Abstract
The presence in human nuclear chromosomes of multiple sequences that are highly similar to human mitochondrial tRNAs (tRNA-lookalikes) raises intriguing questions about the possible functionality of these genomic loci. In this perspective, we explore the significance of the mitochondrial tRNA-lookalikes based on a series of properties that argue for their non-accidental nature. We particularly focus on the possibility of transcription as well as on potential functional roles for these sequences that can range from their acting as DNA regulatory elements to forming functional mature tRNAs or tRNA-derived fragments. Extension of our analysis to other simians (chimp, gorilla, rhesus, and squirrel monkey), 2 rodents (mouse and rat), a marsupial (opossum) and 3 invertebrates (fruit-fly, worm, and sponge) revealed that mitochondrial tRNA-lookalikes are prevalent in primates and the opossum but absent from the other analyzed organisms.
Keywords: mitochondrion, tRNA, tRNA fragment, tRF, tRNA-half, tRNA-lookalike
Introduction
tRNA (tRNAs, Fig. 1A) have long been believed to be housekeeping molecules. It can be argued that their well-established and well-understood participation in the process of translation has contributed to the general scientific studies remaining relatively away from possible non-translational roles. In recent years however, tRNA molecules are again in the spotlight. Their demonstrated roles in diseases and cellular/molecular processes,1-4 directly or through tRNA-derived fragments (tRFs),5,6 have revived scientific interest in these ancient molecules. To further add to this emerging complexity, the distinction of nuclear and mitochondrial tRNAs should also be considered.
Figure 1.
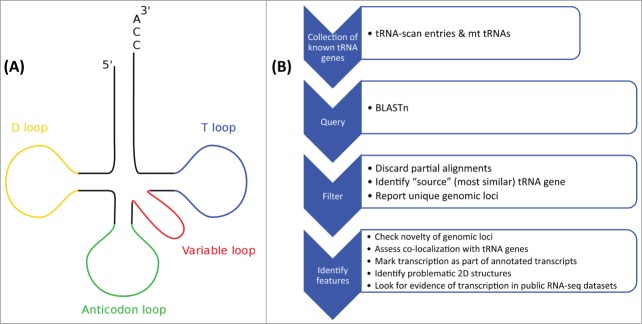
(A) The typical 2D (cloverleaf) structure of a tRNA with each loop (D, T, variable and anticodon loop) colored differently. The post-transcriptionally added CCA-trinucleotide at the 3’ end of the mature molecule is also indicated. (B) The general workflow of our previous study7 that identified the tRNA-lookalikes and their properties.
Owing to the prokaryotic provenance of the mitochondrion, it has long been believed that there is a structural and functional distinction between mitochondrial and nuclear tRNAs. This was compounded by the fact that these 2 tRNA collections exist and operate in 2 distinct cellular compartments. However, our recent findings of numerous mitochondrial tRNA-lookalikes in the nuclear genome7 suggest that the boundaries separating these 2 different “tRNA worlds” may not be so sharp. In this point-of-view we review the possible biological mechanisms that may hinder or allow the expression of tRNA-lookalikes and discuss their potential participation in tRNA-related functions.
Several Notable Properties
Our recent investigation of the human nuclear genome has revealed a large number (497) of genomic loci that resemble known nuclear and mitochondrial tRNA genes – we refer to these sequences as “tRNA-lookalikes”7; a graphical summary of our earlier study is provided in Figure 1B. In fact, the sequences of 8 of these nuclear genome tRNA-lookalikes are identical to the sequences of 7 mitochondrial tRNAs (mtAlaTGC has 2 copies in the nuclear genome). Perhaps surprisingly, the majority of these 497 tRNA-lookalike loci had not previously been annotated as such. Of the 497 loci, 351 best resemble one or other of the 22 known mitochondrial tRNAs. An additional 103 tRNA-lookalikes resemble one of the 508 bona fide nuclear tRNAs. The remaining 43 tRNA-lookalikes resemble pseudo-tRNAs.8
From a sequence standpoint, the collection of tRNA-lookalikes that we have uncovered doubles the number of genomic loci (508) in the human genome that have been linked to bona fide tRNAs. We note that this is not an idle observation. Indeed, using the publicly available ENCODE datasets9 we were able to show evidence of transcription for more than 2 dozen of the lookalikes: the sequenced transcripts matched perfectly the endpoints of the computationally-identified lookalikes. Intriguingly, several of the corresponding loci were transcribed in a cell-type dependent manner. Also supporting their potential participation in cellular processes was our finding that the tRNA-lookalikes are not randomly distributed across the human genome but tend to co-localize with the currently known tRNAs. The fact that the majority of the tRNA-lookalikes resemble mitochondrial sequences, of which there are only 22, further adds to the possibility that these are functionally relevant sequences.
