Abstract
Variants in BCL11A were previously associated with fetal hemoglobin (HbF) levels among Cameroonian sickle cell disease (SCD) patients, however explaining only ∼2% of the variance. In the same patients, we have investigated the relationship between HbF and two SNPs in a BCL11A erythroid-specific enhancer (N = 626). Minor allele frequencies in rs7606173 and rs1427407 were 0.42 and 0.24, respectively. Both variants were significantly associated with HbF levels (p = 3.11e-08 and p = 6.04e-06, respectively) and explained 8% and 6.2% variations, respectively. These data have confirmed a stronger effect on HbF of genomic variations at the BCL11A erythroid-specific enhancer among patients with SCD in Cameroon, the first report on a West African population. The relevance of these findings is of prime importance because the disruption of this enhancer would alter BCL11A expression in erythroid precursors and thus HbF expression, while sparing the induced functional challenges of any alterations on the expression of this transcription factor in non-erythroid lineages, thus providing an attractive approach for new treatment strategies of SCD.
Introduction
Adult fetal hemoglobin (HbF) levels alter the morbidity of sickle cell disease (SCD) (Platt et al., 1994), by inhibiting sickle hemoglobin (HbS) polymerization and therefore reducing hemolytic anemia and tissue injury. The HbF levels are highly variable and inheritable, primarily through genetic variants at three principal loci, BCL11A, HBS1L-MYB, and HBB cluster, accounting for 10%–20% of HbF variation among SCD patients (Thein and Menzel, 2009). Specifically, variations in BCL11A have been shown to be amenable to therapeutic manipulation, leading to the reversal of SCD symptoms in mice models (Xu et al., 2011).
Functional studies have shown that the expression of the transcriptional repressor BCL11A is regulated by erythroid-specific enhancers that contain 3′ DNase hypersensitive sites (DHS) located +62, + 58, and +55 kb from the transcription initiation site of BCL11A (Bauer et al., 2013). Two SNPs of the enhancer element had a stronger association with HbF levels among African American SCD patients: rs1427407 in DHS +62 and rs7606173 in DHS +55. This is consistent with the hypothesis that multiple functional SNPs act in combination to influence BCL11A regulation (Bauer et al 2013; Sebastiani et al., 2015).
We previously reported that the minor allele frequencies (MAFs) of selected variants in BCL11A were similar to those detected in African Americans (Lettre et al., 2008) and the relationships with HbF were significant, but explained less of the variance in expression (∼2% vs. 10%) (Wonkam et al., 2014). Other genomic polymorphisms at this locus could better tag the relation with HbF level in some West African SCD patients. In the present study, we have investigated the relationship between HbF levels and rs1427407 and rs7606173 in the BCL11A erythroid-specific enhancer among 626 patients with SCD (HbSS) from Cameroon.
Methods
Study population and HbF measurement
The study was performed with the approval of the National Ethical Committee Ministry of Public Health, Republic of Cameroon (No 033/CNE/DNM/07), and the University of Cape Town, Faculty of Health Sciences Human Research Ethics Committee (HREC REF: 132/2010). Patients' recruitment was conducted at the Yaoundé Central Hospital and Laquintinie Hospital in Douala, Cameroon. Only clinically stable patients, 5 years or older, with no history of blood transfusion, hydroxyurea treatment or hospitalisation in the preceding 6 weeks were included and clinical events were prospectively collected (Table 1).
Table 1.
Cohort Description
| Variables | Mean ± SD | Value range | Number of observations | |
|---|---|---|---|---|
| Age (years) | 17.3 ± 9.9 | 5–53 | 628 | |
| RBC (1012/L) | 2.3 ± 3.1 | 1.1–2-6.5 | 626 | |
| Hematological indices | Hb (g/dl) | 7.8 ± 1.5 | 3.5–14.5 | 626 |
| MCV(fl) | 84 ± 10.1 | 59–117 | 626 | |
| MCHC (g/dl) | 34 ± 3.7 | 21.5–54.3 | 626 | |
| WBC (109/l) | 14 ± 5.9 | 2.9–49.8 | 626 | |
| Lymphocytes (109/l) | 5.9 ± 2.9 | 1.9–22.6 | 626 | |
| Monocytes (109/l) | 1.5 ± 1.0 | 0.1–11.9 | 626 | |
| Platelets (109/l) | 373 ± 122 | 97–837 | 626 | |
| HbA2 (%) | 3.7 ± 1.8 | 0–9.7 | 626 | |
| HbF (%) | 10.1 ± 8.6 | 0–26.8 | 626 | |
| Clinical events | Vaso-occlusive crisis (No. /year) | 2.9 ± 3.4 | 0–30 | 593 |
| Consultations (No. /year) | 2.8 ± 3.5 | 0–29 | 591 | |
| Hospitalisation (No. /year) | 1.4 ± 2.1 | 0–15 | 591 | |
| Stroke | 4.2% | 25/599 | ||
| 3.7del Alpha-globin gene genotypes | αα/αα | 59.3% | 310/524a | |
| αα/α3.7 | 29.6% | 156/524a | ||
| α3.7/α3.7 | 11.1% | 58/524a | ||
| HBB haplotypes | Benin/Benin | 57% | 307/541a | |
| Benin/Cameroon | 25% | 137/541a | ||
| Benin/atypical | 8% | 45/541a | ||
| Cameroon/Cameroon | 5% | 2/541a |
Number of individuals and not alleles.
