Abstract
Mutations in genes that encode tRNAs, aminoacyl-tRNA syntheases, tRNA modifying enzymes and other tRNA interacting partners are associated with neuropathies, cancer, type-II diabetes and hearing loss, but how these mutations cause disease is unclear. We have hypothesized that levels of tRNA decoding error (mistranslation) that do not fully impair embryonic development can accelerate cell degeneration through proteome instability and saturation of the proteostasis network. To test this hypothesis we have induced mistranslation in zebrafish embryos using mutant tRNAs that misincorporate Serine (Ser) at various non-cognate codon sites. Embryo viability was affected and malformations were observed, but a significant proportion of embryos survived by activating the unfolded protein response (UPR), the ubiquitin proteasome pathway (UPP) and downregulating protein biosynthesis. Accumulation of reactive oxygen species (ROS), mitochondrial and nuclear DNA damage and disruption of the mitochondrial network, were also observed, suggesting that mistranslation had a strong negative impact on protein synthesis rate, ER and mitochondrial homeostasis. We postulate that mistranslation promotes gradual cellular degeneration and disease through protein aggregation, mitochondrial dysfunction and genome instability.
Keywords: tRNA, mRNA mistranslation, proteotoxic stress, protein aggregation, ROS, zebrafish
Introduction
TRNAs (tRNAs) are normally viewed as highly stable and robust cellular adaptors that faithfully translate genes into proteins. However, they also sense amino acid availability, regulate global protein synthesis rate, participate in biosynthesis of various metabolites, protein degradation, apoptosis, initiation of reverse transcription in retroviruses and are a source for a variety of small interfering RNAs known as tRNA fragments.1-5 These functions are dependent on a complex network of interactions between tRNAs and multiple tRNA modifying enzymes, aminoacyl-tRNA synthetases (aaRSs), tRNA 3′and 5′- end processing enzymes, kinases, translation initiation and elongation factors, the ribosome and many other proteins.1 The tRNAs are also one of the most actively transcribed classes of RNAs - transcribed by RNA polymerase III – and a strong correlation between tRNA abundance and codon usage fine tunes translation efficiency and accuracy.6 Therefore, it is not surprising that transcriptional deregulation of tRNAs and mutations in tRNA genes and in genes that encode tRNA interacting partners are associated with human diseases. Indeed, mutations that affect the aminoacylation kinetics of the mitochondrial tRNALeu(UUR) by the leucyl-tRNA synthetase (LARS), cause myopathy, encephalopathy, lactic acidosis, hearing loss, stroke-like episodes (MELAS) and other neurological and non-neurological symptoms,7 while mutations in the mitochondrial tRNA pseudouridine synthase 1 cause myopathy, lactic acidosis and sideroblastic anemia (MLASA).8 Several other mitochondrial tRNA mutations, for example the 8344A > G, mutation of the tRNALys and the 7511T > C mutation of the tRNASer(UCN), that affect protein synthesis efficiency, cause hearing loss.9-11 Similarly, mutations in nuclear encoded alanyl (AARS), glycyl (GARS), tyrosyl (YARS) and lysyl (KARS) - tRNA synthetases are associated with peripheral neurodegenerative Charcot-Marie-Tooth disease12-14 and a 2201C > A mutation in the editing domain of the mouse AARS that promotes mischarging of alanine tRNAs with glycine and serine causes neurodegeneration through rapid loss of Purkinje neurons.15 The human tRNA modifying enzyme TRMT12 and many tRNAs are also overexpressed in breast cancer16,17 and mutations in small and large ribosomal proteins (especially RSP19) cause Diamond-Blackfan anemia (DBA), which is an inherited bone marrow failure syndrome characterized by proapoptotic hematopoiesis, birth defects and predisposition to cancer.18 How these mutations cause such a wide spectrum of diseases in unclear, but it is likely that alterations in tRNA decoding efficiency lower protein synthesis rate and affect cell growth and survival, while tRNA alterations that affect decoding accuracy through altered codon-anticodon interactions lead to protein aggregation and saturation of the protein quality control (PQC) networks that act downstream of mRNA translation.19 Indeed, overloading of the molecular chaperones, autophagy and the ubiquitin proteasome pathway (UPP) lead to protein aggregation, while saturation of the endoplasmic reticulum (ER) with misfolded proteins triggers the unfolded protein response (UPR), phosphorylation of the translation initiation factor eIF2-α, transient attenuation of protein synthesis rate, calcium release from the ER and activation of pro-apoptotic pathways.20 Since saturation of the proteostasis network (PN) is a hallmark of cancer and various neurodegenerative and metabolic diseases,21 we have hypothesized that mutations that affect tRNA decoding accuracy cause disease through deregulation of the proteostasis network (PN). To clarify this hypothesis we have engineered a series of tRNAs that misincorporate serine (Ser) at non-cognate protein sites and produced embryos with variable levels of proteome instability. The transgenic embryos exhibited increased protein aggregation accompanied by downregulation of protein synthesis, disruption of the mitochondrial network, sharp increase in oxidative stress, and severe damage of both mitochondrial and nuclear DNA. The data suggest that mistranslation causes disease through deregulation of protein synthesis and mitochondrial function, proteotoxic stress and destabilization of mitochondrial and nuclear genomes.
Results
Impact of mistranslation on early embryo development
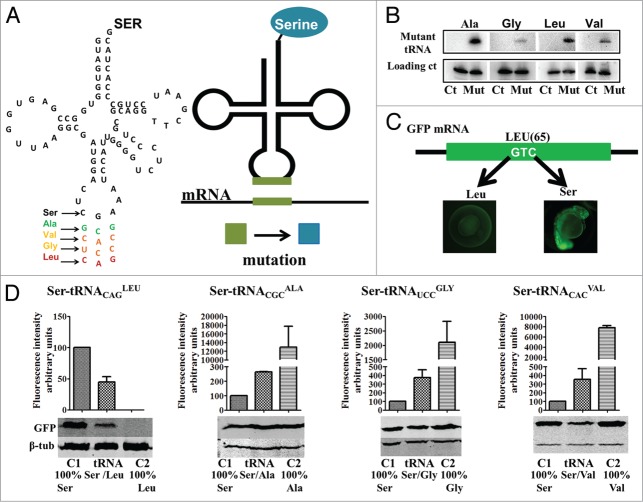
In order to study the cellular consequences of mRNA mistranslation we have generated mutant zebrafish Ser-tRNAs that misincorporate Ser at non-cognate codons alanine (GCG), glycine (GGA), valine (GTG) and leucine (CTG) (Fig. 1A). Since the eukaryotic seryl-tRNA synthetase (SerRS) does not interact with the anticodon of Ser-tRNAs,22,23 mutations in the anticodon of the Ser-tRNA do not interfere with serylation by the SerRS, but alter decoding properties of the Ser-tRNAs (Table S1).
Figure 1.