Our initial study aimed at addressing a specific question: “are there genomic loci that resemble the known nuclear and mitochondrial tRNAs?” To answer it, we formed a tRNA-reference space by combining source sequences from both the nuclear and mitochondrial genomes and simultaneously sought to identify lookalikes elsewhere in the nuclear genome. In terms of starting point and goal, our study is orthogonal to earlier efforts that searched for mitochondrial sequences in the nuclear genome10-12 and discovered what are now referred to as NUMTs (Nuclear Mitochondrial DNA). Just like the tRNA-lookalikes, NUMTs are arranged across the chromosomes in a non-random manner having been associated with specific chromatin structures and with retrotransposons.13,14
In the case of mitochondrial tRNAs it is known that several of them are placed immediately adjacent to one another on the mitochondrial genome. However, what we discovered is that there is a considerable level of granularity when such tRNAs migrate to the nuclear genome. As a matter of fact, we found that immediately adjacent tRNA genes in the mitochondrial chromosome “migrate” independently of one another. As an example we mention that the mitochondrial LeuTAG has 3 times as many nuclear lookalikes as its immediately adjacent SerGCT. Observations such as this could increase the likelihood that selection mechanisms which are not currently understood may be behind (mitochondrial and nuclear) tRNA-lookalikes.
Compartmentalization of cellular processes has provided eukaryotic cells with unprecedented possibilities, as compared to prokaryotic cells, and with highly dynamic interactions and cross-talk between the nuclear and the mitochondrial genome.15 In light of this, in what follows we will focus largely on the mitochondrial tRNA lookalikes alone and discuss possibilities that arise from their presence in the nuclear genome.
Transcription of the Mitochondrial tRNA-Lookalikes
Important differences characterize the transcription of mitochondrial and nuclear tRNAs. The mitochondrial genome is transcribed as a polycystronic RNA and tRNAs are recognized and cleaved (punctuation model).16 On the other hand, expression of nuclear tRNAs relies on the recruitment of RNA Pol III by a specific scaffolding mechanism that is mainly guided by the TFIIIC factor.17 TFIIIC identifies the A and B boxes that coincide with the D and T loops respectively of the mature tRNA molecule and act as promoters of the tDNA regions. The B box (analogous to an enhancer-like element) has been shown to be more important for the binding of TFIIIC than the A-box (promoter-like).17 Whether TFIIIC binding occurs in tRNA-lookalikes remains to be investigated but at least one exact tRNA-lookalike (of the mitochondrial tRNA SerTGA, even if it has an atypical secondary structure18) contains the minimal consensus sequence (GGTTCGAnnCC) that has been proposed for B boxes19: indeed, GTTCGATTC spans positions 48 through 58 inclusive of the lookalike sequence in question and presumed mature tRNA. Therefore we ought to consider the possibility that perhaps a mechanism of transcription of the tRNA-lookalikes exists which is similar to that of the nuclear tRNAs.
In our analysis, we also showed that a number of lookalikes are transcribed as part of other transcripts. As mentioned above, this already happens in the mitochondrion where the mitochondrial tRNAs are excised from a single polycistronic transcript. It is thus possible, in principle, that such nascent molecules acquire the necessary secondary and tertiary structures that would permit recognition of the embedded tRNA-lookalike by nucleases and its subsequent appropriate cleaving from the longer transcripts.
The requirement for proper cleaving is probably not going to be a hindrance for the expression of the lookalikes as tRNAs: as a matter of fact, RNase P and RNase Z, the enzymes cleaving the 5´ and 3´ additional sequences are active in the nucleus as well as in the cytoplasm.20 However, in light of the complexity of these enzymes in the 2 compartments20,21 additional focused experimental investigations will be required to address this matter.
Potential Functional Roles of the Mitochondrial tRNA-Lookalikes
Had it been for the mere presence in the nuclear chromosomes of sequences that resemble mitochondrial tRNAs it would have been difficult to conjecture that they are functionally relevant. Fueling the possibility of functional roles for these tRNA-lookalikes are several specific properties that characterize them and argue for their non-accidental nature.
