Abstract
Ubiquitin-like with PHD and RING Finger domains 1 (UHRF1) is an important nuclear protein that is mutated and aberrantly expressed in many tumors. The protein integrates different chromatin modifications and is essential for their maintenance throughout the cell cycle. Separate chromatin-binding modules of UHRF1 have been studied on a functional and structural level. The unmodified N-terminus of histone H3 is recognized by a PHD domain, while a TTD domain specifically interacts with histone H3 Lysine 9 trimethylation. A SRA region binds hemimethylatd DNA. Emerging evidence indicates that the modules of UHRF1 do not act independently of each other but establish complex modes of interaction with patterns of chromatin modifications. This multivalent readout is regulated by allosteric binding of phosphatidylinositol 5-phosphate to a region outside the PHD, TTD and SRA domains as well as by phosphorylation of one of the linker regions connecting these modules. Here, we summarize the current knowledge on UHRF1 chromatin interaction and introduce a novel model of conformational transitions of the protein that are directed by the flexible and highly charged linker regions. We propose that these are essential in setting up defined structural states of the protein where different domains or combinations thereof are available for binding chromatin modifications or are prevented from doing so. Lastly, we suggest that controlled tuning of intramolecular linker interactions by ligands and posttranslational modifications establishes a rational framework for comprehending UHRF1 regulation and putatively the working mode of other chromatin factors in different physiological contexts.
Keywords: allosteric, chromatin, cooperative, DNA methylation, domains, epigenetic, histone, modifications, multivalent, multiple protein conformations, UHRF1
Introduction
DNA methylation is an important epigenetic modification to establish and maintain cell-type-specific gene expression profiles as well as to ensure genome stability. In mammals, the majority of genomic DNA methylation occurs on the 5-position of cytosine bases within CpG dinucleotides. Within the short palindromic sequence DNA nucleotide methyltransferases (DNMTs) modify both strands (so called symmetric or full methylation). CpG methylation (meCpG) does not operate in isolation but is closely interconnected with specific repressive histone Lysine methylation marks and in particular histone H3 Lysine 9 trimethylation (H3K9me3). Histone modifications seem to be involved in targeting DNMTs to specific regions of the genome for regional methylation of DNA. In turn, DNA sequence and methylation state have a great influence on the Lysine methylation status of histones. The interplay between both epigenetic marks and how these are established is depending on the physiological and developmental cellular context. Since both modifications are associated with transcriptional repression and heterochromatin formation, their faithful duplication and transmission during cell division is crucial to preserve genome integrity (reviewed in ref.1)
Ubiquitin-like with PHD and RING Finger domains 1 (UHRF1) is a nuclear factor essential for the maintenance of meCpG and putatively H3K9me3 patterns during replication. The protein is preferentially localized to pericentric heterochromatin but can also be found in euchromatic regions.2-4 Via multiple chromatin binding domains UHRF1 targets enzymes catalyzing meCpG and H3K9me3, DNMT1, DNMT3a/b and G9a, to genomic loci.5-8 Since the protein besides maintenance of epigenetic marks is also implicated in gene expression regulation, particularly in silencing of tumor suppressor genes, understanding how it affects chromatin regulatory processes through histone and DNA methylation is of great interest.2,9,10
Based on results of functional and structural studies, we recently proposed a role of phosphatidylinositol 5-phosphate (PI(5)P) in allosteric regulation of UHRF1 binding to H3K9me3.11 Here, we summarize findings that indicate multiple conformational states of UHRF1 that expose different domains providing distinct overall chromatin-binding properties. We suggest these different states of UHRF1 are dependent on conserved linker regions. Lastly, we provide rationale how the different interaction modes of the protein with chromatin might be regulated by the interplay of allosteric ligands, posttranslational modifications and protein binding partners.
Domain structure of UHRF1
Human UHRF1 is also referred to as ICBP90, while the mouse ortholog is known as Np95. Most vertebrates also contain a highly similar factor, UHRF2 (mNp97). Since hUHRF1 has been most extensively studied, we will, unless otherwise stated, refer to this protein in our discussions. The five conserved domains of UHRF1 (Fig. 1) comprise an N-terminal ubiquitin-like domain (UBL), which is also referred to as NIRF_N (novel Np95/ICBP90-like RING finger protein N-terminus). This domain has the classic α/β ubiquitin fold and contains conserved surface Lysines K31 and K50, which are putative targets of mono- or poly-ubiquitination (pdb entry 2FAZ, unpublished structure). While UHRF1 protein function and/or proteasomal protein turnover might be regulated by the UBL domain, the exact biological role of this region is not yet fully understood.12
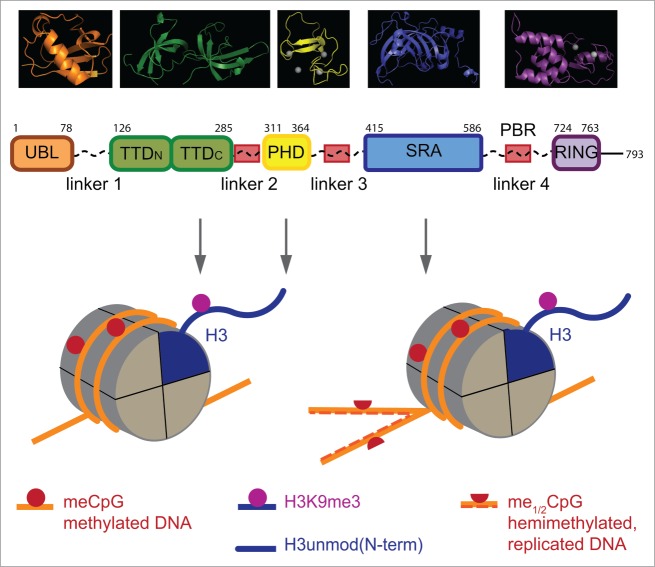
Figure 1.
UHRF1 is a multi-domain factor with several conserved protein motifs connected by linker regions. Schematic representation of the domain structure of human UHRF1 isoform 1 is shown (UniProtKB: Q96T88). Top: The structures of the individual domains of UHRF1 as determined by X-ray crystallography and NMR spectroscopy visualized and rendered using MacPyMOL v 1.7.0.3 (pymol.org). Middle: Domain boundaries are given by respective starting and ending amino acid positions. Conserved stretches of low complexity and high content of basic amino acids within the linker regions are indicated (red boxes). Bottom: Chromatin ligands of the different domains of UHRF1; schematic representation of a single nucleosome in interphase (left) and immediately after replication in S-phase (right). For simplicity only one H3-tail is shown. UBL, ubiquitin like; TTD, tandem tudor domain; PHD, plant homeodomain; SRA, SET and RING associated; PBR, polybasic region; RING, really interesting and new gene. pdb entries are: UBL, 2FAZ; TTDN:TTDC, 3DB3; PHD, 3SHB; SRA, 2PB7; RING, 3FL2.