Proteome mutagenesis using misreading tRNAs (A) A Ser-tRNA gene was mutated in the anticodon region by site directed mutagenesis to produced tRNAs that insert Ser at alanine (GCG), valine (GTG), glycine (GGA) and leucine (CTG) codons. Our method takes advantage of the fact that the seryl-tRNA-synthetase (SerRS) does not rely on the anticodon sequence to recognize the Ser tRNAs, thus allowing the mutant tRNA to be charged with Ser. These mutant tRNAs incorporate Ser at non-cognate Ala, Gly, Val and Leu sites producing mutant proteins. (B) Northern blot analysis showing that all mutant tRNAs are expressed and processed. (C) A GFP reporter was used to quantify the level of Ser misincorporation at non-cognate sites. The residue Ser 65 is critical for GFP fluorescence, insertion of another amino acid at residue 65 shifts overall fluorescence. (D) The mutant tRNAs were able to partially restore wild type GFP fluorescence levels. Western blot analysis of GFP expression was used to normalize fluorescence levels; it also shows that Ser insertion at position 65 is able the restore GFP synthesis or stability in the case of the Leu mutant. Data are presented as the mean ± SD (n = 4). Fluorescence units are relative to Control 1. Control 1 refers to 100% insertion of serine at position 65. Control 2 refers to insertion of 100% insertion of the mutated amino acids tested.
For simplicity, the mutant tRNAs were designated Ser-tRNACGCAla, Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu, indicating that they incorporate Ser at the amino acid sites corresponding to the indicated anticodons. The mutant anticodons were chosen on the basis of the chemical differences between Ser and Ala, Gly, Val and Leu, namely polarity and hydrophobicity. Ser is polar and hydrophilic while the replaced amino acids are non-polar and hydrophobic and aliphatic, as is the case of Ala and Leu.24-26 Importantly, hydrophobic amino acids, such as Ala, Val and Leu are commonly buried in hydrophobic protein cores while hydrophilic amino acids, such as Gly and Ser, are usually located on protein surfaces.25,27 These chemical variables, blossom scores and codon usage patterns were taken into consideration to produce transgenic zebrafish embryos with increasing levels of proteome instability (Table S2). For example, the blossom score for Ser-Ala replacements is 1, Ser-Gly is 0 and Ser-Val or Ser-Leu is -2.28,29 Therefore, Ala replacements with Ser were expected to produce minor proteome destabilization, but Leu replacement with Ser were expected to produce major proteome instability.25
The mutant tRNA genes were microinjected into one cell zebrafish embryos after fertilization and their expression was confirmed by northern blot analysis in 24hpf embryos (Fig. 1B). Decoding activity was monitored using a GFP reporter system (Fig. 1C and 1D) containing non-cognate codons targeted by each mutant tRNA at position 65, replacing the wild type (WT) Ser TCT codon. Since Ser 65 (S65) is essential for chromophore formation and consequently GFP fluorescence, the mutant GFPs had no fluorescence or had significantly altered (increased or decreased) fluorescence when expressed in wild type embryos. However, WT fluorescence levels of the mutant GFPs could be partially restored in embryos co-expressing the mutant tRNAs if they inserted Ser at position 65.30,31
Quantification of the relative GFP fluorescence showed that the mutant tRNAs were functional as they were able to partially restore WT GFP fluorescence levels that correspond to the incorporation of serine at position 65 (Fig. 1C and 1D). In control embryos, Leu incorporation at position 65 mediated by the CTG codon abolished production or stability of the mutant GFP protein and consequently fluorescence, likely due to the sharp differences between serine (polar) and leucine (hydrophobic). Co-expression of the mutant Ser-tRNACAGLeu and GFP(Leu65) codon restored 45% of the WT fluorescence. Conversely, insertion of glycine, alanine and valine at position 65 by the corresponding endogenous zebrafish tRNAs in control embryos, increased GFP fluorescence sharply producing a hyperfluorescent protein. But, incorporation of serine by the mutant heterologous mutant heterologous Ser-tRNACGCAla, Ser-tRNAUCCGly, Ser-tRNACACVal when co-expressed with the corresponding mutant GFPs, brought fluorescence levels back to nearly normal levels (Fig. 1D), indicating that these mutant tRNAs were also active. These data are in line with similar experiments performed in yeast, mammalian cells and chick embryos32 and support the hypothesis that serine tRNAs provide a powerful tool to mutagenize and destabilize the proteome of eukaryotic model systems.
Expression of the mutant tRNAs in embryos reduced their viability (24hpf), only 5% of the embryos expressing the Ser-tRNACAGLeu survived while the embryos expressing Ser-tRNACGCAla, Ser-tRNAUCCGly, Ser-tRNACACVal had a reduction on viability by up to 50%, 16% and 35% respectively (Fig. 2A). Embryo viability was determined at 24hfp for wild type embryos, embryos injected with empty plasmid and with the endogenous serine tRNA plasmid (Fig. S1); no significant difference was observed between these three controls subsequently the embryos injected with the endogenous serine tRNA were selected as the control for subsequent experiments. The reduction of embryo viability was accompanied by an increase in developmental malformations (Fig. 2B), in particular in the case of the embryos expressing the Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu. These malformations involved undeveloped head and spinal region, tail shortening, longitudinal symmetry loss and also developmental arrest (Fig. 2C), further indicating that the mutant tRNAs were fully functional.
Figure 2.
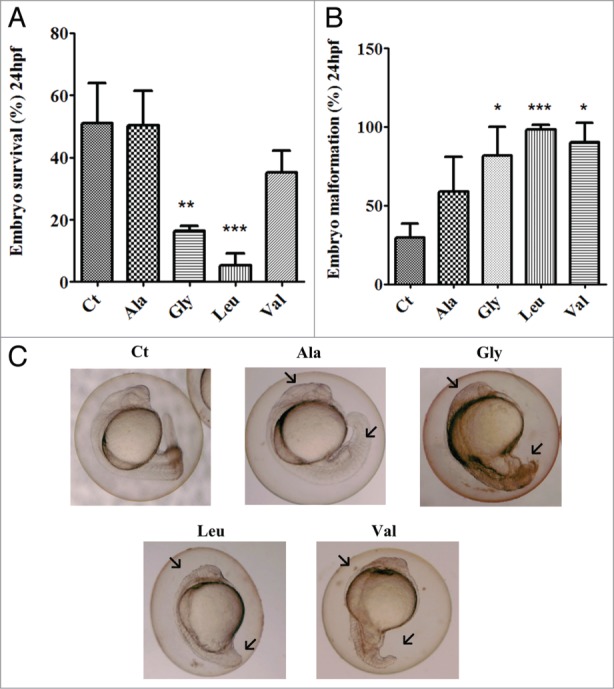
Impact of mRNA mistranslation on early zebrafish development.(A) Mutant tRNAs strongly decreased embryos survival at 24hpf. Errors bars represent the standard deviation of three independent experiments. (Student's unpaired t test, **P < 0.05, ***P < 0.0001) (B) Mutant tRNAs have a strong effect on embryo development and cause morphological alterations. This graph illustrates the percentage of embryos displaying morphological alterations Data are presented as the mean ± SD (n = 3) (Student's unpaired t test against control sample, *P < 0.05, **P < 0.01). (C) Morphological alterations caused by mutant tRNAs. Control embryos (microinjected with endogenous serine tRNA) show normal development. Embryos expressing the mutant misreading tRNAs show various morphological alterations, namely: longitudinal symmetry loss, undeveloped head and spinal region, and also developmental arrest (data not shown).