Complete routine blood counts of patients and hemoglobin electrophoresis were conducted upon arrival at the hospital. Two methods of HbF measurement were employed in this study for successive groups of patients; initially the alkali denaturation test (ADT) in 55.5% (n = 347) of the cohort, and subsequently high performance liquid chromatography (HPLC), when it became available.
In a previous report, we disaggregated the SCD patients sample, based on the HbF assessment technique (ADT vs. HPLC) and found that the significant associations with HbF levels, examined independently, were present in both subgroups in rs11886868 (BCL11A), rs4671393 (BCL11A), rs28384513 (HMIP 1), and rs9494142 (HMIP 2) (Wonkam et al., 2014). To further avoid any major influence on the outcomes of the present study, the association analyses were also corrected for the electrophoresis techniques.
Genotyping
HbS mutation and HBB haplotypes
DNA samples were extracted from peripheral blood following the manufacturer's instructions (Puregene Blood Kit, Qiagen®, Alameda, CA, USA). Molecular analysis for the presence of the sickle mutation was carried out on 200 ng DNA by PCR to amplify a 770 bp segment of the β-globin gene, followed by Dde I restriction analysis of the PCR product (Saiki et al., 1985).
Five restriction fragment length polymorphism (RFLP) regions in the β-globin gene cluster were amplified using published primers and methods (Steinberg et al., 1998) to analyze the XmnI (5'Gγ), HindIII (Gγ), HindIII (Aγ), HincII (Ψ'Ψβ), and HinfI (5'β) for the HBB haplotype background (Bitoungui et al., 2015).
Detection of 3.7 kb α-globin gene deletions
The 3.7 kb alpha-globin gene deletions were successfully screened using the expand-long template PCR (Roche®, UK) following the instructions reported by Tan et al. (2001) with some modifications previously published (Rumaney et al., 2014).
SNPs
For the 626 samples, multiplex SNaPshot PCR and capillary electrophoresis were used to genotype both rs1427407 and rs7606173 at the BCL11A erythroid-specific enhancer loci. Quality control of the capillary electrophoresis results was assessed by direct cycle sequencing in a subset of 10% randomly selected samples that confirmed the SNaPshot results. Details of the methods were previously reported (Wonkam et al., 2014).
Data analysis
Descriptive statistics were obtained for all quantitative data using SPSS (IBM, USA version 21.0). A chi-squared test, with one degree of freedom, was used to perform the Hardy-Weinberg Equilibrium (HWE) test on the SNPs genotype with the following results: rs7606173 HWE p-value was 0.534 (X2 = 0.845) and rs1427407 HWE p-value was 0.0042 (X2 = 12.7). To exclude the ascertainment bias due to technical genotyping reasons, about 10% (N = 60) of the samples were Sanger-sequenced by an independent laboratory (Central Analytical Facilities, University of Stellenbosch) and confirmed all the SNP genotyping results.
The observed frequencies of the rs1427407 variant could be biased by the presence of a third T allele (MAF = 0.048) in 92 samples, suggesting the presence of copy number variations (CNVs) that could be involved in the regulation of BCL11A in some individuals. Alternatively, the violation of HWE for rs1427407 could possibly be due to a selective effect of this HbF-promoting SNP that could potentially influence the course of SCD and ultimately the survival of some patients. These various observations and hypotheses deserve further investigations.
Owing to the skewed distribution of HbF level, we log10-transformed HbF values to approximate normal distribution, and then used these transformed values in the association analysis after correcting for age; gender, electrophoresis technique, history of transfusion, HBB haplotypes, and the 3.7 kb alpha-globin gene deletion genotypes. Using an additive genetic model, under a generalized linear regression framework, we investigated the relationship between the SNPs (rs1427407 and rs7606173) and HbF levels, using the R statistical package version 3.0.3 (The R Foundation for statistical computing, Vienna, Austria). Significance was set at 5%.