The first relevant observation in this regard relates to the fact that the lookalikes favor specific chromosomes where they are located in close proximity to the 508 known nuclear tRNAs. In the human genome, several of the nuclear tRNAs form 2 big clusters on chromosomes 1 and 6 respectively.22,23 These two clusters comprise multiple anticodon families and exhibit expression that is considerably higher compared to the other genomic regions that harbor the remaining nuclear tRNAs.
Genomic clustering can facilitate a local microenvironment where all factors that are required for translation are concentrated. Such an arrangement, which is reminiscent of bacterial operons,24 has the potential to simplify transcriptional strategies and enable efficient recycling of the resulting molecules without the need to coordinate matters over large distances. Possibly related to this are the genomic sequences that code for tRNAs (tDNAs) and have been shown to act as genomic insulators mainly by being bound by TFIIIC.25 As has already been reported, a single tDNA region is not able to induce insulation by itself: co-operation among multiple such regions is often required before chromatin structure can be significantly affected.25,26 It is possible that the tRNA-lookalikes act in a similar way, especially those whose sequence has sufficiently diverged to the point that it does not allow proper cloverleaf folding.
The characteristics of the tRNA-lookalike loci suggest that they may be transcribed either as individual units or processed from longer transcripts. Preliminary evidence that we generated from public RNA-seq data shows that at least some of the tRNA-lookalikes are transcribed and that, conspicuously, the resulting molecules are as long as the typical mature bona fide tRNAs. Of course, these resulting RNAs could function either as tRNAs or tRNA-related molecules. It is important to note that based on the available public data it is unlikely that the tRNA-lookalikes are aberrant transcripts: in fact, the ENCODE repository indicates that they are transcribed in a cell-type specific manner.
Can tRNA-lookalikes participate in the process of translation? This is an important question given that the majority of the lookalikes are mitochondrial. For translation to happen a series of enzymatic reactions need to take place after the tRNA precursor has been transcribed and before the mature tRNA can participate in the process of translation.
Some of the enzymes catalyzing these reactions are able to identify both nuclear and mitochondrial tRNAs whereas other cannot.27,28 For example, the CCA-adding enzymes are the same for the nuclear and the mitochondrial tRNAs. The first exon of these messenger RNAs (mRNAs) codes for the mitochondrial localization signal (MLS) and when translated (as a consequence of choice of a different translational starting site) the enzyme is transported inside the mitochondrion.27 Therefore, if the enzyme came across a mitochondrial tRNA-lookalike in the nucleus, it would be expected to add the CCA tail to it.
On the other hand, amino-acyl tRNA synthetases (aaRS), the enzymes that charge the tRNAs with the respective amino acids, are encoded by different genes for the cytoplasmic and the mitochondrial tRNAs.18,27 The mitochondrial aaRS have a MLS that allows them to enter the mitochondrion whereas nuclear/cytoplasmic aaRS cannot aminoacylate mitochondrial tRNAs. Two exceptions are worth noting here: the aaRS for Lys and Gly can process both mitochondrial and cytoplasmic tRNAs.
An additional crucial element for mitochondrial tRNAs is the need for proper modifications. If the nascent tRNAs are not modified at specific nucleotides, they can easily get unfolded and lose their ability to function as true tRNAs.27 Although modification maps are now becoming available for mammalian mitochondrial tRNAs,29 the enzymes/factors that are responsible for these modifications remain to be determined and characterized in order to investigate possible non-mitochondrial localization. Elongation factors and ribosomomal subunits further complicate a potential translation function,18,27 which is not obvious but not impossible either.
The best known function of tRNAs known to date is indeed their participation in the process of translation. In recent years, other novel aspects of tRNA biology have begun to be uncovered. One important such development is the existence of tRNA fragments (tRFs). TRFs are quickly attracting the attention of researchers as they suggest a whole new level of regulation of cellular and molecular processes.5,6 There are 2 basic types of tRFs: tRNA halves that originate from cleavage of the tRNAs at the anticodon loop and tRFs that arise from either the 5´ or the 3´ end of the mature tRNA molecule.5,30 tRNA halves have been considered to be stress-induced but physiological roles have also been attributed to them, while the shorter tRFs regulate gene expression via multifaceted ways, e.g. due to loading on Argonaute (Ago).5,30
Mitochondrial mature tRNAs can give rise to tRNA halves31 and, possibly, to other currently unknown categories of fragments. Under the prism of the possible biogenesis of the tRFs, tRNA-lookalikes could be potentially important. Unreasoned hypotheses of mitochondrial-specific functions of tRNAs that happen to have exact tRNA-lookalikes in the nuclear chromosome will unavoidably lead to biased and erroneous results as the tRF or tRNA half can conceivably have a non-mitochondrial biogenesis. Even if the biogenesis occurred in the mitochondrion the origin of the template at hand would remain ambiguous.