Mistranslation increases protein ubiquitination and aggregation
As mentioned above, our transgenic embryos co-expressed the mutant tRNAs and wild type (WT) tRNACGCAla, tRNAUCCGly, tRNACACVal and tRNACAGLeu, whose tDNA genes copy number in zebrafish are 112, 47, 227 and 300, respectively. These WT tRNAs ensure production of WT polypeptides and embryo viability. In other words, the mutant Ser-tRNACGCAla, Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu competed with these WT tRNAs for the decoding of the above mentioned codons, producing statistical subpopulations of polypeptides of each protein (statistical proteins). Since the fate of these mutant polypeptides is dependent on the destabilizing nature of the mutations; i.e., certain mutations may result in protein unfolding, ubiquitination, degradation or aggregation, while others may induce low level of protein destabilization compatible with refolding by molecular chaperones, we expected that Ser misincorporation would saturate PQC systems, leading to increased protein ubiquitination and aggregation. Indeed, protein ubiquination was increased in all mutants, with the highest levels occurring in the embryos expressing the Ser-tRNAUCCGly (60%), Ser-tRNACACVal (40%) and Ser-tRNACAGLeu (60%) (Fig. 3A). Insoluble protein aggregates were also detected and the highest levels were present in the embryos expressing the Ser-tRNACAGLeu (70%) (Fig. 3B). In line with these data the carbonyl content of proteins, which measure oxidative damages, was increased by 490% in the embryos expressing the Ser-tRNACAGLeu, respectively (Fig. 3C). We used the oxidative stress generators 3-nitropropionic acid (NPA) and rotenone as positive controls for protein aggregation and observed 120% (20mM NPA) and 90% (15μg/l Rotenone) (Fig. 3D) increase in protein aggregation in WT embryos, showing that oxidative stress has the potential to increase protein aggregation. Taken together these data indicate that mistranslated proteins are more susceptible to oxidative damage and aggregation, that ROS also contributes to protein aggregation and that the chemical differences between Ser and the other amino acids tested determines the intensity of the proteotoxic stress generated by the misreading tRNAs.
Figure 3.
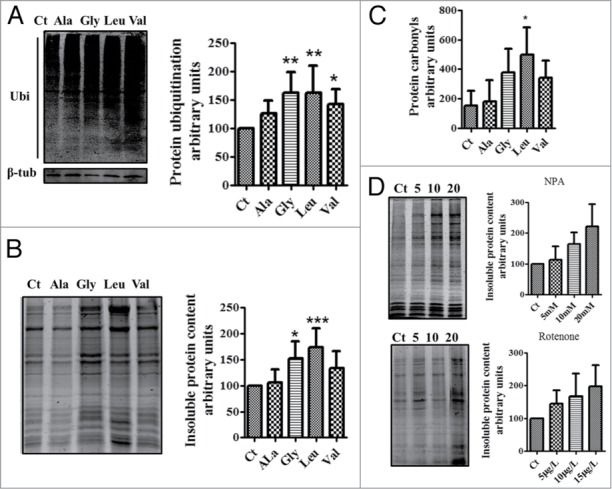
Mistranslation increases the levels of protein ubiquitination and aggregation. (A) western blot analysis showing that mistranslated proteins are poliubiquitinated. Data showed are the mean ± SD (n = 6). (Data analysis was performed using one-way ANOVA followed by a Dunnet test with CI 95% relative to Ct; ***P < 0.001, **P < 0.01, *P < 0.05). (B) Mutant proteins produced by Ser misincorporation accumulate in insoluble aggregates, especially in embryos expressing the Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu. Representative gel showing the insoluble protein fraction of control and mistanslating zebrafish embryos. Data are presented as the mean ± SD (n = 5). (Data analysis was performed using one-way ANOVA followed by a Dunnet test with CI 95% relative to Ct; ***P < 0.001, **P < 0.01, *P < 0.05). (C) Gels showing that mistranslation increases the levels of protein carbonylation. Errors bars represent the standard deviation of four independent experiments (Student's unpaired t test, *P ˂ 0.05). (D) Gels showing that oxidative stress generates protein aggregation in zebrafish. Embryos incubated with the mitochondrial electron transport chain disruptors 3-Nitropropionic acid (NPA) and Rotenone had high levels of insoluble proteins. Data are presented as the mean ± SD (n = 3). Units are relative to control sample in percentage. Western blot and SDS-page samples were normalized for the gel control sample.
The production of protein aggregates in mistranslating embryos was further analyzed using a chimeric Hspb1-mCherry reporter protein (Fig. 4A). Since Hspb1 is an oligomeric small heat shock protein that binds unfolded proteins and keeps them in a folding competent state it can be used to monitor protein aggregation in vivo if fused to a fluorescent probe.33,34 Expression of the Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu produced fluorescent foci localized near the nucleus and also scattered throughout the cytoplasm (Fig. 4B), confirming the increase in protein aggregation in the mistranslating embryos. Ubiquitin immunofluorescence analysis of 5dpf embryos expressing Ser-tRNACGCAla and Ser-tRNACAGLeu confirmed accumulation of polyubiquitinated proteins in the perinuclear foci in the latter (Fig. 4C), indicating that misfolded proteins produced by tRNA misreading are ubiquitinated and aggregate in the perinuclear region. Therefore, we can suggest that mRNA mistranslation leads to the production of aberrant polypeptides that are differentially tagged for degradation and accumulated at differentially locations in the cell.
Figure 4.
