Abstract
More than one third of the cellular proteome is destined for incorporation into cell membranes or export from the cell. In all domains of life, the signal recognition particle (SRP) delivers these proteins to the membrane and protein traffic falls apart without SRP logistics. With the aid of a topogenic transport signal, SRP retrieves its cargo right at the ribosome, from where they are sorted to the translocation channel. Mammalian SRP is a ribonucleoprotein complex consisting of an SRP RNA of 300 nucleotides and 6 proteins bound to it. Assembly occurs in a hierarchical manner mainly in the nucleolus and only SRP54, which recognizes the signal sequence and regulates the targeting process, is added as the last component in the cytosol. Here we present an update on recent insights in the structure, function and dynamics of SRP RNA in SRP assembly with focus on the S domain, and present SRP as an example for the complex biogenesis of a rather small ribonucleoprotein particle.
Keywords: Ribonucleoprotein complex (RNP), RNA folding, RNA structure, RNP assembly, RNA-RNA tertiary interactions, protein-RNA interactions, signal recognition particle (SRP)
The signal recognition particle (SRP) is universally conserved and plays a central role in co-translational protein transport.1,2 The discovery of SRP as a ribonucleoprotein complex presents an example of the serendipity of discovery. Already in 1970, studies on oncornavirus described a 7S RNA (later 7SL RNA) that was derived from the infected host cell rather than from the virus.3 This 7S RNA associated with polyribosomes in a sub-stoichiometric fashion and somehow seemed to participate in the translation process.4 In the 1980s, SRP was first reported as an 11S protein complex in canine pancreatic tissue that is involved in the translocation process of secretory proteins across the endoplasmic reticulum membrane5 and recognizes the signal sequence.6 With the discovery of the 7S RNA as part of the protein complex, SRP was then defined as the signal recognition particle.7
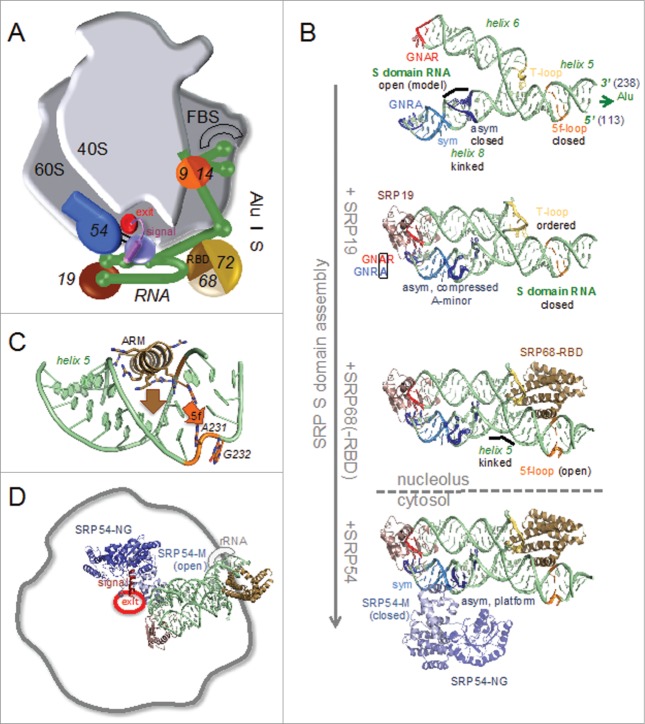
While in the past 30 y the analysis of SRP structure and function has come a long way, the biogenesis of mammalian SRP remained enigmatic. The mammalian SRP consists of a single copy of SRP RNA with about 300 nucleotides that forms the assembly platform for 6 proteins (named according molecular mass: SRP9/14, SRP19, SRP54, SRP68/72)5 (Fig. 1A). SRP assembly starts in the nucleolus,8 but is completed with the addition of SRP54 in the cytosol.9 Here, we focus on the structure, function, and dynamics of mammalian SRP RNA in respect to ribonucleoprotein particle (RNP) assembly. SRP RNA has been found in all domains of life, but the size and secondary structure elements vary significantly between the phylogenetic groups, however with a number of generally conserved features.10,11 Long SRP RNA (7S RNA, in contrast to short bacterial 4.5S RNA) folds into a terminal Alu domain and an S domain of about equal sizes of 150 nucleotides.12,13 The S domain harboring the 4 larger proteins recognizes SRP targets through the presence of an N-terminal signal sequence as soon as it emerges from the ribosomal tunnel exit.14-16 The Alu domain (including SRP9/14) imposes an elongation arrest or retardation of translation17-19 by binding into the elongation factor binding site. The structure of mammalian SRP has been determined by cryo-EM studies in the context of the ribosome – nascent chain complex (RNC),20,21 but is not available on its own. In the past 15 years, sub-structures of mammalian SRP have been determined, but it was only this year that the mechanism of the S domain remodeling RNA-binding domain of the SRP68 protein was described,22 which we will now integrate in the current scheme of SRP assembly.
