Abstract
Understanding the multifaceted nature of microRNA (miRNA) function in mammalian cells is still a challenge. Commonly accepted principles of cooperativity and multiplicity of miRNA function imply that individual mRNAs can be targeted by several miRNAs whereas a single miRNA may concomitantly regulate a subset of different genes. However, there is a paucity of information whether multiple miRNAs regulate critical cellular events and thereby acting redundantly. To gain insight into this notion, we conducted an unbiased high-content miRNA screen by individually introducing 1139 miRNA mimics into Chinese hamster ovary (CHO) cells. We discovered that 66% of all miRNAs significantly impacted on proliferation, protein expression, apoptosis and necrosis. In summary, we provide evidence for a substantial degree of redundancy among miRNAs to maintain cellular homeostasis.
Keywords: chinese hamster ovary, microRNA, pathway regulation, redundancy, screen
Abbreviations
- 3'UTR
3'-untranslated region
- AF647
AlexaFluor→647
- bp
base pair
- cDNA
cDNA
- CH
Chinese hamster
- CHO
Chinese hamster ovary
- cgr
Cricetulus griseus
- CO2
carbon dioxide
- miR
mature microRNA
- MIR
precursor microRNA
- miRNA
microRNA, mRNA, mRNA, nc, non-coding
- PI
propidium iodide,
- pre-miR
precursor microRNA
- puro
puromycin
- qRT-PCR
quantitative reverse-transcriptase real-time PCR
- RNAi
RNA interference
- rpm
rounds per minute
- SEAP
secreted alkaline phosphatase
- siRNA
small-interfering RNA
Introduction
MicroRNAs are endogenous non-protein coding (nc) small RNAs that modulate global gene expression in eukaryotic cells at the post-transcriptional level and are highly conserved across species.1 miRNAs are critically involved in virtually all cellular processes and aberrant miRNA expression is associated with a variety of diseases.2 Thus, miRNAs have gained much attention both as therapeutic target and biomarker for medical applications, as well as in the field of biotechnology as powerful cell engineering tools.3,4
In recent years the question of how miRNAs exert their multiple cellular functions has gained increasing attention. It has become clear, that miRNAs employ certain principles or modes of action which are characterized by the following principles: (i) cooperativity in miRNA function describes the targeting of a single mRNA by several miRNAs;5,6 This cooperative binding has been shown to be important for miRNA-mediated function and to facilitate more effective target repression.7 (ii) Multiplicity of miRNA function takes into account that a single miRNA can target more than one gene, due to the short and imperfect binding mode of miRNAs to their target mRNAs.6 The interplay between cooperativity and multiplicity enables a single miRNA to fine-tune protein biosynthesis from thousands of genes and to form complex regulative networks within the cell.8,9 In the past few years the number of known miRNAs placed in the miRBase repository has risen to a total number of >2800 in humans.10 Considering these vast numbers of miRNAs in the light of their cooperative and multiplying effects on target genes, the interconnected and multilayered network character of their mechanism of action on cellular pathways becomes apparent. Current concepts investigate the regulatory action mostly centered on a single miRNA and its cooperative and multiplying effects on different but distinct cellular scenarios. However, the question whether multiple miRNAs regulate crucial cellular pathways and their mode of action has not been adequately addressed. Regulation of single or multiple pathways by multiple miRNAs, however, may add an additional layer of complexity to the network operation and introduce a possible redundancy of miRNA function.
The present study aimed at assessing the impact of all known cellular miRNAs on cell proliferation, protein expression/secretion, apoptotic and necrotic cell death. We recently conducted an unbiased functional miRNA screen in a recombinant CHO cell line assessing the impact of 1139 miRNAs on crucial cellular functions.11 In this report, we demonstrate that each of these vital cellular pathways could be controlled by an unexpectedly large number of miRNAs, which may indicate a redundancy in pathway regulation guaranteeing survival of the cell. Since the pathways are to some degree interrelated, we found that subsets of miRNAs have both specific as well as overlapping functions. Our results show that >750 miRNAs (66% of library) had an effect on essential cellular pathways such as proliferation, protein expression, apoptosis and necrosis, which supports the idea that many, if not all miRNAs are critically involved to a certain extent in most cellular pathways keeping the cell in homeostasis.
Materials and Methods
Cell culture and transfection
CHO-SEAP suspension cells stably expressing the human secreted alkaline phosphatase (SEAP),12 were cultured in ProCHOTM5 culture medium (Lonza, Vervier, Belgium) with 4 mM L-Glutamine (Lonza) and 0.1% anti-clumping agent (Life Technologies, Carlsbad, CA, USA). Cells were maintained at 37°C, 5% CO2 and 85% humidity with agitation at 140 rpm in an orbital shaker incubator (Kuehner, Birsfelden, Switzerland). Transfections of miRNA mimics and small interfering RNAs (siRNAs) were carried out essentially as described previously.11 An entire murine miRNA mimics library (miRBase release 18) comprising 1139 different miRNAs was used for the transient miRNA screening (Qiagen, Hilden, Germany) (see also Supplementary Table S1). An Alexa Fluor® 647-labeled non-targeting siRNA, a non-labeled, scrambled siRNA (NT siRNA), an anti-SEAP siRNA (siSEAP) (all Qiagen) and a cell death control siRNA (siDeath) (kindly provided by Dr. Eric Lader, Qiagen) served as functional transfection controls.
Flow-cytometry based high-content cell analysis
Transfected cells were analyzed for cell concentration, viability, necrosis, apoptosis and transfection efficiency by quantitative high-throughput flow cytometry employing a MACSQuant® Analyzer (Miltenyi Biotech, Bergisch-Gladbach, Germany). Cell viability was determined by means of Calcein-Violet450-AM (eBioscience, Frankfurt, Germany) staining and necrosis/late apoptosis was detected by propidium iodide (PtdIns) (Roth, Karlsruhe, Germany) exclusion. miRNA transfection efficiency was assessed by analyzing cells for Alexa Fluor® 647 fluorescence. Analysis of apoptosis was performed by means of Nicoletti staining13 and analyzed by flow cytometry. Transfected cells were incubated for 30 min at 4°C in a modified Nicoletti staining solution composed of 0.1% sodium citrate (Roth), 0.05% Triton X-100 (Roth), 10 µg/mL PI (Roth) and 1 U/µL RNase A (Thermo Fisher Scientific, Schwerte, Germany) in phosphate buffered saline.
Analysis of protein expression
Differing SEAP concentrations in the culture supernatant of CHO-SEAP cells were indicative of changes in protein expression/secretion. Changes in SEAP protein levels were determined using a SEAP reporter gene assay (Roche Diagnostics, Mannheim, Germany) according to the vendor's protocol and measured on a SpectraMax® M5e microplate reader (Molecular Devices, Sunnyvale, CA, USA). A standard curve from purified SEAP was used to calculate SEAP concentrations in the culture supernatant.
