptomic study of exosomes released from another melanoma cell line using array technology,14 a relatively close correlation was reported, but it is unclear whether microvesicles were included in that analysis due to important differences in isolation procedures.
A significant number of the miRNAs in exosomes were unique, and not detected in apoptotic bodies or microvesicles, which again is supporting the concept that specific RNA species are loaded into exosomes. These miRNAs include miR-199a-3p, miR-150-5p, miR-142-3p, miR-486-5p, miR-223-3p, previously shown to be associated with uveal melanoma,45 primary cutaneous melanoma,40 melanoma tissues46 and blood samples from melanoma patients47,48 respectively. We also found expression of miR-214-3p that is associated with melanoma metastasis.36,49 Interestingly, miR-214 and miR-199a, both related to melanoma progression, are found in a cluster of miRNAs that are regulated by the transcription factor TWIST-1.50 This transcription factor has been associated with neural cell development, which may be important since melanocytes are known to originate from the neural crest. Further, this miRNA cluster is also related to hypoxia by targeting myocardial PPARδ and impairing mitochondrial fatty acid oxidation.37 Taken together, the most expressed miRNA in exosomes from the MML-1 cells may have a potential role in melanoma progression.
Pathways analysis of the miRNA enriched in group 1 are known to regulate certain genes in a melanoma network, which supports the concept that exosomal miRNA are important regulators in tumorigenesis in melanoma (Supplementary Fig. 4). Of special interest was the finding that pathway analysis of the exosomal miRNAs was targeting pathways closely related to BRAF and NRAS, both kinases involved in melanoma, and often mutated in this disease.
When comparing the specific miRNAs in exosomes from the MML-1 cell line with databases of microRNA from melanoma, and performed a cluster analysis, we found several microRNAs that were overlapping between the 2 types of samples. The GEO accession database of validated miRNA showed similarity of 7 miRNAs which include hsa-miR-142-3p, hsa-miR-150-5p, hsa-miR-155-5p, hsa-miR-223-3p, hsa-miR-486-5p, hsa-miR-335-5p and hsa-miR-494-3p, which may imply clinical relevance of the exosomal miRNA transcriptome. The list of the mRNA targets is shown in the Supplementary File S2. There is no documented evidence for miR-335-5p and miR-494-3p that is associated with melanoma. All other 5 miRNA are associated with melanoma.40,46,48,51-53
In summary, our results show the expression of miRNAs and other RNA types in melanoma cells and their released extracellular vesicle subsets including apoptotic bodies, microvesicles and exosomes. The RNA species significantly differ between cells and among the extracellular vesicle subsets, which may have vast importance to understand melanoma development and progression. Furthermore, the differences in RNA cargo in the different subsets strongly support the notion that the cells release distinct extracellular vesicle subsets, which are likely to be produced by different pathways. Beyond the miRNAs, there are also many other ncRNA subsets present in the different vesicles, which increases the level of complexity of extracellular vesicle RNA cargo and function, and is important to acknowledge in the development of future extracellular RNA melanoma EV-based biomarker development.
Material and Methods
Cell cultures
The following malignant melanoma cell lines were used; A375 (ATCC, Manassas, VA, USA) which was cultured in DMEM (Sigma-Aldrich, St. Louis, MO, USA), MML-1 (CLS, Eppelheim, Germany) which was cultured in RPMI-1640 (Sigma-Aldrich) and SK-MEL-28 (ATCC) which was cultured in DMEM (Lonza, Basel, Switzerland). All culture media contained 10% fetal bovine serum (Sigma-Aldrich), 100 U/ml penicillin (HyClone, Logan, UT, USA), 100 µg/ml streptomycin (HyClone), and 2 mM L-glutamine (HyClone). In addition, A375 cell culture media also contained 1% non-essential amino acids (PAA Laboratories, Pasching, Austria) and 5 mM β-mercaptoethanol (Sigma-Aldrich). Prior to use, the fetal bovine serum was ultracentrifuged at 120,000 × g for 18 hours using a Type 45 Ti rotor (Beckmann Coulter, Brea, CA, USA) to pellet the bovine exosomes. All cells were maintained at 37°C in 5% CO2. Cell viability was assessed by trypan blue exclusion assay.