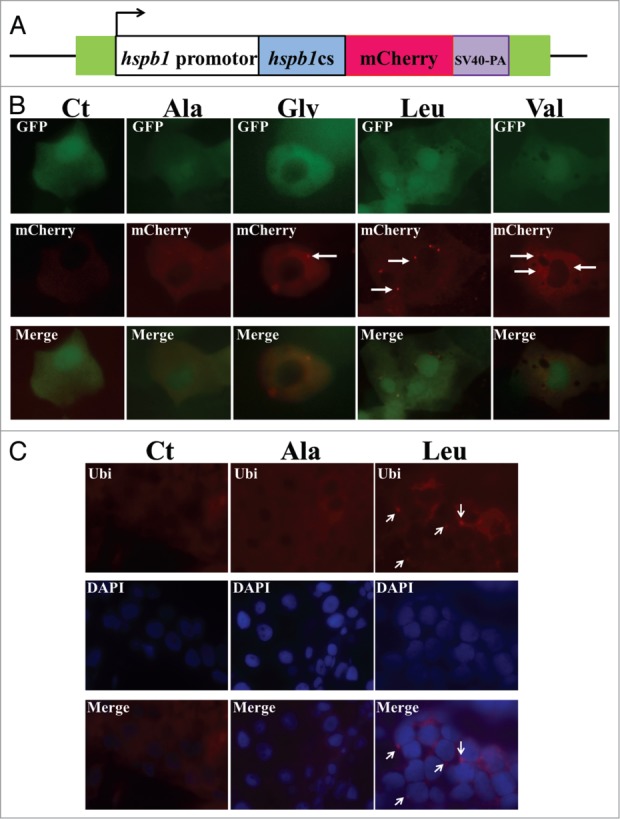
Mistranslation induces the formation of protein aggregates. (A) A fluorescence reporter constructed by fusing the zebrafish HSPB1 protein with mCherry was used to monitor the formation of protein aggregates. (B) Mistranslating embryos expressing the HSPB1-mCherry chimeric protein were observed using fluorescence microscopy. The accumulation of protein aggregates was observed by formation of HSPB1-mCherry fluorescent foci which were more visible in embryos expressing the Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu. (C) Immunofluorescence analysis of 5dpf embryos expressing the Ser-tRNACGCAla and the Ser-tRNACAGLeu show that the ubiquitin foci accumulated in the perinuclear region of the cytoplasm. Nuclei were labeled with DAPI.
Global transcriptional responses to proteome instability induced by mistranslation
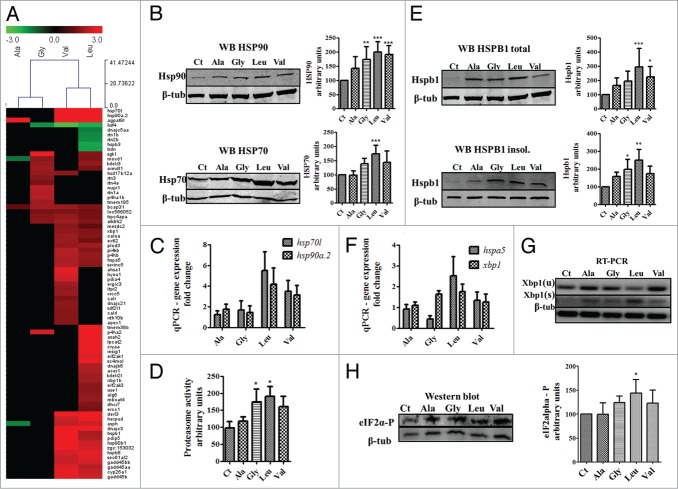
The transcriptomes of the mistranslating embryos were then profiled using DNA microarrays to better understand the nature of the proteotoxic stress response induced by aggregation of the mistranslated proteins. Significant gene expression deregulation was observed in a mutant dependent manner. The Ser-tRNACAGLeu showed the highest level of gene deregulation, with a marked trend of gene upregulation (Fig. 5A). These data (Table 1) supported the activation of the stress response since molecular chaperones, in particular hsp70l was 34.2 and 38.3-fold upregulated in embryos expressing the Ser-tRNACACVal and Ser-tRNACAGLeu, and hsp90a.2 was 7.5-fold and 9.4-fold upregulated in the same embryos. The hsf4 transcription factor which regulates heat shock protein expression through hsf1 repression was downregulated.35
Figure 5.
Induction of stress responses by mRNA mistranslation. (A) Cluster representation of stress response and Endoplasmic Reticulum (ER) genes that are deregulated in mistranslation embryos. These genes are mainly upregulated, in particular in embryos misincorporating Ser at Leu and Val codons. (B) Western blot analysis of HSP90 and HSP70 showing that these chaperones are induced by mRNA mistranslation. Data shown are the mean ± SD (n = 4). (Data statistical analysis was performed using one-way ANOVA followed by a Dunnet test with CI 95% relative to Ct; ***P < 0.001, **P < 0.01, *P < 0.05). Intensity units are relative to control. Units are relative to control sample in percentage. Western blot samples were normalized for the gel control sample. (C) qRT-PCR was performed to check the levels of HSP70L and HSP90A.2 mRNAs in mistranslating embryos. (D) Proteasome activity was quantified using the fluorogenic chymotrypsin-like substrate s-LLVY-MCA. Data shown are the mean ± SD (n = 4). (Student's unpaired t test, *P ˂ 0.05 was used). Activity units are relative to control. Units are relative to control sample in percentage. (E) western blot analysis of HSPB1 in total and insoluble fractions show that this small heat shock protein is induced by mRNA mistranslation and that it accumulates in the insoluble fraction. Data are presented as the mean ± SD (n = 3). (Data statistical analysis was performed using one-way ANOVA followed by a Dunnet test with CI 95% relative to Ct; ***P < 0.001, **P < 0.01, *P < 0.05). Units are relative to control sample in percentage. Western blot samples were normalized for the gel control sample. (F) Induction of the ER stress response by mRNA mistranslation. qRT-PCR was performed to quantify the levels of HSPA5/BIP mRNA in mistranslating embryos. BIP acts as an early ER stress sensor. It recognizes and links protein hydrophobic exposed regions, thus signaling misfolded/unfolded polypeptides. (G) Alternative splicing of XBP1 mRNA was monitored by RT-PCR in zebrafish embryos during ER stress induced by mRNA mistranslation. β-tubulin was used as internal control for RT-PCR. (H) Increased phosphorylation levels of eIF2α are consistent with downregulation of translation in mistranslating embryos. Data shown are the mean ± SD (n = 4). (statistical analysis was performed using one-way ANOVA followed by a Dunnet test with CI 95% relative to Ct; ***P < 0.001, **P < 0.01, *P < 0.05). Total eIF2α levels (Fig.S3) are stable in embryos expressing the mutant tRNAs. Units are relative to control sample in percentage. Western blot samples were normalized for the gel control sample.
Table 1.