At the C-terminus of UHRF1 is a Really Interesting and New Gene (RING) domain that has E3 ubiquitin ligase activity. It is composed of 2 zinc-fingers and a novel, unique α-helix bundle structure, which is formed by 3 helices localized upstream and one helix localized downstream of the Zn-fingers (pdb entry 3FL2, unpublished structure). According to a recent report the RING domain establishes histone H3 Lysine 23 (H3K23) monoubiquitination during S-phase. It was proposed that this mark serves as a prerequisite for recruitment of the maintenance DNA methyltransferase DNMT1 to target sites of UHRF1.13
Three central domains of UHRF1 are implicated in specific recognition of chromatin modifications, the unmodified N-terminal region of histone H3 (H3unmod(N-term)), the H3K9me3 mark and hemi-methylated DNA (me1/2CpG) (Fig. 1). The tandem tudor domain (TTD) is composed of 2 subdomains (TTDN:TTDC) that are tightly packed together and that both have a typical tudor family 5-stranded β-barrel fold.14 An aromatic cage that is built by residues F152, Y188 and Y191 of TTDN recognizes di- and tri-methylated Lysine residues. Specificity for binding H3K9me3 is provided by a peptide-binding groove formed between the 2 individual tudor domains establishing specific and tight contacts to the residues upstream and downstream of the methylated Lysine. Binding of the TTD to an H3K9me3 peptide is not sensitive to adjacent Serine 10 phosphorylation (H3S10ph).14,15 In contrast, H3 Lysine 4 trimethylation (H3K4me3) and Threonine 6 phosphorylation (H3T6ph) impair affinity for H3K9me3. These post-translational modifications disrupt the important interactions of the K4 and T6 side chain and backbone residues with the peptide-binding groove between TTDN and TTDC.14
The adjacent Plant Homeo Domain (PHD) is a Zn-finger domain; 3 zinc atoms coordinate its rod-shape structure. It recognizes the N-terminus of the H3-tail solely when unmodified (H3unmod(N-term)).2 Accordingly, the crystal structure of the PHD with bound H3-tail peptide revealed that Alanine 1 (H3A1) and Arginine 2 (H3R2) make hydrogen bond-contact to specific residues of the domain.16 Remarkably, the first zinc atom coordinates a loop, the so-called prePHD that precedes the canonical PHD-fold. This structural feature was first identified in the UHRF1 PHD domain and its detailed function still needs to be determined.16,17 It was suggested that the prePHD might be essential for the right orientation of residue C316, which makes contact with H3K4.16 However, if analyzed in isolation the PHD does not much discriminate the modification status of H3K4. In contrast, phosphorylation of H3 Threonine 3 (H3T3ph), symmetric as well as asymmetric dimethylation of H3R2 (R2me2s/a) and H3A1 N-terminal acetylation (H3A1ac) strongly interfere with binding of the PHD to the H3-tail.2
C-terminal to the PHD is a SET and RING-associated domain (SRA). It consists of a β-barrel flanked by α-helical elements forming a half moon-like structure with a basic inner surface.18 Two loops sticking out of this structure grasp into the major and the minor groove of DNA. R491 is part of the so-called N-K-R finger, which specifically forms hydrogen bonds with CpG sequences. These contacts flip out a cytosine from the double helix placing it in a binding pocket that is tailored for the recognition of 5-methylcytosine (5mC).19 In this pocket 5mC is sandwiched by stacking interactions with 2 aromatic residues (Y478, Y466). The SRA domain exhibits significant specificity for hemi-methylated DNA (me1/2CpG). This is facilitated by N489, which is a part of the N-K-R finger and makes contacts to the non-methylated adverse cytosine on the second DNA strand. Methylation of this cytosine disturbs positioning of the N-K-R finger and therefore impairs SRA binding.18
Interplay of UHRF1 chromatin-binding domains
Given that UHRF1 contains 3 separate domains that can recognize different chromatin modifications, the question arises whether these work independently or in concert. The combinatorial interaction of different domains of one protein with a complex target containing multiple ligands is generally referred to as multivalent binding. Several chromatin factors such as TRIM24 and BPTF have been shown to recognize patterns of histone modifications in a multivalent mode.20,21 In these proteins different binding domains simultaneously engage in interaction with separate histone modifications Such cross-talk in interaction on the same or distinct histone tails is considered a fundamental element of the so-called histone code theory that postulates that patterns of chromatin marks constitute a complex signaling system.22 To this point, multivalent chromatin interactions have been rationalized in a static manner, with recognition domains acting independently of each other.23,24 Several observations indicate that multivalent binding of UHRF1 to chromatin is more complex.
TTD-PHD interplay in recognizing the H3-tail
Different biophysical studies have quantified the interaction strength of the isolated TTD of UHRF1 with H3K9me3 peptides. Depending on the experimental conditions the dissociation constant (KD) was determined within a range of 1.0 µM to 2.5 µM.11,15,25,26 Similarly, binding of the isolated PHD domain to H3unmod(N-term) was mapped at a KD of 0.7 µM to 2.5 µM.2,17,26,27 In contrast, a UHRF1 TTD-PHD cassette showed around 5-fold enhanced binding (KD between 0.15 µM and 0.5 µM) to a histone H3 peptide containing both, an unmodified N-terminus and the K9me3 mark, implying a multivalent binding modus.11,25,26,28
Since the H3-tail is embedded in the peptide-binding groove of the TTD in the isolated structure of this complex, how can the PHD get access to the ultimate N-terminus of H3 in the context of the TTD-PHD cassette? Fluorescence/Förster Resonance Energy Transfer (FRET) experiments revealed a conformational shift induced in the TTD-PHD cassette upon interaction with the H3-tail.25 In agreement, structural studies using cocrystalization as well as NMR measurements indicate that the H3-binding mode of the TTD-PHD cassette is different from that of the isolated TTD (Fig. 2A). Obviously, the short region connecting TTD and PHD (linker 2, Fig. 1) replaces the H3-tail from the peptide-binding groove of the TTD and itself occupies this interface. The resulting arrangement has the H3-tail connecting the PHD, which binds the ultimate N-terminus, with the TTD, which binds K9me3. Two Arginine and a Lysine residue (R295-R296-K297) in linker 2 are crucial for stabilizing this TTD-PHD conformation. Indeed, mutation of R295 and R296 results in loss of multivalent binding.26,28 In contrast, dynamic NMR studies indicate multiple modes of PHD linkage in relation to linker 2 and TTD.28 Also, the TTD-PHD cassette only crystalized in presence of the H3K9me3-tail peptide.26 The findings imply that the 2 domains do not directly interface but that their relative localization is variable without ligand.
Figure 2.
