Abstract
Animal sex is determined by the number of X chromosomes in many species, creating unequal gene dosage (aneuploidy) between sexes. Dosage Compensation mechanisms equalize this dosage difference by regulating X-linked gene expression. In the nematode C. elegans the current model suggests that DC is achieved by a 2-fold transcriptional downregulation in hermaphrodites mediated by the Dosage Compensation Complex (DCC), which restricts access to RNA Polymerase II by an unknown mechanism. Taking a nuclear organization point of view, we showed that the male X chromosome resides in the pore proximal subnuclear compartment whereas the DCC bound to the X, inhibits this spatial organization in the hermaphrodites. Here we discuss our results and propose a model that reassigns the role of DCC from repression of genes to inhibition of activation.
Keywords: C. elegans, chromatin compaction, dosage compensation, nuclear organization, nuclear pores, X chromosome
Abbreviations
- DC
dosage compensation
- DCC
dosage compensation complex
- SMC
structural maintenance of chromosomes.
Introduction
Sex chromosome based sex determination leads to difference in sex chromosome copy number between the sexes.1 In mammals, female animals carry 2 X chromosomes (XX) whereas males have only a single X copy and a Y chromosome (XY). To compensate this X-linked gene dosage difference, one of the X chromosomes in females is almost completely silenced, thereby equalizing the expression between the 2 sexes. In Drosophila, the single X chromosome in males (XY) is hyper transcribed to equalize expression with females (XX). In C. elegans, hermaphrodites (XX) reduce transcription from both X chromosomes by half to match transcriptional output from the single X chromosome in the males (XO). These different strategies that compensate for the gene dosage imbalance between sexes are collectively referred to as Dosage Compensation (DC).2 Failure to compensate dosage imbalance leads to embryonic lethality as documented by mutations crippling the components of dosage compensation machinery, independently of the compensation mechanism in question.3-5
Sex determination in mammals is dependent on the presence or absence of a Y chromosome. In worms as in Drosophila, the sex of the animal is determined by the ratio between X and autosome (X:A).1 Careful genetic dissection in C. elegans revealed that although both controlled by the same upstream transcription factor XOL-1 (XO lethal), sex determination and dosage compensation are separable phenomena.6 In hermaphrodites, the X:A ratio leads to the repression of xol-1 transcription. This leads to the activation of DC and repression of the masculinizing gene her-1 (hermaphrodization of XO animals). In males, the X:A ratio allows xol-1 expression; XOL-1 represses DC and activates her-1, leading to male development.7 Forward genetic screens for mutations leading to hermaphrodite specific phenotypic defects characterized a number of mutations wherein X-linked genes are overexpressed, sign of absence of dosage compensation (Dumpy genes: dpy-21, −26, −27, −28, −30).3,4 Another set of mutants showed Sex Determination and Dosage Compensation Deficiency (sdc-1, −2, −3).8-10 Remarkably, all proteins of the Dpy and Sdc mutant gene classes interact and form a single complex, the Dosage Compensation Complex (DCC).11 The DCC is structurally similar to a condensin I complex, with 2 SMC-like components, DPY-27 and MIX-1, the latter being also part of the mitotic and meiotic condensin complexes.11 Loading of the DCC on the X chromosome is nucleated at rex (recruitment element on X) sites characterized by a 12 bp consensus sequence called MEX (Motif Enriched on X).12,13 Interestingly, these sites also seem to recruit the mitotic and meiotic Condensin I/II complexes in an sdc-2 dependent manner.14 Current estimates suggest the presence of 100 to 300 rex sites on the X chromosome, although only 38 have been formally characterized.12 This imprecise estimation is due to the fact that the DCC is able to spread along the X chromosome from its nucleation sites and accumulate at promoters upstream of transcription start sites.12,13,15
How the loading of a condensin-like complex represses transcription remains unclear. A wealth of genome-wide studies has shown that dosage compensated X chromatin is different from autosomal chromatin in various manners. At the sequence level, the X chromosome has a slightly higher GC content in promoter sequences compared to autosomes.13 As DNA sequence dictates in part nucleosome binding, X-linked gene promoters have higher nucleosome occupancy, independently of DC.16 At the chromatin level, the dosage compensated X chromosomes are depleted for the histone variant H2A.Z (HTZ-1) and H4K16 acetylation while they carry high H4K20 monomethylation.17-20 The first two might be a consequence of lower transcription levels, while the latter is a consequence of DCC binding as the methylation mark spreads with the DCC.18 The function of this methylation marks remains however elusive. Animals deficient for set-4, the methyltransferase carrying di- and tri-methylation of H4K20, in which the entire genome is marked with H4K20 monomethyl have no dosage compensation related phenotypes. Finally, X-linked genes appear transcriptionally over-expressed compared to autosomal genes in males and dosage compensation deficient animals,21-23 suggesting an X-specific transcription activation mechanism. Genome-wide run-on experiments coupled to RNA polymerase II chromatin immunoprecipitation have shown that the DCC globally reduces transcription from the X chromosomes by evicting the polymerase from X-linked genes.22,24 The precise mechanism by which polymerase loading is impaired by the DCC still remains mysterious.25,26
The current model of DC, in which transcription is repressed in hermaphrodites by the DCC leaves many caveats. For example, DCC binding to a gene is not a proxy for its transcriptional downregulation. Across the X chromosome, there are many examples of compensated genes not bound by DCC and conversely, DCC bound genes that do not undergo compensation.12 Secondly, end to end chromosomal fusions between X and autosomes show DCC spreading on autosomal regions for few kilobases but with little change in the transcriptional status of the autosomal genes.13,18 Moreover, autosomal regions bound by DCC do not show a significant change in transcription.12 The exact mechanism of DC in hermaphrodites remains unclear, in particular the molecular path linking DCC loading onto the X chromosome and X-linked transcriptional gene regulation by impairment of polymerase loading.