Stress response and ER genes deregulated by mistranslation in zebrafish embryos. (Microarray data analysis was done with MEV software.85 Significantly deregulated genes for each mutant tRNA were obtained with a t test analysis (p0.05) and a 1.5-fold change deregulation cut off. Fold change values are displayed.)
| Gene symbol | Ala | Gly | Val | Leu | Function |
|---|---|---|---|---|---|
| hsp70l | 34.17 | 38.32 | Heat shock protein | ||
| hsp90a.2 | 7.54 | 9.40 | Heat shock protein | ||
| cryaa | 3.54 | Small heat shock protein | |||
| hsp90b1 | 3.85 | 2.39 | ER chaperone | ||
| hspa5 | 2.02 | 1.72 | ER chaperone | ||
| hspb1 | 3.08 | 1.99 | Small heat shock protein | ||
| hspb3 | −1.93 | Small heat shock protein | |||
| hspb8 | 2.46 | 2.69 | Small heat shock protein | ||
| dnajb6 | 2.08 | DNAJ chaperone | |||
| dnajc3 | 3.35 | 3.33 | ER DNAJ chaperone | ||
| dnajc5aa | −1.50 | DNAJ chaperone | |||
| dnajc21 | 1.68 | DNAJ chaperone | |||
| ahsa1 | 2.69 | HSP90 ATPase activation | |||
| nupr1 | 2.21 | Target of ATF4 | |||
| hsf4 | −1.70 | −2.64 | −2.01 | Regulator of HSP expression | |
| xbp1 | 1.70 | 1.54 | Unfolded Protein Response | ||
| eif2ak1 | 2.61 | eif2alpha kinase | |||
| eif2ak3 | 1.65 | eif2alpha kinase | |||
| derl3 | 3.64 | 4.90 | ER stress response ERAD | ||
| herpud | 2.39 | 5.13 | ER stress response ERAD | ||
| bcap31 | 1.25 | 1.93 | 1.82 | 1.49 | ER stress response ERAD |
| sec61al2 | 2.34 | 2.62 | ER stress response ERAD | ||
| sdf2l1 | 1.65 | ER chaperone | |||
| hyou1 | 2.57 | ER chaperone | |||
| ergic3 | 1.91 | Response to ER stress | |||
| pdia4 | 2.41 | ER chaperone | |||
| pdip5 | 3.50 | 2.07 | ER chaperone | ||
| asph | −1.55 | 2.24 | 2.43 | Calcium homeostasis | |
| calr | 1.52 | ER calcium binding | |||
| calrl | 1.58 | CALR like protein | |||
| itpr2 | 1.76 | IP(3)-sensitive Ca(2+) channels | |||
| rtn1a | 2.12 | ER shaping protein | |||
| trdn | −1.87 | ER calcium release | |||
| rtn1b | −1.60 | ER shaping protein | |||
| rtn2b | −1.64 | ER shaping protein | |||
| rtn3 | 1.62 | ER shaping protein | |||
| rtn4a | 1.46 | ER shaping protein | |||
| ercc1 | 1.47 | DNA repair | |||
| alkbh2 | 1.63 | 1.87 | 1.55 | DNA repair | |
| xrcc5 | 1.80 | DNA repair | |||
| apex1 | 1.60 | DNA repair | |||
| gadd45aa | 2.60 | 2.14 | DNA-damage response | ||
| gadd45bb | 2.35 | 2.50 | DNA-damage response | ||
| gadd45b | 2.67 | 2.41 | DNA-damage response |
The ER resident chaperones hsp90b1/grp94 and hspa5/bip36,37 were also deregulated. Grp94 was 3.8 and 2.5-fold upregulated in embryos expressing the Ser-tRNACACVal and the Ser-tRNACAGLeu, respectively, while bip was upregulated 2.0 fold and 1.7-fold, respectively. Several other genes related to stress response processes were induced (Table 1), namely eif2ak1, eif2ak3/perk which are involved in translation attenuation through eIF2α phosphorylation . The major UPR regulator xbp1 was induced, and overexpression of hspa5/bip, hyou1, copb, zgc:103652, sec61a, gtpbp1, zgc:92245 and chchd2l were well correlated with the results obtained for the overexpression of Xbp1 s.38 The sec61a gene, which encodes a membrane ER protein involved in the import of newly synthesized polypeptides that integrates the ERAD machinery providing export capacity to misfolded and non-ubiquitinated proteins for degradation,39 was upregulated. Sec61a is also a calcium channel that allows Ca2+ leakage from the ER under stress conditions. Under normal conditions Bip seals Sec61a channels, but it translocates to unfolded/misfolded polypetides leaving the Sec61a channels open during stress.40 The data suggested, therefore, that ER stress induced by mistranslation may trigger calcium release to the cytoplasm increasing cytoplasmic calcium levels. Accordingly, calcium homeostatic control genes, namely asph, calr, calrl and itpr2 and also ER shaping genes were deregulated as well (Table 1). Additionally, the ERAD mediators derl3,41 herpud1,42 and bcap3143 (Table 1), the ER co-chaperone dnajc3,44 the co-chaperones dnajb645 and dnajc2146 and the dnaj5aa47 were also deregulated (Table 1). ER protein transport genes and many ER metabolism related genes, mainly lipid metabolism, were in most cases upregulated (Table S4), confirming that mistranslation induces the UPR.
Finally the apex1 gene which is a component of the base excision repair mechanism,48 xrcc5 which repairs double strand breaks in zebrafish embryos,49 and the alkbh2 gene which is involved in repairing DNA etheno adducts,50 were upregulated (Table 1). Gadd45 proteins are involved in cell-cycle regulation and are normally induced by genome damage and stress conditions, and regulate somite formation in zebrafish.51 The gadd45al, gadd45bl and gadd45b genes were upregulated 2.6, 2.3 and 2.6 in embryos expressing the Ser-tRNACACVal and 2.1, 2.5 and 2.4 in embryos expressing the Ser-tRNACAGLeu, supporting the hypothesis that mistranslation affects the genome stability.
Stress responses induced by mistranslation
The effects of mistranslation on the stress response pathways was further analyzed, with particular focus on the heat-shock response and ubiquitin proteasome pathway. Expression of HSP70 and HSP90 was upregulated at both the mRNA and protein levels, especially in the embryos expressing the Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu (Fig. 5C and 5C, Table S5). In the case of the embryos expressing the Ser-tRNAUCCGly Hsp70 was induced 3.2-fold at the mRNA level and 1.4-fold at the protein level, Hsp90 was 2.8-fold induced at the mRNA level and 1.7-fold at the protein level. Embryos expressing the Ser-tRNACACVal upregulated Hsp70 11.2-fold at the mRNA level and 1.4-fold at the protein level, Hsp90 was 8.8-fold induced at the mRNA level and 1.9-fold at the protein level. Finally, in the case of the Ser-tRNACAGLeu, Hsp70 was upregulated 44.8-fold at the mRNA level and 1.7-fold at the protein level and Hsp90 was upregulated 18.2-fold at the mRNA level and 2-fold at the protein level. The ubiquitin proteasome pathway was strongly induced (Fig. 3A and 5D) in particular in the embryos expressing the most destabilizing Ser-tRNACAGLeu, Ser-tRNAUCCGly and Ser-tRNACACVal. In other words, the mistranslations with the biggest chemical differences between misincorporated and WT amino acids resulted in differential UPP activation, in line with the results described above. The small heat shock protein HSPB1 was also induced at the mRNA and protein levels (Fig. 5E) and was accumulated in the insoluble fraction (Fig. 5E), which is in line with its binding to misfolded and aggregated proteins. These data are in line with the strong upregulation of the UPP observed above (Fig. 3A and 5D) and is also consistent with upregulation of the UPR since hspa5/bip and xbp1 were upregulated 2.5 and 1.8-fold respectively, in the Ser-tRNACAGLeu (Fig. 5F, Table S4). We have also detected splicing of the transcription factor Xbp1 in all four mutants, with the highest level in the embryos expressing the Ser-tRNACAGLeu (Fig. 5G).