Results
Patients
Table 1 summarizes the description of the cohort. All patients were homozygous for the HbS mutation. Among them, 50.3% (n = 316) were female. After the analysis of 1082 chromosomes, Benin (74%; n = 799) and Cameroon haplotypes (19%; n = 207) were the most prevalent. In combination, the Benin/Benin haplotype represented 57% (n = 307) and the Benin/Cameroon 25% (n = 137) of the cohort (Table 1). Among 524 patients successfully genotyped for the 3.7 kb alpha-globin gene deletion, 40.7% (n = 214) had co-inherited alpha-thalassaemia (Table 1).
SNPs in BCL11A hypersensitivity sites and HbF levels
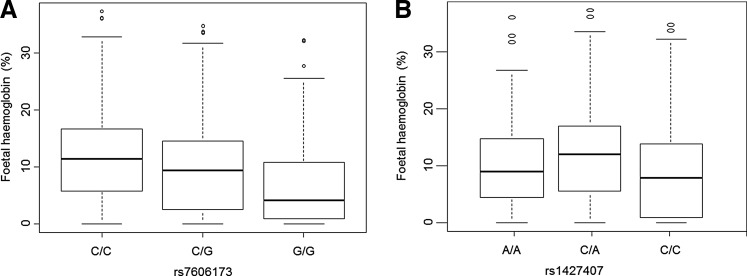
In the present study, MAFs in both rs7606173 and rs1427407 variants were 0.42 and 0.24, respectively (Table 2). Both genetic variants were significantly associated with HbF levels (p = 3.11e-08 and p = 6.04e-06) (Fig. 1A) and explained 8% and 6.2% variations in HbF levels, respectively (Table 2). The relationship of both SNPs with HbF was not influenced by the electrophoresis techniques, alpha-thalassaemia genotypes, or HBB haplotypes. There were no significant associations between the two SNPs and other various hematological indices or clinical events, but rs7606173 that was associated with hemoglobin levels (p = 0.02) and consultation rates (p = 0.04).
Table 2.
Differential HbF Association Results for Selected SNPs at the BCL11A Specific Erythroid Enhancer in Cameroonian SCD HbSS Patients (N = 626)
| Locus Chr. 2 | SNP | Position& | Allele change | MAF$ | Effect Size# (SE) | Variance explained (%) | P values |
|---|---|---|---|---|---|---|---|
| BCL11A enhancer | rs1427407 (DHS +62) | 60718043 | C>A | 0.26 | 0.119 (0.024) | 6.2 | 6.04e-06 |
| BCL11A enhancer | rs7606173 (DHS +55) | 60725451 | C>G | 0.42 | 0.1 (0.025) | 8 | 3.11e-08 |
Chr, chromosome; Position on NCBI Build 36.1; $MAF, minor allele frequency; #Effect sizes and standard errors are given in standard deviation units for the minor allele.
FIG. 1.
Box-plots showing the distribution of foetal haemoglobin levels conditioned on SNP genotypes. (A) Fetal hemoglobin levels, conditioned on the rs7606173 SNP genotypes, and (B) conditioned on the rs1427407 SNP genotypes. Boxes have lines at the lower quartile, median, and upper quartile. The plot whiskers extend up and down from the median a distance 1.5 times the interquartile range of the boxes (truncated at zero where necessary). Outliers are the points outside the whiskers indicated as circles.
Discussion
The present article is the first report on the importance of the SNPs rs1427407 and rs7606173 among West African SCD patients. The results are consistent with those reported among African Americans (Bauer et al., 2013) and patients from Saudi Arabia and India (Sebastiani et al., 2015) and Tanzania for rs1427407 (Ntatiro et al., 2015). The results of this study are of particular importance for the local and global SCD scientific communities for the following reasons. First, nearly two-thirds of SCD patients globally are born and reside in Africa (Piel et al., 2013) and any potentially important SCD research performed on other populations should be replicated in African populations to assess and confirm their relevance for future care and therapeutic interventions.
Second, it well known that Africans have the highest genetic variations and diversities (Gurdasani et al., 2015), implying some regional genetic specificities. For instance, we have previously shown that rs9399137, which acts as a tagging HbF-promoting SNP for the HMIP-2 sub-locus in European populations (Thein et al., 2007), occurs at a low frequency in this Cameroonian cohort (Wonkam et al., 2014). In addition, a 3 bp deletion in perfect linkage disequilibrium (LD) with rs9399137 in Chinese, European, and some African American populations was reported to be the candidate functional motif for the HMIP-2 region (Farell et al., 2011).