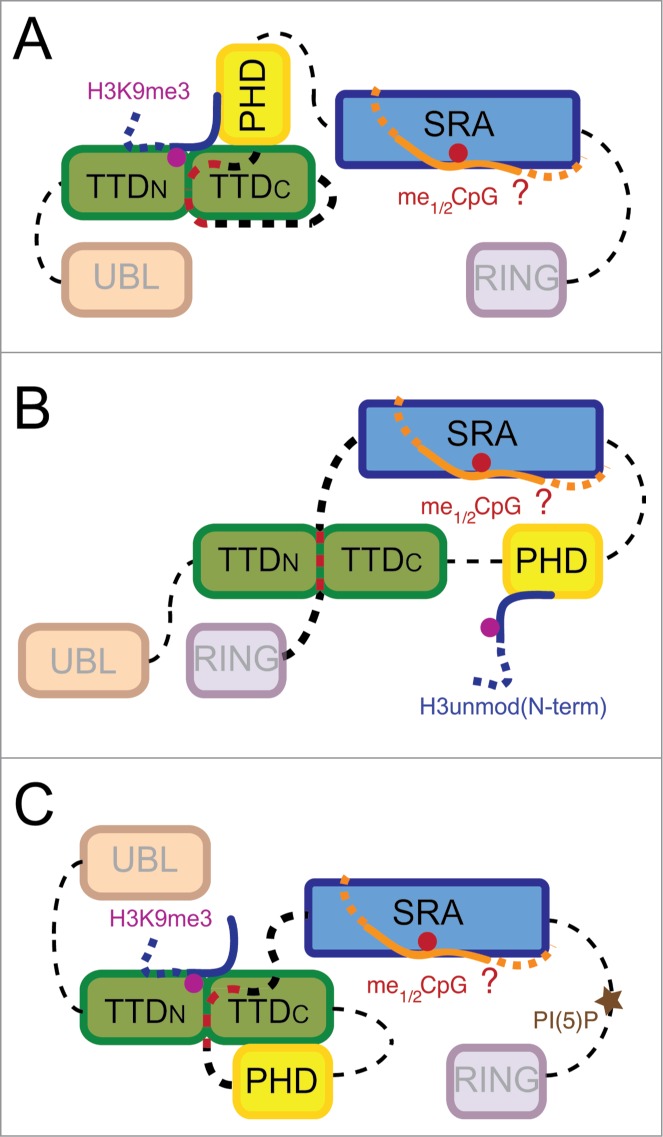
Conformational transition model for UHRF1 binding to chromatin marks. The three conformational states shown (A, B, C) correspond to binding modes deduced by different studies. While some parameters of domain and ligand interaction are known, others are hypothetical. For example, the exact coupling of H3unmod(N-term), H3K9me3, and me1/2CpG binding is not known. Also, the mechanism of PHD inhibition in state (C) has not been resolved. Additional other binding states and relative orientations of the domains might exist. For representation purposes, linker regions are not drawn to scale. Linker regions that are involved in setting up different conformational states via interaction with the peptide-binding groove of the TTD are highlighted. Note that the role of linker 3 has not been determined. Binding of the allosteric regulator PI(5)P to the PBR region releases linker 4 from the TTD peptide-binding groove.
The reduced binding surface of the TTD with the H3-tail in the TTD-PHD/H3K9me3 complex as well as the flexible linkage of the ligand and between the domains explain the observed relative weak enhancement of interaction by the multivalent binding mode. Indeed, the PHD domain is dominating the interaction of the TTD-PHD cassette with the H3K9me3-tail. If binding of the PHD to the H3-tail is abolished by mutation of D334 or modification of the N-terminus of the H3 peptide (A1ac, T3phos), the binding of the neighboring TTD to methylated K9 is drastically weakened or completely lost. Conversely, PHD interaction with the unmodified N-terminus of H3 in the context of the PHD-TTD cassette is largely unaffected by mutation of the TTDN aromatic cage (F152, Y188) or histone modifications such as H3K4me3, which abolish the binding of the isolated TTD to the H3-tail.15,25,26,28
Control of TTD/H3K9me3 interaction by a C-terminal polybasic region
While reductionistic approaches studying isolated domains and combinations thereof have provided detailed insights into the chromatin interaction potential of UHRF1, other studies looked directly at the full-length protein. Here, striking differences in interaction specificity for the H3-tail were observed. While native UHRF1 analyzed in the context of mammalian cell extracts shows clear preferences for H3K9me3, the recombinant protein expressed in bacteria or insect cells binds similarly to unmodified H3 and H3K9me3 peptides.2-4,11,29 We recently showed that this discrepancy is due to intramolecular interaction of the TTD with a polybasic region (PBR) located between the SRA and RING domains of UHRF1 (Fig. 2B).11 Our studies imply that in the absence of ligands (i.e. the recombinant purified protein) the PBR but not linker 2 occupies the peptide-binding groove of the TTD. This prevents interaction with H3K9me3, whereas the PHD is unaffected and can bind the ultimate unmodified H3-tail.
NMR experiments have revealed that a K-R-K motif (K648-R649-K650) is essential for placing the PBR in the peptide-binding groove of the TTD surface. Competition experiments with isolated domains indicate that this interaction is stronger than the similar TTD-linker 2 interplay. It also fully blocks the binding of the H3K9me3-tail. In the context of the recombinant, full-length protein mutagenesis of the K-R-K motif is necessary to release the PBR from the TTD.11 The resulting mutant UHRF1 protein appears to be in an intermediate state as the TTD and PHD both can bind their respective K9me3 and H3unmod(N-terminus) ligands.
Dialysis of recombinant UHRF1 against nuclear extract isolated from HeLa cells induced yet another UHRF1 binding state. Interestingly, this form resembles the native cellular protein in preference for H3K9me3 over H3K9me0 (Fig. 2C).11 The results not only infer an allosteric regulatory mode of UHRF1, but also indicate that states of the protein exist where the PHD domain is prevented from binding H3unmod(N-term).
Multivalent binding to histone H3 and DNA methylation
The isolated SRA domain of UHRF1 binds me1/2CpG with a dissociation constant of around 200 nM.30 Of the 3 chromatin-binding domains it therefore has the tightest interaction with a ligand. While life cell imaging and fluorescence recovery after photobleaching have revealed that the SRA domain dominates targeting of mNp95 to pericentromeric heterochromatin in vivo, mutagenesis studies imply that the PHD and/or TTD domains are also required in this context.29 Indeed, in vitro binding experiments with mNp95 protein isolated from cells show that presence of histone H3K9me3 peptide promotes interaction of the SRA with hemi-methylated DNA. Conversely, interaction with histone peptides is enhanced in presence of un-/methylated DNA.31 While the mechanistic details of this interplay have not yet been unveiled, the findings suggest that the SRA domain cooperates with the TTD and/or PHD in binding to multiple-modified chromatin targets.
A conformational transition model for UHRF1 chromatin binding
It has not been possible to deduce a simple, coherent picture of UHRF1 multivalent chromatin binding on the basis of the research on cassettes composing more than one of the chromatin binding domains, our molecular analysis of the full-length protein as well as based on the multiple studies of deletion and point mutants of the protein in recombinant form, extracted from cells, or in different cellular context (see for example refs.3,29). A putative explanation might come from the idea that the TTD, PHD, and SRA (and possibly UBL and RING) domains do not work independently. Indeed, we favor the view that engagement of the different binding domains of UHRF1 with ligands influences the interaction properties of each other. Since the structural analysis of the isolated domains has not indicated any conformational changes of the binding pockets induced by ligand, the cooperative mode of interaction must be mediated on another level. We suggest that UHRF1 exists in multiple protein conformations where different, structurally invariable binding domains or combinations thereof are either exposed and available for interaction with chromatin marks or where these are occluded and prevented from ligand binding (Fig. 2). We postulate that these conformational states are in constant exchange with each other and that the actual equilibrium between the distinct forms determines the apparent binding properties of UHRF1.
Conserved linker regions likely establish different UHRF1 conformational states
How are different conformational states of UHRF1 established? The regions connecting the conserved and easily recognized chromatin modification-binding domains might play a major role (Figs. 1 and 2). These contain the linker 2 between the TTD and PHD (26 aa in hUHRF1), linker 3 between the PHD and SRA (51 aa in hUHRF1), and the PBR containing region between the SRA and RING (linker 4, 138 aa in hUHRF1). Algorithms that predict secondary structures fail to assign particular folds to these regions. The linkers might therefore form random, intrinsically disordered structures.