Mitochondrial tRNA-Lookalikes in Other Organisms
To determine whether the concept of nuclear copies of mitochondrial tRNA-lookalikes is unique to the human genome, we extended our previous analyses7 to 10 additional organisms: primates (Chimpanzee, Gorilla, Macaca and the Squirrel Monkey), rodents (Mouse, Rat), invertebrates (fruit fly, worm), a marsupial (opossum), and a poriferan (the sponge Reniera). The species’ official names, the genome assembly numbers and the results of these analyses are shown in Table 1. We find that the nuclear genomes of primates hosted the highest number of mitochondrial tRNA-lookalikes (between 200 and 500). Rodents had a much lower number of lookalikes (∼50) despite the fact that the mouse and rat genomes have lengths nearly identical to that of the human genome. On the other hand, very few lookalikes could be found in D. melanogaster, C. elegans, and the sponge A. queenslandica (aka Reniera). We should note that the somewhat high number of lookalikes in the sponge as compared to Drosophila and the worm could be an overestimate: the genome for this organism has not yet been assembled in chromosomes but exists as scaffolds many of which likely overlap. Nonetheless, our results indicate that the high number of tRNA-lookalikes is an attribute of primates. The relative absence of lookalikes in rodents and their presence in the opossum suggests 2 possibilities: either tRNA lookalikes represent independent evolutionary events in some animals (primates and marsupials), or they were present in the common ancestor of animals but selectively retained in some and/or lost in others. There is evidence that argues for the first hypothesis, like the fact that the rate of mitochondrial genome change is greater than the respective rate of the nuclear chromosomes,32 as well as the low sequence conservation among mitochondrial tRNA genes among mammals which results in mitochondrial genes that are organism-specific.33 However, these findings in conjunction with the known diversity of tRNAs in both sequence and structure further highlight the observation that answering whether tRNA-lookalikes are functional requires organism-specific considerations. This is particularly true of mitochondrial tRNAs that may, for example, lack the T and the D loop.18,34 Addressing all such considerations will necessitate additional and likely lengthy studies that go beyond the scope of the perspective put forth in this study.
Table 1.
Number of tRNA-lookalikes of mitochondrial tRNA genes in different organisms in the animal kingdom
| Organism | Species name | Genome Assembly | Number of MT tRNAs | Number of exact mt tRNA-lookalikes | Number of all MT tRNA- lookalikes |
|---|---|---|---|---|---|
| Human | Homo sapiens | GRCh37 | 22 | 8 | 497 |
| Chimanzee | Pan troglodytes | CHIMP2.1 | 22 | 7 | 388 |
| Gorilla | Gorilla gorilla | gorGor2.1 | 22 | 0 | 255 |
| Macaca | Macaca mulatta | MMUL 1.0 | 22 | 5 | 419 |
| Squirrel Monkey | Saimiri boliviensis | saiBol1 | 22 | 2 | 272 |
| Mouse | Mus musculus | GRCm38.p2 | 22 | 12 | 53 |
| Rat | Rattus norvegicus | RGSC6.0 | 22 | 7 | 46 |
| Opossum | Monodelphis domestica | monDom5 | 21 | 39 | 543 |
| Drosophila | Drosophila melanogaster | BDGP6 | 21 | 0 | 1 |
| Worm | Caenorhabditis elegans | WS246 | 22 | 0 | 2 |
| Sponge | Amphimedon queenslandica | 1 | 17 | 1 | 16 |
Mitochondrial tRNA-Lookalikes as a Potential Source of Intact tRNAs
As with many nuclear tRNAs that have been shown to enter the mitochondrion in a wide range of taxa,35,36 the same could hold true, in principle, for tRNA-lookalikes. In such an event, tRNA-lookalikes would be contributing to intra-mitochondrial tRNA pools to various extents, and possibly in a cell-dependent manner. Such a hypothesis is supported by the observation that some organisms completely lack tRNAs in their mitochondrial genome and rely on nuclear production.35,36 Additionally, it has been reported that marsupial mitochondrial genomes lack a functional Lys tRNA gene37: instead, the tRNA that is charged with Lys has been shown to be of nuclear origin.37 Our search for mitochondrial tRNA-lookalikes in the opossum (M. domestica) revealed 543 such lookalikes in its nuclear genome. All 21 mitochondrial tRNAs are represented among these lookalikes. Of the 543 lookalikes 13 are nearly identical (82%–100%) copies of the non-functional mitochondrial Lys tRNA gene. This is a relatively low number compared to the average of 26 lookalikes per mitochondrial tRNA gene in this species.
