Abstract
Termination of protein synthesis is not 100% efficient. A number of natural mechanisms that suppress translation termination exist. One of them is STOP codon readthrough, the process that enables the ribosome to pass through the termination codon in mRNA and continue translation to the next STOP codon in the same reading frame. The efficiency of translational readthrough depends on a variety of factors, including the identity of the termination codon, the surrounding mRNA sequence context, and the presence of stimulating compounds. Understanding the interplay between these factors provides the necessary background for the efficient application of the STOP codon suppression approach in the therapy of diseases caused by the presence of premature termination codons.
Keywords: translational readthrough, termination codon, STOP codon suppression, premature termination codon, near-cognate tRNA, mRNA
Abbreviations
- nc-tRNA
near-cognate tRNA
- eRF1/3
eukaryotic release factor 1/3
- NTC
normal termination codon
- PABP
poly(A)-binding proteins
- RT
STOP codon readthrough
- PTC
premature termination codons
- PTC-RT
premature termination codons readthrough
- eIF3
eukaryotic translation initiation factor
- AAGs
aminoglycoside antibiotics
Termination of Translation
Termination of translation is one of the most complex stages in protein biosynthesis.1 It occurs when the STOP codon in mRNA (UAA-ochre, UAG-amber or UGA-opal) enters the A-site on the small ribosomal subunit, and leads to the release of a nascent polypeptide chain from the peptidyl-tRNA positioned at the ribosomal P-site. Both in eukaryotes and bacteria, 2 classes of release factors (I and II) mediate this process, although these mechanisms are distinct.
In bacteria, there are 2 class-I release factors, RF1 or RF2; they recognize STOP codons through the direct action of a “peptide anti-codon” sequence (RF1 interacts with UAG and UAA, RF2 with UGA and UAA) and mediate the hydrolysis of the peptidyl-tRNA ester bond.2,3 Subsequently, a class-II release factor, RF3, which possesses GTPase activity, mediates recycling of class-I factors by ejecting them from post-termination complexes.4-6
Eukaryotic translation is terminated by a protein heterodimer consisting of 2 release factors, eRF1 (class-I) and eRF3 (class-II).7 In contrast to bacteria, where 2 class-I release factors are required, in eucaryotes, eRF1 alone recognizes all 3 STOP codons,8-10 although its interaction with UGA differs from interactions with UAA or UAG.11 eRF1 is a tRNA-shaped protein, containing 3 domains: N-terminal, middle and C-terminal.12 Recognition of STOP codons occurs through a complex, 3-dimensional network formed by conserved residues in the N-terminal domain of eRF1.13-15 The highly conserved GlyGlyGln motif in the middle domain of eRF1 is responsible for the release of a peptide from the ribosome.9 The middle domain as well as the C-terminal domain of eRF1 interact with eRF3, and the C-terminal domain of eRF3 binds GTP. Formation of a ternary eRF1•eRF3•GTP complex16-18 leads to GTP hydrolysis.5,19 Activation of the eRF3 GTPase activity additionally requires the interaction of the ternary complex with poly(A)-binding proteins (PABPs) present at the 3′-UTR of mRNA.20 Hydrolysis of GTP leads to the correct positioning of eRF1 in the ribosomal peptidyl transferase center, which catalyzes the cleavage of the peptidyl–tRNA bond and releases a polypeptide chain from the ribosome.21
To summarize, effective termination of translation in eukaryotes requires the presence of a STOP codon in the ribosomal A-site, its interaction with 2 release factors, eRF1 and eRF3, and a close distance between the terminating ribosome and 3′UTR-bound PABPs (Fig. 1).
Figure 1.
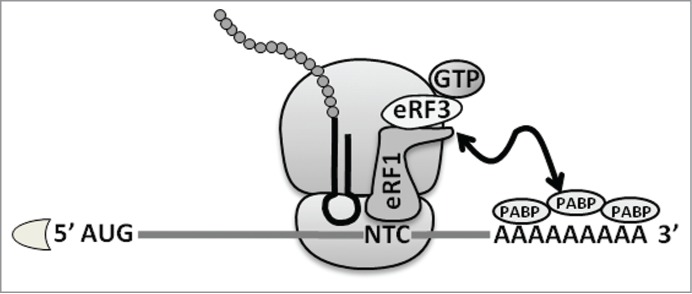
The major factors involved in translation termination in Eukaryota. NTC – normal termination codon; PABP – poly(A)-binding proteins; eRF1 and eRF3 – termination factors.
Translational readthrough of natural STOP codons
The process of protein synthesis termination, although effective, is not 100% efficient. Several natural mechanisms of termination suppression exist, including ribosomal frameshifting,22 suppressor tRNAs (aminoacylated tRNAs with anticodons complementary to STOP codons in mRNA)23 and STOP codon readthrough (RT).24,25
In this review, we will focus on the latter mechanism. STOP codon RT relies on competition between 2 distinct phenomena: the recognition of a termination codon by eRF1, which triggers the proper termination of translation, and accommodation of a near-cognate tRNA (nc-tRNA) in the A-site of the ribosome, which leads to erroneous decoding of the STOP codon. Nc-tRNAs are able to pair with STOP codons at 2 of the 3 positions of a codon–anticodon sequence. This interaction may compete with the STOP codon recognition by the release factor eRF1, and thereby inhibit the process of protein synthesis termination. As a result, an amino acid is erroneously incorporated into the polypeptide chain and the ribosome continues translation to the next STOP codon in the same reading frame (Fig. 2).
Figure 2.
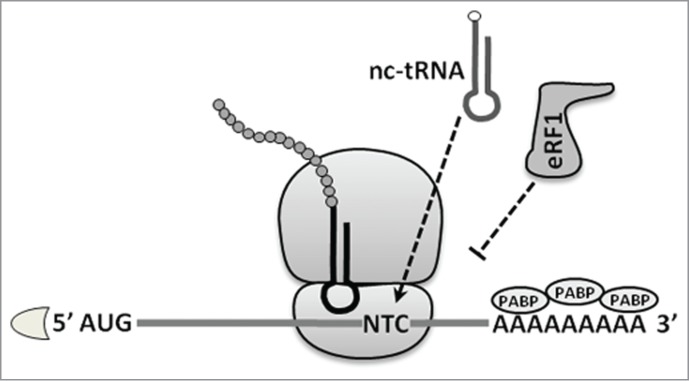
The principle of termination codon readthrough. Near-cognate tRNA (nc-tRNA) successfully competes with eRF1 and leads to the decoding of a STOP codon.
Spontaneous suppression of the translation termination, called basal RT, has been best studied in viruses. So far, prediction of eukaryote genes undergoing RT is mostly based on phylogenetic comparisons and ribosome profiling studies,26-28 and only few genes have been studied experimentally.29,30 Nevertheless, it has been estimated that the level of basal RT of STOP codons in mammalian cells ranges from 0.01 to 0.1%.31
Efficiency of Basal RT
Type of the STOP codon
The potential efficiency of basal RT is different for each of 3 STOP codons. Experiments in mammalian cell lines, using an overexpressed dual luciferase reporter vector carrying each of the STOP codons, have demonstrated that UGA has the highest basal RT potential and thus the lowest fidelity. UAG is less “leaky,” and UAA has the highest fidelity.30,32-34
Although the identity of a particular STOP codon is crucial for the efficiency of basal RT, this process also depends on other factors. Experimental studies in a number of organisms have shown that both downstream and upstream sequence context plays an essential role in determining the RT potential of STOP codons.