A Nuclear Organization Perspective to Understand DC
Dosage compensation has been investigated from the nuclear organization point of view in different model organisms, exploring the interaction between chromosome organization and chromosome-wide gene regulation.27 In mammals, DC is initiated by the expression of the long noncoding RNA Xist from one of the X copies in females. Xist spreads along the entire X chromosome and represses transcription by multiple mechanisms.28 Xist binding changes the chromatin conformation of the X chromosome29 and its spreading depends on the 3D organization of the X chromosome. Chromosome Conformation Capture techniques demonstrated that the first regions bound by Xist are in close proximity to the Xist locus in the nuclear space.30 Moreover, binding of the Xist RNA leads to local chromatin conformation changes correlated with gene expression regulation.31
Another line of evidence supporting the role of nuclear organization in chromosome-wide gene regulation came from studies of the Drosophila DCC. In flies, X-linked genes are upregulated 2-fold in males; a consequence of the recruitment of the fly DCC, the male-specific lethal (MSL) complex onto the X chromosome.32 Consequent to this loading, the male X chromosome shows a specific 3D conformation.33 Fly DCC activates X-linked gene transcription via H4K16 acetylation34 and co-purifies with 2 nucleoporins, MTOR/TPR and NUP153. These nucleoporins interact broadly with the male X chromosome and are important for transcriptional upregulation and spatial positioning,35,36 suggesting a functional link between dosage compensation and nuclear organization.
In comparison, very little is known about sex-specific nuclear organization of the X chromosome in worms. All autosomes arrange as an Omega (Ω) shape inside the nuclear space, with both arms anchored at the nuclear lamina. On the opposite, the X chromosome in hermaphrodites is largely internal and only loosely interacting with the periphery at telomeres.37,38 This difference in organization was suggested early to be a consequence of dosage compensation in hermaphrodite animals.37 Given the similarity between the DCC and condensins, a specific higher order structure of the X chromosomes has been suggested as a model for years,39 but never tested directly. In males, no specific chromosome organization or X-specific chromatin marks have been described owing to the technical roadblock of obtaining a pure male population in large quantities. Additionally, addressing the X organization in males can provide insights into the yet unexplored mechanisms of the suggested male X chromosome upregulation.21-23 Here, we discuss a new model of DC in worms based on the latest findings. We describe an X specific organization in C. elegans and the possible links between tridimensional folding of the chromosome inside the nucleus and its transcriptional regulation.
The DCC Condenses the X Chromosome in Hermaphrodites
Since the DCC shares subunits with Condensin I and II and is structurally similar to these complexes, it was proposed early that the DCC could promote X chromosome compaction during interphase.39 To test this hypothesis directly, we and others made use of DNA-fluorescent in situ hybridization to measure actual physical distances between different loci separated by a given genomic distance. These distances were measured in both sexes in either embryos post establishment of DC (between 50–150 cell stages) or in tail tip hypodermal cells of adult animals. In both systems X chromosome distances were significantly shorter in hermaphrodites compared to males, irrespective of their position along the chromosome (Fig. 1A–D). Whole chromosome paints in adult intestinal or tail tip nuclei led to the same conclusion that the X chromosome is more compact in hermaphrodites than in males.40 Finally, formally demonstrating that increased compaction is a consequence of DCC loading onto the X, non-compensating XO hermaphrodites9 show similar X compaction rates as males41 while knock-down of DCC subunits leads to increased X chromosome volume.40 Together, these studies demonstrate that the DCC indeed condenses the X chromosome. Surprisingly however, 2-points measurements in males showed that the X chromosome appears significantly more compact than autosomes (Fig. 1E).41
Figure 1.