Figure 1.
SRP mediated co-translational protein targeting. (A) Scheme of human SRP bound to the ribosome – nascent chain complex (RNC). The S domain recognizes the signal at the ribosomal tunnel exit and the Alu domain binds to the factor binding site (FBS) causing “elongation arrest." (B) RNA gymnastics in SRP S domain assembly. SRP19 (pdb code 3ktv (42) and SRP68-RBD (4p3e (22); and remainder of SRP68/72) are attached in the nucleolus, while the functional particle with SRP54 (1qzw; structure from an archaeal complex57) is completed in the cytosol. (C) SRP RNA remodeling of helix 5 and the 5f-loop (orange) by the insertion of an ARM motif (brown) of SRP68-RBD. (D) The assembled SRP S domain bound to the RNC.20 The signal is recognized by SRP54-M (open conformation) and SRP54-NG is flipped to the apex of the particle. The opened SRP RNA 5f-loop is in contact with rRNA.
SRP RNA is transcribed by RNA polymerase III and is only slightly processed. Its 3’ end consists of a modifiable oligo(U) tract like other RNA polymerase III transcripts. In yeast this region was recently shown to bind to the stabilizing La protein homolog and to the nuclear RNA quality machinery including TRAMP-stimulated exosomal degradation.23 The 3’ end of human SRP RNA is shortened to a single uridine and an adenine is added.24 While the cellular control mechanisms for particle assembly are basically unknown, the structural analysis of the relevant folding intermediates is well advanced. The Alu domain RNA consists of 2 folding units, the 5’ and the 3’ region. For the human Alu domain, it has been shown that the 5’ region is flexible in respect to the 3’ region via a hinge.25 The SRP9/14 heterodimer induces and stabilizes a compact Alu RNA structure with the 5’ region folded back onto the 3’ region and tertiary interactions between 4 RNA helices (helices 2 to 5, helix 1 is absent in human SRP).26 Recently, we reported the structure of the complete Alu RNA from B. subtilis, which adopts a stable fold in the absence of proteins due to prokaryote specific, inbuilt stabilizing elements.27 The Alu domain RNA is the ancestor of the Alu elements, which are retrotransposable DNA elements that comprise more than 10% of the primate genome.28 Alu RNP structures are not only part of the SRP and the SRP9/14 heterodimer can also assemble with transcripts of the Alu elements29 underlining the high conservation of the Alu RNA fold.