3′ context
In both bacteria and eukaryotes, the base immediately following the STOP codon (position +4, with the first nucleotide of the termination codon marked as +1) exerts the strongest influence on RT efficiency.35-39 This has led to the hypothesis that an actual translation termination signal consists of a tetranucleotide sequence, not only the STOP codon itself.40,41
While different studies are consistent in demonstrating that the termination efficiency depends on the +4 nucleotide, it has been shown both in yeast,24,42 and in higher eukaryotes that this effect depends greatly on the identity of the STOP codon. Which tetranucleotide is the most efficient in eliciting the RT in eukaryotes remains controversial (see Table 1). For example, the level of basal UGA-C readthrough in mammalian cells (3–4%) was shown to be 3–6 times higher than for the remaining UGA-N tetranucleotides. However, for other STOP codons, the presence of C at the +4 position did not significantly affect the efficiency of suppression: for UAG (1–2%) or for UAA (≤0 .5%).33
Table 1.
Basal RT potential: the influence of the STOP codon identity and of the +4 nucleotide context
Until recently, STOP codon RT was documented in only few genes in higher eukaryotes: syn, kelch and hdc in Drosophila melanogaster,26,43,44 and β-globin gene in rabbits.45 However, the recent comparative phylogenetic analyses of Drosophila and other metazoan genomes identified over 280 genes undergoing RT.26,46 32% percent of these genes contained UGA codon followed by cytosine (UGA-C); these genes were nearly 10 times more likely to undergo RT than genes with other nucleotide contexts. Importantly, these results confirmed that the order of STOP codon “leakiness” in eukaryotes was UGA>UAG>UAA, and the influence of +4 nucleotide on RT was C>U>G>A.26 The common conclusion of the studies in eukaryotes was that cytosine at position +4 promoted higher levels of basal RT (equivalent to the lower efficiency of termination); this was especially evident for the “leakiest” UGA codon. This conclusion is consistent with the earlier observation that leaky STOP codons in combination with the +4 cytosine (UGA-C and UAG-C) are rarely used termination contexts in mammals.39 Nonetheless, the identity of a nucleotide immediately downstream of the STOP codon is still not sufficient to predict the RT efficiency for a given nonsense mutation.
The variability in the impact of the +4 nucleotide position on the efficiency of STOP codons RT can be, at least partially, explained by the influence of other surrounding sequences. Again, an important insight into the broader sequence context of a basal RT has come from studies on viruses, where 3 main types of motifs stimulating RT have been identified.23 Type I involves the UAG-CAAUYA motif, type II – UGA-CGG or UGA-CUA motif, and type III consists of the UAG-G motif plus a downstream stimulatory RNA pseudoknot structure.23,47-50
Motifs similar to those found in viruses have also been identified in eukaryotes. The CAA sequence downstream of the STOP codon was associated with the high RT level in yeast.24,51 A more recent study of the leaky context in yeast revealed that at least 6 nucleotides (−CAAUUA) downstream of the STOP codon were involved in promoting RT.52 Tests performed in the human kidney epithelial cell line (HEK-293T) revealed a high level of RT (7–31%) in 4 genes featuring a highly conserved -CUAG sequence downstream of the UGA codon;30 experimental deletion of this 3′-motif almost completely abolished RT, clearly demonstrating the importance of the downstream -CUAG sequence in the UGA readthrough in mammals. The significance of the 3 sequence context was also confirmed in another study, which revealed that the C-terminally extended peroxisomal isoforms of 2 human proteins, LDHB and MDH1, result from translational RT of the respective genes containing the UAG-CU motif at the 3′end of their regular coding sequences.53
An interesting example of the interplay between the adjacent sequence context and STOP codon RT was reported in a study on a patient with junctional epidermolysis bullosa, the terminal recessive disease caused by mutations in the gene LAMA3.54 The patient's condition was surprisingly good, in spite of the presence of 2 nonsense mutations in LAMA3 (R943X/R1159X). The study revealed that expected deficiency of the full length laminin-332 protein was rescued by spontaneous RT of the R943X allele. The authors suggested that the sequence surrounding the 943X codon (AGU-UGA-CUA) was a crucial factor in the induction of premature STOP suppression.
5′context
The evolutionary conservation of the 5′-sequence context of termination codons in Escherichia coli and humans led to the postulate that the upstream sequence adjacent to the STOP codon also plays a role in the efficiency of translation termination.55 Indeed, it was shown that the penultimate and/or ultimate positions could modulate the level of translational RT in bacteria and yeast.51 Experiments on yeast demonstrated that the presence of adenine at 2 positions immediately upstream of the termination codon stimulates RT of UAG, and most probably of other STOP codons as well.42,56 This is consistent with the observation that adenines 5′-adjacent to STOP codons are evolutionarily conserved in genes regulated by RT: among 91 plant and animal viral RNAs subjected to RT, 65 sequences had adenine in position −1, 69 in position −2, and 50 carried adenines both in positions −1 and −2.42,56 The importance of the upstream sequence context in the STOP codon RT is also supported by studies on mammalian cells, although the RT potential of specific 5′-adjacent sequences is still not clear. In experiments performed on mouse fibroblasts (NIH3T3) and human HEK293T cell lines, the highest RT was observed when the −1 position was occupied by adenine or generally purine; in these models, uracil at the −1 position was always associated with the lowest RT level.30,57
In any case, the influence of the 5′-adjacent sequence on the efficiency of the STOP codon RT is considered to be more subtle compared to the effect of the 3′-sequence context.58
Mechanisms explaining the impact of sequence context on the RT efficiency
In spite of intensive studies, molecular mechanisms explaining the influence of the sequence context upon the RT efficiency remain unclear.
The effect of a nucleotide following the termination codon (position +4) appears to be linked to interactions of mRNA with the translational machinery rather than to interactions of the STOP codon with nc-tRNAs.59 Crosslinking experiments have demonstrated that the +4 nucleotide in mRNA interacts with eRF1.60 One of the ways by which the sequence downstream of the termination codon could affect RT is the formation of secondary structures (stem–loops, pseudoknots), which can interact with the ribosome.61,62 Such interaction of the mRNA pseudoknot with the ribosome was suggested to favor the binding of nc-tRNAs at the A-site over binding of eRF1.25 It was even suggested that structures formed by the 3′-end of mRNA, acting as components of efficient RT cassettes in eukaryotes may in fact be the norm rather than an exception.62
Regarding the downstream sequence context, it has been proposed that in yeast, the sequence 5′ to the STOP codon could stimulate RT through the direct interaction with the tRNA at the ribosomal P-site. Alternatively, ultimate amino acids of the nascent polypeptide could elicit RT through interaction with the release factors and the ribosome.51 The latter mechanism was, however, excluded in higher eukaryotes. Instead, it was suggested that adenine at position −1 or −2 could induce RT by modifying the structure of mRNA at the P-site; this would distort the structure of the ribosome, and modulate the competition between release factors and natural suppressor tRNAs.56
Recently, it has been proposed that eIF3, the eukaryotic translation initiation factor, could be involved in promoting programmed RT of all 3 STOP codons set in the unfavorable termination context.63 In this mechanism, which appears to be evolutionary conserved, eIF3 interacts with the pre-termination complex, where it prevents eRF1 from recognizing the third/wobble position of the STOP codon. As a consequence, nc-tRNAs with a mismatch in the same position can decode the STOP codon, allowing the protein synthesis to be continued.