However, in these Cameroonian patients, rs9389269 was not linked to rs9399137 (r2 = 0; D’ = 0.138) (Wonkam et al., 2014). Moreover, we reported, in the HMIP-2 sub-locus, a much higher frequency of rs9389269 (0.18) in Cameroonians as compared to Tanzanian SCD patients (0.03), an observation that could indicate a high degree of variation in the MAF of this SNP amongst SCD patients from various African population groups (Wonkam et al., 2014). These results emphasize the need to report on multiple African populations to fully capture the importance of various disease-modifying SNPs.
Lastly, Sebastiani et al. (2015) have reported on the association between variants at BCL11A erythroid-specific enhancer and HbF levels in African American SCD patients. However, among self-identified African Americans, analysis of genomic admixture indicates that the median proportion of European ancestry is 18.5% with very large variation, ranging from an estimate of over 99% West African ancestry less than 1% (Bryc et al., 2010). Therefore, any results of genomic studies performed on African Americans are not always appropriate proxies of what could be expected among sub-Saharan Africans. The importance of replicating findings of HbF-promoting SNPs at BCL11A in various populations is thus crucial to identifying possible targets for therapeutic intervention to increase HbF levels in patients with SCD.
Nevertheless, a major clinical caution of exploring the decreased expression of BCL11A, in order to promote HbF expression, rests on the other deleterious functional implications of this transcription factor, whose reduced expression could have serious developmental and neurological consequences. Indeed, patients described with microdeletions encompassing BCL11A display neurodevelopmental abnormalities including intellectual disability and brain malformation (Balci et al., 2015; Funnell et al., 2015; Peter et al., 2014). This emphasises the importance of the BCL11A erythroid-specific enhancer being subject to genetic variations that influence HbF levels as reported in the present study.
Bioinformatics analysis suggests that rs7606173 in DHS +55 changes a binding site for MZF1, a gene actively transcribed in hematopoietic cells (Sebastiani et al., 2015) and most importantly, functional analysis have shown that the disruption of this enhancer would impair BCL11A expression in erythroid precursors and thus HbF expression, while conserving the “undruggable” nature of this transcription factor in non-erythroid lineages (Bauer et al., 2013).
It is also possible that, in the future, genome editing could allow the introduction of beneficial SNPs at the BCL11A erythroid-specific enhancer locus to improve the clinical course of SCD patients. Preliminary proof of concept was shown by experiments that introduced the −175> point mutation and successfully elevated HbF into erythroid cell lines (Wienert et al., 2015). In addition, using a specific guide, RNA and Cas9, authors recently corrected one allele of the SCD HBB gene in human erythroid cells (Huang et al., 2015).
A limitation of the present research is the number of SNPs that were studied. Additional genotyping of SNPs will allow the definition of various haplotypes that will best capture the combinatorial importance of BCL11A elements (Bae et al., 2012).
Conclusion and Perspectives
The present study validates the relationship between HbF levels and SNPs in a BCL11A erythroid-specific enhancer among SCD patients in Cameroon, the first report on a West African cohort. The relevance of this finding is of prime importance because the disruption of this enhancer would impair BCL11A expression in erythroid precursors and thus HbF expression, while sparing the induced various functional challenges of any alterations of this transcription factor expression in non-erythroid lineages, thus providing an attractive approach for future treatment of SCD.
Acknowledgments
Conceived and designed the experiments: GP, AW. Performed the experiments: GP, VJNB AW. Analyzed the data: AW, APK, GL, SEA. Contributed reagents/materials/analysis tools: VJNB, BCC, APK, AW. Wrote the article: GP, AW. Revised and approved the manuscript: GP, VJNB, BCC, APK, SEA, AW.
Funding. The molecular experiments of the study were funded by the National Health Laboratory Services (NHLS), South Africa; and the NIH, USA, Grant Number 1U01HG007459-01. The students' bursaries were supported by the Third World Academy of Sciences (TWAS), Oppenheimer Memorial Trust, the National Research Foundation, and FirstRand Laurie Dippenaar Scholarship, South Africa. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author Disclosure Statement
The authors have declared that no competing financial interests exist.