The Dosage Compensation Complex compacts the (X)chromosome in hermaphrodites. (A) Schematic map of the chromosome showing the positions of the genomic segments analyzed. Overlay represents the LEM-2 (lamina-associated protein) ChIP-chip profile with high (red) and low (blue) enrichment on the chromosomes, adapted from Ikegami et al. 2010 (B) Partial projection of a z-stack obtained after FISH with 2 probes separated by 1 Mb on X chromosome as shown in A. Images were acquired in hermaphrodite (top panel) or male (bottom panel) embryos. FISH probes are shown in red and green, nuclei counterstained with DAPI (blue) (C) Box plot representation for the nuclear diameter normalized distance distribution for the 1 Mb region on the autosome (green) and the X chromosome (white) as shown in A, in hermaphrodite embryos (solid line) or male (dashed lines) embryos. (D) Cumulative distribution representation for the data in C. (E) Model of chromatin compaction summarizing the observed chromosome compaction levels of chromosomes according to sex and DCC presence.
Another question arising from these observations was whether the DCC mediates this compaction by enhanced interaction between specific sites (for example rex sites) giving rise to rosette structures or whether DCC loading led to sequence-independent global compaction? To answer this we scored the frequency of ‘rex/rex’ and ‘rex/non-rex’ colocalization in both sexes. In hermaphrodites, rex sites colocalized with each other more frequently compared to rex/non-rex (∼18% compared to ∼8%), irrespective of the genomic distance between them. In males, although the overall frequency of interaction was low, the trend remained the same. This suggests that the observed higher frequency of interaction in hermaphrodites is not rex specific but an outcome of DCC mediated overall chromatin compaction. However, our observations are limited to a small subset of rex sites. Owing to the large number of these motifs on the X chromosome, there might be a possibility of random rex sites clustering together or a more dynamic interaction between these sites (Fig. 1E). Chromosome Conformation Capture experiments will likely provide insights into whether and how rex sites impact on genome folding.
The Male X Chromosome Demonstrates a Spatial Preference for the Nuclear Periphery
Gene expression regulation is controlled at multiple levels, ranging from transcription factor binding to the position of the locus inside the nuclear space. Over the last years, it became increasingly clear that subnuclear positioning can have a profound effect on gene expression in many model systems.42 In C. elegans, 3 distinct nuclear domains have been described.43 The nuclear interior was shown to be enriched with active developmentally regulated genes, reminiscent of the internal euchromatic regions seen using electron microscopy.44 The nuclear periphery is a contrasted environment with silent chromatin anchored at the nuclear lamina37,38,45 and active domains located close to nuclear pores by transcription-dependent mechanisms.46,47 We asked whether the X chromosome would show a sex-specific spatial positioning inside the nucleus. Using both FISH and live imaging, we scored the position of multiple loci across the X chromosome in embryos. In hermaphrodites, our observations agreed with the published reports that the X remains largely randomly positioned except the extremities of the arms that interact with the nuclear periphery (Fig. 2B). In males however, all loci scored on the X chromosome showed perinuclear positioning, demonstrating a sex-specific X chromosome conformation (Fig. 2A). Moreover, knocking down DCC subunits in hermaphrodites led to male-like peripheral positioning of an otherwise randomly located X locus. Together, these experiments suggested that default positioning of the X chromosome is at the nuclear periphery and that DCC loading led to internal (re-)positioning of the chromosome.
Figure 2.
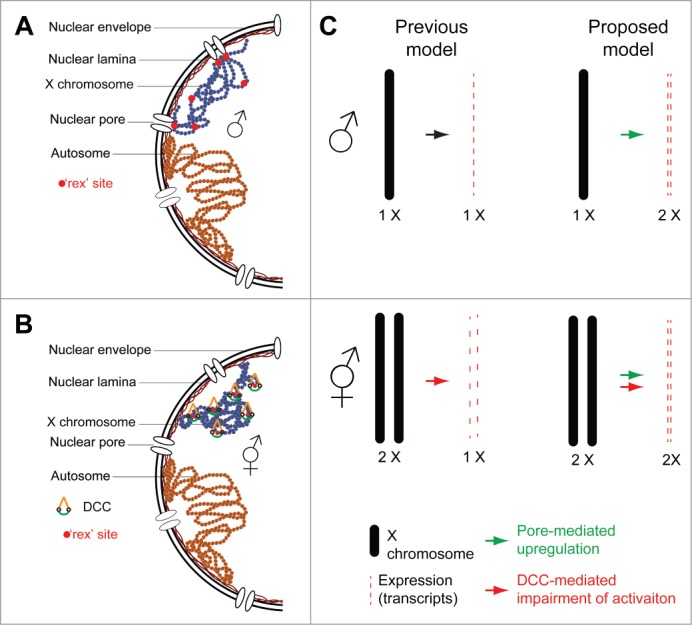
Interplay between Nuclear Organization and Dosage Compensation. (A) Schematic representation of the chromosome organization inside a male nucleus. rex sites direct the interaction of the X chromosome with the nuclear periphery, which in turn leads to pore interactions, promoting transcriptional upregulation. (B) Same as A in a hermaphrodite nucleus. DCC binding to rex sites and spreading on X chromatin impairs perinuclear localization and interaction with nuclear pore proteins, thereby impairing activation of X-linked transcripts and promoting chromosomal compaction. (C) Comparison between the previous and proposed models of Dosage Compensation in C. elegans. In the proposed model, the final transcriptional output of the X chromosome is a consequence of pore mediated activation in males and impairment of this activation by the DCC in hermaphrodites.