Last but not least, a study of the S. cerevisiae genome, searching for adjacent open reading frames separated only by a unique STOP codon (called SORFs),64,65 identified several “leaky” STOP codons surrounded by sequences different from those believed to promote RT. This implies that our understanding of the role of 5′- and 3′-sequence context in the STOP codon suppression is still incomplete, and suggests that mechanisms other than RT might be involved in allowing ribosomes to bypass STOP codons.65,66
Other factors influencing the efficiency of termination codon RT
The STOP codon identity and the surrounding sequence context are not the only factors that influence the efficiency of termination codon suppression. The RT of STOP codons can be also enhanced by the increased level of mRNA that is subjected to termination suppression67 or the depletion of termination machinery components like eRF1 and/or eRF3.68-70 Such mechanisms involve changes in the cellular level of gene expression and are not dependent on STOP codon identity and mRNA sequence context.
It has been shown that post-translational modification of some ribosomal proteins can also influence the RT process. Hydroxylation of the proline residue at position 64 of RPS23 protein in the 40S ribosomal subunit is essential for the accurate translation process. The hydroxylation site is located in the ribosomal decoding center and can affect the termination codon recognition. Inhibition of the hydroxylases involved in RPS23 modification could stimulate the production of full-length proteins from sequences containing nonsense mutations.71,72
Alternatively, translational RT can be stimulated by externally provided chemical compounds, such as low molecular weight drugs. Aminoglycoside antibiotics (AAGs) are among the most important representatives of this group.58
STOP codon RT stimulated by AAGs
AAGs are oligosaccharides consisting of streptidine or 2-deoxystreptidine as the molecular core and a variable number of sugar rings and ammonium groups.73 Early papers indicating the RT potency of AAGs were already published in the 1960s.74,75 AAGs, commonly used to treat Gram-negative bacterial infections, efficiently bind to the 7 nucleotide sequence in 16S rRNA and inhibit the function of the bacterial ribosome.11 In eukaryotes, a one-base difference (A->G at position 1408) in the 7 nucleotide motif in 18S rRNA significantly lowers the efficiency of its interaction with AAGs (e.g. a ∼25–50-fold decrease in binding affinity for paromomycin).76,77 Nonetheless, this low efficiency is sufficient to reduce discrimination between cognates and nc-tRNAs in eukaryotes, thereby stimulating translational RT.
Our current understanding does not allow unambiguous prediction of the sequence context's influence upon the susceptibility of STOP codons to AAG-mediated suppression in mammalian cells.31-33 Nevertheless, the majority of evidence indicates that, as in basal RT, UGA is the “leakiest” termination codon and cytosine (or pyrimidine) residue at the +4 position correlates with the highest level of AAG-induced RT (see Table 2).
Table 2.
RT potential in the presence of RT-stimulating compounds; the influence of the STOP codon identity and of the +4 nucleotide context
| STOP CODON |
||||||
|---|---|---|---|---|---|---|
| UGA | > | UAG | > | UAA | RT-STIMULATING DRUG | REFERENCE |
| C>U>A>G | C>U∼G∼C | C>U>G∼A | G418 | Howard et al.32 | ||
| C>U>A∼G | C>U∼G∼C | C ∼U∼G∼C | Gentamicin | |||
| C>U∼A>G | C ∼U∼G∼C | C ∼U∼G∼C | Paromomycin | |||
| C>A∼G>U | U>C∼G>A | C>U>G>A | G418 | Manuvakhova et al.33 | ||
| C>A=G >U | U>C=G >A | C>G>A=U | Gentamicin | |||
| C>A≥G>U | C>A=G =U | G=U >A=C | Paromomycin | |||
| C>A=G =U | G>C=U >A | U>C>A=G | Neomycin | |||
| C>A>G>U | U>C>A=G | U>A=C =G | Sisomycin | |||
| C>U>A=G | G>C>A>U | A>U>C>G | Lividomycin | |||
| C>A∼G∼U | C>A∼G∼U | C>A∼G∼U | Gentamicin | Floquet et al.31 | ||
Statistical analysis of the RT potential, based on the study of cultured mouse cells (NIH3T3) transfected with 66 sequences containing a variety of termination codon contexts, suggested that the combination of UGA-C with uracil in position −1 (U-UGA-C) had the highest potential of AAG-stimulated RT. In the presence of AAGs, all codons studied with this sequence context systematically underwent RT, resulting in a full-length protein expression exceeding 0.5% of wild-type sequence expression.31
Biological consequences of the STOP codon RT
The process of STOP codons RT may have important biological consequences, ranging from those essential for the cell life (as in programmed RT) to detrimental ones.
Viruses have long been known to use RT to increase their coding capacity.36,65,78,79 Some pathogenic fungi that infect plants utilize RT in order to include additional signaling sequences targeting the synthesized proteins to a specific cellular compartment, like peroxisomes.80 In higher organisms, the biological function of programmed STOP codon RT is less clear, although recent studies have led to the identification of some functional, RT-derived extended proteins. Examples include regulated RT in a number of Drosophila genes,28 peroxisomal isoforms of metabolic enzymes in diverse model organisms53 or programmed RT in mammals.29,30,45,53,81
On the other hand, the RT-mediated C-terminal extension of polypeptides may cause a dominant-negative effect of mistranslated proteins, which may interfere with normal, cellular functions or lead to the gaining a new function harmful to the cell. Synthesis of useless polypeptides is also a waste of resources. There are mechanisms in the cell to prevent detrimental effects of the RT process. One of them is the existence of tandem STOPs, which are secondary in-frame STOP codons present downstream from the primary STOP codon. Tandem STOPs have been purported to limit the level of STOP codon suppression by providing a second chance for the translation termination apparatus to stop protein synthesis.66,82 Tandem STOPs in yeast are preferentially located at the third codon after the primary STOP; the six preceding nucleotides act as a sequence context favoring translation termination at the first STOP.83 According to studies in yeast and in ciliated species (Paramecium tetraurelia and Tetrahymena thermophila), tandem STOP codons are more frequent in highly expressed genes, where translation termination occurs more frequently.83,84
Decoding of STOP codons
There are several nc-tRNAs, which can recognize each of the termination codons, but their usage in the process of translational RT does not appear to be random (see Table 3). According to a number of studies, mainly in viral and yeast mRNAs, UGA is the most frequently decoded as tryptophan, but it can also be misread by cysteine or arginine tRNAs.85-88 UAA is decoded as glutamine.85,86,88 Depending on the study, UAG has been reported to be decoded as either glutamine or tryptophan,85,86,88 or as tryptophan, tyrosine or lysine.24 A recent study, using a novel mass spectrometry-based approach for identifying amino acids incorporated at the STOP codon in the in vivo reporter system in yeast, has confirmed that the UGA codon is decoded as tryptophan, cysteine or arginine, but indicated that both UAA and UAG can be decoded as glutamine, tyrosine or lysine.15 In addition, the latter study suggested that the sequence surrounding the STOP codon had no impact on the identity or proportion of amino acids incorporated during the RT process.
Table 3.
Near-cognate codons for the eukaryotic STOP codons. *Near-cognate codons reported to be most frequently involved in the translational RT of each of the STOP codons are shown in bold (in vitro studies) or underlined (in vivo reporter studies)
| TERMINATION CODONS | EFFECT ON TRANSLATION | ||
|---|---|---|---|
| UAA |
UAG |
UGA |
STOP |
| |
POSSIBLE NEAR-COGNATE CODONS* |
|
AMINO ACID INSERTED |
| GAA | GAG | Glu E | |
| AAA | AAG | Lys K | |
| CAA | CAG | Gln Q | |
| AGA, CGA | Arg R | ||
| GGA | Gly G | ||
| UCA | UCG | UCA | Ser S |
| UUA | UUG | UUA | Leu L |
| UGG | UGG | Trp W | |
| UGC, UGU | Cys C | ||
| UAC, UAU | UAC, UAU | Tyr Y | |
Inconclusive results obtained in a variety of studies and studied organisms suggest that the identity of the amino acid inserted at the STOP codon is likely to be irrelevant. This is related to the fact that the purpose of the STOP codon suppression in the programmed RT is to access another ORF. This feature distinguishes translational RT from, for example, selenocysteine insertion at UGA codons, where character of accommodated tRNA is precisely determined.89
In conclusion, in spite of the observed bias in the decoding of STOP codons, it is currently impossible to predict theoretically which amino acid will be incorporated in the synthesized peptide during the process of termination codon RT.