Extracellular vesicles (EVs) isolation
EVs were isolated from the supernatant of 3 cell lines A375, MML-1 and SK-MEL-28 (80–90% confluence) using differential centrifugation. The protocol has previously been described as “the modified protocol-2B” by Crescitelli, et al.6 Briefly, cells were pelleted by centrifugation at 300 × g for 10 minutes and the supernatant was harvested to pellet down apoptotic bodies at 2,000 × g for 20 minutes and microvesicles at 16,500 × g for 20 minutes. The supernatant harvested from the 16,500 × g centrifugation was filtered (0.2 µm pore size, Sarstedt, Nümbrecht-Rommelsdorf, Germany) to remove particles greater than 200 nm. Exosomes were pelleted from the filtered supernatant by ultracentrifugation at 120,000 × g for 70 minutes. All the EVs were re-suspended in the appropriate buffers depending on downstream analyses (PBS or lysis buffer).
Total RNA isolation and analysis
Vesicular RNA and cellular RNA were isolated using miRCURY™ RNA Isolation Kit (Exiqon, Vedbaek, Denmark) following the manufacturer's protocol. All the RNA samples were stored at −80°C until used. The quality, size and concentration of the isolated RNA was determined using capillary electrophoresis (Agilent 2100 Bioanalyzer, Agilent Technologies®, Santa Clara, CA, USA) with total RNA 6000 Nano Chip and Small RNA chip following the manufacturer's instructions.
RNase treatment and particle number measurement
Extracellular vesicles suspended in PBS were used for the RNAse treatment. A part of the vesicles 5 μl was used for the particle count analysis using ZetaView® (a Nanotracking analysis (NTA) device: Particle Matrix, Meerbusch, Germany). Before RNase treatment cellular RNA was used as control for the RNase treatment. Equal volume of samples were divided in 2 parts: non-treated and RNase treated. The RNase treated (5 μg/ml) samples were incubated at 37°C for 20 mins while the non-treated were kept on ice, and immediately RNA was extracted from these samples using the Total RNA isolation kit (miRCURY™). The samples were later analyzed by Agilent 2100 Bioanalyzer using total RNA Nano chip. The RNA concentration was used as a normalizing factor for the RNA load per particle analysis.
Small RNA sequencing library generation and data analysis
For each library, 50 ng of RNA was ligated to adapters containing a unique index barcode (Ion Xpress™ RNA-Seq Barcode 1–16 Kit, Life Technologies, Carlsbad, CA, USA). RNA samples were reverse transcribed to cDNA using adaptor specific primers, especially designed for short RNA sequencing. Using the Magnetic Bead Purification Module (Life Technologies), cDNA samples were size-selected from 94 to 200 nt that includes the adaptor sequences around 25bp. PCR amplification was then performed followed by a library clean-up step using nucleic acid beads (Life Technologies). The yield and size distribution of the small RNA libraries were assessed using the Agilent 2100 Bioanalyzer™ instrument with the High sensitivity DNA chip (Agilent). Equally pooled libraries were clonally amplified onto Ion Sphere™ Particles (ISPs) supplied by the Ion OneTouch™ 200 Template Kit v2 DL kit (Life Technologies) and enriched using the One Touch 2 ES system (Life Technologies). Enriched ISPs loaded with libraries were sequenced on the Ion Torrent PGM™ using Ion™ 318 V2 chips (Life Technologies) and the Ion PGM™ 200 V2 Sequencing Kit (Life Technologies). Two libraries were pooled per chip. Pre-processing of reads, removal of adapters and barcodes were performed using the Torrent Suite (v.3.4.1). Sequences were analyzed for quality control (FASTQC) and aligned to the Human genome (HG19) using the Torrent Suite. Output files (.bam) were uploaded, mapped to miRBase V.20 and further analyzed using Partek Genomic Suite (Partek Inc., Singapore). Reads were normalized to reads per million reads (RPM).
miRNAs identified with at least 2 reads were used for further analysis. Sequences were also mapped to Ensembl Release 74 to identify other non-coding and coding RNA species.