Despite the lack of conserved folds, short (ca. 20 aa) sequence stretches are highly conserved within linker 2, linker 3 and PBR (Fig. 3A). For example, the region between aa 372 and 391 of linker 3 shows sequence identity of around 90% in all analyzed UHRF1 proteins.12 Besides the conserved sequences, these stretches of the linker regions are of relative low complexity, enriched in basic amino acids (i.e., Lysine and Arginine residues). This seems to be functionally important as for hUHRF1 the R-R-K element in linker 2 and the K-R-K element in the PBR have been shown to be crucial for binding of these regions to the peptide-binding groove of the TTD.11,26,28 Since the interacting polar residues on the surface of the TTD domain also exhibit conservation throughout all analyzed species (Fig. 3B), the competition of linker 2 and PBR for binding the TTD seems to constitute a major element for establishing different UHRF1 conformational states. Due to the high sequence conservation and a central K-K-K element we further hypothesize that linker 3 might also interface with the peptide-binding groove of the TTD (Fig. 2C). We think that depending on which linker associates with the TTD, different overall conformations of UHRF1 are established. Lastly, we theorize that other regions of UHRF1 might contain polar surfaces similar to the peptide-binding groove of the TTD for accommodating linker 2, linker 3 or PBR in the different conformational states.
Figure 3.
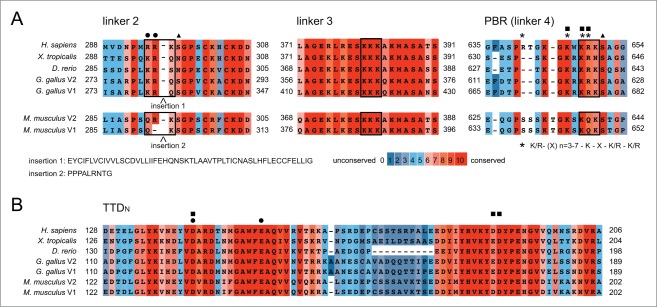
The different linker regions and the peptide binding-groove of the TTD are highly conserved. Multiple sequence alignments of sequence stretches within the linker regions (A) and the peptide-binding groove of the TTD (B) of UHRF1. Alignments were prepared using the PRALINE alignment tool at http://www.ibi.vu.nl/programs/pralinewww/. Numbers correspond to amino acid positions at the beginning and end of the respective protein sequences of the different species. Sequences of insertions found in splicing variants of mouse and chicken proteins are given. Symbols indicate residues implied in the interaction of the TTD peptide-binding groove with linker 2 (dots) or the PBR region within linker 4 (squares). Triangles mark known sites of phosphorylation. Asterisks mark positions corresponding to a putative PIP binding consensus motif within the PBR. Boxes highlight elements of basic residues found in linkers 2, 3 and 4. UHRF1 amino acid sequences according to NCBI: Homo sapiens, NP_001276981.1; Mus musculus v1, NP_001104550.1, Mus musculus v2, NP_001104548.1, Gallus gallus v1, XP_418269.4, Gallus gallus v2, XP_004949013.1; Xenopus laevis, F6UA42.2; Danio rerio, NP_998242.1.
As linker 2 and its interaction with the peptide-binding groove of the TTD might be elementary in setting up distinct conformational states of UHRF1, it is interesting to note that in mouse as well as in chicken splicing variants of the protein exist that only differ in this region. Even more perplexing is the fact that both insertions occur right at the R-R-K element important for the interaction with the peptide-binding groove of the TTD (Fig. 3A). While the linker 2 of mNp95 variant 2 is very similar to the corresponding region of the single human UHRF1 protein, variant 1 contains an insertion of 9 aa, which eliminates the first Arginine of the motif. And whereas the chicken sequences are only predicted but lack experimental validation, variant 1 of this organism has a 54 aa insertion just ahead of the R-R-K element.
Unfortunately, work on mNP95 did so far not discriminate between variants 1 and 2 (note that chicken UHRF1 has not yet been studied). Nevertheless, careful reevaluation of the available data indicates that the linker insertion in variant 1 might block multivalent binding of the TTD-PHD cassette to the H3K9me3-tail.31 We therefore think that the combinatorial binding of both domains is strongly dependent on the sequence and length of linker 2 that could alter the spacing and orientation of the TTD and PHD. On the basis of our conformational transition model of UHRF1, we predict that certain states are not or less populated by the different splicing variants of UHRF1 in mouse and chicken. These should therefore have non-overlapping roles. Detailed analysis of the expression and function of the splicing variants in mouse and/or chicken might be a fruitful avenue for further dissecting the molecular working mode and regulation of UHRF1.
Regulation of UHRF1 conformational states
While intrinsically disordered, the linker regions might nevertheless adopt a series of limited, defined transitional states enabling interaction with different polar surfaces of UHRF1 and determining different conformational and binding states. The kinetic barrier for these transitions could be relatively low due to the intrinsic linker flexibility. As consequence, the protein might be able to rapidly change conformations in response to external stimuli and structural perturbations. This in turn would facilitate changes in the population of certain states therefore affecting UHRF1 target binding and cellular function.32
Different events might affect the equilibrium of the conformational states of UHRF1. If forms of the protein exist that enable more than one of the chromatin binding domains to interact with their ligand (Fig. 2), engagement of one of the exposed regions with its target will facilitate a secondary binding event of the other available domain in a cooperative fashion. While we do not (yet) know how many different forms and binding states of UHRF1 might exist, such interpretation is in full agreement with the observed enhancement of H3K9me3-tail binding of mNP95 by DNA and vice versa.31
Allosteric ligands provide another level of regulating UHRF1 conformational states. Based on our observations that recombinant UHRF1 behaved differently from cellular protein but could be converted to a state with similar H3K9me3-binding properties after dialysis against nuclear extract, we biochemically defined the cofactor and regulator as phosphatidylinositol 5-phosphate (PI(5)P).11 PIPs are amphiphilic glycerophospholipids that consist of a polar inositol head group linked by a phosphodiester bridge to a glycerol backbone branching 2 non-polar fatty acids (Fig. 4). PI(5)P is a low abundance PIP present primarily in mammalian cytoplasmic cell membranes with a smaller population in the cell nucleus.33 While its nuclear functions are not fully clear, its presence in this compartment is highly conserved throughout eukaryotes.34-36 We could show that PI(5)P binding to UHRF1 specifically requires the PBR region and that this interaction releases the latter from the peptide-binding groove of the TTD. In consequence, a conformation of UHRF1 is established that allows the TTD to bind H3K9me3 independent of the PHD.11
Figure 4.
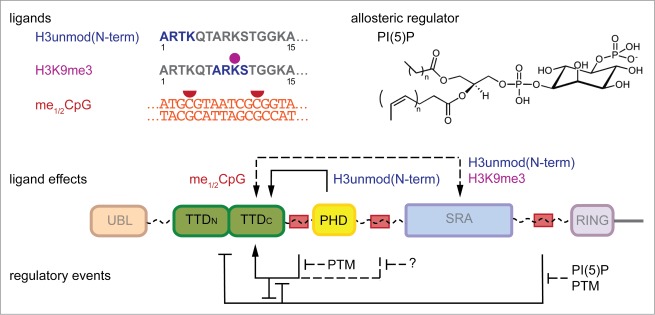
Regulation of the conformational transitions of UHRF1. The interplay of the chromatin-binding domains of UHRF1 and therefore the conformational states of the protein are affected by binding of different ligands, the allosteric regulator phosphatidylinositol 5-phosphate (PI(5)P) as well as posttranslational modifications (PTM) within the linker regions. Enhancing as well as suppressing effects on chromatin modification-binding properties of the TTD, PHD and SRA domains have been described. Dashed lines indicate domain interplay that has not been fully resolved. The regulation of linker 3 is hypothetical (question mark).