Translational RT of premature termination codons
STOP codon RT stimulated by chemical compounds such as AAG is a promising approach to restore protein translation from pathogenic alleles containing premature termination codons (PTC). It is estimated that 20% of all genetic human diseases are caused by single base-pair substitutions, which introduce PTCs into the mRNA.90 The idea of the RT-driven correction of PTC mutations has been tested for many inherited human diseases, especially in the context of using AAGs as RT-stimulating agents.
The first report on the PTC-RT induced by AAGs was published already in 1985. In this study, PTC-containing sequences cloned in a reporter gene overexpressed in COS-7 cells were used to demonstrate that paromomycin and G418 could restore almost 20% of the wild-type protein activity.91 The first in vivo proof of the therapeutic potential of AAGs was demonstrated in mdx mice, an animal model of Duchenne muscular dystrophy caused by premature PTC in the dystrophin gene.92 Following treatment with gentamicin, up to 10–20% of the normal level expression of the full-length dystrophin was observed. Since then, the PTC-RT stimulating potential in the therapy of human genetic diseases has been tested for many aminoglycosides, their mimetics and derivatives. A variety of different models have been used in these studies, including in vitro transcription and translation systems,33 cell lines,34,93-99 and animal models.100-104 Some of the compounds have shown promising results and have been tested in clinical trials.105-114
Although clinical improvement has been observed in some of the studies, the efficiency of PTC-readthrough therapies is not always satisfactory. Essentially, the same factors which determine RT of natural termination codons may also influence the efficiency of PTC suppression. The lessons from the RT of natural STOP codons provide important clues in this respect: the identity of a STOP codon and its surrounding sequence remain the most important factors. Until recently, neither of these could be changed without invasive genome modifications. While the emerging technologies based on the CRISPR/Cas9 system115 promise a change in the therapeutic potential related to the correction of PTC at the genomic level, it remains important to know how PTCs and their surrounding sequence context influence the potential therapeutic use of translational RT. This knowledge is necessary to make an informed choice of a particular approach, best suited to a given mutation, or to apply additional measures that can influence the RT level. Even more importantly, the efficiency of various AAGs or their derivatives in promoting the suppression of natural STOP codons has to be taken into account in efforts to establish the most effective therapeutic protocol for a given PTC mutation. Finally, further studies are required to better understand and systematize the variety of sequence-unrelated factors that can modulate the suppression of translation termination.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Science Center, grants no. 2011/01/B/NZ4/04840 (EZ) and no. 2013/09/D/NZ4/01692 (ZBB).
References
- 1.Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol 2012; 4:a013706; PMID:22751155; http://dx.doi.org/ 10.1101/cshperspect.a013706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scolnick E, Tompkins R, Caskey T, Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad Sci U S A 1968; 61:768-74; PMID:4879404; http://dx.doi.org/ 10.1073/pnas.61.2.768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura Y, Ito K. A tripeptide discriminator for stop codon recognition. FEBS Lett 2002; 514:30-3; PMID:11904176; http://dx.doi.org/ 10.1016/S0014-5793(02)02330-X [DOI] [PubMed] [Google Scholar]
- 4.Zavialov AV, Buckingham RH, Ehrenberg M. A posttermination ribosomal complex is the guanine nucleotide exchange factor for peptide release factor RF3. Cell 2001; 107:115-24; PMID:11595190; http://dx.doi.org/ 10.1016/S0092-8674(01)00508-6 [DOI] [PubMed] [Google Scholar]
- 5.Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell 2006; 125:1125-36; PMID:16777602; http://dx.doi.org/ 10.1016/j.cell.2006.04.035 [DOI] [PubMed] [Google Scholar]
- 6.Gao H, Zhou Z, Rawat U, Huang C, Bouakaz L, Wang C, Cheng Z, Liu Y, Zavialov A, Gursky R, et al.. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 2007; 129:929-41; PMID:17540173; http://dx.doi.org/ 10.1016/j.cell.2007.03.050 [DOI] [PubMed] [Google Scholar]
- 7.Bertram G, Bell HA, Ritchie DW, Fullerton G, Stansfield I. Terminating eukaryote translation: domain 1 of release factor eRF1 functions in stop codon recognition. RNA N Y N 2000; 6:1236-47; http://dx.doi.org/ 10.1017/S1355838200000777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frolova L, Le Goff X, Rasmussen HH, Cheperegin S, Drugeon G, Kress M, Arman I, Haenni AL, Celis JE, Philippe M. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 1994; 372:701-3; PMID:7990965; http://dx.doi.org/ 10.1038/372701a0 [DOI] [PubMed] [Google Scholar]
- 9.Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D. The crystal structure of human eukaryotic release factor eRF1-mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 2000; 100:311-21; PMID:10676813; http://dx.doi.org/ 10.1016/S0092-8674(00)80667-4 [DOI] [PubMed] [Google Scholar]
- 10.Kisselev L, Ehrenberg M, Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J 2003; 22:175-82; PMID:12514123; http://dx.doi.org/ 10.1093/emboj/cdg017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan-Minogue H, Bedwell DM. Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity. RNA N Y N 2008; 14:148-57; http://dx.doi.org/ 10.1261/rna.805208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frolova LY, Merkulova TI, Kisselev LL. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA N Y N 2000; 6:381-90; http://dx.doi.org/ 10.1017/S135583820099143X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolosov P, Frolova L, Seit-Nebi A, Dubovaya V, Kononenko A, Oparina N, Justesen J, Efimov A, Kisselev L. Invariant amino acids essential for decoding function of polypeptide release factor eRF1. Nucleic Acids Res 2005; 33:6418-25; PMID:16282590; http://dx.doi.org/ 10.1093/nar/gki927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng Z, Saito K, Pisarev AV, Wada M, Pisareva VP, Pestova TV, Gajda M, Round A, Kong C, Lim M, et al.. Structural insights into eRF3 and stop codon recognition by eRF1. Genes Dev 2009; 23:1106-18; PMID:19417105; http://dx.doi.org/ 10.1101/gad.1770109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blanchet S, Rowe M, Von der Haar T, Fabret C, Demais S, Howard MJ, Namy O. New insights into stop codon recognition by eRF1. Nucleic Acids Res 2015; 43:3298-308; PMID:25735746; http://dx.doi.org/ 10.1093/nar/gkv154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitkevich VA, Kononenko AV, Petrushanko IY, Yanvarev DV, Makarov AA, Kisselev LL. Termination of translation in eukaryotes is mediated by the quaternary eRF1*eRF3*GTP*Mg2+ complex. The biological roles of eRF3 and prokaryotic RF3 are profoundly distinct. Nucleic Acids Res 2006; 34:3947-54; PMID:16914449; http://dx.doi.org/ 10.1093/nar/gkl549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kononenko AV, Mitkevich VA, Dubovaya VI, Kolosov PM, Makarov AA, Kisselev LL. Role of the individual domains of translation termination factor eRF1 in GTP binding to eRF3. Proteins 2008; 70:388-93; PMID:17680691; http://dx.doi.org/ 10.1002/prot.21544 [DOI] [PubMed] [Google Scholar]