Bioinformatics and statistics
Bioinformatics-based examination of putative target genes for miRNAs exhibiting related functions was performed by a comprehensive in silico target prediction analysis using 5 different miRNA target prediction algorithms (miRanda, TargetScan, DIANA-mT, miRDB and miRWalk). Potential target genes that were commonly predicted by at least 3 of the mentioned algorithms were further considered for gene clustering analysis using the PANTHER classification system (www.pantherdb.org),14 followed by a comparative analysis.
For statistical analysis of the primary screening results, biological triplicate data points of each effector miRNA were normalized to the mean value of the respective on-plate non-targeting control miRNA, to allow for inter-plate comparisons between different 96-well screen plates and to identify impactful miRNAs. Significant miRNA-mediated changes on cellular readout parameters were determined by applying a one-way analysis of variances (ANOVA) combined with a Dunnett's multiple comparison post-test (against the non-targeting control miRNA; * p<0.05, ** p<0.01, *** p<0.001).
A detailed expression analysis of impactful miRNAs in CHO cells was carried out by first comparing all miRNA hits from the high-content screening with miRNAs already annotated for Cricetulus griseus (C. griseus) in the publically available miRBase repository (release 21). In a next step, miRNAs which were not annotated yet for C. griseus, but have already been detected in different small RNA sequencing approaches of CHO cell lines, were also considered to be expressed. Finally, all remaining mouse miRNA sequences were aligned to the recently published Chinese hamster (CH) genome to examine if the miRNA hairpin is at least genomically present.
Results and Discussion
miRNA-mediated impact on mammalian cellular behavior may be studied by transient gain-of-function experiments in which miRNA mimics are introduced into cultured cells resulting in an elevation of the cytosolic abundance of a given miRNA. CHO cells are frequently applied as a cell system of choice, e.g. for studying cytogenetics and DNA repair mechanisms but also as host cells for the production of therapeutic proteins.15,16 In order to investigate which miRNAs significantly affect fundamental cellular functions such as proliferation, apoptosis, necrosis or protein expression/secretion, we have separately introduced an entire murine miRNA mimics library into a recombinant CHO cell line constitutively expressing SEAP as model protein. Three days following transfection, cells were subjected to a high-content analysis to determine miRNA-induced changes in crucial cellular processes. Functional transfection controls reproducibly confirmed highly efficient and functional intracellular transfer of the miRNAs (Fig. 1A, B). Functionally relevant miRNAs were ascertained as screening hits if they were found to induce a statistically significant change (one-way analysis of variances (ANOVA); p<0.05) in at least one of the observed phenotypes as compared to cells transfected with a scrambled siRNA.
Figure 1:
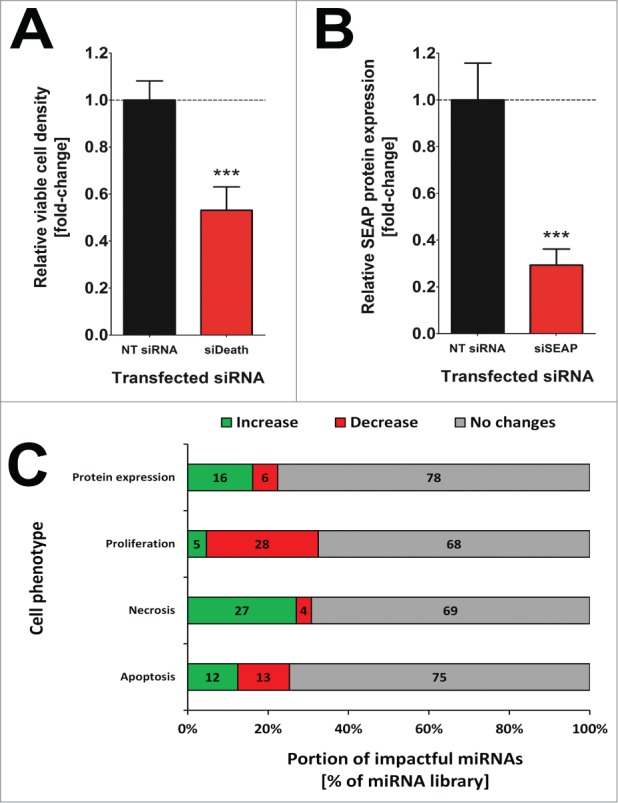
Confirmation of functional small ncRNA transfection on all miRNA screen plates (n=73) in the primary miRNA screen. Normalized mean values of (A) viable cell density and (B) SEAP protein expression level of siDeath and siSEAP siRNA transfected CHO-SEAP cells, respectively. Sample values were normalized to the respective NT siRNA transfected on-plate control cells. Data are presented as mean ± s.e.m. and for statistical analysis an unpaired, 2-tailed t-test was applied (*** p <0.001). (C) Overview on how many miRNAs significantly affected crucial cellular processes such as protein expression, cell proliferation, necrosis and apoptosis in CHO-SEAP cells. Data are presented as percentages of the entire miRNA library comprising 1139 murine miRNA mimics and the amount of miRNAs increasing (green) or decreasing (red) observed phenotypes are indicated. miRNAs were considered as impactful if the change in phenotype was statistically significant against the negative control cells.
miRNA-mediated effects on protein expression
Strikingly, we found that an unexpectedly large proportion of miRNAs significantly influenced each of the investigated cellular parameters (Fig. 1C). Notably, miRNAs that influenced protein expression levels due to acute apoptotic or necrotic cell death were excluded from the above presented numbers. Therefore, 255 miRNAs (22.4%) of all miRNAs influenced overall protein expression, whereof 184 miRNAs (16.2%) increased protein expression while 71 miRNAs (6.2%) reduced functional SEAP protein concentrations in the culture supernatant, suggesting a decreased protein expression, secretion or improper folding. The reasons behind these changes in protein concentration in the culture supernatant might be diverse and certainly rely on the target genes (and pathways), which were affected. Basically, observed alterations might be due to an involvement in a variety of cellular processes including gene transcription, protein translation/modification, unfolded protein response (UPR), ubiquitination/proteasomal degradation, metabolism, intracellular trafficking, cytoskeleton dynamics or secretion/exocytosis.
Modulating the expression of transcription factors such as the X-box binding protein (XBP1) might be causative for observed alterations in SEAP concentrations since overexpression of XBP1 has previously been shown to support protein expression and enhance exocytosis in mammalian cells.17 Thus, miRNA-mediated post-transcriptional silencing of transcription factors like XBP1 might be responsible for reduced SEAP protein levels following miRNA transfection in the presented screen. To support this hypothesis, XBP1 has been demonstrated to be a direct target of miR-214–3p in human hepatocellular carcinoma cells,18 and intriguingly, our data suggest that miR-214–3p significantly decreased SEAP protein expression in CHO cells (Fig. 2A).