While we do not yet know how exactly PI(5)P is bound by UHRF1 – the protein has to accommodate both, the polar head group as well as the hydrophobic acyl chains – it is interesting to note that the PBR resembles in sequence a previously defined consensus PIP interaction motif (R/K-(Xn = 3–7)-K-X-K/R-K/R) shared by several PIP interacting factors.37 Given that the linker 2 and linker 3 are also highly conserved and enriched in basic amino acids, we are wondering whether other ligands modulate the behavior of these regions. We think this is an idea worth entertaining, particularly since synthetic PI(5)P did not fully recuperate all changes of recombinant UHRF1 observed after dialysis against cellular extract.11
The transitions between different conformational states of UHRF1 l conveyed by the linker regions might be further regulated by posttranslational modifications (PTM). These could impair binding to the peptide-binding groove of the TTD or putatively other regions of the protein (Fig. 4). Indeed, it was shown previously that phosphorylation of a Serine residue (S298) adjacent to R295-R296-K297, decreases the binding affinity of the TTD-PHD cassette for the H3K9me3-tail. The altered binding stoichiometry of the complex indicates that the coupling of TTD and PHD mediated by linker 2 is impaired by the PTM.26
A number of additional sites of Serine phosphorylation and other posttranslational modification have been identified in UHRF1 by proteomics studies. Some of these map to highly conserved residues within the linker 2, linker 3 and PBR regions (Fig. 3A).38-43 These events have the potential to regulate UHRF1 conformational states. For example, phosphorylation of S651 that is found during human embryonic stem cell differentiation and is next to the K648-R649-K650 element40 might impair the interaction of the PBR with the peptide-binding groove of the TTD thereby enhancing a particular UHRF1 chromatin-binding mode.
Finally, we point out that binding of UHRF1 to other proteins such as DNMTs, G9a, USP7, etc. will affect the equilibrium of the various forms of the protein. These interactions require distinct, specific surfaces that will or will not be available in the different conformational states of UHRF1. Conversely, it is expected that engagement of a particular protein partner will stabilize a certain conformation and accompanying chromatin mark-binding mode of UHRF1 (Fig. 4).
Cellular role of the different chromatin binding states of UHRF1
Many studies on UHRF1 have emphasized the importance of the 3 chromatin-binding domains in facilitating specific chromatin association and multiple functions in cellular context. We think that the fact that defined functions could not be pinpointed exclusively to particular domains agrees with extensive collaboration and interplay of these regions. It was shown that the TTD, PHD and SRA domains are all participating in targeting of UHRF1 to heterochromatin,3,28 a process that might be dependent on H3K9me3 and meCpG. The TTD, PHD, and SRA regions are also necessary for recruitment of DNMT1 to replicating foci and thus for proper maintenance of DNA-methylation.3,13,28 However, it was also found that UHRF1 binds to promoter regions of euchromatic genes and regulates their expression. Interestingly, these loci lack H3K9me3 but exhibit DNA methylation paired with H3unmod(N-term) (i.e. absence of H3R2 symmetric dimethylation).2 It is conceivable that UHRF1 binds to these genomic regions by multivalent interaction of the PHD and SRA domains with the unmodified N-terminal H3-tail and methylated DNA, respectively.
Additionally, it was found that UHRF1 interacts with the de novo DNA methyltransferases DNMT3a and DNMT3b and that this complex mediates epigenetic silencing in a histone H3 Lysine 9 methyltransferase (i.e., G9a, Suv39h1/2) dependent manner.8 Depletion of Np95 in mouse embryonic stem cells results in severe global DNA hypomethylation.44 A similar DNA methylation defect is caused by depletion of all 3 DNMTs45 or of histone methyltransferases (G9a, Suv39h1/2).46,47 Hence, there is circumstantial evidence that UHRF1 is not only involved in replication-coupled maintenance of DNA methylation, but also in de novo DNA methylation in an H3K9me3-dependent manner. Since de novo methylation has no hemimethylated DNA template, UHRF1 targeting to these chromatin regions might be dependent on interaction of the TTD or TTD-PHD cassette with H3K9me3.
Our work showed that manipulation of cellular PI(5)P levels affects UHRF1 subnuclear localization.11 Several links between the reported functions of UHRF1 and nuclear PI(5)P levels support the idea of allosteric control of the protein. It was shown previously that nuclear PI(5)P levels are tightly regulated during the cell cycle and in response to certain cellular signaling cascades. PI(5)P levels increase up to 20-fold during transition of G1- to S-phase.34,48-50 Cell cycle analysis of UHRF1 localization in relation to heterochromatic foci indicates changed distribution at the onset of S-phase,51 which might be caused by regulation of TTD/H3K9me3 interaction by PI(5)P. Also, UHRF1 interacts with several factors associated with the DNA damage response such as Eme1, TIP60, Ku70, and PARP1.44,52-54 PI(5)P levels are increased after irradiation and exposure to oxidative stress.34,49,50,55 Since H3K9me3 has an important role in repair mechanisms of DNA damage,56 we speculate that UHRF1 might in a specific context target the repair factors to H3K9me3 enriched loci (Fig. 5).
Figure 5.
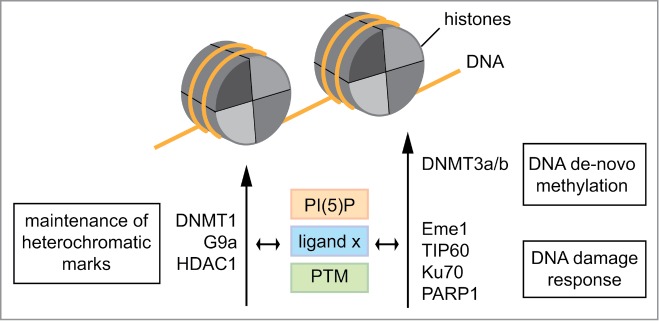
Regulation of interaction of UHRF1 with chromatin and multiple interaction partners. The allosteric regulator phosphatidylinositol 5-phosphate (PI(5)P), different chromatin marks as ligands and posttranslational modifications regulate the postulated different conformations of UHRF1. This impacts on interaction with different protein partners in distinct physiological pathways of chromatin control. Conversely, the protein-binding partners of UHRF1 putatively affect interaction with ligands, allosteric regulators as well as establishment of post-translational modifications.