A ‘rex’ Site Motif is Sufficient to Target the X Chromosome to the Nuclear Periphery in Males
These observations raised a fundamental question: which sequences target the X chromosome to the periphery in males? Very few DNA motifs were characterized that are sufficient to autonomously direct an external locus to a specific sub-nuclear position. In yeasts, 2 motifs known as gene recruitment sequence elements (GRSI and GRSII) have been reported to target ectopic loci to the nuclear pores.48,49 More recently, mammalian Lamina associating sequences (LAS) binding several transcriptional repressors target different loci to the nuclear lamina for repression.50,51 In C. elegans, heat shock-induced promoters were shown to be recruited to nuclear pores in a similar manner as yeast GRS.46
Prime candidates for X-specific sequences that may direct male-specific targeting of the chromosome to the nuclear periphery are rex sites. rex sites are enriched on the X chromosome and in hermaphrodites, they are sufficient for loading of the DCC.52 Since our FISH analysis showed an enrichment of rex sites at the periphery in males, we hypothesized that these sites could be sufficient to target the chromosome in males. To test this, we generated transgenic lines with a 33 bp rex sequence52 ectopically integrated in the middle of an autosome. FISH analysis with embryos in both sexes revealed that the locus is randomly located in hermaphrodite nuclei whereas it shows significant enrichment at the nuclear rim in males. Moreover, insertion at the same site of the same construct with a mutated rex sequence, unable to load the DCC, is no longer able to target this locus to the periphery in males. Given the specificity and density of rex sites on the X chromosome, these sites would be sufficient to target the entire X chromosome to the periphery, although there might be other sequences involved in the process.
The X Chromosome in Males Shows Widespread Interaction With a Nuclear Pore Protein
If rex sites are sufficient to target the X chromosome to the nuclear periphery in males, which perinuclear compartment is the X chromosome targeted to? Two major subcompartments have been described at the nuclear rim: the nuclear lamina, which in C. elegans as in other metazoans clusters repressed chromatin; and nuclear pores, which was shown to be a hub for active genes. To answer this question, we made use of a genome-wide mapping protein-DNA interactions in vivo, DNA adenine methyltransferase identification (DamID). In brief, the protein of interest (in our case, lamin/LMN-1 and a nucleoporin/MEL-28) is fused with a DNA Adenine Methyltransferase (DAM) which methylates adenine residues in close proximity with the protein. These methylated fragments can then be amplified and sequenced to identify binding sites of the protein of interest. By using a single-tube protocol, we could generate genome-wide interaction profiles starting from 20 adult animals, hermaphrodites or males, for LMN-1 and MEL-28, although at low resolution (100 kb). Genome interaction with the nuclear lamina was similar in males and hermaphrodites. However for MEL-28, the X chromosome showed a broad interaction compared to autosomes, specifically in males. The male X chromosome therefore resides at the nuclear periphery and interacts with nuclear pore proteins, likely in proximity of these given the observed location of the X chromosome (Fig. 2A). However, we cannot rule out that the interaction with a pore protein occurs inside the nucleoplasm.
Interaction with nuclear pore proteins has been shown to facilitate transcriptional upregulation in C. elegans and other species.53,54 Therefore, it would be interesting to see whether nuclear pore proteins are involved in the upregulation of the single X copy in males. Secondly, whether DCC knockdown leads to enhanced interaction with the nuclear pore proteins in hermaphrodites remains to be tested. Since XX embryos that fail to dosage compensate, die due to excessive transcription from the 2 X copies, interaction with nucleoporins would serve as a general mechanism to upregulate transcription in both sexes.
A Structural Model for Dosage Compensation in C. elegans
As discussed above, how DCC mediates transcriptional downregulation remains unclear. Based on our results, we propose a “structural” model of dosage compensation, based on the subnuclear localization of the X chromosome. Our observations suggest that the previously reported upregulation of the X chromosome occurs in males at the nuclear pores via interaction with nucleoporins. Targeting to the nuclear periphery is achieved by rex sites, which leads to perinuclear positioning of the male X chromosome (Fig. 2A and C). However, owing to the low resolution of our DamID datasets, it is impossible to determine whether rex sites preferentially interact with nuclear pores. Using a modeling approach, we showed that interaction of a polymer with discrete anchoring sites such as nuclear pores could explain the observed increased compaction of the X compared to autosomes in males (in the absence of DCC). In hermaphrodites, recruitment of the DCC on ‘rex’ sites masks the interaction of the X chromosome with perinuclear anchors and nucleoporins, thereby modifying nuclear positioning and impairing transcriptional activation (Fig. 2B and C). This model could explain a number of previous observations. First, it would provide a mechanism for the male-specific overexpression of X-linked genes. Second, it would explain why DCC spreading onto the autosomal moiety of X to autosome fusions (up to 2 Mb)13,24 does not correlate with transcriptional downregulation of autosomal genes.13 If rex site anchoring activates transcription and DCC impairs this activation, in the absence of rex site (in case of autosomes) the loading of the DCC should have little transcriptional impact.