Since the activation of the stress response results in downregulation of protein synthesis we have quantified its rate using the SUnSET technology, which measures the incorporation of puromycin into nascent polypeptides.52 Western blot analysis of total protein extracts labeled with puromycin showed 30% and 40% decrease in protein synthesis rate in embryos expressing the Ser-tRNACACVal and Ser-tRNACAGLeu (Fig.S2). As before, the severity of downregulation was correlated with the differences in the chemical properties between Ser and the other amino acids; especially in the cases of Ser-tRNACACVal and Ser-tRNACAGLeu where the decrease in protein synthesis was stronger. This prompted us to monitor the phosphorylation status of the translation initiation factor 2 (eIF2α), which is a major regulator of protein synthesis. There was increased phosphorylation of eIF2α and stable levels of total eIF2α, in line with the differences observed for the rate of protein synthesis. EIF2α-P levels reached 40% in embryos expressing the Ser-tRNACAGLeu (Fig. 5H and S3) and there was a small increase in phosphorylation in embryos expressing the Ser-tRNAUCCGly and Ser-tRNACACVal.
Mistranslation leads to the accumulation of reactive oxygen species
Stress induced production of reactive oxygen species (ROS) in the ER and subsequent Ca2+ release from the ER increases mitochondrial ROS production in several model systems.20,53-55 To clarify whether our misreading tRNAs would impact on oxidative stress we have quantified ROS using DHE and Amplex® Red Hydrogen Peroxide kit (Molecular Probes®). DHE analysis showed an increased production of the superoxide anion in embryos expressing Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu (Fig. 6A). Production of hydrogen peroxide (H2O2), especially in the embryos expressing the Ser-tRNACACVal and Ser-tRNACAGLeu (Fig. 6B), was increased by 200 and 270%, respectively, in line with increased activity of catalase (50% for Ser-tRNACACVal and 180% for Ser-tRNACAGLeu), which converts H2O2 to H2O and O2 (Fig. 6C and 6D). The gene expression profiling also showed deregulation of several redox maintenance and antioxidant defense genes (Fig. 6D, Table 2), namely the antioxidant cat gene56 whose expression was upregulated 1.9-fold in Ser-tRNACAGLeu expressing embryos. Several selenoproteins and other antioxidant defense related genes, such as glrx, mgst1 and gstp1 were also induced (Table 2). Similar results were obtained for the peroxissomal pex13, lonp2, zgc:114079 and pmp34 genes (Table 2), indicating that our proteome instability model recapitulated the oxidative stress phenotype observed in protein conformational diseases.57
Figure 6.
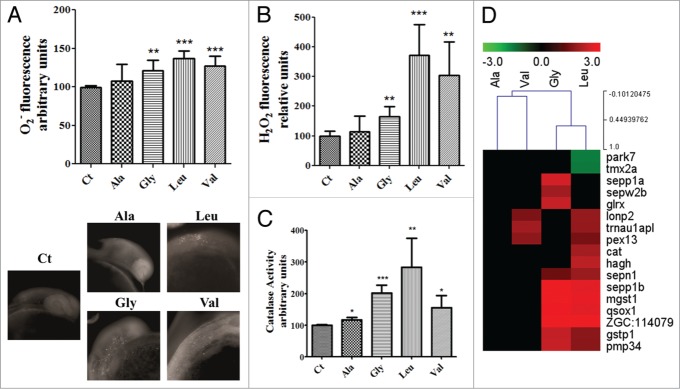
Oxidative stress is one of the endpoints of mistranslation. (A) The detection of the superoxide anion with dihydroethidium by fluorimetry (a) and microscopy (b) showed a discrete increase in its levels in the mutant embryos expressing the mutant tRNAs. Data are presented as the mean ± SD (n = 5) (Student's unpaired t test, **P ˂ 0.005, ***P ˂ 0.0001 was used for data statistical analysis). (B) Hydrogen peroxide levels were highly increased in the mistranslating embryos. Data are presented as the mean ± SD (n = 4) (Student's unpaired t test, **P ˂ 0.01, ***P ˂ 0.001). (C) Catalase activity was also induced by mistranslation, balancing the increase in the production of hydrogen peroxide. Data are presented as the mean ± SD (n = 4) (Student's unpaired t test, *P ˂ 0.05, **P ˂ 0.01, ***P ˂ 0.0005). (D) Cluster representation of redox maintenance and antioxidant defense genes deregulated in mistranslating embryos, showing that these genes are mainly upregulated, with the strongest upregulation in embryos expressing the Ser-tRNAUCCGly and the Ser-tRNACAGLeu. Units are relative to control sample in percentage.
Table 2.
Antioxidant defense genes deregulated by mistranslation in zebrafish embryos. (Microarray data analysis was done with MEV software.85 Significantly deregulated genes for each mutant tRNA were obtained with a t test analysis (p0.05) and a 1.5-fold change deregulation cut off. Fold change values are displayed.)
| Gene symbol | Ala | Gly | Val | Leu | Function |
|---|---|---|---|---|---|
| park7 | −1.48 | Oxidative stress response, apoptosis activator | |||
| sepp1a | 2.49 | Selenoprotein | |||
| sepp1b | 3.26 | 2.66 | Selenoprotein | ||
| sepn1 | 1.32 | 1.81 | Selenoprotein | ||
| sepw2b | 1.90 | Selenoprotein | |||
| trnau1apl | 1.93 | 1.75 | Protein associated with selenocysteine tRNA | ||
| qsox1 | 2.88 | 2.66 | Cell redox homeostasis | ||
| glrx | 2.33 | Antioxidant defense | |||
| gstp1 | 2.30 | 1.65 | Antioxidant defense | ||
| mgst1 | 3.17 | 2.74 | Antioxidant defense | ||
| cat | 1.94 | Antioxidant defense peroxissomal | |||
| pex13 | 1.69 | 1.45 | Antioxidant defense peroxissomal | ||
| lonp2 | 1.50 | 1.75 | Antioxidant defense peroxissomal | ||
| zgc:114079 | 5.50 | 4.91 | Antioxidant defense peroxissomal | ||
| zgc:162641 | 2.33 | 1.64 | Antioxidant defense peroxissomal |
Mistranslation impacts mitochondrial organization and function
The increase in oxidative stress and upregulation of DNA repair genes prompted us to investigate whether mistranslation increased DNA damage. We focused on the integrity of both the mitochondrial and nuclear genomes in the mistranslating zebrafish embryos using qPCR.58 Mitochondrial damages accumulated in response to Ser-tRNACGCAla (0.39 lesions/10Kb), Ser-tRNAUCCGly (0.82 lesions/10Kb), Ser-tRNACACVal (0.77 lesions/10Kb) and Ser-tRNACAGLeu (1.62 lesions/10Kb). Nuclear genome damage was also observed in response to Ser-tRNACGCAla (0.16 lesions/10Kb), Ser-tRNAUCCGly (0.36 lesions/10Kb), Ser-tRNACACVal (0.34 lesions/10Kb) and Ser-tRNACAGLeu (0.52 lesions/10Kb), (Fig. 7A). The higher level of damage of the mitochondrial DNA is likely related to reduced DNA repair capacity relative to repair of nuclear DNA and higher exposure to ROS.59-61
Figure 7.