The S domain RNA (human: nucleotides 101 to 250) comprises the distal part of helix 5 as well as helices 6 and 8 (helix 7 forms a structured loop) and a number of conserved internal bulges and apical tetranucleotide loops (tetraloops) (Fig. 1B, first panel). The connection between the Alu domain and the S domain by helix 5 (parts 5d and 5e (10)) is not stabilized by protein as indicated by the cryo-EM reconstruction of human SRP bound to the RNC.20 The connection seems to form a flexible hinge necessary for adapting SRP to the ribosomal surface. S domain RNA folding depends on the SRP19 and SRP68 proteins. SRP19 comprises a single, monomeric RNA binding domain (RBD), while SRP68 comes as a large solenoidal heterodimer together with SRP72, the structure of which so far is unknown. A significant portion of SRP68/72 seems to be flexible, as it does not give rise to defined electron density when bound to the RNC.20,21 Chemical probing data and mutational analyses revealed the primary binding site for SRP19 to involve the distal end of helix 6 and its closing GNAR tetraloop with an unusual conservation of an adenine at the third position.30 SRP68 localizes to the 3-way junction connecting helices 5, 6 and 8.20,31-34 SRP72 binds to helix 5 adjacent to SRP68 and was described to stabilize an RNA “kink-turn” at the 5e-loop,35 however, no structure of this interaction is yet available. Interestingly, SRP68 and SRP72 have been implied in SRP export36 and also SRP independent functions.37-39
Mammalian S domain RNA by itself is flexible and its structure could not be determined until now. First atomic insights in RNA structure and in the process of SRP assembly came from the structure of human SRP19 bound to helix 6. SRP19 is a flexible protein with αβ topology that adopts a stable fold upon RNA binding. It binds to a widened major groove and the phosphoribose backbone of the GNAR tetraloop (Fig. 1B, second panel) leaving the conserved adenine solvent exposed.40 However, crystal packing immediately suggested a plausible model for its strict conservation by the formation of RNA-RNA tertiary interactions, which could be subsequently confirmed by all structures including the complete S domain RNA (for human SRP22,41,42). Binding of SRP19 exposes the GNAR adenine for the formation of a non-canonical A-A base pair with the conserved adenine in the classical GNRA-type tetraloop closing helix 8. This interaction clamps the apices of helices 6 and 8 together inducing the typical closed S domain RNA structure. S domain closure results in remarkable further structural consolidations. The internal asymmetric bulge-loop within helix 8 is compressed and while the long strand bulges out to form a binding platform for subsequent SRP54 assembly (see below), 2 adenines of the short strand insert in the minor groove of helix 6 by classical A-minor motifs, a recurrent and highly relevant RNA-RNA interaction.43 The connection of helices 5, 6, and 8 folds into a 3-way junction similar to the hammerhead ribozyme,44 with helix 7 adopting a so-called T-loop structure45 including a U-turn motif, which again is fixed to helix 5 by A-minor tertiary interactions.
The overall structure of the S domain RNA is shaped by SRP19. However, SRP reconstituted without SRP68/72 lacks elongation retardation activity and is inactive in translocation.46,47 How does the large SRP68/72 heterodimer execute these crucial functions? The N-terminal half of SRP68 contains an RBD48 that has been shown by chemical probing to induce manifold changes especially in the RNA 3-way junction and at the distal part of helix 5 including the conserved 5f-loop.31-33 Despite these data, SRP68/72 has long remained the “terra incognita” in SRP research. Some of its mysteries could be resolved recently by structure determination of the ternary complex of human SRP68-RBD, S domain RNA and SRP19.22 The purely α-helical SRP68-RBD has similarity to a tetratricopeptide repeat (TPR) fold (Fig. 1B, third panel). It comprises an extended positively charged patch for RNA binding but does not resemble any classical RNA-binding domain.49 SRP68-RBD binds as a rigid body to the 3-way junction of SRP RNA helices 5, 6, and 8 mainly via the phosphoribose backbone.19 Three α-helices constitute a concave surface with one α-helix being accommodated in the T-loop structure and one α-helix protruding deeply into the major groove of helix 5 at the 5f-loop, thus significantly stabilizing the RNA 3-way junction. Like for SRP19, SRP68-RBD binding remodels the complete S domain RNA. SRP68-RBD induces a kink of helix 5 in respect to the coaxially stacked helix 8 by about 20° away from helix 6 with the hinge point locating proximal to the asymmetric loop of helix 8. It is important to note, that the SRP68-mediated kink in SRP RNA is maintained upon interaction of SRP with the RNC and that the kink is essential for establishing a specific contact between SRP RNA and rRNA (Fig. 1D). This interaction provides an explanation for the deleterious effect of SRP68 depletion on translocation.47 In addition to these long-range RNA rearrangements, the insertion of an α-helix (a so-called arginine-rich motif (ARM)50) into the major groove causes a groove opening that coincides with the remodeling of the 5f-loop (Fig. 1C) (see also51). Like the asymmetric loop in helix 8, the asymmetric 5f-loop switches from an protein-free inward “closed” conformation41 to a protein-bound “open” conformation exposing 2 conserved purines bases (human: A231, G232). Intriguingly, the corresponding region in bacterial 4.5S RNA is known to be responsible for SRP GTPase activation and thus is critically involved in the control of the targeting process.52,53 Therefore, the principle of SRP GTPase stimulation by exposed purine bases might be conserved also in mammalian SRP. However, this has not been shown so far and further experiments are necessary.