Open Questions and Considerations with the Model
Although we demonstrate that a single rex site is able to autonomously target a randomly positioned locus toward the nuclear periphery in males specifically, we do now know whether rex sites interact with nuclear pores or other perinuclear structures. Similarly, although the X chromosome interacts broadly with a nuclear pore subunit, the low resolution obtained with DamID did not allow to identify which regions on the X chromosome interact with the nucleoporins in males. In particular, the X chromosome is highly enriched for non coding RNA genes, some of which have been reported to be transcribed at the nuclear pores in hermaphrodites.47 Second, the X-linked gene activation machinery remains largely unknown but is likely to be similar to the nucleoporin containing activating complex acting on autosomes. In particular, the SAGA/TREX complex has been shown in both mammalian cells, Drosophila and nematodes to mediate transcriptional activation at the pores via interaction with nuclear basket proteins in a number of other organisms.46,55,56 Third, since the X chromosome in hermaphrodites is 2-fold downregulated and not completely silent, two mechanisms linking DCC loading and transcriptional downregulation could be envisioned. In a first static scenario, the DCC remains stably associated with the X chromosome in hermaphrodites and does not allow association with nuclear pores, impairing transcriptional upregulation while allowing transcription. In a second, dynamic scenario, DCC binding could be a highly dynamic process due to the sliding properties of the condensin-like complex. Regions not masked by the DCC would get intermittent interaction with pore proteins, initiating transcriptional activation. This creates open chromatin, attracting the DCC which in turns restricts transcription. Sliding of the DCC would then re-initiate the activation/repression cycle. Setting up conditional tools will help addressing the dynamics of DCC loading and pore mediated activation.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The Meister laboratory is funded by the Swiss National Foundation (SNF assistant professor grant PP00P3_133744) and the University of Bern.
References
- 1.Disteche CM. Dosage Compensation of the Sex Chromosomes. Annu Rev Genet 2011; 46:120913144909001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ercan S. Mechanisms of x chromosome dosage compensation. J genomics [Internet] 2015. [cited 2015 Apr 1] 3:1-19. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4303597&tool=pmcentrez&rendertype=abstract; PMID:25628761; http://dx.doi.org/ 10.7150/jgen.10404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hodgkin J. X chromosome dosage and gene expression in Caenorhabditis elegans: Two unusual dumpy genes. MGG Mol Gen Genet [Internet] 1983. [cited 2015 Apr 30]; 192:452-8. Available from: http://link.springer.com/10.1007/BF00392190; http://dx.doi.org/ 10.1007/BF00392190 [DOI] [Google Scholar]
- 4.Plenefisch JD, Delong L, Meyer BJ. Genes That Implement. 1989; 121:57-76; PMID:2917714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belote J, Lucchesi J. Male-specific lethal mutations of Drosophila melanogaster. Genetics [Internet] 1980; (1):165-86. Available from: http://www.genetics.org/content/96/1/165.short. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgkin J. Sex Determination and Dosage Compensation in Caenorhabditis Elegans. Annu Rev Genet [Internet] 1987; 21:133-54. Available from: http://dx.doi.org/ 10.1146/annurev.ge.21.120187.001025 [DOI] [PubMed] [Google Scholar]
- 7.Zarkower D. Somatic sex determination. WormBook 2006. :1-12; PMID:18050479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nusbaum C, Meyer BJ. The Caenorhabditis elegans gene sdc-2 controls sex determination and dosage compensation in XX animals. Genetics 1989; 122:579-93; PMID:2759421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis TL, Meyer BJ. SDC-3 coordinates the assembly of a dosage compensation complex on the nematode X chromosome. Development [Internet] 1997; 124:1019-31. Available from: http://www.ncbi.nlm.nih.gov/pubmed/9056777; PMID:9056777 [DOI] [PubMed] [Google Scholar]
- 10.Villeneuve A M, Meyer BJ. sdc-1: a link between sex determination and dosage compensation in C. elegans. Cell 1987; 48:25-37; PMID:3791412; http://dx.doi.org/ 10.1016/0092-8674(87)90352-7 [DOI] [PubMed] [Google Scholar]