On a related note, in humans, the mitochondrial LysRS has been reported to be released from mitochondria and be included in HIV viral particles.38 This suggests a subsequent use of the mitochondrial LysRS in the cytoplasm of newly infected cells and, thus, a concomitant potential interaction with transcribed nuclear lookalikes of the mitochondrial Lys tRNA. Whether the virion-packaged LysRS participates in such an interaction or whether such an interaction has any relevance in the HIV infection cycle39,40 are currently open questions.
The above observations make it conceivable that tRNA lookalikes, among other possible functional roles, serve as a repository of intact, “backup copies” in the organisms where they are found, especially since, as we have shown, the tRNA-lookalikes are depleted in mutations.7 These backup nuclear tRNA-lookalikes would thus be able to “annul” the impact of any mutations that might occur in their original mitochondrial counterparts, in a manner analogous to the marsupial tRNA(Lys). Under such a scenario, the tRNA-lookalikes would be transcribed in the nucleus and, after being imported in the mitochondrion, would participate in local translation thereby alleviating any functional shortcomings that might result from mutations acquired by their mitochondrial tRNA counterpart. Exploring this possibility further we note that in humans the only mitochondrial tRNA that does not have nuclear lookalikes is LeuTAA (even when searching at the most tolerant setting7). In this regard, and unlike its 21 counterparts, the mitochondrial LeuTAA would not have the ability to be “rescued” by a potential nuclear “backup” copy. It so happens, that mutations in the mitochondrial LeuTAA sequence have been associated with many diseases.4 The absence of a nuclear tRNA lookalike and the association of mitochondrial LeuTAA with disease make the “backup copy” scenario an intriguing possibility.
Another interesting mitochondrial anticodon is the TrpTCA. The UGA codon in the nucleus is a stop codon whereas in mitochondria it is translated to tryptophan. It is important to note that the mitochondrial tRNA gene for TrpTCA has 25 tRNA-lookalikes in nuclear chromosomes with 80.0% to 100.0% similarity to the mitochondrial tRNA gene.7 Many questions arise from this observation, as the genetic code could be affected.
Conclusion
New knowledge about tRNAs has been accumulating at a quick pace in recent years. These emerging findings are noteworthy and have been helping to expand the collective understanding of the spectrum of roles that tRNAs play. At the same time, the discovery of numerous conspicuously-placed human mitochondrial tRNA-lookalikes in the human nuclear genome open the door to many intriguing possibilities and novel functions, which, currently, are neither characterized nor understood. Lending support to these possibilities is the fact that additional searches revealed that non-human genomes include mitochondrial tRNA-lookalikes in their nuclear genomes. In particular, we found such lookalikes in other old world and new world monkeys and in a marsupial. Surprisingly, mitochondrial tRNA-lookalikes were found to be relatively absent from rodents, and absent from the fruit-fly, the worm, and a sponge. Their overwhelming and uneven representation among the nuclear tRNA-lookalikes has now cast a different light on mitochondrial tRNAs and raises the distinct possibility of a molecular cross-talk that transcends the boundaries of the mitochondrial and nuclear compartments.
Data Availability
The data that were used to generate the entries of Table 1 are available for download through our website at: https://cm.jefferson.edu/data-tools-downloads/trna-lookalikes-beyond-homo-sapiens/.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like the thank the anonymous reviewers for valuable comments that helped strengthen the presentation.
Funding
The work was supported in part by a W. M. Keck Foundation grant (IR), NIH grant GM106047 (YK), and by institutional funds.