With the incorporation of SRP9/14, SRP19, and SRP68/72, nucleolar assembly is complete and the pre-SRP is exported (not well investigated) to the cytosol where SRP54 is added as the last component. SRP54 is the key-player of SRP and comprises a methionine-rich M domain for signal sequence and RNA binding, and an SRP GTPase domain (SRP54-NG) for regulation (Fig. 1B, fourth panel). Binding of the M-domain (SRP54-M; “closed” conformation without signal54) to helix 8 of the SRP RNA induces the formation of a stacked platform by the asymmetric loop and involves the read-out of the modified minor groove of the most highly conserved SRP RNA sequence formed by 5 non-canonical base pairs of the symmetric loop.55 SRP is now ready to bind to an RNC, and to accommodate a hydrophobic signal sequence emerging from the ribosomal exit tunnel (“open” conformation of the M domain20) (Fig. 1D). RNC binding again results in rearrangements, e.g. involving a conserved hinge region in SRP54.16 Protein targeting proceeds by the interaction of SRP54-NG with its twin in the SRP receptor (SRα-NG, not shown).52,56 The gymnastics described here for the SRP RNA during SRP assembly is then continued in an elaborate choreography involving the RNC-SRP complex, the membrane associated SRP receptor (SR) and the translocation machinery.
Disclosure of Potential Conflicts of Interest
I.S. is an investigator of the Cluster of Excellence:CellNetworks.
Acknowledgments
We thank the ESRF and Jürgen Kopp from the BZH/CellNetworks crystallization platform for continuous, excellent support.
Funding
We are grateful for support by the Deutsche Forschungsgemeinschaft (DFG) (SFB638, FOR967, and GRK1188).
References
- 1. Akopian D, Shen K, Zhang Shan SO. Signal recognition particle: an essential protein-targeting machine. Annu Rev Biochem 2013; 82:693-721; PMID:23414305; http://dx.doi.org/ 10.1146/annurev-biochem-072711-164732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cross BC, Sinning I, Luirink J, High S. Delivering proteins for export from the cytosol. Nat Rev Mol Cell Biol 2009; 10:255-64; PMID:19305415; http://dx.doi.org/ 10.1038/nrm2657 [DOI] [PubMed] [Google Scholar]
- 3. Bishop JM, Levinson WE, Sullivan D, Fanshier L, Quintrell N, Jackson J. The low molecular weight RNAs of Rous sarcoma virus. II. The 7 S RNA. Virology 1970; 42:927-37; PMID:4321311; http://dx.doi.org/ 10.1016/0042-6822(70)90341-7 [DOI] [PubMed] [Google Scholar]
- 4. Walker TA, Pace NR, Erikson RL, Erikson E, Behr F. The 7S RNA common to oncornaviruses and normal cells is associated with polyribosomes. Proc Natl Acad Sci U S A 1974; 71:3390-94; PMID:4530311; http://dx.doi.org/ 10.1073/pnas.71.9.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Walter P, Blobel G. Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum. Proc Natl Acad Sci U S A 1980; 77:7112-6; PMID:6938958; http://dx.doi.org/ 10.1073/pnas.77.12.7112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum. II. Signal recognition protein (SRP) mediates the selective binding to microsomal membranes of in-vitro-assembled polysomes synthesizing secretory protein. J Cell Biol 1981; 91:551-6; PMID:7309796; http://dx.doi.org/ 10.1083/jcb.91.2.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walter P, Blobel G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 1982; 299:691-8; PMID:6181418; http://dx.doi.org/ 10.1038/299691a0 [DOI] [PubMed] [Google Scholar]