While nothing is known about the in vivo significance of the PTMs of UHRF1 within the linker region, we note that this essential effector protein is incorporated into various complexes involved in different chromatin regulating processes (i.e. maintenance of DNA methylation, de novo DNA methylation, histone H3 methylation, and histone H4 deacetylation).7,8,44,51,57,58 Thus, it was shown to be an important factor to facilitate DNA replication, cell cycle progression, transcriptional repression, and DNA damage response.2,53,59-61 This variety of functions argues for different states of the protein that provide distinct binding interfaces not only for histone and DNA modifications but also for specific protein interaction partners (Fig. 5).52,53,57,62
Concluding Remarks
Here, we discussed in detail the modification binding behavior of UHRF1 that reveals novel features of a chromatin binding protein. First of all, the different regions of the protein appear not to function in isolation. We expect that further functional and structural analysis of the full-length protein or of cassettes containing more than one of the TTD, PHD and SRA chromatin-binding domains will uncover that these generally do not interact with their targets independent of each other. We think their interplay induces complex binding modes that cannot simply be explained by static multivalent and simultaneous engagement of different protein surfaces with complex ligands (i.e., key and lock behavior). Instead, UHRF1 might exist in multiple, distinct but constantly exchanging conformations. We propose that these are brought about by flexible linkers that intramolecularly compete for a limited number of different sites thereby inducing and stabilizing folds of the protein that expose or occlude the different chromatin-binding interfaces. Besides cooperativity in interaction with chromatin marks, a major consequence of these considerations is a general rationale for comprehending regulation of UHRF1 by allosteric ligands such as PI(5)P and posttranslational modifications that both target linker regions outside of the actual chromatin-binding modules.
The transitions between the different conformations and binding states of UHRF1 are dynamically regulated in a physiological context and in response to external stimuli. This in turn controls specific localization of UHRF1 to defined target loci of the genome as well as interaction with different protein partners such as chromatin modifying enzymes. Any perturbation of these regulatory processes will have profound consequences on chromatin structural organization and subsequently lead to developmental aberrations, chromosome instability and oncogenesis – a major outcome of UHRF1 mutation or abnormal expression.63,64
UHRF1 might be a paradigm for comprehending chromatin interaction and regulation of other factors in response to cellular internal or external stimuli. We note that other related multi-domain proteins as for example DNMT1 and PARP1 also comprise flexible linkers between their functional domains. Interestingly, these might putatively also interact with PIPs via polybasic regions.37 We expect that future studies will not only reveal a more general influence of small cellular metabolites such as PIPs onto chromatin binding factors but also provide a general framework for the role of interchanging, transient conformational states in protein regulation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Ron Finn for help with protein alignment.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, FI 1513/2–1) and the Max Planck Society.
References
- 1.Rose NR, Klose RJ. Understanding the relationship between DNA methylation and histone lysine methylation. Biochim Biophys Acta 2014; 1839:1362–72; PMID:24560929; http://dx.doi.org/ 10.1016/j.bbagrm.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajakumara E, Wang Z, Ma H, Hu L, Chen H, Lin Y, Guo R, Wu F, Li H, Lan F, et al.. PHD finger recognition of unmodified histone H3R2 links UHRF1 to regulation of euchromatic gene expression. Mol Cell 2011; 43:275–84; PMID:21777816; http://dx.doi.org/ 10.1016/j.molcel.2011.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun 2013; 4:1563; PMID:23463006; http://dx.doi.org/ 10.1038/ncomms2562 [DOI] [PubMed] [Google Scholar]
- 4.Karagianni P, Amazit L, Qin J, Wong J. ICBP90, a novel methyl K9 H3 binding protein linking protein ubiquitination with heterochromatin formation. Mol Cell Biol 2008; 28:705–17; PMID:17967883; http://dx.doi.org/ 10.1128/MCB.01598-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esteve PO, Chin HG, Smallwood A, Feehery GR, Gangisetty O, Karpf AR, Carey MF, Pradhan S. Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 2006; 20:3089–103; PMID:17085482; http://dx.doi.org/ 10.1101/gad.1463706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharif J, Koseki H. Recruitment of Dnmt1 roles of the SRA protein Np95 (Uhrf1) and other factors. Prog Mol Biol Transl Sci 2011; 101:289–310; PMID:21507355; http://dx.doi.org/ 10.1016/B978-0-12-387685-0.00008-1 [DOI] [PubMed] [Google Scholar]
- 7.Kim JK, Esteve PO, Jacobsen SE, Pradhan S. UHRF1 binds G9a and participates in p21 transcriptional regulation in mammalian cells. Nucleic Acids Res 2009; 37:493–505; PMID:19056828; http://dx.doi.org/ 10.1093/nar/gkn961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meilinger D, Fellinger K, Bultmann S, Rothbauer U, Bonapace IM, Klinkert WE, Spada F, Leonhardt H. Np95 interacts with de novo DNA methyltransferases, Dnmt3a and Dnmt3b, and mediates epigenetic silencing of the viral CMV promoter in embryonic stem cells. EMBO Rep 2009; 10:1259–64; PMID:19798101; http://dx.doi.org/ 10.1038/embor.2009.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronner C, Fuhrmann G, Chedin FL, Macaluso M, Dhe-Paganon S. UHRF1 Links the Histone code and DNA Methylation to ensure Faithful Epigenetic Memory Inheritance. Genet Epigenet 2010; 2009:29–36; PMID:21643543 [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Yang YZ, Shi CZ, Zhang P, Moyer MP, Zhang HZ, Zou Y, Qin HL. UHRF1 promotes cell growth and metastasis through repression of p16(ink(4)a) in colorectal cancer. Ann Surg Oncol 2012; 19:2753–62; PMID:22219067; http://dx.doi.org/ 10.1245/s10434-011-2194-1 [DOI] [PubMed] [Google Scholar]
- 11.Gelato KA, Tauber M, Ong MS, Winter S, Hiragami-Hamada K, Sindlinger J, Lemak A, Bultsma Y, Houliston S, Schwarzer D, et al.. Accessibility of different histone H3-binding domains of UHRF1 is allosterically regulated by phosphatidylinositol 5-phosphate. Mol Cell 2014; 54:905–19; PMID:24813945; http://dx.doi.org/ 10.1016/j.molcel.2014.04.004 [DOI] [PubMed] [Google Scholar]
- 12.Bronner C, Achour M, Arima Y, Chataigneau T, Saya H, Schini-Kerth VB. The UHRF family: oncogenes that are drugable targets for cancer therapy in the near future? Pharmacol Ther 2007; 115:419–34; PMID:17658611; http://dx.doi.org/ 10.1016/j.pharmthera.2007.06.003 [DOI] [PubMed] [Google Scholar]
- 13.Nishiyama A, Yamaguchi L, Sharif J, Johmura Y, Kawamura T, Nakanishi K, Shimamura S, Arita K, Kodama T, Ishikawa F, et al.. Uhrf1-dependent H3K23 ubiquitylation couples maintenance DNA methylation and replication. Nature 2013; 502:249–53; PMID:24013172; http://dx.doi.org/ 10.1038/nature12488 [DOI] [PubMed] [Google Scholar]