Figure 2:
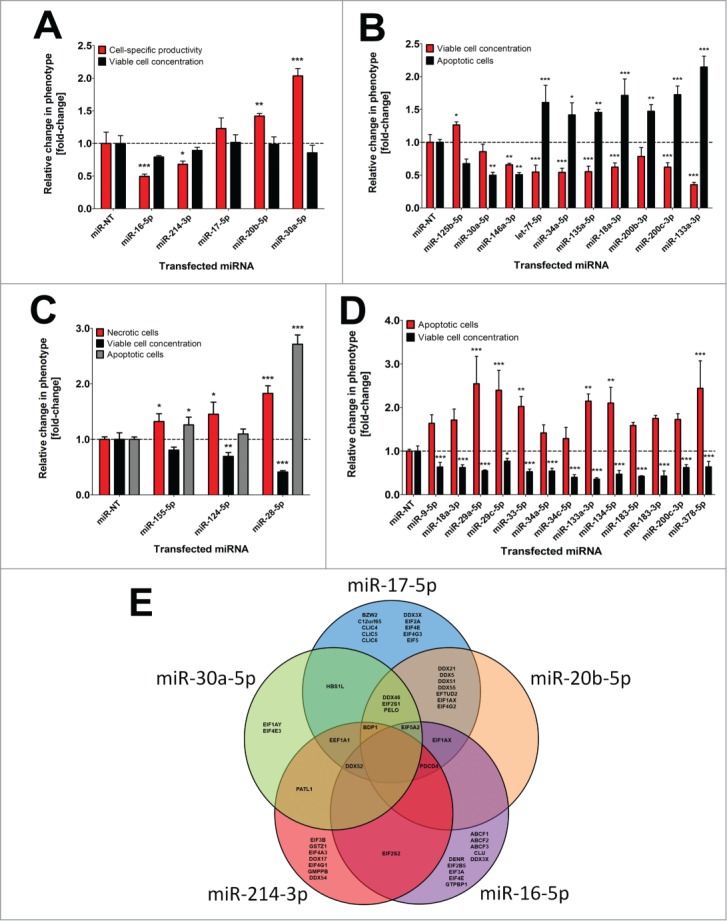
Influence of single miRNAs on different cell phenotypes in CHO-SEAP cells. (A) Effects of individual pro- and anti-productive miRNAs on SEAP protein level (black) and viable cell concentration (gray) 72 h following transfection in the primary miRNA screen. (B) Effects of individual pro- and anti-proliferative miRNAs on viable cell concentration (black) and apoptosis (gray) 72 h following transfection in the primary miRNA screen. (C) Effects of individual pro-necrotic miRNAs on necrotic (black) and apoptotic (white) cell death as well as viable cell concentration (gray) 72 h following transfection in the primary miRNA screen. (D) Effects of individual pro-apoptotic miRNAs on apoptosis (black) and viable cell concentration (gray) 72 h following transfection in the primary miRNA screen. Triplicate sample values of each miRNA were normalized to the mean value of the miR-NT transfected control cells. Error bars represent the standard deviation (SD) of 3 independent transfections. Statistics: One-way ANOVA (* p <0.05; ** p<0.01; *** p<0.001). (E) VENN diagram of bioinformatically predicted target genes for miR-17–5p, miR-20b-5p, miR-30a-5p, miR-214–3p and miR-16–5p in humans involved in translational control. The five different prediction algorithms miRanda, TargetScan, DIANA-mT, miRDB or miRWalk were used and overlaps indicate shared target genes.
Hammond and Lee reported that the siRNA-mediated knock-down of cofilin (CFL), an actin-modulating protein involved in cytoskeleton dynamics, increased protein expression in CHO cells.19 Down-regulation of CFL diminishes actin stress fibers and a destabilized actin cytoskeleton has been shown to enhance heterologous protein expression.20 CFL2 is assumed to be regulated by miR-17–5p and miR-20b-5p in CHO cells.21 Furthermore, another interesting actin binding protein, twinfilin-1 (TWF1), has been shown to be targeted by miR-30a-5p.22 Notably, we observed that transient introduction miR-17–5p, miR-20b-5p and miR-30a-5p into CHO-SEAP cells resulted in markedly increased SEAP expression levels in the miRNA screen (Fig. 2A), suggesting that CFL and TWF might presumably be a target of those miRNAs in CHO cells, too.
Other conceivable targets might be for instance the ubiquitin pathway which represents a key interface to be modulated by miRNAs in order to control protein abundance. In this conjunction, miR-16 has been shown to regulate several effector genes of the ubiquitin pathway (e.g., UBE4A, UBE2V1 and UBE2S) and thus seems to be involved in protein synthesis control in humans.22 Ectopic overexpression of miR-16–5p severely decreased SEAP expression in the presented miRNA screen (Fig. 2A) supporting the idea that miR-16 might be an important regulator of protein expression in mammalian cells.
In this conjunction, it is also conceivable that miRNA-mediated downregulation of particular genes involved in ubiquitination might result in elevated levels of specific proteins. Notably, stringent bioinformatics target prediction for miR-17, miR-20b, miR-30a, miR-214 and miR-16 yielded a reasonable high number of genes exhibiting translational regulatory activity (Fig. 2E). Common putative target genes specific for pro-productive miR-17, miR-20b and miR-30a comprised DDX46, EIF2S1 and PELO. These genes were jointly predicted by at least 4 of the 5 different target recognition algorithms (miRanda, TargetScan, DIANA-mT, miRDB or miRWalk) which we applied for analysis. Furthermore, HBS1L is supposed to be a shared target of miR-17 and miR-30a according to our target prediction (Fig. 2E). Excessive translation of particular mRNAs in mammalian cells, e.g. during production of therapeutic proteins, might trigger listerin-dependent nascent protein ubiquitination to keep protein homeostasis.23 This process, however, relies on ribosome subunit dissociation which is directed by the ribosome splitting factors PELO/HBS1/ABCE1.23 Disrupting ribosome splitting by post-transcriptional down-regulation of one of these critical factors might prevent this special form of ubiquitination and could thus be responsible for the observed increase in SEAP abundance following miR-17, miR-20b or miR-30a introduction in CHO cells. These examples, together with the many novel miRNAs we have identified to affect SEAP protein levels, strongly argument for a highly redundant regulation of the underlying processes in mammals.
Control of cell proliferation by miRNAs
Our data suggest that a total of 370 miRNAs (33.4%) influence cellular proliferation of CHOs, whereof 53 miRNAs (4.7%) significantly increased while 317 (28.7%) decreased viable cell numbers. Influencing the expression of genes involved in cell cycle regulation such as checkpoint kinases entails severe changes in cell proliferation. Oncogenes require circumventing cell cycle arrest which could occur once a host cell senses intra- or extracellular abnormalities such as DNA damage or infection. Aberrant expression of cell cycle checkpoint or loss-of-function of tumor suppressor genes thus ultimately leads to cancer and uncontrolled cell growth.