- 8. Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci U S A 1998; 95:7981-6; PMID:9653126; http://dx.doi.org/ 10.1073/pnas.95.14.7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci U S A 2000; 97:55-60; PMID:10618370; http://dx.doi.org/ 10.1073/pnas.97.1.55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rosenblad MA, Gorodkin J, Knudsen B, Zwieb C, Samuelsson T. SRPDB: Signal Recognition Particle Database. Nucleic Acids Res 2003; 31:363-4; PMID:12520023; http://dx.doi.org/ 10.1093/nar/gkg107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rosenblad MA, Larsen N, Samuelsson T, Zwieb C. Kinship in the SRP RNA family. RNA biology 2009; 6:508-16; PMID:19838050; http://dx.doi.org/ 10.4161/rna.6.5.9753 [DOI] [PubMed] [Google Scholar]
- 12. Ullu E, Murphy S, Melli M. Human 7SL RNA consists of a 140 nucleotide middle-repetitive sequence inserted in an alu sequence. Cell 1982; 29:195-202; PMID:6179628; http://dx.doi.org/ 10.1016/0092-8674(82)90103-9 [DOI] [PubMed] [Google Scholar]
- 13. Gundelfinger ED, Krause E, Melli M, Dobberstein B. The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res 1983; 11:7363-74; PMID:6196719; http://dx.doi.org/ 10.1093/nar/11.21.7363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lutcke H, Dobberstein B. Structure and function of signal recognition particle (SRP). Molecular biology reports 1993; 18:143-7; PMID:8232296; http://dx.doi.org/ 10.1007/BF00986769 [DOI] [PubMed] [Google Scholar]
- 15. Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol 1994; 10:87-119; PMID:7888184; http://dx.doi.org/ 10.1146/annurev.cb.10.110194.000511 [DOI] [PubMed] [Google Scholar]
- 16. Wild K, Halic M, Sinning I, Beckmann R. SRP meets the ribosome. Nat Struct Mol Biol 2004; 11:1049-53; PMID:15523481; http://dx.doi.org/ 10.1038/nsmb853 [DOI] [PubMed] [Google Scholar]
- 17. Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J Cell Biol 1981; 91:557-61; PMID:7309797; http://dx.doi.org/ 10.1083/jcb.91.2.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Siegel V, Walter P. Removal of the Alu structural domain from signal recognition particle leaves its protein translocation activity intact. Nature 1986; 320:81-84; PMID:2419765; http://dx.doi.org/ 10.1038/320081a0 [DOI] [PubMed] [Google Scholar]
- 19. Thomas Y, Bui N, Strub K. A truncation in the 14 kDa protein of the signal recognition particle leads to tertiary structure changes in the RNA and abolishes the elongation arrest activity of the particle. Nucleic Acids Res 1997; 25:1920-9; PMID:9115358; http://dx.doi.org/ 10.1093/nar/25.10.1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 2004; 427:808-14; PMID:14985753; http://dx.doi.org/ 10.1038/nature02342 [DOI] [PubMed] [Google Scholar]
- 21. Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature 2006; 444:507-11; PMID:17086193; http://dx.doi.org/ 10.1038/nature05326 [DOI] [PubMed] [Google Scholar]
- 22. Grotwinkel JT, Wild K, Segnitz B, Sinning I. SRP RNA remodeling by SRP68 explains its role in protein translocation. Science 2014; 344:101-4; PMID:24700861; http://dx.doi.org/ 10.1126/science.1249094 [DOI] [PubMed] [Google Scholar]
- 23. Leung E, Brown JD. Biogenesis of the signal recognition particle. Biochemical Society transactions 2010; 38:1093-8; PMID:20659010; http://dx.doi.org/ 10.1042/BST0381093 [DOI] [PubMed] [Google Scholar]
- 24. Sinha KM, Gu J, Chen Y, Reddy R. Adenylation of small RNAs in human cells. Development of a cell-free system for accurate adenylation on the 3'-end of human signal recognition particle RNA. J Biol Chem 1998; 273:6853-9; PMID:9506988; http://dx.doi.org/ 10.1074/jbc.273.12.6853 [DOI] [PubMed] [Google Scholar]
- 25. Weichenrieder O, Stehlin C, Kapp U, Birse DE, Timmins PA, Strub K, Cusack S. Hierarchical assembly of the Alu domain of the mammalian signal recognition particle. RNA 2001; 7:731-40; PMID:11350037; http://dx.doi.org/ 10.1017/S1355838201010160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weichenrieder O, Wild K, Strub K, Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature 2000: 408:167-73; PMID:11089964; http://dx.doi.org/ 10.1038/35041507 [DOI] [PubMed] [Google Scholar]