- 18.Des Georges A, Hashem Y, Unbehaun A, Grassucci RA, Taylor D, Hellen CUT, Pestova TV, Frank J. Structure of the mammalian ribosomal pre-termination complex associated with eRF1.eRF3.GDPNP. Nucleic Acids Res 2014; 42:3409-18; PMID:24335085; http://dx.doi.org/ 10.1093/nar/gkt1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eurwilaichitr L, Graves FM, Stansfield I, Tuite MF. The C-terminus of eRF1 defines a functionally important domain for translation termination in Saccharomyces cerevisiae. Mol Microbiol 1999; 32:485-96; PMID:10320572; http://dx.doi.org/ 10.1046/j.1365-2958.1999.01346.x [DOI] [PubMed] [Google Scholar]
- 20.Uchida N, Hoshino S-I, Imataka H, Sonenberg N, Katada T. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/Poly(A)-dependent translation. J Biol Chem 2002; 277:50286-92; PMID:12381739; http://dx.doi.org/ 10.1074/jbc.M203029200 [DOI] [PubMed] [Google Scholar]
- 21.Salas-Marco J, Bedwell DM. GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 2004; 24:7769-78; PMID:15314182; http://dx.doi.org/ 10.1128/MCB.24.17.7769-7778.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Ribosomal frameshifting from −2 to +50 nucleotides. Prog Nucleic Acid Res Mol Biol 1990; 39:159-83; PMID:2247607; http://dx.doi.org/ 10.1016/S0079-6603(08)60626-1 [DOI] [PubMed] [Google Scholar]
- 23.Beier H, Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res 2001; 29:4767-82; PMID:11726686; http://dx.doi.org/ 10.1093/nar/29.23.4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fearon K, McClendon V, Bonetti B, Bedwell DM. Premature translation termination mutations are efficiently suppressed in a highly conserved region of yeast Ste6p, a member of the ATP-binding cassette (ABC) transporter family. J Biol Chem 1994; 269:17802-8; PMID:7517933 [PubMed] [Google Scholar]
- 25.Bertram G, Innes S, Minella O, Richardson J, Stansfield I. Endless possibilities: translation termination and stop codon recognition. Microbiol Read Engl 2001; 147:255-69 [DOI] [PubMed] [Google Scholar]
- 26.Jungreis I, Lin MF, Spokony R, Chan CS, Negre N, Victorsen A, White KP, Kellis M. Evidence of abundant stop codon readthrough in Drosophila and other metazoa. Genome Res 2011; 21:2096-113; PMID:21994247; http://dx.doi.org/ 10.1101/gr.119974.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E, et al.. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 2011; 478:476-82; PMID:21993624; http://dx.doi.org/ 10.1038/nature10530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn JG, Foo CK, Belletier NG, Gavis ER, Weissman JS. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife 2013; 2:e01179; PMID:24302569; http://dx.doi.org/ 10.7554/eLife.01179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yamaguchi Y, Hayashi A, Campagnoni CW, Kimura A, Inuzuka T, Baba H. L-MPZ, a novel isoform of myelin P0, is produced by stop codon readthrough. J Biol Chem 2012; 287:17765-76; PMID:22457349; http://dx.doi.org/ 10.1074/jbc.M111.314468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loughran G, Chou M-Y, Ivanov IP, Jungreis I, Kellis M, Kiran AM, Baranov PV, Atkins JF. Evidence of efficient stop codon readthrough in four mammalian genes. Nucleic Acids Res 2014; 42:8928-38; PMID:25013167; http://dx.doi.org/ 10.1093/nar/gku608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Floquet C, Hatin I, Rousset J-P, Bidou L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet 2012; 8:e1002608; PMID:22479203; http://dx.doi.org/ 10.1371/journal.pgen.1002608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howard MT, Shirts BH, Petros LM, Flanigan KM, Gesteland RF, Atkins JF. Sequence specificity of aminoglycoside-induced stop condon readthrough: potential implications for treatment of Duchenne muscular dystrophy. Ann Neurol 2000; 48:164-9; PMID:10939566; http://dx.doi.org/ 10.1002/1531-8249(200008)48:2%3c164::AID-ANA5%3e3.0.CO;2-B [DOI] [PubMed] [Google Scholar]
- 33.Manuvakhova M, Keeling K, Bedwell DM. Aminoglycoside antibiotics mediate context-dependent suppression of termination codons in a mammalian translation system. RNA N Y N 2000; 6:1044-55; http://dx.doi.org/ 10.1017/S1355838200000716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bidou L, Hatin I, Perez N, Allamand V, Panthier J-J, Rousset J-P. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther 2004; 11:619-27; PMID:14973546; http://dx.doi.org/ 10.1038/sj.gt.3302211 [DOI] [PubMed] [Google Scholar]
- 35.Pedersen WT, Curran JF. Effects of the nucleotide 3′ to an amber codon on ribosomal selection rates of suppressor tRNA and release factor-1. J Mol Biol 1991; 219:231-41; PMID:2038055; http://dx.doi.org/ 10.1016/0022-2836(91)90564-M [DOI] [PubMed] [Google Scholar]
- 36.Skuzeski JM, Nichols LM, Gesteland RF, Atkins JF. The signal for a leaky UAG stop codon in several plant viruses includes the two downstream codons. J Mol Biol 1991; 218:365-73; PMID:2010914; http://dx.doi.org/ 10.1016/0022-2836(91)90718-L [DOI] [PubMed] [Google Scholar]
- 37.Li G, Rice CM. The signal for translational readthrough of a UGA codon in Sindbis virus RNA involves a single cytidine residue immediately downstream of the termination codon. J Virol 1993; 67:5062-7; PMID:8331741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tate WP, Poole ES, Horsfield JA, Mannering SA, Brown CM, Moffat JG, Dalphin ME, McCaughan KK, Major LL, Wilson DN. Translational termination efficiency in both bacteria and mammals is regulated by the base following the stop codon. Biochem Cell Biol Biochim Biol Cell 1995; 73:1095-103; http://dx.doi.org/ 10.1139/o95-118 [DOI] [PubMed] [Google Scholar]
- 39.McCaughan KK, Brown CM, Dalphin ME, Berry MJ, Tate WP. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc Natl Acad Sci U S A 1995; 92:5431-5; PMID:7777525; http://dx.doi.org/ 10.1073/pnas.92.12.5431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown CM, Stockwell PA, Trotman CN, Tate WP. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res 1990; 18:6339-45; PMID:2123028; http://dx.doi.org/ 10.1093/nar/18.21.6339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tate WP, Mannering SA. Three, four or more: the translational stop signal at length. Mol Microbiol 1996; 21:213-9; PMID:8858577; http://dx.doi.org/ 10.1046/j.1365-2958.1996.6391352.x [DOI] [PubMed] [Google Scholar]