Western Blotting
Cells and extracellular vesicles were lysed using RIPA buffer (Thermo Scientific) and were sonicated for 5 minutes 3 times with intermittent vortexing in between to extract all the proteins from the EVs. The protein concentrations of different EVs samples were determined using the BCA Protein assay kit (Pierce, Thermo Scientific, Rockford, IL, USA). Thirty micrograms of protein lysates were loaded on to a 10% SDS-PAGE gel. Samples were transferred to nitrocellulose membrane (Bio-Rad laboratories, Hercules, CA, USA) and blocked in 5% Non-Fat Dry Milk (Bio-Rad Laboratories) for one hour. Membranes were incubated with primary antibodies against Flotillin-1 (1:1,000), Calnexin (1:1,000), Nucleoporin-p62 (1:1,000), BCL-2 (1:1,000; all above from Santa Cruz Biotechnology, Santa Cruz, CA, USA) and TSG101 (1:1,000; clone 4A10; Abcam, Cambridge, UK) at 4°C overnight. The membranes were washed 3 times in TBST buffer and incubated with secondary antibodies for one hour at room temperature. Secondary antibodies used were for Flotillin-1, Calnexin, BCL-2 and Nucleoporin p62: (1:10,000) ECL anti-rabbit IgG horseradish peroxidase-linked F(ab')2 fragment (donkey-anti-rabbit); and for TSG101: (1:2,000) ECL anti-mouse IgG horseradish peroxidase-linked F(ab’)2 fragment (sheep-anti-mouse); (GE Healthcare, Buckinghamshire, UK). The membranes were washed again 3 times for 5 minutes in TBST buffer and detected and analyzed with ECL Prime Western Blotting Detection (GE HealthCare) and a VersaDoc 4000 MP (Bio-Rad Laboratories).
Transmission electron microscopy and cytospins
Microvesicles and exosomes (10 μg) were dissolved in PBS and was loaded onto Formvar/Carbon-coated grids (Ted Pella Inc., Redding, CA, USA). Samples were incubated onto grids, previously UV treated for 15 minutes; and then fixed in 2.5% glutaraldehyde and contrasted in 2% uranyl acetate. Samples were examined in a LEO 912AB Omega electron microscope (Carl Zeiss NTS, Jena, Germany). EVs were measured with the iTEM® software (Olympus-SiS, Münster, Germany).
Apoptotic bodies (50 μg) were dissolved in 100 μl of PBS. The filters for the cytospins were pre-wet with PBS and all the sample was loaded on to the fixed filters in the bucket of the CYTOSPIN 3 rotor. The samples were run at maximum speed for 25 minutes in CYTOSPIN 3 for pelleting the samples on the glass slide. The samples were fixed in methanol and Giemsa-stained for 20 minutes. The samples were then air dried and assessed by Axioplan 2 microscope (Carl Zeiss Jena GmbH, Eching, Germany) by using oil immersion magnification of 100X.
Validation of miRNAs
Individual miRNA primer assay (Exiqon) was performed on MML-1 cells and their EVs according to the manufactures instructions. Briefly, total RNAs were isolated from the cells and EVs and 5 ng/μl of total RNA were converted to cDNA using universal cDNA synthesis kit to make the final reaction volume up to 10 μl (Exiqon). Then 1 μl of cDNA solution was diluted with 79 μl of nuclease free water. From this reaction, 4 μl of diluted cDNA was used for the qRT-PCR using the PCR starter kit (Exiqon). UniSp6 RNA Spike-in template provided in the kit was also converted to cDNA and amplified using UniSp6 RNA Spike-in control primer set that was used as internal control. Furthermore, miR-103a-3p was used as a positive control for cells provided in the starter kit. The plate was run on Bio-Rad CFX96 (Bio-Rad Laboratories, Inc.) real time detection system for 40 cycles following the provided protocol. Data are analyzed by using the Ct values exported from the CFX manager software.