- 11.Csankovszki G, Collette K, Spahl K, Carey J, Snyder M, Petty E, Patel U, Tabuchi T, Liu H, McLeod I, et al.. Three Distinct Condensin Complexes Control C. elegans Chromosome Dynamics. Curr Biol [Internet] 2009; 19:9-19. Available from: http://dx.doi.org/ 10.1016/j.cub.2008.12.006; PMID:19119011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jans J, Gladden JM, Ralston EJ, Pickle CS, Michel AH, Pferdehirt RR, Eisen MB, Meyer BJ, Michel H, Pferdehirt RR, et al.. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev [Internet] 2009. [cited 2014 Mar 21]; 23:602-18. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2658519&tool=pmcentrez&rendertype=abstract; PMID:19270160; http://dx.doi.org/ 10.1101/gad.1751109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ercan S, Dick LL, Lieb JD. The C. elegans dosage compensation complex propagates dynamically and independently of X chromosome sequence. Curr Biol [Internet] 2009. [cited 2014 Mar 21]; 19:1777-87. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2783177&tool=pmcentrez&rendertype=abstract; PMID:19853451; http://dx.doi.org/ 10.1016/j.cub.2009.09.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kranz A-L, Jiao C-Y, Winterkorn LH, Albritton SE, Kramer M, Ercan S. Genome-wide analysis of condensin binding in Caenorhabditis elegans. Genome Biol [Internet] 2013. [cited 2014 Mar 21]; 14:R112. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24125077; PMID:24125077; http://dx.doi.org/ 10.1186/gb-2013-14-10-r112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csankovszki G, McDonel P, Meyer BJ. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science 2004; 303:1182-5; PMID:14976312; http://dx.doi.org/ 10.1126/science.1092938 [DOI] [PubMed] [Google Scholar]
- 16.Ercan S, Lubling Y, Segal E, Lieb JD. High nucleosome occupancy is encoded at X-linked gene promoters in C. elegans. Genome Res 2011; 21:237-44; PMID:21177966; http://dx.doi.org/ 10.1101/gr.115931.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petty E, Laughlin E, Csankovszki G. Regulation of DCC localization by HTZ-1/H2A.Z and DPY-30 does not correlate with H3K4 methylation levels. PLoS One [Internet] 2011. [cited 2014 Mar 21]; 6:e25973. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3187824&tool=pmcentrez&rendertype=abstract; PMID:21998734; http://dx.doi.org/ 10.1371/journal.pone.0025973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vielle A, Lang J, Dong Y, Ercan S, Kotwaliwale C, Rechtsteiner A, Appert A, Chen QB, Dose A, Egelhofer T, et al.. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet [Internet] 2012. [cited 2014 Mar 21]; 8:e1002933. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3441679&tool=pmcentrez&rendertype=abstract; PMID:23028348; http://dx.doi.org/ 10.1371/journal.pgen.1002933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wells MB, Snyder MJ, Custer LM, Csankovszki G. Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol Cell Biol [Internet] 2012. [cited 2014 Mar 21]; 32:1710-9. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3347233&tool=pmcentrez&rendertype=abstract; PMID:22393255; http://dx.doi.org/ 10.1128/MCB.06546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genet [Internet] 2008. [cited 2014 Mar 21]; 4:e1000187. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2522285&tool=pmcentrez&rendertype=abstract; PMID:18787694; http://dx.doi.org/ 10.1371/journal.pgen.1000187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, et al.. Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet [Internet] 2011. [cited 2014 Mar 19]; 43:1179-85. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3576853&tool=pmcentrez&rendertype=abstract; PMID:22019781; http://dx.doi.org/ 10.1038/ng.948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife [Internet] 2013. [cited 2014 Mar 21]; 2:e00808. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3687364&tool=pmcentrez&rendertype=abstract; PMID:23795297; http://dx.doi.org/ 10.7554/eLife.00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B. Global analysis of X-chromosome dosage compensation. J Biol 2006; 5:3; PMID:16507155; http://dx.doi.org/ 10.1186/jbiol30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pferdehirt RR, Kruesi WS, Meyer BJ. regulation of gene expression An MLL / COMPASS subunit functions in the C. elegans dosage compensation complex to target X chromosomes for transcriptional regulation of gene expression. Genes Dev 2011; 25:499-515; PMID:21363964; http://dx.doi.org/ 10.1101/gad.2016011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csankovszki G, Petty EL, Collette KS. The worm solution: a chromosome-full of condensin helps gene expression go down. Chromosome Res [Internet] 2009. [cited 2014 Mar 21]; 17:621-35. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2992697&tool=pmcentrez&rendertype=abstract; PMID:19802703; http://dx.doi.org/ 10.1007/s10577-009-9061-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferrari F, Alekseyenko AA, Park PJ, Kuroda MI. Transcriptional control of a whole chromosome: emerging models for dosage compensation. Nat Struct Mol Biol [Internet] 2014. [cited 2014 Mar 21]; 21:118-25. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24500429; PMID:24500429; http://dx.doi.org/ 10.1038/nsmb.2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chow JC, Heard E. Nuclear organization and dosage compensation. Cold Spring Harb Perspect Biol [Internet] 2010. [cited 2014 Mar 21]; 2:a000604. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid= 964184&tool=pmcentrez&rendertype=abstract; PMID:20943757; http://dx.doi.org/ 10.1101/cshperspect.a000604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nora EP, Heard E. Chromatin Structure and Nuclear Organization Dynamics during X-Chromosome Inactivation. Cold Spring Harb Symp Quant Biol [Internet] 2011; 485:LXXV Available from: http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=21447823&retmode=ref&cmd=prlinks\npapers2://publication/doi/10.1101/sqb.2010.75.032 [DOI] [PubMed] [Google Scholar]
- 29.Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJG, Zhu Y, Kaaij LJT, van Ijcken W, Gribnau J, Heard E, et al.. The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 2011; 25:1371-83; PMID:21690198; http://dx.doi.org/ 10.1101/gad.633311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al.. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science [Internet] 2013; 341:1237973. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3778663&tool=pmcentrez&rendertype=abstract; PMID:23828888; http://dx.doi.org/ 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al.. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature [Internet] 2012. [cited 2014 Mar 20]; 485:381-5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3555144&tool=pmcentrez&rendertype=abstract; PMID:22495304; http://dx.doi.org/ 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad T, Akhtar A. Dosage compensation in Drosophila melanogaster: epigenetic fine-tuning of chromosome-wide transcription. Nat Rev Genet [Internet] 2012; 13:123-34. PMID:22251873; Available from: http://dx.doi.org/ 10.1038/nrg3124 [DOI] [PubMed] [Google Scholar]
- 33.Grimaud C, Becker PB. The dosage compensation complex shapes the conformation of the X chromosome in Drosophila. Genes Dev [Internet] 2009. [cited 2014 Mar 21]; 23:2490-5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2779748&tool=pmcentrez&rendertype=abstract; PMID:19884256; http://dx.doi.org/ 10.1101/gad.539509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith ER, Allis CD, Lucchesi JC. Linking Global Histone Acetylation to the Transcription Enhancement of X-chromosomal Genes in Drosophila Males. J Biol Chem 2001; 276:31483-6; PMID:11445559; http://dx.doi.org/ 10.1074/jbc.C100351200 [DOI] [PubMed] [Google Scholar]
- 35.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al.. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 2006; 21:811-23; PMID:16543150; http://dx.doi.org/ 10.1016/j.molcel.2006.02.007 [DOI] [PubMed] [Google Scholar]
- 36.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet [Internet] 2010. [cited 2014 Mar 21]; 6:e1000846. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2820533&tool=pmcentrez&rendertype=abstract; PMID:20174442; http://dx.doi.org/ 10.1371/journal.pgen.1000846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ikegami K, Egelhofer TA, Strome S, Lieb JD. Caenorhabditis elegans chromosome arms are anchored to the nuclear membrane via discontinuous association with LEM-2. Genome Biol [Internet] 2010. [cited 2014 Mar 21]; 11:R120. Available from: http://genomebiology.com/2010/11/12/R120; PMID:21176223; http://dx.doi.org/ 10.1186/gb-2010-11-12-r120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Towbin BD, González-Aguilera C, Sack R, Gaidatzis D, Kalck V, Meister P, Askjaer P, Gasser SM. Step-wise methylation of histone H3K9 positions heterochromatin at the nuclear periphery. Cell [Internet] 2012. [cited 2014 Mar 20]; 150:934-47. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22939621; PMID:22939621; http://dx.doi.org/ 10.1016/j.cell.2012.06.051 [DOI] [PubMed] [Google Scholar]
- 39.Chuang PT, Albertson DG, Meyer BJ. DPY-27:a chromosome condensation protein homolog that regulates C. elegans dosage compensation through association with the X chromosome. Cell [Internet] 1994; 79:459-74. Available from: http://www.ncbi.nlm.nih.gov/pubmed/7954812; PMID:7954812; http://dx.doi.org/ 10.1016/0092-8674(94)90255-0 [DOI] [PubMed] [Google Scholar]