One of the most prominent tumor suppressors is p53, a transcription factor exhibiting a variety of crucial functions in cell cycle control and apoptosis.24 Aberrant p53 function has been demonstrated to be a hallmark of cancer and post-transcriptional downregulation of tumor suppressors might be responsible for miRNA-mediated cell growth.25 A number of previous studies suggested that miR-125b-5p and miR-30a-5p directly target p53, thereby averting apoptosis and promoting proliferation.26,27 Furthermore, Le and co-workers discovered that miR-125b-5p not only regulates p53 but also affects the expression of a number of other effector genes within the p53 network, which are highly conserved across species.28 In the presented work, inhibition of apoptosis and partially the acceleration of cell proliferation by miR-125b and miR-30a could be confirmed in CHO cells (Fig. 2B). In addition, stable overexpression of pre-miR-30a in CHO cells was shown to enhance cell proliferation.11
Conversely, oncogenes such as c-Myc, KRAS or growth factor receptors, whose overexpression is directly linked to uncontrolled cell growth and tumorigenesis, might have been targeted by miRNAs leading to diminished cell growth. In papillary thyroid cancer cells, let-7f has been detected to be down-regulated, whereas overexpression of let-7 was shown to inhibit cell growth, by targeting c-Myc.29 Furthermore, miR-34a and miR-135a were demonstrated to target c-Myc in renal cell carcinoma leading to growth arrest and apoptosis.30,31 Tumor suppressor function of miR-18a-3p was reported by Tsang et al., who suggested that the oncogene KRAS is directly regulated by miR-18a-3p in different human carcinomas.32 Finally, there is increasing evidence that the down-regulation or loss-of-function of the microRNAs miR-146a, miR-200b/c and miR-133a critically promotes cell growth and metastasis of various cancers. The common function of these miRNAs relies on the direct target interaction with either the epidermal (EGFR) or vascular-endothelial growth factor receptor (VEGFR).33,34 Collectively, these findings are in keeping with our observations that introduction of let-7f-5p, miR-34a-5p, miR-135a-5p, miR-18a-3p, miR-146a-3p, miR-200b-3p, miR-200c-3p and miR-133a-3p decreased CHO proliferation and induced cell death (Fig. 2B). Considering that we were able to substantially expand the known pool of anti-proliferative miRNAs by our comprehensive screening approach, results from this study might be a good resource to identify novel miR-based therapeutic targets for manipulating tumor growth and metastasis.
miRNA-mediated modulation of necrosis
Interestingly, cell proliferation data was inversely correlated with the number of miRNAs influencing necrotic cell death. Hence, a total of 351 miRNAs (31%) influenced necrosis, whereof 3.7% (42) of the miRNAs decreased whereas 308 miRNAs (27.0%) increased susceptibility for necrotic cell death. Necrotic cells are morphologically characterized by an enhanced cell volume, organelle swelling, lack of intranucleosomal DNA fragmentation and plasma membrane rupture.35 Necrosis usually occurs in response to physico-chemical stress, e.g., by destabilization of the cytoskeleton or loss of membrane integrity, by increased generation of reactive oxygen species or by the disruption of cellular calcium homeostasis.36 A detailed example demonstrating our flow cytometry based cellular analysis of necrosis and apoptosis is illustrated in Supplementary Figure S1, showing specific effects of the pro-necrotic miRNA miR-3097–5p and the pro-apoptotic miRNA miR-466k on CHO cells. Of note, there are only very few reports connecting miRNA function to necrosis. The reason for this might be that for many years necrosis was considered to be caused solely by exterior stimulations. However, in recent years programmed necrosis, also called necroptosis, has gained growing attention in cancer therapy as for its potential to circumvent apoptosis resistance mechanisms, which have been developed by a variety of malignant tissues during treatment.37 Key mediators of necroptosis are the RIPK1/3 genes which can induce necrosis upon receptor stimulation.38 In human cardiomyocyte progenitor cells, miR-155–5p has been shown to prevent necrosis via targeting RIP1.39 In contrast to that, miR-155–5p increased the number of both necrotic and apoptotic cells and diminished proliferation, pointing toward a different function of miR-155 in CHO cells (Fig. 2C). Furthermore, Mucaj et al. recently reported that miR-124 expression is downregulated in glioblastomas and enforced miR-124 expression stops survival and induces cell death of tumor cells.40 In colorectal cancers, miR-124–5p has been shown to regulate expression of the SMC4 gene and reduced miR-124–5p levels are correlated with poor patient prognosis.41 This pro-necrotic effect of miR-124 could also be validated by our screening approach and introduction of miR-124–5p clearly induced necrosis and decreased viable cell numbers (Fig. 2C). Cell death induced by miR-124 in glioblastomas was demonstrated to be caused by the direct downregulation of TEAD1, MAPK14 as well as SERP1.40 However, it still remains to be elucidated if miR-124-mediated induction of necroptosis in CHO cells is also caused by silencing this set of genes.
Finally, miR-28–5p was found as one of the most potent pro-necrotic and pro-apoptotic miRNAs in CHO cells by substantially increasing both parameters compared to control cells (Fig. 2C). miR-28–5p has been shown to directly target MAPK1.42 Inhibition of the MAPK1 pathway is associated with apoptosis and impaired cell proliferation.43 In accordance with our data, miR-28 expression has been demonstrated to be down-regulated in a variety of cancer cells and enforced expression of miR-28–5p blocked proliferation and induced apoptosis in various cancer cells.44 Furthermore, in mouse embryonic fibroblasts overexpression of miR-28 induced apoptosis and inhibited proliferation by post-transcriptional repression of SRSF1.45 Besides its function as a tumor suppressor, miR-28 appears to induce different types of cell death (apoptosis/necroptosis) in a cell type and species specific manner.
miRNA-mediated effects on apoptosis
288 miRNAs (25.3%) influenced apoptosis and a remarkably balanced proportion of 142 (12.5%) and 146 (12.8%) miRNAs increased or decreased apoptosis rate, respectively, indicating that the genes controlling apoptosis appear to be tightly regulated by miRNAs. Evasion of apoptosis has been shown to be a major cause of cancer, thus deregulated miRNA expression can be either oncogenic or tumor suppressive depending on the miRNA function.46 One of the most promising miRNAs for clinical treatment of a variety of cancers is the miR-34 family, consisting of miR-34a, -b and -c since these pro-apoptotic miRNAs have been shown to be downregulated in many tumors.47 Among the 142 pro-apoptotic miRNAs identified in CHO cells, we found functionally homolog candidates corresponding to human pro-apoptotic miRNAs such as miR-9, miR-18a, miR-29a, miR-29c, miR-33, miR-34a, miR-34c, miR-133a, miR-134, miR-183, miR-200c and miR-378 (Fig. 2D). These findings obtained using CHO cells confirm previous observation in other mammalian cells and suggest a high degree of evolutionary conservation for these miRNA-regulated pathways.
However, it has not yet been clarified if the miRNA-mediated phenotypic changes in CHO are directly associated with a down-regulation of the above mentioned genes/pathways similar to human and mouse. Although these genes are prominent examples and key players in a plethora of diseases, there are many more cellular pathways and target genes which may trigger changes in cellular behavior.