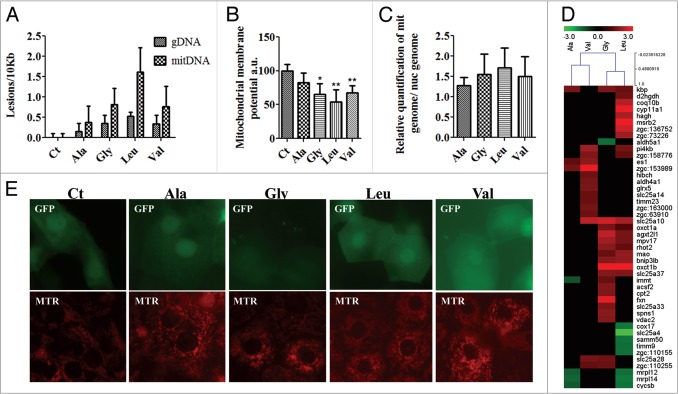
Mistranslation increases damage of mitochondrial and nuclear DNA and alters the mitochondrial morphology. (A) Quantification of DNA damage showing that mistranslation leads to the accumulation of damages in both the nuclear and mitochondrial DNA. DNA damage was determined by qPCR. Values are expressed as lesions/10 kb of DNA. Data are presented as the mean ± SD (n = 3). (B) Relative mtDNA copy number was measured by qRT-PCR, results show a tendency toward an increased mitochondrial abundance. Data are presented as the mean ± SD (n = 3). Units are relative to control sample in percentage. (C) Mitochondrial membrane potential was assessed by JC-1 staining; fluorescence was measured on mitochondria purified from control and mistranslating embryos. Data are presented as the mean ± SD (n = 3) (Student's unpaired t test, *P ˂ 0.05, **P ˂ 0.01). (D) Mistranslating embryos were labeled with Mito Tracker Red and observed with fluorescence microscopy. Mistranslation affected mitochondrial morphology, in particular the tubular structure of mitochondria was lost in mistranslating embryos. These differences in the shape of mitochondria were more pronounced in embryos expressing the Ser-tRNACACVal and the Ser-tRNACAGLeu. (E) Cluster representation of mitochondrial genes deregulated in mistranslating embryos showing a trend toward upregulation. The higher upregulation was observed in embryos expressing the Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu.
The above data indicated that mistranslation likely had a major impact on mitochondrial function. To test this hypothesis we have characterized the mitochondrial membrane potential and mitochondrial morphology. The former was reduced by 35%, 33% and 46% in embryos expressing the Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu (Fig. 7B), indicating that oxidative phosphorylation and ATP production were affected. Real-Time quantitative PCR (Fig. 7C) analysis showed 28%, 55%, 50% and 70% increase in the levels of mitochondrial DNA in embryos expressing the tRNACGCAla, Ser-tRNAUCCGly, Ser-tRNACACVal and Ser-tRNACAGLeu, respectively. And, the morphology of the mitochondrial network showed major alterations in embryos expressing the Ser-tRNACAGLeu. Lower levels of disruption of the mitochondrial network were observed in the case of Ser-tRNACGCAla, Ser-tRNAUCCGly and Ser-tRNACACVal embryos (Fig. 7D), indicating that proteome instability produced by mRNA mistranslation increases the ratio of fission/fusion.
Those alterations in the mitochondrial network were consistent with deregulation of expression of several mitochondrial genes (Fig. 7E, Table 3). For example park2/parkin, which promotes mitochondrial degradation by facilitating their elimination by specific autophagy – mitophagy,62 was 4.8-fold upregulated in embryos expressing the Ser-tRNACAGLeu (Fig. 7E). Conversely, park7/dj-1 which protects against ROS and translocates to the mitochondrial membrane under mitochondrial oxidative stress63 was downregulated 1.5-fold in embryos expressing the Ser-tRNACAGLeu (Fig. 6D, Table 2), indicating increased susceptibility of mitochondria to ROS. Furthermore loss of Park7/Dj-1 elicits mitochondrial membrane potential reduction,64 an effect also observed in our samples. Genes related to mitochondrial network dynamics, namely rhot2, zgc:63910/mtfp1 and mff65-67 were upregulated. The gene zgc:63910 was 1.5-fold upregulated in embryos expressing the Ser-tRNACACVal, mff was 1.2-fold upregulated in embryos expressing the Ser-tRNACAGLeu and rhot2 was upregulated 1.9-fold in embryos expressing the Ser-tRNAUCCGly and 1.4 in Ser-tRNACAGLeu embryos. These three factors are activators of mitochondrial fission and importantly Rhot2/Miro2 is a fission activator that responds to elevated Ca2+ signals in the cytoplasm,65 which is frequent during ER stress.
Table 3.
Mitochondrial genes deregulated in mistranslating embryos. (Microarray data analysis was performed using the MEV software.85 Significantly deregulated genes for each mutant tRNA were obtained with a t test analysis (p0.05) and a 1.5-fold change deregulation cut off. Fold change values are displayed.)
| Gene symbol | Ala | Gly | Val | Leu | Function |
|---|---|---|---|---|---|
| park2 | 4,8 | Mitochondrial turnover | |||
| cox17 | −1,57 | COX assembly | |||
| cycsb | −1,22 | −1,52 | Mitochondrial respiratory chain | ||
| bnip3lb | 1,84 | 1,99 | Apoptosis | ||
| zgc:158776 | 1,76 | 1,42 | Apoptosis | ||
| spns1 | 1,89 | Apoptosis | |||
| mpv17 | 2,15 | 1,64 | Response to ROS | ||
| glrx5 | 1,69 | Biogenesis of iron-sulfur clusters | |||
| msrb2 | 4,88 | Response to ROS | |||
| mrpl12 | −1,26 | −1,47 | Mitochondrial ribosomal protein | ||
| mrpl14 | −1,36 | −1,58 | Mitochondrial ribosomal protein | ||
| zgc:110255 | 1,51 | 1,43 | Mitochondrial ribosomal protein | ||
| zgc:63910 | 1,52 | Mitochondrial fission | |||
| mff | 1,2 | Mitochondrial fission | |||
| rhot2 | 1,88 | 1,43 | Mitochondrial dynamics | ||
| samm50 | −1,53 | Ultrastructure organization | |||
| atg10 | 1,5 | 1,5 | Autophagy | ||
| atg4b | 1,8 | Autophagy | |||
| becn1 | 1,6 | 1,9 | 1,9 | Autophagy | |
| gabarap | 1,9 | Autophagy | |||
| map1lc3b | 2,4 | 1,4 | Autophagy |
Several apoptosis and autophagy related genes were also upregulated (Table 3). For example zgc:158776/smac/diablo which is a mitochondrial protein related to apoptosis and caspase activation68 was upregulated by Ser-tRNACACVal and by Ser-tRNACAGLeu expressing embryos. The bnip3lb gene encoding a mitochondrial outer membrane protein involved in autophagy and autophagic cell death69 was also upregulated in Ser-tRNAUCCGly and Ser-tRNACAGLeu expressing embryos. Interestingly, the major zebrafish autophagic markers and autophagy related genes were coordinately expressed in our mistranslating embryos namely lc3/map1lc3b, gabarap, becn1, atg10 and atg4b (Table 3).70 The lc3/map1lc3b gene was upregulated both in Ser-tRNAUCCGly and Ser-tRNACAGLeu, becn1 was upregulated in the three mutants, specifically in Ser-tRNAUCCGly, Ser-tRNACACVal and the Ser-tRNACAGLeu (Table S6), further supporting the strong negative effect of mistranslation on mitochondrial function.