- 42.Bonetti B, Fu L, Moon J, Bedwell DM. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J Mol Biol 1995; 251:334-45; PMID:7650736; http://dx.doi.org/ 10.1006/jmbi.1995.0438 [DOI] [PubMed] [Google Scholar]
- 43.Klagges BR, Heimbeck G, Godenschwege TA, Hofbauer A, Pflugfelder GO, Reifegerste R, Reisch D, Schaupp M, Buchner S, Buchner E. Invertebrate synapsins: a single gene codes for several isoforms in Drosophila. J Neurosci Off J Soc Neurosci 1996; 16:3154-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steneberg P, Samakovlis C. A novel stop codon readthrough mechanism produces functional Headcase protein in Drosophila trachea. EMBO Rep 2001; 2:593-7; PMID:11463742; http://dx.doi.org/ 10.1093/embo-reports/kve128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chittum HS, Lane WS, Carlson BA, Roller PP, Lung FD, Lee BJ, Hatfield DL. Rabbit beta-globin is extended beyond its UGA stop codon by multiple suppressions and translational reading gaps. Biochemistry (Mosc) 1998; 37:10866-70; http://dx.doi.org/ 10.1021/bi981042r [DOI] [PubMed] [Google Scholar]
- 46.Lin MF, Carlson JW, Crosby MA, Matthews BB, Yu C, Park S, Wan KH, Schroeder AJ, Gramates LS, St Pierre SE, et al.. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res 2007; 17:1823-36; PMID:17989253; http://dx.doi.org/ 10.1101/gr.6679507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alam SL, Wills NM, Ingram JA, Atkins JF, Gesteland RF. Structural studies of the RNA pseudoknot required for readthrough of the gag-termination codon of murine leukemia virus. J Mol Biol 1999; 288:837-52; PMID:10329183; http://dx.doi.org/ 10.1006/jmbi.1999.2713 [DOI] [PubMed] [Google Scholar]
- 48.Harrell L, Melcher U, Atkins JF. Predominance of six different hexanucleotide recoding signals 3′ of read-through stop codons. Nucleic Acids Res 2002; 30:2011-7; PMID:11972340; http://dx.doi.org/ 10.1093/nar/30.9.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brierley I, Pennell S, Gilbert RJC. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat Rev Microbiol 2007; 5:598-610; PMID:17632571; http://dx.doi.org/ 10.1038/nrmicro1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Napthine S, Yek C, Powell ML, Brown TDK, Brierley I. Characterization of the stop codon readthrough signal of Colorado tick fever virus segment 9 RNA. RNA N Y N 2012; 18:241-52; http://dx.doi.org/ 10.1261/rna.030338.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mottagui-Tabar S, Tuite MF, Isaksson LA. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur J Biochem FEBS 1998; 257:249-54; http://dx.doi.org/ 10.1046/j.1432-1327.1998.2570249.x [DOI] [PubMed] [Google Scholar]
- 52.Namy O, Hatin I, Rousset JP. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2001; 2:787-93; PMID:11520858; http://dx.doi.org/ 10.1093/embo-reports/kve176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stiebler AC, Freitag J, Schink KO, Stehlik T, Tillmann BAM, Ast J, Bölker M. Ribosomal readthrough at a short UGA stop codon context triggers dual localization of metabolic enzymes in Fungi and animals. PLoS Genet 2014; 10:e1004685; PMID:25340584; http://dx.doi.org/ 10.1371/journal.pgen.1004685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pacho F, Zambruno G, Calabresi V, Kiritsi D, Schneider H. Efficiency of translation termination in humans is highly dependent upon nucleotides in the neighbourhood of a (premature) termination codon. J Med Genet 2011; 48:640-4; PMID:21693480; http://dx.doi.org/ 10.1136/jmg.2011.089615 [DOI] [PubMed] [Google Scholar]
- 55.Arkov AL, Korolev SV, Kisselev LL. 5′ contexts of Escherichia coli and human termination codons are similar. Nucleic Acids Res 1995; 23:4712-6; PMID:8524665; http://dx.doi.org/ 10.1093/nar/23.22.4712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tork S, Hatin I, Rousset J-P, Fabret C. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res 2004; 32:415-21; PMID:14736996; http://dx.doi.org/ 10.1093/nar/gkh201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cassan M, Rousset JP. UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol Biol 2001; 2:3; PMID:11242562; http://dx.doi.org/ 10.1186/1471-2199-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee H-LR, Dougherty JP. Pharmaceutical therapies to recode nonsense mutations in inherited diseases. Pharmacol Ther 2012; 136:227-66; PMID:22820013; http://dx.doi.org/ 10.1016/j.pharmthera.2012.07.007 [DOI] [PubMed] [Google Scholar]
- 59.Phillips-Jones MK, Hill LS, Atkinson J, Martin R. Context effects on misreading and suppression at UAG codons in human cells. Mol Cell Biol 1995; 15:6593-600; PMID:8524224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bulygin KN, Repkova MN, Ven'yaminova AG, Graifer DM, Karpova GG, Frolova LY, Kisselev LL. Positioning of the mRNA stop signal with respect to polypeptide chain release factors and ribosomal proteins in 80S ribosomes. FEBS Lett 2002; 514:96-101; PMID:11904189; http://dx.doi.org/ 10.1016/S0014-5793(02)02304-9 [DOI] [PubMed] [Google Scholar]
- 61.Wills NM, Gesteland RF, Atkins JF. Evidence that a downstream pseudoknot is required for translational read-through of the Moloney murine leukemia virus gag stop codon. Proc Natl Acad Sci U S A 1991; 88:6991-5; PMID:1871115; http://dx.doi.org/ 10.1073/pnas.88.16.6991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Firth AE, Wills NM, Gesteland RF, Atkins JF. Stimulation of stop codon readthrough: frequent presence of an extended 3′ RNA structural element. Nucleic Acids Res 2011; 39:6679-91; PMID:21525127; http://dx.doi.org/ 10.1093/nar/gkr224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beznosková P, Wagner S, Jansen ME, von der Haar T, Valášek LS. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res 2015; 43:5099-111; PMID:Can't; http://dx.doi.org/ 10.1093/nar/gkv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Namy O, Duchateau-Nguyen G, Rousset J-P. Translational readthrough of the PDE2 stop codon modulates cAMP levels in Saccharomyces cerevisiae. Mol Microbiol 2002; 43:641-52; PMID:11929521; http://dx.doi.org/ 10.1046/j.1365-2958.2002.02770.x [DOI] [PubMed] [Google Scholar]