- 40.Lau AC, Nabeshima K, Csankovszki G. The C. elegans dosage compensation complex mediates interphase X chromosome compaction. Epigenetics Chromatin [Internet] 2014; 7:31. Available from: http://www.epigeneticsandchromatin.com/content/7/1/31; PMID:25400696; http://dx.doi.org/ 10.1186/1756-8935-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharma R, Jost D, Kind J, Gómez-Saldivar G, van Steensel B, Askjaer P, Vaillant C, Meister P. Differential spatial and structural organization of the X chromosome underlies dosage compensation in C. elegans. Genes Dev [Internet] 2014. [cited 2015 Apr 1]; 28:2591-6. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=4248290&tool=pmcentrez&rendertype=abstract; PMID:25452271; http://dx.doi.org/ 10.1101/gad.248864.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrai C, de Castro IJ, Lavitas L, Chotalia M, Pombo A. Gene positioning. Cold Spring Harb Perspect Biol 2010; 2:a000588; PMID:20484389; http://dx.doi.org/ 10.1101/cshperspect.a000588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma R, Meister P. Nuclear organization in the nematode C. elegans. Curr. Opin. Cell Biol. 2013; 25:395-402; PMID:23481208; http://dx.doi.org/ 10.1016/j.ceb.2013.02.002 [DOI] [PubMed] [Google Scholar]
- 44.Meister P, Towbin BD, Pike BL, Ponti a, Gasser SM. The spatial dynamics of tissue-specific promoters during C-elegans development. Genes Dev [Internet] 2010; 24:766-82. Available from: ://000276730300006; PMID:20395364; http://dx.doi.org/ 10.1101/gad.559610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.González-Aguilera C, Ikegami K, Ayuso C, de Luis A, Iñiguez M, Cabello J, Lieb JD, Askjaer P. Genome-wide analysis links emerin to neuromuscular junction activity in Caenorhabditis elegans. Genome Biol [Internet] 2014; 15:R21. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24490688; PMID:24490688; http://dx.doi.org/ 10.1186/gb-2014-15-2-r21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohner S, Kalck V, Wang X, Ikegami K, Lieb JD, Gasser SM, Meister P. Promoter- and RNA polymerase II-dependent hsp-16 gene association with nuclear pores in Caenorhabditis elegans. J Cell Biol [Internet] 2013. [cited 2014 Mar 21]; 200:589-604. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3587839&tool=pmcentrez&rendertype=abstract; PMID:23460676; http://dx.doi.org/ 10.1083/jcb.201207024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ikegami K, Lieb J. Integral Nuclear Pore Proteins Bind to Pol III-Transcribed Genes and Are Required for Pol III Transcript Processing in C. elegans. Mol Cell 2013; 51:840-9; PMID:24011592; http://dx.doi.org/ 10.1016/j.molcel.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol [Internet] 2010. [cited 2014 Mar 19]; 12:111-8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2835469&tool=pmcentrez&rendertype=abstract; PMID:20098417; http://dx.doi.org/ 10.1038/ncb2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell [Internet] 2010. [cited 2014 Mar 19]; 40:112-25. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2953765&tool=pmcentrez&rendertype=abstract; PMID:20932479; http://dx.doi.org/ 10.1016/j.molcel.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zullo JM, Demarco IA, Piqué-Regi R, Gaffney DJ, Epstein CB, Spooner CJ, Luperchio TR, Bernstein BE, Pritchard JK, Reddy KL, et al.. DNA sequence-dependent compartmentalization and silencing of chromatin at the nuclear lamina. Cell [Internet] 2012. [cited 2014 Mar 20]; 149:1474-87. Available from: http://www.ncbi.nlm.nih.gov/pubmed/22726435; PMID:22726435; http://dx.doi.org/ 10.1016/j.cell.2012.04.035 [DOI] [PubMed] [Google Scholar]
- 51.Harr JC, Luperchio TR, Wong X, Cohen E, Wheelan SJ, Reddy KL. Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. 2015; 208:33-52; PMID:25559185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature 2006; 444:614-8; PMID:17122774; http://dx.doi.org/ 10.1038/nature05338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-Bound Nuclear Pore Components Regulate Gene Expression in Higher Eukaryotes. Cell [Internet] 2010; 140:372-83. PMID:20144761; Available from: http://dx.doi.org/10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kalverda B, Pickersgill H, Shloma V V, Fornerod M. Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes Inside the Nucleoplasm. Cell [Internet] 2010; 140:360-71. PMID:20144760; Available from: http://dx.doi.org/ 10.1016/j.cell.2010.01.011; [DOI] [PubMed] [Google Scholar]
- 55.Umlauf D, Bonnet J, Waharte F, Fournier M, Stierle M, Fischer B, Brino L, Devys D, Tora L. The human TREX-2 complex is stably associated with the nuclear pore basket. J Cell Sci [Internet] 2013; 126:2656-67. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23591820; PMID:23591820; http://dx.doi.org/ 10.1242/jcs.118000 [DOI] [PubMed] [Google Scholar]
- 56.Kurshakova MM, Krasnov AN, Kopytova DV, Shidlovskii YV, Nikolenko JV, Nabirochkina EN, Spehner D, Schultz P, Tora L, Georgieva SG. SAGA and a novel Drosophila export complex anchor efficient transcription and mRNA export to NPC. EMBO J 2007; 26:4956-65; PMID:18034162; http://dx.doi.org/ 10.1038/sj.emboj.7601901 [DOI] [PMC free article] [PubMed] [Google Scholar]