- 14.Nady N, Lemak A, Walker JR, Avvakumov GV, Kareta MS, Achour M, Xue S, Duan S, Allali-Hassani A, Zuo X, et al.. Recognition of multivalent histone states associated with heterochromatin by UHRF1 protein. J Biol Chem 2011; 286:24300–11; PMID:21489993; http://dx.doi.org/ 10.1074/jbc.M111.234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rothbart SB, Krajewski K, Nady N, Tempel W, Xue S, Badeaux AI, Barsyte-Lovejoy D, Martinez JY, Bedford MT, Fuchs SM, et al.. Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nat Struct Mol Biol 2012; 19:1155–60; PMID:23022729; http://dx.doi.org/ 10.1038/nsmb.2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lallous N, Legrand P, McEwen AG, Ramon-Maiques S, Samama JP, Birck C. The PHD finger of human UHRF1 reveals a new subgroup of unmethylated histone H3 tail readers. PLoS One 2011; 6:e27599; PMID:22096602; http://dx.doi.org/ 10.1371/journal.pone.0027599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu L, Li Z, Wang P, Lin Y, Xu Y. Crystal structure of PHD domain of UHRF1 and insights into recognition of unmodified histone H3 arginine residue 2. Cell Res 2011; 21:1374–8; PMID:21808300; http://dx.doi.org/ 10.1038/cr.2011.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avvakumov GV, Walker JR, Xue S, Li Y, Duan S, Bronner C, Arrowsmith CH, Dhe-Paganon S. Structural basis for recognition of hemi-methylated DNA by the SRA domain of human UHRF1. Nature 2008; 455:822–5; PMID:18772889; http://dx.doi.org/ 10.1038/nature07273 [DOI] [PubMed] [Google Scholar]
- 19.Hashimoto H, Horton JR, Zhang X, Bostick M, Jacobsen SE, Cheng X. The SRA domain of UHRF1 flips 5-methylcytosine out of the DNA helix. Nature 2008; 455:826–9; PMID:18772888; http://dx.doi.org/ 10.1038/nature07280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al.. TRIM24 links a non-canonical histone signature to breast cancer. Nature 2010; 468:927–32; PMID:21164480; http://dx.doi.org/ 10.1038/nature09542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruthenburg AJ, Li H, Milne TA, Dewell S, McGinty RK, Yuen M, Ueberheide B, Dou Y, Muir TW, Patel DJ, et al.. Recognition of a mononucleosomal histone modification pattern by BPTF via multivalent interactions. Cell 2011; 145:692–706; PMID:21596426; http://dx.doi.org/ 10.1016/j.cell.2011.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000; 403:41–5; PMID:10638745; http://dx.doi.org/ 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 23.Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol 2007; 8:983–94; PMID:18037899; http://dx.doi.org/ 10.1038/nrm2298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du J, Patel DJ. Structural biology-based insights into combinatorial readout and crosstalk among epigenetic marks. Biochim Biophys Acta 2014; 1839:719–27; PMID:24747177; http://dx.doi.org/ 10.1016/j.bbagrm.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng J, Yang Y, Fang J, Xiao J, Zhu T, Chen F, Wang P, Li Z, Yang H, Xu Y. Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein. J Biol Chem 2013; 288:1329–39; PMID:23161542; http://dx.doi.org/ 10.1074/jbc.M112.415398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arita K, Isogai S, Oda T, Unoki M, Sugita K, Sekiyama N, Kuwata K, Hamamoto R, Tochio H, Sato M, et al.. Recognition of modification status on a histone H3 tail by linked histone reader modules of the epigenetic regulator UHRF1. Proc Natl Acad Sci U S A 2012; 109:12950–5; PMID:22837395; http://dx.doi.org/ 10.1073/pnas.1203701109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie S, Jakoncic J, Qian C. UHRF1 double tudor domain and the adjacent PHD finger act together to recognize K9me3-containing histone H3 tail. J Mol Biol 2012; 415:318–28; PMID:22100450; http://dx.doi.org/ 10.1016/j.jmb.2011.11.012 [DOI] [PubMed] [Google Scholar]
- 28.Rothbart SB, Dickson BM, Ong MS, Krajewski K, Houliston S, Kireev DB, Arrowsmith CH, Strahl BD. Multivalent histone engagement by the linked tandem Tudor and PHD domains of UHRF1 is required for the epigenetic inheritance of DNA methylation. Genes Dev 2013; 27:1288–98; PMID:23752590; http://dx.doi.org/ 10.1101/gad.220467.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res 2010; 38:1796–804; PMID:20026581; http://dx.doi.org/ 10.1093/nar/gkp1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian C, Li S, Jakoncic J, Zeng L, Walsh MJ, Zhou MM. Structure and hemimethylated CpG binding of the SRA domain from human UHRF1. J Biol Chem 2008; 283:34490–4; PMID:18945682; http://dx.doi.org/ 10.1074/jbc.C800169200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pichler G, Wolf P, Schmidt CS, Meilinger D, Schneider K, Frauer C, Fellinger K, Rottach A, Leonhardt H. Cooperative DNA and histone binding by Uhrf2 links the two major repressive epigenetic pathways. J Cell Biochem 2011; 112:2585–93; PMID:21598301; http://dx.doi.org/ 10.1002/jcb.23185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma B, Tsai CJ, Haliloglu T, Nussinov R. Dynamic allostery: linkers are not merely flexible. Structure 2011; 19:907–17; PMID:21742258; http://dx.doi.org/ 10.1016/j.str.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pendaries C, Tronchere H, Racaud-Sultan C, Gaits-Iacovoni F, Coronas S, Manenti S, Gratacap MP, Plantavid M, Payrastre B. Emerging roles of phosphatidylinositol monophosphates in cellular signaling and trafficking. Adv Enzyme Regul 2005; 45:201–14; PMID:16023705; http://dx.doi.org/ 10.1016/j.advenzreg.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 34.Clarke JH, Letcher AJ, D'Santos CS, Halstead JR, Irvine RF, Divecha N. Inositol lipids are regulated during cell cycle progression in the nuclei of murine erythroleukaemia cells. Biochem J 2001; 357:905–10; PMID:11463365; http://dx.doi.org/ 10.1042/0264-6021:3570905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balla T, Wymann M, York JD. Phosphoinositides II: The Diverse Biological Functions. Dordrecht: Springer Netherlands; 2012. [Google Scholar]
- 36.Barlow CA, Laishram RS, Anderson RA. Nuclear phosphoinositides: a signaling enigma wrapped in a compartmental conundrum. Trends Cell Biol 2010; 20:25–35; PMID:19846310; http://dx.doi.org/ 10.1016/j.tcb.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lewis AE, Sommer L, Arntzen MO, Strahm Y, Morrice NA, Divecha N, D'Santos CS. Identification of nuclear phosphatidylinositol 4,5-bisphosphate-interacting proteins by neomycin extraction. Mol Cell Proteomics 2011; 10:M110 003376; PMID:21048195; http://dx.doi.org/ 10.1074/mcp.M110.003376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trotzier MA, Bronner C, Bathami K, Mathieu E, Abbady AQ, Jeanblanc M, Muller CD, Rochette-Egly C, Mousli M. Phosphorylation of ICBP90 by protein kinase A enhances topoisomerase IIalpha expression. Biochem Biophys Res Commun 2004; 319:590–5; PMID:15178447; http://dx.doi.org/ 10.1016/j.bbrc.2004.05.028 [DOI] [PubMed] [Google Scholar]