Bioinformatics
The clean sequences generated from the Ion Torrent sequencer were used. Files were trimmed using PRINSEQ-lite (minimum fragment length was set to 10 and Q = 20)54 and quality of the trimmed reads were assessed by FASTQC.55 Reads were then aligned using the STAR 2-pass alignment procedure.56 Read groups were added to the alignments; then sorted; duplicates removed and indexed using picard tools.57 The reads were counted using HTSeq58 using the Gencode V19 annotation.59 Differential expression was analyzed using DESeq2.60 To compare the miRNA profile among EV types and cell, the RPM was normalized by quantile normalization with MATLAB (version 2010a). All the scatter plots and their correlations were visualized and calculated by MATLAB. For a uniform cut-off, the distribution of gene expression in the transcriptomic data was modeled into a 2-component Gaussian mixture distribution61,62 using gmdistribution function of MATLAB. Among two Gaussian density functions, one with lower mean intensity was annotated as “absent” peak, and the other as “present” peak. Whether each gene was “absent” or “present” were determined based on expression values with a 1% false positive rate in this model.
To test the confidence of this experiment, principal component analysis was performed with 4 samples (2 biological replicates for each sample) by MATLAB. P-values between samples were calculated by an integrative statistical hypothesis testing method34 that combined adjusted P-values from paired two-tailed t-tests and the median ratio test using MATLAB. Heatmap was drawn by “clustergram” function of MATLAB to catalog the miRNAs. To build the hypothetical pathways analysis between the miRNAs and their mRNA targets, mRNA-miRNA pairs with at most −0.3 context+ scores was used. To perform the GO enrichment analysis with mRNA targets, we used DAVID.39 To draw the miRNA networks, we adopted the miRNA-mRNA target database of miRDB.63 The miRNA-mRNA target pair were considered as confident only if the target score is over 80 as suggested in miRDB. The number of sharing mRNA targets between 2 miRNAs was then calculated by the code generated with Python 2.7.3. The number of sharing mRNA targets between 2 miRNAs is proportional to the width of the edge.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Erik Larsson for graciously providing server space. We are also grateful to Martin Bergö at Sahlgrenska Cancer Center, University of Gothenburg, Sweden, for providing the A375 melanoma cell line.
Funding
This work was funded by the Swedish Research Council - K2014-85x-22504-01-3 (for 2014), K2011-56K-20676-04-6 (for 2013), VBG Group Herman Krefting Foundation for Asthma and Allergy Research - 20131212 (for 2014), 20121218 (for 2013), the Swedish Heart and Lung Foundation - 20120528 (for 2013 and 2014) and the Swedish Cancer Foundation - CAN 2012/690(for 2013 and 2014). AFH is supported by the National Health Medical Research Council of Australia, [Program Grant 628946 to A.F.H.]; and the Australian Research Council [ARC Future Fellowship FT100100560]. This work was also supported by grants from the Swedish Cancer Society, the Sahlgrenska Academy, and BioCARE - a National Strategic Cancer Research Program at University of Gothenburg (to JN), and Assar Gabrielsson's and W.M. Lundgren's Foundations (to TRL and JB).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Jerant AF, Johnson JT, Sheridan CD, Caffrey TJ. Early detection and treatment of skin cancer. Am Fam Phys 2000; 62:357-68, 75–6, 81–2; PMID:10929700 [PubMed] [Google Scholar]
- 2.Ascierto PA, Kirkwood JM, Grob JJ, Simeone E, Grimaldi AM, Maio M, Palmieri G, Testori A, Marincola FM, Mozzillo N. The role of BRAF V600 mutation in melanoma. J Transl Med 2012; 10:85; PMID:22554099; http://dx.doi.org/ 10.1186/1479-5876-10-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature 2007; 445:851-7; PMID:17314971; http://dx.doi.org/ 10.1038/nature05661 [DOI] [PubMed] [Google Scholar]