Vast numbers of miRNAs regulate crucial cellular pathways
Given the fact that each of the examined cellular pathways was regulated by 20–30% of the whole miRNA library, we became interested in how many impactful miRNAs have the capacity to significantly affect at least one of the observed phenotypes. Interestingly, 66% of the entire miRNA library (754 miRNAs) significantly impacted on protein expression/secretion, proliferation, apoptosis or necrosis, which was far above our initial assumptions (Fig. 4A). Such a finding might not be surprising for well-studied cellular processes such as apoptosis, however, considering the comparably high numbers of impactful miRNAs in other processes, the outcome of our studies clearly provide deeper insights into the multifaceted and redundant nature of miRNA-mediated pathway regulation. In addition, although we investigated merely 4 different cellular phenotypes, we determined significant changes for >750 miRNAs suggesting that the selected cellular processes are tightly regulated by vast numbers of different miRNAs.
Figure 4:
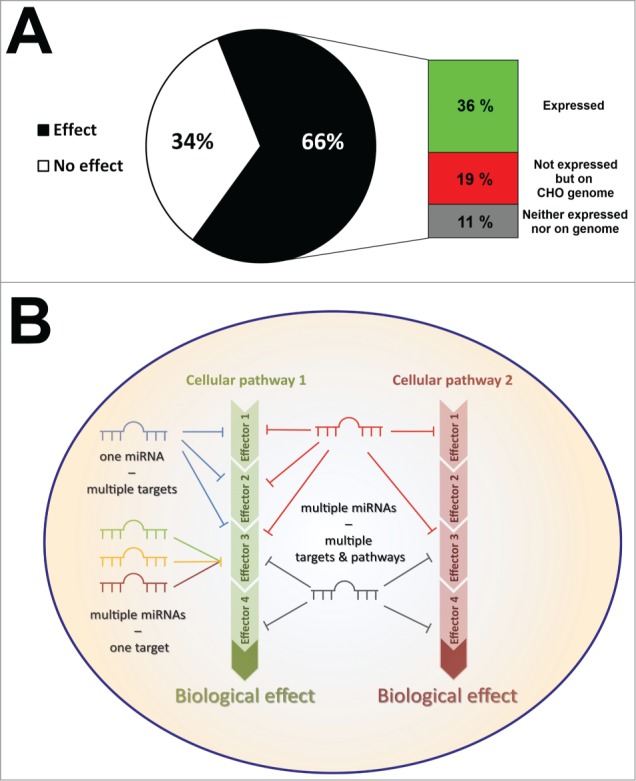
(A) Impactful murine miRNAs from the primary miRNA screen were found to be either expressed in CHO cells (green), not expressed but present as mature miRNA sequence on the CHO genome (red), or did not have any equivalent in CHO cells (gray). (B) Schematic overview on the main principles of miRNA-mediated regulation of cellular pathways in mammals. A single miRNA can regulate various effector genes of a particular pathway since miRNAs are capable of post-transcriptionally regulating hundreds of different mRNAs due to the imperfect nature of target recognition. This principle is known as multiplicity of miRNA function. Furthermore, as most mRNAs have been found to possess multiple binding sites for miRNAs, individual mRNAs can be regulated by different miRNAs in parallel which known as cooperative miRNA function. Finally, vast miRNAs are capable of regulating multiple cellular processes by targeting various effector genes of different pathways thereby ensuring cellular homeostasis. This phenomenon is called redundancy in miRNA function.
Generally, each cell type of an organism usually exhibits a particular miRNA expression pattern depending on the distinct function of a cell or tissue. In the latest miRBase version (release 21), 1982 murine mature miRNAs have been annotated. RNA deep sequencing data of different CHO lines indicated that approximately 400 miRNAs are expressed in CHO cells.48 Although Chinese hamsters possess a considerably lower number of chromosomes (n=22) compared to mice (n=40), the genomic information is highly overlapping.49 Thus, the presence of only 400 different miRNAs appears to be underestimated and recently reported computational re-analysis of CHO genomic data identified hundreds of additional pre-miR sequences. This suggests that other CH tissue types might take advantage of different miRNA pools to selectively control their transcriptome. However, this postulate does not inevitably mean that those miRNAs not expressed in CHO cells, are incapable of inducing any phenotypic changes once they are introduced, as there might be target transcripts still available for these miRs. Among all impactful miRNAs identified in the presented screening, 55% (36% of the entire library) are assumed to be expressed in CHO cells while 29% (19%) and 16% (11%) are only present on the CH genome or even completely absent, respectively (Fig. 4A). This might suggest that the CH might have lost some miRNA genes but still expresses mRNAs harboring conserved miRNA binding sites in their 3'UTRs which could be targeted by murine miRNAs.
By detailed data analysis it became apparent that vast numbers of miRNAs influence multiple cellular processes (Fig. 3A). More specifically, 471 miRNAs were found to affect more than one of the examined cellular phenotypes in CHO cells (Supplementary Figure S2). In addition, we provide detailed results of some multifunctional miRNAs influencing 2 or 3 obviously non-interrelated cellular states. For example, the miRNAs miR-5100, miR-543–3p, miR-30a-5p, miR-30d-5p and miR-20b-3p exhibited anti-apoptotic effects but were also found to influence SEAP protein expression or secretion (Fig. 3B). Interestingly, among anti-apoptotic miRNAs were also candidates that additionally decreased cell proliferation, e.g. miR-24–2–5p, miR-146a-3p or miR-362–3p (Fig. 3C). The combination of anti-apoptosis and anti-proliferation appears to be contradictory since inhibition of apoptosis is often in conjunction with increased survival. Decreasing the susceptibility to apoptosis is further not directly linked to necrotic cell death. Nonetheless, we could identify miRNAs decreasing both necrosis and apoptosis in parallel as depicted for miR-194–1–3p, miR-200c-5p, miR-5626–5p and miR-33–3p (Fig. 3D). These additional effects all seem to be contrary to an anti-apoptotic function and are likely not a consequence thereof. Furthermore, our high-content screening approach also enabled the identification of miRNAs influencing more than two different cell functions. For instance, miR-501–3p, miR-5615–3p, miR-493–3p, miR-1960 and miR-196a-1–3p decreased apoptotic cell death and cell proliferation, and surprisingly increased cell-specific SEAP expression (Fig. 3E).