- 39.Ma H, Chen H, Guo X, Wang Z, Sowa ME, Zheng L, Hu S, Zeng P, Guo R, Diao J, et al.. M phase phosphorylation of the epigenetic regulator UHRF1 regulates its physical association with the deubiquitylase USP7 and stability. Proc Natl Acad Sci U S A 2012; 109:4828–33; PMID:22411829; http://dx.doi.org/ 10.1073/pnas.1116349109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rigbolt KT, Prokhorova TA, Akimov V, Henningsen J, Johansen PT, Kratchmarova I, Kassem M, Mann M, Olsen JV, Blagoev B. System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci Signal 2011; 4:rs3; PMID:21406692; http://dx.doi.org/ 10.1126/scisignal.2001570 [DOI] [PubMed] [Google Scholar]
- 41.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A 2008; 105:10762–7; PMID:18669648; http://dx.doi.org/ 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 2006; 127:635–48; PMID:17081983; http://dx.doi.org/ 10.1016/j.cell.2006.09.026 [DOI] [PubMed] [Google Scholar]
- 43.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, Gnad F, Cox J, Jensen TS, Nigg EA, et al.. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal 2010; 3:ra3; PMID:20068231; http://dx.doi.org/ 10.1126/scisignal.2000475 [DOI] [PubMed] [Google Scholar]
- 44.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al.. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 2007; 450:908–12; PMID:17994007; http://dx.doi.org/ 10.1038/nature06397 [DOI] [PubMed] [Google Scholar]
- 45.Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al.. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cell 2006; 11:805–14; PMID:16824199; http://dx.doi.org/ 10.1111/j.1365-2443.2006.00984.x [DOI] [PubMed] [Google Scholar]
- 46.Lehnertz B, Ueda Y, Derijck AAHA, Braunschweig U, Perez-Burgos L, Kubicek S, Chen TP, Li E, Jenuwein T, Peters AHFM. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 2003; 13:1192–200; PMID:12867029; http://dx.doi.org/ 10.1016/S0960-9822(03)00432-9 [DOI] [PubMed] [Google Scholar]
- 47.Dong KB, Maksakova IA, Mohn F, Leung D, Appanah R, Lee S, Yang HW, Lam LL, Mager DL, Schubeler D, et al.. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J 2008; 27:2691–701; PMID:18818693; http://dx.doi.org/ 10.1038/emboj.2008.193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Divecha N, Banfic H. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor ( for IGF-1 ) in the plasma membrane , and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kin. EMBO J 1991; 1:3207–14; PMID:1655412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, Heck AJ, D'Santos CS, Divecha N. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol Cell 2006; 23:685–95; PMID:16949365; http://dx.doi.org/ 10.1016/j.molcel.2006.07.014 [DOI] [PubMed] [Google Scholar]
- 50.Jones DR, Foulger R, Keune WJ, Bultsma Y, Divecha N. PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J 2013; 27:1644–56; PMID:23241309; http://dx.doi.org/ 10.1096/fj.12-218842 [DOI] [PubMed] [Google Scholar]
- 51.Papait R, Pistore C, Negri D, Pecoraro D, Cantarini L, Bonapace IM. Np95 is implicated in pericentromeric heterochromatin replication and in major satellite silencing. Mol Biol Cell 2007; 18:1098–106; PMID:17182844; http://dx.doi.org/ 10.1091/mbc.E06-09-0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Achour M, Fuhrmann G, Alhosin M, Ronde P, Chataigneau T, Mousli M, Schini-Kerth VB, Bronner C. UHRF1 recruits the histone acetyltransferase Tip60 and controls its expression and activity. Biochem Biophys Res Commun 2009; 390:523–8; PMID:19800870; http://dx.doi.org/ 10.1016/j.bbrc.2009.09.131 [DOI] [PubMed] [Google Scholar]
- 53.Mistry H, Gibson L, Yun JW, Sarras H, Tamblyn L, McPherson JP. Interplay between Np95 and Eme1 in the DNA damage response. Biochem Biophys Res Commun 2008; 375:321–5; PMID:18692478; http://dx.doi.org/ 10.1016/j.bbrc.2008.07.146 [DOI] [PubMed] [Google Scholar]
- 54.De Vos M, El Ramy R, Quenet D, Wolf P, Spada F, Magroun N, Babbio F, Schreiber V, Leonhardt H, Bonapace IM, et al.. Poly(ADP-ribose) polymerase 1 (PARP1) associates with E3 ubiquitin-protein ligase UHRF1 and modulates UHRF1 biological functions. J Biol Chem 2014; 289:16223–38; PMID:24782312; http://dx.doi.org/ 10.1074/jbc.M113.527424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Divecha N, Banfic H, Irvine RF. The polyphosphoinositide cycle exists in the nuclei of Swiss 3T3 cells under the control of a receptor (for IGF-I) in the plasma membrane, and stimulation of the cycle increases nuclear diacylglycerol and apparently induces translocation of protein kinase C to the nucleus. EMBO J 1991; 10:3207–14; PMID:1655412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y, Xu C, Price BD. Mechanistic links between ATM and histone methylation codes during DNA repair. Prog Mol Biol Transl Sci 2012; 110:263–88; PMID:22749149; http://dx.doi.org/ 10.1016/B978-0-12-387665-2.00010-9 [DOI] [PubMed] [Google Scholar]
- 57.Unoki M, Nishidate T, Nakamura Y. ICBP90, an E2F-1 target, recruits HDAC1 and binds to methyl-CpG through its SRA domain. Oncogene 2004; 23:7601–10; PMID:15361834; http://dx.doi.org/ 10.1038/sj.onc.1208053 [DOI] [PubMed] [Google Scholar]
- 58.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, Jacobsen SE. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 2007; 317:1760–4; PMID:17673620; http://dx.doi.org/ 10.1126/science.1147939 [DOI] [PubMed] [Google Scholar]
- 59.Arima Y, Hirota T, Bronner C, Mousli M, Fujiwara T, Niwa S, Ishikawa H, Saya H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cell 2004; 9:131–42; PMID:15009091; http://dx.doi.org/ 10.1111/j.1356-9597.2004.00710.x [DOI] [PubMed] [Google Scholar]
- 60.Jeanblanc M, Mousli M, Hopfner R, Bathami K, Martinet N, Abbady AQ, Siffert JC, Mathieu E, Muller CD, Bronner C. The retinoblastoma gene and its product are targeted by ICBP90: a key mechanism in the G1/S transition during the cell cycle. Oncogene 2005; 24:7337–45; PMID:16007129; http://dx.doi.org/ 10.1038/sj.onc.1208878 [DOI] [PubMed] [Google Scholar]
- 61.Mistry H, Tamblyn L, Butt H, Sisgoreo D, Gracias A, Larin M, Gopalakrishnan K, Hande MP, McPherson JP. UHRF1 is a genome caretaker that facilitates the DNA damage response to gamma-irradiation. Genome Integr 2010; 1:7; PMID:20678257; http://dx.doi.org/ 10.1186/2041-9414-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Berkyurek AC, Suetake I, Arita K, Takeshita K, Nakagawa A, Shirakawa M, Tajima S. The DNA methyltransferase Dnmt1 directly interacts with the SET and RING finger-associated (SRA) domain of the multifunctional protein Uhrf1 to facilitate accession of the catalytic center to hemi-methylated DNA. J Biol Chem 2014; 289:379–86; PMID:24253042; http://dx.doi.org/ 10.1074/jbc.M113.523209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Papait R, Pistore C, Grazini U, Babbio F, Cogliati S, Pecoraro D, Brino L, Morand AL, Dechampesme AM, Spada F, et al.. The PHD domain of Np95 (mUHRF1) is involved in large-scale reorganization of pericentromeric heterochromatin. Mol Biol Cell 2008; 19:3554–63; PMID:18508923; http://dx.doi.org/ 10.1091/mbc.E07-10-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bronner C, Krifa M, Mousli M. Increasing role of UHRF1 in the reading and inheritance of the epigenetic code as well as in tumorogenesis. Biochem Pharmacol 2013; 86:1643–9; PMID:24134914; http://dx.doi.org/ 10.1016/j.bcp.2013.10.002 [DOI] [PubMed] [Google Scholar]