- 4.Yang C, Kim SH, Bianco NR, Robbins PD. Tumor-derived exosomes confer antigen-specific immunosuppression in a murine delayed-type hypersensitivity model. PloS One 2011; 6:e22517; PMID:21829629; http://dx.doi.org/ 10.1371/journal.pone.0022517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robbins PD, Morelli AE. Regulation of immune responses by extracellular vesicles. Nat Rev Immunol 2014; 14:195-208; PMID:24566916; http://dx.doi.org/ 10.1038/nri3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crescitelli R, Lasser C, Szabo TG, Kittel A, Eldh M, Dianzani I, Buzas EI, Lotvall J. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J Extracell Vesicles 2013; 2; PMID:24223256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sluijter JP, Verhage V, Deddens J, van den Akker F, Doevendans PA. Microvesicles and exosomes for intracardiac communication. Cardiovasc Res 2014; 102:302-11; PMID:24488559; http://dx.doi.org/ 10.1093/cvr/cvu022 [DOI] [PubMed] [Google Scholar]
- 8.Waldenstrom A, Ronquist G. Role of exosomes in myocardial remodeling. Circ Res 2014; 114:315-24; PMID:24436427; http://dx.doi.org/ 10.1161/CIRCRESAHA.114.300584 [DOI] [PubMed] [Google Scholar]
- 9.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007; 9:654-9; PMID:17486113; http://dx.doi.org/ 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 10.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, Rak J. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nature cell biology 2008; 10:619-24; PMID:18425114; http://dx.doi.org/ 10.1038/ncb1725 [DOI] [PubMed] [Google Scholar]
- 11.Sharghi-Namini S, Tan E, Ong LL, Ge R, Asada HH. Dll4-containing exosomes induce capillary sprout retraction in a 3D microenvironment. Sci Rep 2014; 4:4031; PMID:24504253; http://dx.doi.org/ 10.1038/srep04031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang XL, Zhang Y, Tan B, Luo CL, Wu XH. Renal tumor-derived exosomes inhibit hepaCAM expression of renal carcinoma cells in ap-AKT-dependent manner. Neoplasma 2014; 61:416-23; PMID:24645843; http://dx.doi.org/ 10.4149/neo_2014_051 [DOI] [PubMed] [Google Scholar]
- 13.Tauro BJ, Mathias RA, Greening DW, Gopal SK, Ji H, Kapp EA, Coleman BM, Hill AF, Kusebauch U, Hallows JL, et al.. Oncogenic H-ras reprograms Madin-Darby canine kidney (MDCK) cell-derived exosomal proteins following epithelial-mesenchymal transition. Mol Cell Proteomics 2013; 12:2148-59; PMID:23645497; http://dx.doi.org/ 10.1074/mcp.M112.027086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao D, Ohlendorf J, Chen Y, Taylor DD, Rai SN, Waigel S, Zacharias W, Hao H, McMasters KM. Identifying mRNA, microRNA and protein profiles of melanoma exosomes. PloS One 2012; 7:e46874; PMID:23056502; http://dx.doi.org/ 10.1371/journal.pone.0046874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, Garcia-Santos G, Ghajar C, et al.. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012; 18:883-91; PMID:22635005; http://dx.doi.org/ 10.1038/nm.2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skog J, Wurdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry WT Jr., Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 2008; 10:1470-6; PMID:19011622; http://dx.doi.org/ 10.1038/ncb1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res 2004:D109-11; PMID:14681370; http://dx.doi.org/ 10.1093/nar/gkh023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell 2005; 122:6-7; PMID:16009126; http://dx.doi.org/ 10.1016/j.cell.2005.06.036 [DOI] [PubMed] [Google Scholar]
- 19.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010; 11:597-610; PMID:20661255 [DOI] [PubMed] [Google Scholar]
- 20.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004; 5:522-31; PMID:15211354; http://dx.doi.org/ 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 21.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al.. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065-70; PMID:16103053; http://dx.doi.org/ 10.1158/0008-5472.CAN-05-1783 [DOI] [PubMed] [Google Scholar]
- 22.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res 2012; 40:10937-49; PMID:22965126; http://dx.doi.org/ 10.1093/nar/gks832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nolte-'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, t Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res 2012; 40:9272-85; PMID:22821563; http://dx.doi.org/ 10.1093/nar/gks658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed W, Philip PS, Tariq S, Khan G. Epstein-Barr virus-encoded small RNAs (EBERs) are present in fractions related to exosomes released by EBV-transformed cells. PloS One 2014; 9:e99163; PMID:24896633; http://dx.doi.org/ 10.1371/journal.pone.0099163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, van Eijndhoven MA, Sadek P, Sie D, Zini N, Middeldorp JM, Ylstra B, de Menezes RX, et al.. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 2014; 8:1649-58; PMID:25242326; http://dx.doi.org/ 10.1016/j.celrep.2014.08.027 [DOI] [PubMed] [Google Scholar]