Figure 3:
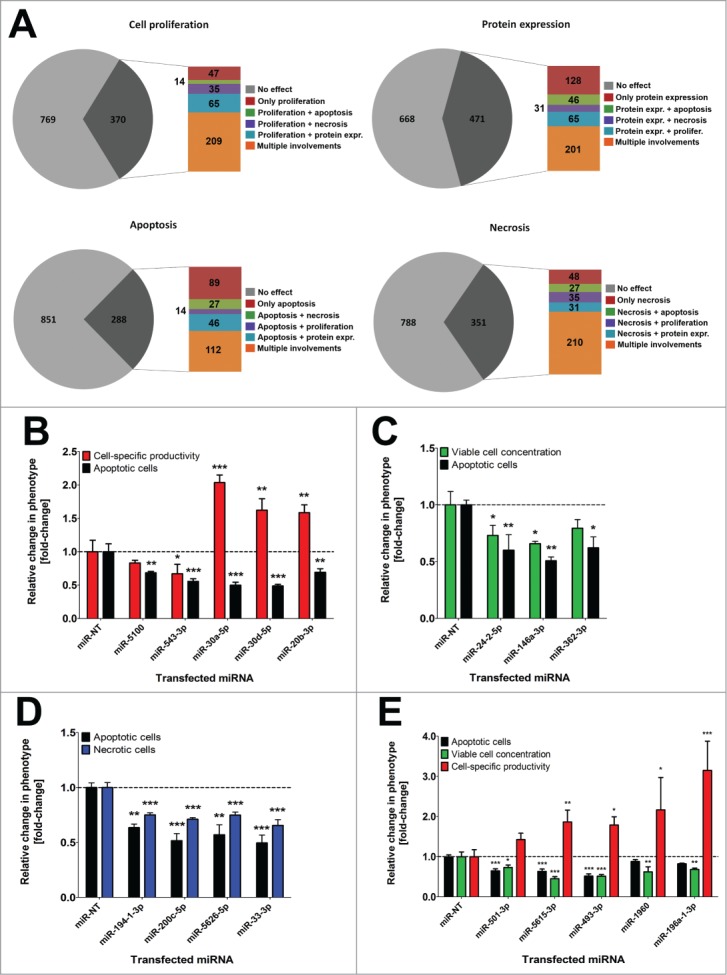
Vast numbers of miRNAs redundantly regulate multiple cellular pathways in CHO-SEAP cells. (A) Phenotypic distribution of all impactful miRNAs significantly affecting cell proliferation, protein expression, apoptosis and necrosis. Subdivisions indicate the total number of miRNAs regulating only a single (red), 2 (green, violet and blue) or multiple (orange) cellular processes. (B) Influence of anti-apoptotic (black bars) miRNAs on cell-specific SEAP productivity (red bars). (C) Influence of anti-apoptotic miRNAs on cell proliferation (green bars). (D) Influence of anti-apoptotic miRNAs on necrotic cell death (blue bars). (E) Influence of anti-apoptotic miRNAs on cell proliferation and cell-specific SEAP productivity. Data are presented as mean ± SD for 3 independent transfections. Statistics: One-way ANOVA (* p <0.05; ** p<0.01; *** p<0.001).
Taken together, our findings underscore the hypothesis that miRNA regulation is highly redundant in mammals, and crucial cellular processes seem to be controlled by a plethora of different miRNAs in parallel. Advantages which could arise from such a regulatory redundancy might be that cells can maintain their homeostasis even if specific miRNA species are lost, e.g., due to genomic DNA mutations. In summary, many miRNAs were found to influence one particular cellular process presumably due to the regulation of one or more genes involved in a specific pathway. Intriguingly, a much greater number of miRNAs was identified to modulate more than one phenotype presumably by controlling multiple pathways.
Concluding remarks
In recent years, the widespread functionality of miRNAs to post-transcriptionally fine-tune gene expression in eukaryotic cells has been the focus of various research projects. Valuable reports enabled deeper insights into the complex miRNA:mRNA relationship and its downstream target regulation activity. This resulted in different hypotheses addressing specific miRNA actions, e.g. the multiplicity of target recognition of a single miRNA resulting in efficient regulation of molecular pathways.6 Cooperativity describes another feature by which different miRNAs target the same mRNA at different positions (binding sites) within the 3'UTR.7,50 However, the principle of functional redundancy of miRNAs has not yet been adequately addressed for mammalian cells. Miska and colleagues already introduced the term redundancy in regard to miRNAs when they reported that knockout of 83% of the miRNA repertoire in Caenorhabditis elegans did not affect viability and developmental timing.51 Their findings already pointed toward a putative redundant function of miRNAs since residual miRNAs might take over regulatory activity of deleted miRNAs to buffer transcriptomic balance. Hence, our functional screening data clearly support the results of Miska et al. and strongly promote the hypothesis of a similar redundant regulation of multiple cellular pathways in mammals. A schematic overview on the 3 major principles of miRNA function – multiplicity, cooperativity and redundancy – is depicted inFigure 4B.
Finally, screening of miRNA mimics has previously been performed by several groups to identify target miRNAs but was generally limited to a distinct phenotype in a particular disease model. Taking advantage of an unbiased gain-of-function miRNA screening in conjunction with a high-content cell analysis greatly helped to decipher multifunctional miRNAs in mammalian cells. Our results might provide new basis for further examinations of single miRNAs or miRNA families, which had been identified to control specific cell behavior. Our results highlight the presence of multifunctional miRNAs and support the idea that miRNAs can act redundantly to maintain function of conserved crucial cell functions. Further testing of miRNA hits from the presented screening approach in other organisms will help to gain deeper insights into conserved miRNA target interactions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
Acknowledgments address the International Graduate School in Molecular Medicine of Ulm University, Germany, for scientific encouragement and support. We thank Fabian Stiefel and Dr. Matthias Hackl for bioinformatics support.