References
- 1. Giege R. Toward a more complete view of tRNA biology. Nat Struct Mol Biol 2008; 15:1007-14; PMID:18836497; http://dx.doi.org/ 10.1038/nsmb.1498 [DOI] [PubMed] [Google Scholar]
- 2. Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/ 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Raina M, Ibba M. tRNAs as regulators of biological processes. Front Genet 2014; 5:171; PMID:24966867; http://dx.doi.org/ 10.3389/fgene.2014.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abbott JA, Francklyn CS, Robey-Bond SM. Transfer RNA and human disease. Front Genet 2014; 5:158; PMID:24917879; http://dx.doi.org/ 10.3389/fgene.2014.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shigematsu M, Honda S, Kirino Y. Tranfer RNA as a source of small functional RNA. J Mol Biol Mol Imaging 2014; 1(2):8. [PMC free article] [PubMed] [Google Scholar]
- 6. Sobala A, Hutvagner G. Transfer RNA-derived fragments: origins, processing, and functions. Wiley Interdiscip Rev RNA 2011; 2:853-62; PMID:21976287; http://dx.doi.org/ 10.1002/wrna.96 [DOI] [PubMed] [Google Scholar]
- 7. Telonis AG, Loher P, Kirino Y, Rigoutsos I. Nuclear and mitochondrial tRNA-lookalikes in the human genome. Front Genet 2014; 5:344; PMID:25339973; http://dx.doi.org/ 10.3389/fgene.2014.00344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 2009; 37:D93-7; PMID:18984615; http://dx.doi.org/ 10.1093/nar/gkn787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57-74; PMID:22955616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bensasson D, Feldman MW, Petrov DA. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J Mol Evol 2003; 57:343-54; PMID:14629044; http://dx.doi.org/ 10.1007/s00239-003-2485-7 [DOI] [PubMed] [Google Scholar]
- 11. Yao YG, Kong QP, Salas A, Bandelt HJ. Pseudomitochondrial genome haunts disease studies. J Med Genet 2008; 45:769-72; PMID:18611982; http://dx.doi.org/ 10.1136/jmg.2008.059782 [DOI] [PubMed] [Google Scholar]
- 12. Leister D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet 2005; 21:655-63; PMID:16216380 [DOI] [PubMed] [Google Scholar]
- 13. Ramos A, Barbena E, Mateiu L, del Mar Gonzalez M, Mairal Q, Lima M, Montiel R, Aluja MP, Santos C. Nuclear insertions of mitochondrial origin: Database updating and usefulness in cancer studies. Mitochondrion 2011; 11:946-53; PMID:21907832; http://dx.doi.org/ 10.1016/j.mito.2011.08.009 [DOI] [PubMed] [Google Scholar]
- 14. Tsuji J, Frith MC, Tomii K, Horton P. Mammalian NUMT insertion is non-random. Nucleic Acids Res 2012; 40:9073-88; PMID:22761406; http://dx.doi.org/ 10.1093/nar/gks424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat Rev Genet 2008; 9:383-95; PMID:18368053; http://dx.doi.org/ 10.1038/nrg2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981; 290:470-4; PMID:7219536; http://dx.doi.org/ 10.1038/290470a0 [DOI] [PubMed] [Google Scholar]
- 17. Orioli A, Pascali C, Pagano A, Teichmann M, Dieci G. RNA polymerase III transcription control elements: themes and variations. Gene 2012; 493:185-94; PMID:21712079; http://dx.doi.org/ 10.1016/j.gene.2011.06.015 [DOI] [PubMed] [Google Scholar]
- 18. Watanabe Y, Suematsu T, Ohtsuki T. Losing the stem-loop structure from metazoan mitochondrial tRNAs and co-evolution of interacting factors. Front Genet 2014; 5:109; PMID:24822055; http://dx.doi.org/ 10.3389/fgene.2014.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. . Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Sstruct Mol Biol 2010; 17:620-8; PMID:20418882; http://dx.doi.org/ 10.1038/nsmb.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Letters 2010; 584:287-96; PMID:19931535; http://dx.doi.org/ 10.1016/j.febslet.2009.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rossmanith W. Of P and Z: mitochondrial tRNA processing enzymes. Biochimi Biophy Acta 2012; 1819:1017-26; PMID:22137969; http://dx.doi.org/ 10.1016/j.bbagrm.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Craig LC, Wang LP, Lee MM, Pirtle IL, Pirtle RM. A human tRNA gene cluster encoding the major and minor valine tRNAs and a lysine tRNA. DNA 1989; 8:457-71; PMID:2766931; http://dx.doi.org/ 10.1089/dna.1.1989.8.457 [DOI] [PubMed] [Google Scholar]