- 27. Kempf G, Wild K, Sinning I. Structure of the complete bacterial SRP Alu domain. Nucleic Acids Res 2014; 2:12284-94; PMID:25270875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deininger P. Alu elements: know the SINEs. Genome Biol 2011; 12:236; PMID:22204421; http://dx.doi.org/ 10.1186/gb-2011-12-12-236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kremerskothen J, Zopf D, Walter P, Cheng JG, Nettermann M, Niewerth U, Maraia RJ, Brosius J. Heterodimer SRP9/14 is an integral part of the neural BC200 RNP in primate brain. Neurosci Lett 1998; 245:123-6; PMID:9605471; http://dx.doi.org/ 10.1016/S0304-3940(98)00215-8 [DOI] [PubMed] [Google Scholar]
- 30. Zwieb C. Recognition of a tetranucleotide loop of signal recognition particle RNA by protein SRP19. J Biol Chem 1992; 267:15650-6; PMID:1379233 [PubMed] [Google Scholar]
- 31. Yin J, Iakhiaeva E, Menichelli E, Zwieb C. Identification of the RNA binding regions of SRP68/72 and SRP72 by systematic mutagenesis of human SRP RNA. RNA biology 2007; 4:154-9; PMID:18347438; http://dx.doi.org/ 10.4161/rna.4.3.5428 [DOI] [PubMed] [Google Scholar]
- 32. Menichelli E, Isel C, Oubridge C, Nagai K. Protein-induced conformational changes of RNA during the assembly of human signal recognition particle. J Mol Biol 2007; 367:187-203; PMID:17254600; http://dx.doi.org/ 10.1016/j.jmb.2006.12.056 [DOI] [PubMed] [Google Scholar]
- 33. Maity TS, Fried HM, Weeks KM. Anti-cooperative assembly of the SRP19 and SRP68/72 components of the signal recognition particle. Biochem J 2008; 415:429-37; PMID:18564060; http://dx.doi.org/ 10.1042/BJ20080569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siegel V, Walter P. Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein-RNA "footprinting". Proc Natl Acad Sci U S A 1988; 85:1801-5; PMID:2450348; http://dx.doi.org/ 10.1073/pnas.85.6.1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Iakhiaeva E, Wower J, Wower IK, Zwieb C. The 5e motif of eukaryotic signal recognition particle RNA contains a conserved adenosine for the binding of SRP72. RNA 2008; 14:1143-53; PMID:18441046; http://dx.doi.org/ 10.1261/rna.979508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van Nues RW, Leung E, McDonald JC, Dantuluru I, Brown JD. Roles for Srp72p in assembly, nuclear export and function of the signal recognition particle. RNA biology 2008; 5:73-83; PMID:18418087; http://dx.doi.org/ 10.4161/rna.5.2.6042 [DOI] [PubMed] [Google Scholar]
- 37. Kirwan M, Walne AJ, Plagnol V, Velangi M, Ho A, Hossain U, Vulliamy T, Dokal I. Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am J Human Genet 2012; 90:888-92; PMID:22541560; http://dx.doi.org/ 10.1016/j.ajhg.2012.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Utz PJ, Hottelet M, Le TM, Kim SJ, Geiger ME, van Venrooij WJ, Anderson P. The 72-kDa component of signal recognition particle is cleaved during apoptosis. J Biol Chem 1998; 273:35362-70; PMID:9857079; http://dx.doi.org/ 10.1074/jbc.273.52.35362 [DOI] [PubMed] [Google Scholar]
- 39. Li J, Zhou F, Zhan D, Gao Q, Cui N, Li J, Iakhiaeva E, Zwieb C, Lin B, Wong J. A novel histone H4 arginine 3 methylation-sensitive histone H4 binding activity and transcriptional regulatory function for signal recognition particle subunits SRP68 and SRP72. J Biol Chem 2012; 287:40641-51; PMID:23048028; http://dx.doi.org/ 10.1074/jbc.M112.414284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wild K, Sinning I, Cusack S. Crystal structure of an early protein-RNA assembly complex of the signal recognition particle. Science 2001; 294:598-601; PMID:11641499; http://dx.doi.org/ 10.1126/science.1063839 [DOI] [PubMed] [Google Scholar]