Funding
This study was supported by the Postgraduate Scholarships Act of the Ministry for Science, Research and Arts of the federal state government of Baden-Württemberg, Germany.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Pasquinelli AE. MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 2012; 13: 271–82; PMID:22411466; http://dx.doi.org/ 10.1038/nrg3162 [DOI] [PubMed] [Google Scholar]
- 2.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell 2012; 148: 1172–87; PMID:22424228; http://dx.doi.org/ 10.1016/j.cell.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KM, Lim SK. Role of miRNAs in bone and their potential as therapeutic targets. Curr Opin Pharmacol 2014; 16C: 133–41; PMID:24907412; http://dx.doi.org/ 10.1016/j.coph.2014.05.001 [DOI] [PubMed] [Google Scholar]
- 4.Hackl M, Borth N, Grillari J. miRNAs–pathway engineering of CHO cell factories that avoids translational burdening. Trend Biotechnol 2012; 30: 405–6; PMID:22673691; http://dx.doi.org/ 10.1016/j.tibtech.2012.05.002 [DOI] [PubMed] [Google Scholar]
- 5.Xu W, Liu M, Peng X, Zhou P, Zhou J, Xu K, Xu H, Jiang S. miR-24-3p and miR-27a-3p promote cell proliferation in glioma cells via cooperative regulation of MXI1. Intl J Oncol 2013; 42: 757–66; PMID:23254855; http://dx.doi.org/ 10.3892/ijo.2012.1742 [DOI] [PubMed] [Google Scholar]
- 6.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature 2011; 469: 336–42; PMID:21248840; http://dx.doi.org/ 10.1038/nature09783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rinck A, Preusse M, Laggerbauer B, Lickert H, Engelhardt S, Theis FJ. The human transcriptome is enriched for miRNA-binding sites located in cooperativity-permitting distance. RNA Biol 2013; 10: 1125–35; PMID:23696004; http://dx.doi.org/ 10.4161/rna.24955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kowarsch A, Marr C, Schmidl D, Ruepp A, Theis FJ. Tissue-specific target analysis of disease-associated microRNAs in human signaling pathways. PloS one 2010; 5: e11154; PMID:20614023; http://dx.doi.org/ 10.1371/journal.pone.0011154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kowarsch A, Preusse M, Marr C, Theis FJ. miTALOS: analyzing the tissue-specific regulation of signaling pathways by human and mouse microRNAs. Rna 2011; 17: 809–19; PMID:21441347; http://dx.doi.org/ 10.1261/rna.2474511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucl Acid Res 2014; 42: D68–73; PMID:24275495; http://dx.doi.org/ 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer S, Buck T, Wagner A, Ehrhart C, Giancaterino J, Mang S, Schad M, Mathias S, Aschrafi A, Handrick R, et al.. A functional high-content miRNA screen identifies miR-30 family to boost recombinant protein production in CHO cells. Biotechnol J 2014; 9(1): 1279-92; PMID:25061012; http://dx.doi.org/ 10.1002/biot.201400306 [DOI] [PubMed] [Google Scholar]
- 12.Fischer S, Wagner A, Kos A, Aschrafi A, Handrick R, Hannemann J, Otte K. Breaking limitations of complex culture media: functional non-viral miRNA delivery into pharmaceutical production cell lines. J Biotechnol 2013; 168: 589–600; PMID:23994267; http://dx.doi.org/ 10.1016/j.jbiotec.2013.08.027 [DOI] [PubMed] [Google Scholar]
- 13.Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C. A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Method 1991; 139: 271–9; PMID:1710634; http://dx.doi.org/ 10.1016/0022-1759(91)90198-O [DOI] [PubMed] [Google Scholar]
- 14.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, et al.. The PANTHER database of protein families, subfamilies, functions and pathways. Nucl Acid Res 2005; 33: D284–8; PMID:15608197; http://dx.doi.org/ 10.1093/nar/gki078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayapal KP, Wlaschin KF, Hu WS, Yap MG. Recombinant protein therapeutics from CHO cells - 20 years and counting. Chem Eng Prog 2007; 103: 40–7 [Google Scholar]
- 16.Mencucci MV, Bravo MV, Bianchi MS, Bolzan AD. Streptonigrin induces delayed chromosomal instability involving interstitial telomeric sequences in chinese hamster ovary cells. Mutat Res 2012; 747: 46–52; PMID:22504371; http://dx.doi.org/ 10.1016/j.mrgentox.2012.03.011 [DOI] [PubMed] [Google Scholar]
- 17.Tigges M, Fussenegger M. Xbp1-based engineering of secretory capacity enhances the productivity of Chinese hamster ovary cells. Metabol Engin 2006; 8: 264–72; PMID:16635796; http://dx.doi.org/ 10.1016/j.ymben.2006.01.006 [DOI] [PubMed] [Google Scholar]
- 18.Duan Q, Wang X, Gong W, Ni L, Chen C, He X, Chen F, Yang L, Wang P, Wang DW. ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PloS one 2012; 7: e31518; PMID:22359598; http://dx.doi.org/ 10.1371/journal.pone.0031518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammond S, Lee KH. RNA interference of cofilin in chinese hamster ovary cells improves recombinant protein productivity. Biotechnol Bioeng 2012; 109: 528–35; PMID:21915848; http://dx.doi.org/ 10.1002/bit.23322 [DOI] [PubMed] [Google Scholar]
- 20.Hayduk EJ, Lee KH. Cytochalasin D can improve heterologous protein productivity in adherent chinese hamster ovary cells. Biotechnol Bioengin 2005; 90: 354–64; PMID:15772946; http://dx.doi.org/ 10.1002/bit.20438 [DOI] [PubMed] [Google Scholar]
- 21.Clarke C, Henry M, Doolan P, Kelly S, Aherne S, Sanchez N, Kelly P, Kinsella P, Breen L, Madden SF, et al.. Integrated miRNA, mRNA and protein expression analysis reveals the role of post-transcriptional regulation in controlling CHO cell growth rate. BMC Genom 2012; 13: 656; PMID:23170974; http://dx.doi.org/ 10.1186/1471-2164-13-656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008; 455: 58–63; PMID:18668040; http://dx.doi.org/ 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- 23.Shao S, von der Malsburg K, Hegde RS. Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Molecular cell 2013; 50: 637–48; PMID:23685075; http://dx.doi.org/ 10.1016/j.molcel.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eliyahu D, Raz A, Gruss P, Givol D, Oren M. Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature 1984; 312: 646–9; PMID:6095116; http://dx.doi.org/ 10.1038/312646a0 [DOI] [PubMed] [Google Scholar]
- 25.Lane DP, Benchimol S. p53: oncogene or anti-oncogene? Genes Dev 1990; 4: 1–8; PMID:2137806; http://dx.doi.org/ 10.1101/gad.4.1.1 [DOI] [PubMed] [Google Scholar]