- 23. Mungall AJ, Palmer SA, Sims SK, Edwards CA, Ashurst JL, Wilming L, Jones MC, Horton R, Hunt SE, Scott CE, et al. . The DNA sequence and analysis of human chromosome 6. Nature 2003; 425:805-11; PMID:14574404; http://dx.doi.org/ 10.1038/nature02055 [DOI] [PubMed] [Google Scholar]
- 24. Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 2008; 91:243-8; PMID:18082363 [DOI] [PubMed] [Google Scholar]
- 25. Van Bortle K, Corces VG. tDNA insulators and the emerging role of TFIIIC in genome organization. Transcription 2012; 3:277-84; PMID:22889843; http://dx.doi.org/ 10.4161/trns.21579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Donze D. Extra-transcriptional functions of RNA Polymerase III complexes: TFIIIC as a potential global chromatin bookmark. Gene 2012; 493:169-75; PMID:21986035; http://dx.doi.org/ 10.1016/j.gene.2011.09.018 [DOI] [PubMed] [Google Scholar]
- 27. Suzuki T, Nagao A, Suzuki T. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 2011; 45:299-329; PMID:21910628; http://dx.doi.org/ 10.1146/annurev-genet-110410-132531 [DOI] [PubMed] [Google Scholar]
- 28. Betat H, Rammelt C, Morl M. tRNA nucleotidyltransferases: ancient catalysts with an unusual mechanism of polymerization. Cell Mol Life Sci 2010; 67:1447-63; PMID:20155482; http://dx.doi.org/ 10.1007/s00018-010-0271-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Suzuki T, Suzuki T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res 2014; 42:7346-57; PMID:24831542; http://dx.doi.org/ 10.1093/nar/gku390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA biology 2013; 10:1798-806; PMID:24351723; http://dx.doi.org/ 10.4161/rna.27177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. Rna 2008; 14:2095-103; PMID:18719243; http://dx.doi.org/ 10.1261/rna.1232808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown WM, George M, Jr., Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci U S A 1979; 76:1967-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Helm M, Brule H, Friede D, Giege R, Putz D, Florentz C. Search for characteristic structural features of mammalian mitochondrial tRNAs. Rna 2000; 6:1356-79; PMID:11073213; http://dx.doi.org/ 10.1017/S1355838200001047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ohtsuki T, Watanabe Y. T-armless tRNAs and elongated elongation factor Tu. IUBMB life 2007; 59:68-75; PMID:17454297; http://dx.doi.org/ 10.1080/15216540701218722 [DOI] [PubMed] [Google Scholar]
- 35. Rubio MA, Hopper AK. Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip Reviews RNA 2011; 2:802-17; PMID:21976284; http://dx.doi.org/ 10.1002/wrna.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Review Biochem 2011; 80:1033-53; PMID:21417719; http://dx.doi.org/ 10.1146/annurev-biochem-060109-092838 [DOI] [PubMed] [Google Scholar]
- 37. Dorner M, Altmann M, Paabo S, Morl M. Evidence for import of a lysyl-tRNA into marsupial mitochondria. Mol Biol Cell 2001; 12:2688-98; PMID:11553708; http://dx.doi.org/ 10.1091/mbc.12.9.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kaminska M, Shalak V, Francin M, Mirande M. Viral hijacking of mitochondrial lysyl-tRNA synthetase. J Virol 2007; 81:68-73; PMID:17050605; http://dx.doi.org/ 10.1128/JVI.01267-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kleiman L, Jones CP, Musier-Forsyth K. Formation of the tRNALys packaging complex in HIV-1. FEBS Letters 2010; 584:359-65; PMID:19914238; http://dx.doi.org/ 10.1016/j.febslet.2009.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pavon-Eternod M, Wei M, Pan T, Kleiman L. Profiling non-lysyl tRNAs in HIV-1. Rna 2010; 16:267-73; PMID:20007329; http://dx.doi.org/ 10.1261/rna.1928110 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that were used to generate the entries of Table 1 are available for download through our website at: https://cm.jefferson.edu/data-tools-downloads/trna-lookalikes-beyond-homo-sapiens/.