- 65.Namy O, Duchateau-Nguyen G, Hatin I, Hermann-Le Denmat S, Termier M, Rousset J-P. Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res 2003; 31:2289-96; PMID:12711673; http://dx.doi.org/ 10.1093/nar/gkg330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Williams I, Richardson J, Starkey A, Stansfield I. Genome-wide prediction of stop codon readthrough during translation in the yeast Saccharomyces cerevisiae. Nucleic Acids Res 2004; 32:6605-16; PMID:15602002; http://dx.doi.org/ 10.1093/nar/gkh1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, Virgilis D, Neu-Yilik G, Kulozik AE, Kerem E, et al.. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest 2007; 117:683-92; PMID:17290305; http://dx.doi.org/ 10.1172/JCI28523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carnes J, Jacobson M, Leinwand L, Yarus M. Stop codon suppression via inhibition of eRF1 expression. RNA N Y N 2003; 9:648-53; http://dx.doi.org/ 10.1261/rna.5280103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chauvin C, Salhi S, Le Goff C, Viranaicken W, Diop D, Jean-Jean O. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol Cell Biol 2005; 25:5801-11; PMID:15987998; http://dx.doi.org/ 10.1128/MCB.25.14.5801-5811.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Diop D, Chauvin C, Jean-Jean O. Aminoglycosides and other factors promoting stop codon readthrough in human cells. C R Biol 2007; 330:71-9; PMID:17241950; http://dx.doi.org/ 10.1016/j.crvi.2006.09.001 [DOI] [PubMed] [Google Scholar]
- 71.Loenarz C, Sekirnik R, Thalhammer A, Ge W, Spivakovsky E, Mackeen MM, McDonough MA, Cockman ME, Kessler BM, Ratcliffe PJ, et al.. Hydroxylation of the eukaryotic ribosomal decoding center affects translational accuracy. Proc Natl Acad Sci U S A 2014; 111:4019-24; PMID:24550462; http://dx.doi.org/ 10.1073/pnas.1311750111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singleton RS, Liu-Yi P, Formenti F, Ge W, Sekirnik R, Fischer R, Adam J, Pollard PJ, Wolf A, Thalhammer A, et al.. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc Natl Acad Sci U S A 2014; 111:4031-6; PMID:24550447; http://dx.doi.org/ 10.1073/pnas.1314482111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.François B, Russell RJM, Murray JB, Aboul-ela F, Masquida B, Vicens Q, Westhof E. Crystal structures of complexes between aminoglycosides and decoding A site oligonucleotides: role of the number of rings and positive charges in the specific binding leading to miscoding. Nucleic Acids Res 2005; 33:5677-90; http://dx.doi.org/ 10.1093/nar/gki862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Davies J, Gilbert W, Gorini L. STREPTOMYCIN, SUPPRESSION, AND THE CODE. Proc Natl Acad Sci U S A 1964; 51:883-90; PMID:14173007; http://dx.doi.org/ 10.1073/pnas.51.5.883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Anderson WF, Gorini L, Breckenridge L. Role of ribosomes in streptomycin-activated suppression. Proc Natl Acad Sci U S A 1965; 54:1076-83; PMID:5327252; http://dx.doi.org/ 10.1073/pnas.54.4.1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Recht MI, Douthwaite S, Dahlquist KD, Puglisi JD. Effect of mutations in the A site of 16 S rRNA on aminoglycoside antibiotic-ribosome interaction. J Mol Biol 1999; 286:33-43; PMID:9931247; http://dx.doi.org/ 10.1006/jmbi.1998.2446 [DOI] [PubMed] [Google Scholar]
- 77.Lynch SR, Puglisi JD. Structural origins of aminoglycoside specificity for prokaryotic ribosomes. J Mol Biol 2001; 306:1037-58; PMID:11237617; http://dx.doi.org/ 10.1006/jmbi.2000.4420 [DOI] [PubMed] [Google Scholar]
- 78.Dreher TW, Miller WA. Translational control in positive strand RNA plant viruses. Virology 2006; 344:185-97; PMID:16364749; http://dx.doi.org/ 10.1016/j.virol.2005.09.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Von der Haar T, Tuite MF. Regulated translational bypass of stop codons in yeast. Trends Microbiol 2007; 15:78-86; PMID:17187982; http://dx.doi.org/ 10.1016/j.tim.2006.12.002 [DOI] [PubMed] [Google Scholar]
- 80.Freitag J, Ast J, Bölker M. Cryptic peroxisomal targeting via alternative splicing and stop codon read-through in fungi. Nature 2012; 485:522-5; PMID:22622582; http://dx.doi.org/ 10.1038/nature11051 [DOI] [PubMed] [Google Scholar]
- 81.Eswarappa SM, Potdar AA, Koch WJ, Fan Y, Vasu K, Lindner D, Willard B, Graham LM, DiCorleto PE, Fox PL. Programmed translational readthrough generates antiangiogenic VEGF-Ax. Cell 2014; 157:1605-18; PMID:24949972; http://dx.doi.org/ 10.1016/j.cell.2014.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nichols JL. Nucleotide sequence from the polypeptide chain termination region of the coat protein cistron in bacteriophage R17 RNA. Nature 1970; 225:147-51; PMID:5409960; http://dx.doi.org/ 10.1038/225147a0 [DOI] [PubMed] [Google Scholar]
- 83.Liang H, Cavalcanti ARO, Landweber LF. Conservation of tandem stop codons in yeasts. Genome Biol 2005; 6:R31; PMID:15833118; http://dx.doi.org/ 10.1186/gb-2005-6-4-r31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Adachi M, Cavalcanti ARO. Tandem stop codons in ciliates that reassign stop codons. J Mol Evol 2009; 68:424-31; PMID:19294453; http://dx.doi.org/ 10.1007/s00239-009-9220-y [DOI] [PubMed] [Google Scholar]
- 85.Feng YX, Copeland TD, Oroszlan S, Rein A, Levin JG. Identification of amino acids inserted during suppression of UAA and UGA termination codons at the gag-pol junction of Moloney murine leukemia virus. Proc Natl Acad Sci U S A 1990; 87:8860-3; PMID:2247457; http://dx.doi.org/ 10.1073/pnas.87.22.8860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zerfass K, Beier H. The leaky UGA termination codon of tobacco rattle virus RNA is suppressed by tobacco chloroplast and cytoplasmic tRNAs(Trp) with CmCA anticodon. EMBO J 1992; 11:4167-73; PMID:1396598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Urban C, Beier H. Cysteine tRNAs of plant origin as novel UGA suppressors. Nucleic Acids Res 1995; 23:4591-7; PMID:8524647; http://dx.doi.org/ 10.1093/nar/23.22.4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nilsson M, Rydén-Aulin M. Glutamine is incorporated at the nonsense codons UAG and UAA in a suppressor-free Escherichia coli strain. Biochim Biophys Acta 2003; 1627:1-6; PMID:12759186; http://dx.doi.org/ 10.1016/S0167-4781(03)00050-2 [DOI] [PubMed] [Google Scholar]
- 89.Labunskyy VM, Hatfield DL, Gladyshev VN. Selenoproteins: molecular pathways and physiological roles. Physiol Rev 2014; 94:739-77; PMID:24987004; http://dx.doi.org/ 10.1152/physrev.00039.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mort M, Ivanov D, Cooper DN, Chuzhanova NA. A meta-analysis of nonsense mutations causing human genetic disease. Hum Mutat 2008; 29:1037-47; PMID:18454449; http://dx.doi.org/ 10.1002/humu.20763 [DOI] [PubMed] [Google Scholar]
- 91.Burke JF, Mogg AE. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res 1985; 13:6265-72; PMID:2995924; http://dx.doi.org/ 10.1093/nar/13.17.6265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Barton-Davis ER, Cordier L, Shoturma DI, Leland SE, Sweeney HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest 1999; 104:375-81; PMID:10449429; http://dx.doi.org/ 10.1172/JCI7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bedwell DM, Kaenjak A, Benos DJ, Bebok Z, Bubien JK, Hong J, Tousson A, Clancy JP, Sorscher EJ. Suppression of a CFTR premature stop mutation in a bronchial epithelial cell line. Nat Med 1997; 3:1280-4; PMID:9359706; http://dx.doi.org/ 10.1038/nm1197-1280 [DOI] [PubMed] [Google Scholar]