References
- Bae HT, Baldwin CT, Sebastiani P, et al. (2012). Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood 120, 1961–1962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balci TB, Sawyer SL, Davila J, Humphreys P, and Dyment DA. (2015). Brain malformations in a patient with deletion 2p16.1: A refinement of the phenotype to BCL11A. Eur J Med Genet 58, 351–354 [DOI] [PubMed] [Google Scholar]
- Bauer DE, Kamran SC, Lessard S, et al. (2013). An erythroid enhancer of BCL11A subject to genetic variation determines fetal hemoglobin level. Science 342, 253–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoungui VJ, Pule GD, Hanchard N, Ngogang J, and Wonkam A. (2015). Beta-globin gene haplotypes among Cameroonians and review of the global distribution: Is there a case for a single sickle mutation origin in Africa? OMICS 19, 171–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryc K, Auton A, Nelson MR, et al. (2010). Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci USA 107, 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell JJ, Sherva RM, Chen ZY, et al. (2011). A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood 117, 4935–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funnell AP, Prontera P, Ottaviani V, et al. (2015). 2p15-p16.1 microdeletions encompassing and proximal to BCL11A are associated with elevated HbF in addition to neurologic impairment. Blood 126, 89–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdasani D, Carstensen T, Tekola-Ayele F, et al. (2015). The African Genome Variation Project shapes medical genetics in Africa. Nature 517, 327–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wang Y, Yan W, et al. (2015). Production of gene-corrected adult beta globin protein in human erythrocytes differentiated from patient iPSCs after genome editing of the sickle point nutation. Stem Cells 33, 1470–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettre G, Sankaran VG, Bezerra MA, et al. (2008). DNA polymorphisms at the BCL11A, HBS1L-MYB, and beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc Natl Acad Sci USA 105, 11869–11874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mtatiro SN, Singh T, Rooks H, et al. (2014). Genome wide association study of fetal hemoglobin in sickle cell anemia in Tanzania. PLoS One 9, e111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter B, Matsushita M, Oda K, and Raskind W. (2014). De novo microdeletion of BCL11A is associated with severe speech sound disorder. Am J Med Genet A 164A, 2091–2096 [DOI] [PubMed] [Google Scholar]
- Piel FB, Patil AP, Howes RE, et al. (2013). Global epidemiology of sickle haemoglobin in neonates: A contemporary geostatistical model-based map and population estimates. Lancet 381, 142–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt OS, Brambilla DJ, Rosse WF, et al. (1994). Mortality in sickle cell disease. Life expectancy and risk factors for early death. N Engl J Med 330, 1639–1644 [DOI] [PubMed] [Google Scholar]
- Rumaney MB, Ngo Bitoungui VJ, Vorster AA, et al. (2014). The co-inheritance of alpha-thalassemia and sickle cell anemia is associated with better hematological indices and lower consultations rate in Cameroonian patients and could improve their survival. PLoS One 9, e100516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, et al. (1985). Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230, 1350–1354 [DOI] [PubMed] [Google Scholar]
- Sebastiani P, Farrell JJ, Alsultan A, et al. (2015). BCL11A enhancer haplotypes and fetal hemoglobin in sickle cell anemia. Blood Cells Mol Dis 54, 224–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg MH, Lu ZH, Nagel RL, et al. (1998). Hematological effects of atypical and Cameroon beta-globin gene haplotypes in adult sickle cell anemia. Am J Hematol 59, 121–126 [DOI] [PubMed] [Google Scholar]
- Tan AS, Quah TC, Low PS, and Chong SS. (2011). A rapid and reliable 7-deletion multiplex polymerase chain reaction assay for alpha-thalassemia. Blood 98, 250–251 [DOI] [PubMed] [Google Scholar]
- Thein SL, and Menzel S. (2009). Discovering the genetics underlying foetal haemoglobin production in adults. Br J Haematol 145, 455–467 [DOI] [PubMed] [Google Scholar]
- Thein SL, Menzel S, Peng X, et al. (2007). Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc Natl Acad Sci USA 104,11346–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienert B, Funnell AP, Norton LJ, et al. (2015). Editing the genome to introduce a beneficial naturally occurring mutation associated with increased fetal globin. Nat Commun 6, 7085. [DOI] [PubMed] [Google Scholar]
- Wonkam A, Ngo Bitoungui VJ, Vorster AA, et al. (2014). Association of variants at BCL11A and HBS1L-MYB with hemoglobin F and hospitalization rates among sickle cell patients in Cameroon. PLoS One 9, e92506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Peng C, Sankaran VG, et al. (2011). Correction of sickle cell disease in adult mice by interference with fetal hemoglobin silencing. Science 334, 993–996 [DOI] [PMC free article] [PubMed] [Google Scholar]