- 26.Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, et al.. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics 2015; 31(6); 933-9; PMID; http://dx.doi.org/ 10.1093/bioinformatics/btu741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DK, Lee J, Kim SR, Choi DS, Yoon YJ, Kim JH, et al. EVpedia: a community web portal for extracellular vesicles research. Bioinformatics 2015; 31(6):933-9; PMID ; http://dx.doi.org/ 10.1093/bioinformatics/btu741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tewari M. A functional extracellular transcriptome in animals? implications for biology, disease and medicine. Genome Biol 2015; 16:47; PMID:25723626; http://dx.doi.org/ 10.1186/s13059-015-0613-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim EL, Trinh DL, Scott DW, Chu A, Krzywinski M, Zhao Y, Robertson AG, Mungall AJ, Schein J, Boyle M, et al.. Comprehensive miRNA sequence analysis reveals survival differences in diffuse large B-cell lymphoma patients. Genome Biol 2015; 16(1):18; PMID:25723320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Gassart A, Geminard C, Fevrier B, Raposo G, Vidal M. Lipid raft-associated protein sorting in exosomes. Blood 2003; 102:4336-44; PMID:12881314; http://dx.doi.org/ 10.1182/blood-2003-03-0871 [DOI] [PubMed] [Google Scholar]
- 31.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, Chalmers RT, Webb DJ, Dear JW. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med 2012; 10:5; PMID:22221959; http://dx.doi.org/ 10.1186/1479-5876-10-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 2010; 107:6328-33; PMID:20304794; http://dx.doi.org/ 10.1073/pnas.0914843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vojtech L, Woo S, Hughes S, Levy C, Ballweber L, Sauteraud RP, Strobl J, Westerberg K, Gottardo R, Tewari M, et al.. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 2014; 42:7290-304; PMID:24838567; http://dx.doi.org/ 10.1093/nar/gku347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hwang D, Rust AG, Ramsey S, Smith JJ, Leslie DM, Weston AD, de Atauri P, Aitchison JD, Hood L, Siegel AF, et al.. A data integration methodology for systems biology. Proc Natl Acad Sci U S A 2005; 102:17296-301; PMID:16301537; http://dx.doi.org/ 10.1073/pnas.0508647102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu SD, Tseng YT, Shrestha S, Lin YL, Khaleel A, Chou CH, Chu CF, Huang HY, Lin CM, Ho SY, et al.. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res 2014; 42:D78-85; PMID:24304892; http://dx.doi.org/ 10.1093/nar/gkt1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pitta C, et al.. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J 2011; 30:1990-2007; PMID:21468029; http://dx.doi.org/ 10.1038/emboj.2011.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, McClellan EA, Poels E, Sluimer JC, van den Hoogenhof MM, et al.. The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell Metab 2013; 18:341-54; PMID:24011070; http://dx.doi.org/ 10.1016/j.cmet.2013.08.009 [DOI] [PubMed] [Google Scholar]
- 38.Xu D, Tan J, Zhou M, Jiang B, Xie H, Nie X, Xia K, Zhou J. Let-7b and microRNA-199a inhibit the proliferation of B16F10 melanoma cells. Oncol Lett 2012; 4:941-6; PMID:23162627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009; 4:44-57; PMID:19131956; http://dx.doi.org/ 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 40.Sand M, Skrygan M, Sand D, Georgas D, Gambichler T, Hahn SA, Altmeyer P, Bechara FG. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res 2013; 351:85-98; PMID:23111773; http://dx.doi.org/ 10.1007/s00441-012-1514-5 [DOI] [PubMed] [Google Scholar]
- 41.Kogure T, Lin WL, Yan IK, Braconi C, Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011; 54:1237-48; PMID:21721029; http://dx.doi.org/ 10.1002/hep.24504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedlander MR, Lizano E, Houben AJ, Bezdan D, Banez-Coronel M, Kudla G, Mateu-Huertas E, Kagerbauer B, Gonzalez J, Chen KC, et al.. Evidence for the biogenesis of more than 1,000 novel human microRNAs. Genome Biol 2014; 15:R57; PMID:24708865; http://dx.doi.org/ 10.1186/gb-2014-15-4-r57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White NM, Cabanski CR, Silva-Fisher JM, Dang HX, Govindan R, Maher CA. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol 2014; 15:429; PMID:25116943; http://dx.doi.org/ 10.1186/s13059-014-0429-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCormick KP, Willmann MR, Meyers BC. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence 2011; 2:2; PMID:21356093; http://dx.doi.org/ 10.1186/1758-907X-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Worley LA, Long MD, Onken MD, Harbour JW. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res 2008; 18:184-90; PMID:18477892; http://dx.doi.org/ 10.1097/CMR.0b013e3282feeac6 [DOI] [PubMed] [Google Scholar]