- 94.Howard MT, Anderson CB, Fass U, Khatri S, Gesteland RF, Atkins JF, Flanigan KM. Readthrough of dystrophin stop codon mutations induced by aminoglycosides. Ann Neurol 2004; 55:422-6; PMID:14991821; http://dx.doi.org/ 10.1002/ana.20052 [DOI] [PubMed] [Google Scholar]
- 95.Zilberberg A, Lahav L, Rosin-Arbesfeld R. Restoration of APC gene function in colorectal cancer cells by aminoglycoside- and macrolide-induced read-through of premature termination codons. Gut 2010; 59:496-507; PMID:19951906; http://dx.doi.org/ 10.1136/gut.2008.169805 [DOI] [PubMed] [Google Scholar]
- 96.Floquet C, Deforges J, Rousset J-P, Bidou L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res 2011; 39:3350-62; PMID:21149266; http://dx.doi.org/ 10.1093/nar/gkq1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Floquet C, Rousset J-P, Bidou L. Readthrough of premature termination codons in the adenomatous polyposis coli gene restores its biological activity in human cancer cells. PloS One 2011; 6:e24125; PMID:21909382; http://dx.doi.org/ 10.1371/journal.pone.0024125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fuchshuber-Moraes M, Carvalho RS, Rimmbach C, Rosskopf D, Carvalho MA, Suarez-Kurtz G. Aminoglycoside-induced suppression of CYP2C19*3 premature stop codon. Pharmacogenet Genomics 2011; 21:694-700; PMID:21934639; http://dx.doi.org/ 10.1097/FPC.0b013e328349daba [DOI] [PubMed] [Google Scholar]
- 99.Ho G, Reichardt J, Christodoulou J. In vitro read-through of phenylalanine hydroxylase (PAH) nonsense mutations using aminoglycosides: a potential therapy for phenylketonuria. J Inherit Metab Dis 2013; 36:955-9; PMID:23532445; http://dx.doi.org/ 10.1007/s10545-013-9602-6 [DOI] [PubMed] [Google Scholar]
- 100.Arakawa M, Shiozuka M, Nakayama Y, Hara T, Hamada M, Kondo S, Ikeda D, Takahashi Y, Sawa R, Nonomura Y, et al.. Negamycin restores dystrophin expression in skeletal and cardiac muscles of mdx mice. J Biochem (Tokyo) 2003; 134:751-8; http://dx.doi.org/ 10.1093/jb/mvg203 [DOI] [PubMed] [Google Scholar]
- 101.Du M, Jones JR, Lanier J, Keeling KM, Lindsey JR, Tousson A, Bebök Z, Whitsett JA, Dey CR, Colledge WH, et al.. Aminoglycoside suppression of a premature stop mutation in a Cftr−/− mouse carrying a human CFTR-G542X transgene. J Mol Med Berl Ger 2002; 80:595-604; http://dx.doi.org/ 10.1007/s00109-002-0363-1 [DOI] [PubMed] [Google Scholar]
- 102.Brendel C, Klahold E, Gärtner J, Huppke P. Suppression of nonsense mutations in Rett syndrome by aminoglycoside antibiotics. Pediatr Res 2009; 65:520-3; PMID:19190538; http://dx.doi.org/ 10.1203/PDR.0b013e31819d9ebc [DOI] [PubMed] [Google Scholar]
- 103.Mattis VB, Ebert AD, Fosso MY, Chang C-W, Lorson CL. Delivery of a read-through inducing compound, TC007, lessens the severity of a spinal muscular atrophy animal model. Hum Mol Genet 2009; 18:3906-13; PMID:19625298; http://dx.doi.org/ 10.1093/hmg/ddp333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gunn G, Dai Y, Du M, Belakhov V, Kandasamy J, Schoeb TR, Baasov T, Bedwell DM, Keeling KM. Long-term nonsense suppression therapy moderates MPS I-H disease progression. Mol Genet Metab 2014; 111:374-81; PMID:24411223; http://dx.doi.org/ 10.1016/j.ymgme.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clancy JP, Bebök Z, Ruiz F, King C, Jones J, Walker L, Greer H, Hong J, Wing L, Macaluso M, et al.. Evidence that systemic gentamicin suppresses premature stop mutations in patients with cystic fibrosis. Am J Respir Crit Care Med 2001; 163:1683-92; PMID:11401894; http://dx.doi.org/ 10.1164/ajrccm.163.7.2004001 [DOI] [PubMed] [Google Scholar]
- 106.Wilschanski M, Yahav Y, Yaacov Y, Blau H, Bentur L, Rivlin J, Aviram M, Bdolah-Abram T, Bebok Z, Shushi L, et al.. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med 2003; 349:1433-41; PMID:14534336; http://dx.doi.org/ 10.1056/NEJMoa022170 [DOI] [PubMed] [Google Scholar]
- 107.Politano L, Nigro G, Nigro V, Piluso G, Papparella S, Paciello O, Comi LI. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results. Acta Myol Myopathies Cardiomyopathies Off J Mediterr Soc Myol Ed Gaetano Conte Acad Study Striated Muscle Dis 2003; 22:15-21 [PubMed] [Google Scholar]
- 108.Hirawat S, Welch EM, Elfring GL, Northcutt VJ, Paushkin S, Hwang S, Leonard EM, Almstead NG, Ju W, Peltz SW, et al.. Safety, tolerability, and pharmacokinetics of PTC124, a nonaminoglycoside nonsense mutation suppressor, following single- and multiple-dose administration to healthy male and female adult volunteers. J Clin Pharmacol 2007; 47:430-44; PMID:17389552; http://dx.doi.org/ 10.1177/0091270006297140 [DOI] [PubMed] [Google Scholar]
- 109.Kerem E, Hirawat S, Armoni S, Yaakov Y, Shoseyov D, Cohen M, Nissim-Rafinia M, Blau H, Rivlin J, Aviram M, et al.. Effectiveness of PTC124 treatment of cystic fibrosis caused by nonsense mutations: a prospective phase II trial. Lancet Lond Engl 2008; 372:719-27; http://dx.doi.org/ 10.1016/S0140-6736(08)61168-X [DOI] [PubMed] [Google Scholar]
- 110.Malik V, Rodino-Klapac LR, Viollet L, Wall C, King W, Al-Dahhak R, Lewis S, Shilling CJ, Kota J, Serrano-Munuera C, et al.. Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann Neurol 2010; 67:771-80; PMID:20517938 [DOI] [PubMed] [Google Scholar]
- 111.Sermet-Gaudelus I, Boeck KD, Casimir GJ, Vermeulen F, Leal T, Mogenet A, Roussel D, Fritsch J, Hanssens L, Hirawat S, et al.. Ataluren (PTC124) induces cystic fibrosis transmembrane conductance regulator protein expression and activity in children with nonsense mutation cystic fibrosis. Am J Respir Crit Care Med 2010; 182:1262-72; PMID:20622033; http://dx.doi.org/ 10.1164/rccm.201001-0137OC [DOI] [PubMed] [Google Scholar]
- 112.Wilschanski M, Miller LL, Shoseyov D, Blau H, Rivlin J, Aviram M, Cohen M, Armoni S, Yaakov Y, Pugatsch T, et al.. Chronic ataluren (PTC124) treatment of nonsense mutation cystic fibrosis. Eur Respir J 2011; 38:59-69; PMID:21233271; http://dx.doi.org/ 10.1183/09031936.00120910 [DOI] [PubMed] [Google Scholar]
- 113.Finkel RS, Flanigan KM, Wong B, Bönnemann C, Sampson J, Sweeney HL, Reha A, Northcutt VJ, Elfring G, Barth J, et al.. Phase 2a study of ataluren-mediated dystrophin production in patients with nonsense mutation Duchenne muscular dystrophy. PloS One 2013; 8:e81302; PMID:24349052; http://dx.doi.org/ 10.1371/journal.pone.0081302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bushby K, Finkel R, Wong B, Barohn R, Campbell C, Comi GP, Connolly AM, Day JW, Flanigan KM, Goemans N, et al.. Ataluren treatment of patients with nonsense mutation dystrophinopathy. Muscle Nerve 2014; 50:477-87; PMID:25042182; http://dx.doi.org/ 10.1002/mus.24332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xiao-Jie L, Hui-Ying X, Zun-Ping K, Jin-Lian C, Li-Juan J. CRISPR-Cas9: a new and promising player in gene therapy. J Med Genet 2015; 52:289-96; PMID:25713109; http://dx.doi.org/ 10.1136/jmedgenet-2014-102968 [DOI] [PubMed] [Google Scholar]