- 41. Kuglstatter A, Oubridge C, Nagai K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nature structural biology 2002; 9:740-4; PMID:12244299; http://dx.doi.org/ 10.1038/nsb843 [DOI] [PubMed] [Google Scholar]
- 42. Wild K, Bange G, Bozkurt G, Segnitz B, Hendricks A, Sinning I. Structural insights into the assembly of the human and archaeal signal recognition particles. Acta Crystallogr D Biol Crystallogr 2010; 66:295-303; PMID:20179341; http://dx.doi.org/ 10.1107/S0907444910000879 [DOI] [PubMed] [Google Scholar]
- 43. Battle DJ, Doudna J.A. Specificity of RNA-RNA helix recognition. Proc Natl Acad Sci U S A 2002; 99:11676-81; PMID:12189204; http://dx.doi.org/ 10.1073/pnas.182221799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scott WG, Horan LH, Martick M. The hammerhead ribozyme: structure, catalysis, and gene regulation. Prog Mol Biol Trans Sci 2013; 120:1-23; PMID:24156940; http://dx.doi.org/ 10.1016/B978-0-12-381286-5.00001-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA 2003; 9:640-3; PMID:12756321; http://dx.doi.org/ 10.1261/rna.2202703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Siegel V, Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: analysis of biochemical mutants of SRP. Cell 1988; 52:39-49; PMID:2830980; http://dx.doi.org/ 10.1016/0092-8674(88)90529-6 [DOI] [PubMed] [Google Scholar]
- 47. Siegel V, Walter P. Elongation arrest is not a prerequisite for secretory protein translocation across the microsomal membrane. J Cell Biol 1985; 100:1913-21; PMID:2581979; http://dx.doi.org/ 10.1083/jcb.100.6.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lutcke H, Prehn S, Ashford AJ, Remus M, Frank R, Dobberstein B. Assembly of the 68- and 72-kD proteins of signal recognition particle with 7S RNA. J Cell Biol 1993; 121:977-85; PMID:8388879; http://dx.doi.org/ 10.1083/jcb.121.5.977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nature reviews. Mol Cell Biol 2007; 8:479-90; PMID:17473849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 1996; 273:1547-51; PMID:8703216; http://dx.doi.org/ 10.1126/science.273.5281.1547 [DOI] [PubMed] [Google Scholar]
- 51. Weeks KM, Crothers DM. Major groove accessibility of RNA. Science 1993; 261:1574-7; PMID:7690496; http://dx.doi.org/ 10.1126/science.7690496 [DOI] [PubMed] [Google Scholar]
- 52. Ataide SF, Schmitz N, Shen K, Ke A, Shan SO, Doudna JA, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science 2011; 331:881-6; PMID:21330537; http://dx.doi.org/ 10.1126/science.1196473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shen K, Wang Y, Hwang Fu YH, Zhang Q, Feigon J, Shan SO. Molecular mechanism of GTPase activation at the SRP RNA distal end. J Biol Chem 2013; 288:36385-97; PMID:24151069; http://dx.doi.org/ 10.1074/jbc.M113.513614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rosendal KR, Sinning I, Wild K. Crystallization of the crenarchaeal SRP core. Acta Crystallogr D Biol Crystallogr 2004; 60:140-3; PMID:14684910; http://dx.doi.org/ 10.1107/S0907444903023734 [DOI] [PubMed] [Google Scholar]
- 55. Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science 2000; 287:1232-9; PMID:10678824; http://dx.doi.org/ 10.1126/science.287.5456.1232 [DOI] [PubMed] [Google Scholar]
- 56. Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science 2006; 312:745-7; PMID:16675701; http://dx.doi.org/ 10.1126/science.1124864 [DOI] [PubMed] [Google Scholar]
- 57. Rosendal KR, Wild K, Montoya G, Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc Natl Acad Sci U S A 2003; 100:14701-6; PMID:14657338; http://dx.doi.org/ 10.1073/pnas.2436132100 [DOI] [PMC free article] [PubMed] [Google Scholar]