- 46.Segura MF, Belitskaya-Levy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, Darvishian F, Berman RS, Shapiro RL, Pavlick AC, et al.. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res 2010; 16:1577-86; PMID:20179230; http://dx.doi.org/ 10.1158/1078-0432.CCR-09-2721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leidinger P, Keller A, Borries A, Reichrath J, Rass K, Jager SU, Lenhof HP, Meese E. High-throughput miRNA profiling of human melanoma blood samples. BMC Cancer 2010; 10:262; PMID:20529253; http://dx.doi.org/ 10.1186/1471-2407-10-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, Berman RS, Shapiro RL, Pavlick AC, Hernando E, et al.. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med 2012; 10:155; PMID:22857597; http://dx.doi.org/ 10.1186/1479-5876-10-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bar-Eli M. Searching for the ‘melano-miRs’: miR-214 drives melanoma metastasis. EMBO J 2011; 30:1880-1; PMID:21593728; http://dx.doi.org/ 10.1038/emboj.2011.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee YB, Bantounas I, Lee DY, Phylactou L, Caldwell MA, Uney JB. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res 2009; 37:123-8; PMID:19029138; http://dx.doi.org/ 10.1093/nar/gkn920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chan E, Patel R, Nallur S, Ratner E, Bacchiocchi A, Hoyt K, Szpakowski S, Godshalk S, Ariyan S, Sznol M, et al.. MicroRNA signatures differentiate melanoma subtypes. Cell cycle 2011; 10:1845-52; PMID:21543894; http://dx.doi.org/ 10.4161/cc.10.11.15777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Philippidou D, Schmitt M, Moser D, Margue C, Nazarov PV, Muller A, Vallar L, Nashan D, Behrmann I, Kreis S. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res 2010; 70:4163-73; PMID:20442294; http://dx.doi.org/ 10.1158/0008-5472.CAN-09-4512 [DOI] [PubMed] [Google Scholar]
- 53.Nana-Sinkam S, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol 2014; 15:445; PMID:25315999; http://dx.doi.org/ 10.1186/s13059-014-0445-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011; 27:863-4; PMID:21278185; http://dx.doi.org/ 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andrews S. Fast QC, A Quality Control tool for High Throughput Sequence Data, 2014; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 56.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15-21; PMID:23104886; http://dx.doi.org/ 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup . The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009; 25:2078-9; PMID:19505943; http://dx.doi.org/ 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Anders S, Pyl PT, Huber W : HTSeq - A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31:166; PMID ; http://dx.doi.org/ 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al.. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 2012; 22:1760-74; PMID:22955987; http://dx.doi.org/ 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014; 15:550; PMID:25516281; http://dx.doi.org/ 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hong BS, Cho JH, Kim H, Choi EJ, Rho S, Kim J, Kim JH, Choi DS, Kim YK, Hwang D, et al.. Colorectal cancer cell-derived microvesicles are enriched in cell cycle-related mRNAs that promote proliferation of endothelial cells. BMC Genomics 2009; 10:556; PMID:19930720; http://dx.doi.org/ 10.1186/1471-2164-10-556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi DS, Yang JS, Choi EJ, Jang SC, Park S, Kim OY, Hwang D, Kim KP, Kim YK, Kim S, et al.. The protein interaction network of extracellular vesicles derived from human colorectal cancer cells. J Proteome Res 2012; 11:1144-51; PMID:22149170; http://dx.doi.org/ 10.1021/pr200842h [DOI] [PubMed] [Google Scholar]
- 63.Wang X. miRDB: a microRNA target prediction and functional annotation database with a wiki interface. Rna 2008; 14:1012-7; PMID:18426918; http://dx.doi.org/ 10.1261/rna.965408 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.