- 26.Li J, Donath S, Li Y, Qin D, Prabhakar BS, Li P. miR-30 regulates mitochondrial fission through targeting p53 and the dynamin-related protein-1 pathway. PLoS Genet 2010; 6: e1000795; PMID:20062521; http://dx.doi.org/ 10.1371/journal.pgen.1000795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 2009; 23: 862–76; PMID:19293287; http://dx.doi.org/ 10.1101/gad.1767609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le MT, Shyh-Chang N, Khaw SL, Chin L, Teh C, Tay J, O'Day E, Korzh V, Yang H, Lal A, et al.. Conserved regulation of p53 network dosage by microRNA-125b occurs through evolving miRNA-target gene pairs. PLoS Genet 2011; 7: e1002242; PMID:21935352; http://dx.doi.org/ 10.1371/journal.pgen.1002242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ricarte-Filho JC, Fuziwara CS, Yamashita AS, Rezende E, da-Silva MJ, Kimura ET. Effects of let-7 microRNA on Cell Growth and Differentiation of Papillary Thyroid Cancer. Transl Oncol 2009; 2: 236–41; PMID:19956384; http://dx.doi.org/ 10.1593/tlo.09151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamada Y, Hidaka H, Seki N, Yoshino H, Yamasaki T, Itesako T, Nakagawa M, Enokida H. Tumor-suppressive microRNA-135a inhibits cancer cell proliferation by targeting the c-MYC oncogene in renal cell carcinoma. Cancer Sci 2013; 104: 304–12; PMID:23176581; http://dx.doi.org/ 10.1111/cas.12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamamura S, Saini S, Majid S, Hirata H, Ueno K, Chang I, Tanaka Y, Gupta A, Dahiya R. MicroRNA-34a suppresses malignant transformation by targeting c-Myc transcriptional complexes in human renal cell carcinoma. Carcinogen 2012; 33: 294–300; PMID:22159222; http://dx.doi.org/ 10.1093/carcin/bgr286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang WP, Kwok TT. The miR-18a* microRNA functions as a potential tumor suppressor by targeting on K-Ras. Carcinogen 2009; 30: 953–9; PMID:19372139; http://dx.doi.org/ 10.1093/carcin/bgp094 [DOI] [PubMed] [Google Scholar]
- 33.Xu B, Wang N, Wang X, Tong N, Shao N, Tao J, Li P, Niu X, Feng N, Zhang L, et al.. MiR-146a suppresses tumor growth and progression by targeting EGFR pathway and in a p-ERK-dependent manner in castration-resistant prostate cancer. Prostate 2012; 72: 1171–8; PMID:22161865; http://dx.doi.org/ 10.1002/pros.22466 [DOI] [PubMed] [Google Scholar]
- 34.Roybal JD, Zang Y, Ahn YH, Yang Y, Gibbons DL, Baird BN, Alvarez C, Thilaganathan N, Liu DD, Saintigny P, et al.. miR-200 Inhibits lung adenocarcinoma cell invasion and metastasis by targeting Flt1/VEGFR1. Mol Cancer Res 2011; 9: 25–35; PMID:21115742; http://dx.doi.org/ 10.1158/1541-7786.MCR-10-0497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laster SM, Wood JG, Gooding LR. Tumor necrosis factor can induce both apoptic and necrotic forms of cell lysis. J Immunol 1988; 141: 2629–34; PMID:3171180 [PubMed] [Google Scholar]
- 36.Vanlangenakker N, Vanden Berghe T, Krysko DV, Festjens N, Vandenabeele P. Molecular mechanisms and pathophysiology of necrotic cell death. Curr Mol Med 2008; 8: 207–20; PMID:18473820; http://dx.doi.org/ 10.2174/156652408784221306 [DOI] [PubMed] [Google Scholar]
- 37.Cho YS, Park SY. Harnessing of programmed necrosis for fighting against cancers. Biomol Ther 2014; 22: 167–75; PMID:25009696; http://dx.doi.org/ 10.4062/biomolther.2014.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK. Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 2009; 137: 1112–23; PMID:19524513; http://dx.doi.org/ 10.1016/j.cell.2009.05.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, van Mil A, Vrijsen K, Zhao J, Gao L, Metz CH, Goumans MJ, Doevendans PA, Sluijter JP. MicroRNA-155 prevents necrotic cell death in human cardiomyocyte progenitor cells via targeting RIP1. J Cell Mol Med 2011; 15: 1474–82; PMID:20550618; http://dx.doi.org/ 10.1111/j.1582-4934.2010.01104.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mucaj V, Lee SS, Skuli N, Giannoukos DN, Qiu B, Eisinger-Mathason TS, Nakazawa MS, Shay JE, Gopal PP, Venneti S, et al.. MicroRNA-124 expression counteracts pro-survival stress responses in glioblastoma. Oncogene 2014; PMID:24954504; http://dx.doi.org/ 10.1038/onc.2014.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jinushi T, Shibayama Y, Kinoshita I, Oizumi S, Jinushi M, Aota T, Takahashi T, Horita S, Dosaka-Akita H, Iseki K. Low expression levels of microRNA-124-5p correlated with poor prognosis in colorectal cancer via targeting of SMC4. Cancer Med 2014; 3(6); 1544-52; PMID:25081869; http://dx.doi.org/ 10.1002/cam4.309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Girardot M, Pecquet C, Boukour S, Knoops L, Ferrant A, Vainchenker W, Giraudier S, Constantinescu SN. miR-28 is a thrombopoietin receptor targeting microRNA detected in a fraction of myeloproliferative neoplasm patient platelets. Blood 2010; 116: 437–45; PMID:20445018; http://dx.doi.org/ 10.1182/blood-2008-06-165985 [DOI] [PubMed] [Google Scholar]
- 43.Dai Y, Chen S, Pei XY, Almenara JA, Kramer LB, Venditti CA, Dent P, Grant S. Interruption of the Ras/MEK/ERK signaling cascade enhances Chk1 inhibitor-induced DNA damage in vitro and in vivo in human multiple myeloma cells. Blood 2008; 112: 2439–49; PMID:18614762; http://dx.doi.org/ 10.1182/blood-2008-05-159392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider C, Setty M, Holmes AB, Maute RL, Leslie CS, Mussolin L, Rosolen A, Dalla-Favera R, Basso K. MicroRNA 28 controls cell proliferation and is down-regulated in B-cell lymphomas. Proc Natl Acad Sci U S A 2014; 111: 8185–90; PMID:24843176; http://dx.doi.org/ 10.1073/pnas.1322466111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verduci L, Simili M, Rizzo M, Mercatanti A, Evangelista M, Mariani L, Rainaldi G, Pitto L. MicroRNA (miRNA)-mediated interaction between leukemia/lymphoma-related factor (LRF) and alternative splicing factor/splicing factor 2 (ASF/SF2) affects mouse embryonic fibroblast senescence and apoptosis. J Biol Chem 2010; 285: 39551–63; PMID:20923760; http://dx.doi.org/ 10.1074/jbc.M110.114736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Othman N, Nagoor NH. The role of microRNAs in the regulation of apoptosis in lung cancer and its application in cancer treatment. BioMed Res Intl 2014; 2014: 318030; PMID:24999473; http://dx.doi.org/ 10.1155/2014/318030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ 2010; 17: 193–9; PMID:19461653; http://dx.doi.org/ 10.1038/cdd.2009.56 [DOI] [PubMed] [Google Scholar]
- 48.Hackl M, Jakobi T, Blom J, Doppmeier D, Brinkrolf K, Szczepanowski R, Bernhart SH, Honer Zu, Siederdissen C, Bort JA, Wieser M, et al.. Next-generation sequencing of the chinese hamster ovary microRNA transcriptome: Identification, annotation and profiling of microRNAs as targets for cellular engineering. J Biotechnol 2011; 153: 62–75; PMID:21392545; http://dx.doi.org/ 10.1016/j.jbiotec.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brinkrolf K, Rupp O, Laux H, Kollin F, Ernst W, Linke B, Kofler R, Romand S, Hesse F, Budach WE, et al.. Chinese hamster genome sequenced from sorted chromosomes. Nat Biotechnol 2013; 31: 694–5; PMID:23929341; http://dx.doi.org/ 10.1038/nbt.2645 [DOI] [PubMed] [Google Scholar]
- 50.Wu S, Huang S, Ding J, Zhao Y, Liang L, Liu T, Zhan R, He X. Multiple microRNAs modulate p21Cip1/Waf1 expression by directly targeting its 3' untranslated region. Oncogene 2010; 29: 2302–8; PMID:20190813; http://dx.doi.org/ 10.1038/onc.2010.34 [DOI] [PubMed] [Google Scholar]
- 51.Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, McGonagle SM, Bartel DP, Ambros VR, Horvitz HR. Most caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet 2007; 3: e215; PMID:18085825; http://dx.doi.org/ 10.1371/journal.pgen.0030215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