Discussion
Mistranslation induces proteome aggregation in zebrafish
The maintenance of a functional proteome is fundamental to life. Translational errors occur naturally at a frequency of 10−4 and are not known to be associated with cell degeneration, however small increases in the level of translational errors are detrimental.71 We have observed accumulation of ubiquitinated protein aggregates in embryos misincorporating Ser at Gly, Val and Leu codons and minor effects on embryos misincorporating Ser at Ala codons where there was only a small increase in ubiquitination, carbonylation and protein aggregation (Figs. 3A, 3B, and 2C). In yeast and mammalian cells the protein aggregates accumulate at two distinct locations; the insoluble protein deposit (IPOD) that mediates the stabilization of terminally aberrant aggregated and insoluble proteins and the juxtanuclear quality control (JUNQ) that concentrates chaperones and the quality control machinery and degrades misfolded proteins.72 Additionally, stress or aging can increase the load of misfolded proteins in the cell, saturating the protein quality control machinery located at the JUNQ, leading to the accumulation of potentially toxic aggregates that are directed to the IPOD for destruction.72 Our results indicate that mRNA mistranslation produces polyubiquitinated protein aggregates in the perinuclear region, protein aggregates in the cytoplasm and high levels of insoluble protein, whose compatibility with the features of JUNQ and IPOD locations raises the possibility that our protein aggregates also accumulated at these protein quality control compartments. The results are consistent with the differences in chemical properties between Ser and the targeted amino acids, i.e., Ser and Ala are chemically similar and the Ser-tRNACGCAla produced mild proteotoxic phenotypes while Ser misincorporations at the other amino acids (Gly < Val < Leu) sites produced increasing levels of protein ubiquitination and aggregation and upregulation of the stress response (Figs. 2 and 4). Mistranslation also activated the ER-unfolded protein response (ER-UPR). Since the UPR coordinates several quality control mechanisms of the secretory pathway and plays a role in several pathologies such as cancer, neurodegenerative, metabolic and inflammatory diseases,73 our results are consistent with the hypothesis that mistranslation causes disease through ER stress. Indeed, ER stress signals extend to the mitochondria, are associated with ROS production and activate JNK and TOR signaling pathways.20 In zebrafish, ER stress is associated with liver disease74 and it will be interesting to investigate in future studies whether mistranslating transgenic fish will develop liver and other protein conformational diseases.
Induction of the UPR triggers eIF2-α phosphorylation through activation of PERK, reducing the delivery of tRNAiMet to the ribosome and consequently decreasing translation rate.75 Our data confirmed the downregulation of protein synthesis rate associated with increasing levels of eIF2-αP, depending on the chemical differences between Ser and the amino acids targeted by the misreading tRNAs. Studies from other laboratories showed that protein aggregation does induce eIF2-αP and long-term downregulation of protein synthesis, rather than toxicity associated with aggregated proteins is the main cause of neuronal cell death.76 We have observed increased levels of apoptosis (data not shown) and it will be interesting to determine whether the latter is a direct result of downregulation of protein synthesis that can be alleviated through inhibition of eIF2-α phosphorylation.
Mistranslation destabilizes the nuclear and mitochondrial genomes and the mitochondrial network
We have observed remarkable negative impacts of mistranslation on both the nuclear and mitochondrial genomes, and upregulation of DNA repair and DNA damage response genes, but further studies are needed to clarify whether these negative phenotypes are a direct consequence of downregulation of protein synthesis and protein aggregation or are associated with oxidative damage or disruption of DNA replication, recombination and repair. In any case, this unexpected deregulation is relevant to understand the role of tRNA mutations in cancer, degenerative diseases and aging.
Mitochondria form highly structured networks which are maintained through dynamic fusion and fission.77,78 The dynamic balance between mitochondrial fusion and fission is crucial for diverse biological processes, namely apoptosis, embryonic development and human diseases.78-80 Our transcriptome profiling data showed that mitochondrial fission processes are upregulated by proteotoxic stress, suggesting that the observed fragmentation of the mitochondrial network is due to increased fission. Since the ER and mitochondria are interconnected at the mitochondria-associated membrane (MAM) and enable lipid, Ca2+ and ATP shuttling, it is likely that mRNA mistranslation affects mitochondrial function through elevated ER stress. Furthermore, association of the ER and mitochondrial membranes plays a role in mitochondrial dynamics and these contacts increase under stress potentiating apoptosis activation.81 Prolonged ER stress increases the calcium flow from the stressed ER into the mitochondrial matrix through the MAM, and the accumulation of Ca2+ in the mitochondrial matrix saturates its buffering capacity resulting in the opening of the mitochondrial permeability transition pore (mtPTP). Modulation of the stress signal from the stressed ER to the mitochondria can also be executed by ROS and high levels of oxidative stress can also deregulate Ca2+ signaling and homeostasis through oxidative damages to Ca2+ channels, namely SERCA and IP3R.82 Therefore, our data suggests that mistranslation may disrupt mitochondrial function by deregulating Ca2+ signaling between the ER and the mitochondria, as suggested before.82 It also show that proteome instability on its own (in presence of a fully functional PN) is sufficient to disrupt the mitochondrial network, increase ROS, saturate the ER and downregulate protein synthesis. Further work will be necessary to address the relative contribution of the ER and of other subcellular compartments to ROS production during mistranslation, but considering the activation of the UPR, mitochondrial instability and the increased ROS production, it is likely that the observed increase in ROS in the mistranslating embryos may result from both ER saturation and mitochondrial dysfunction. In any case, the data demonstrate that mistranslation has a dominant effect on the mitochondrial network and is a major cause of ROS accumulation, mimicking several of the major endpoints of neurodegenerative diseases.
Conclusions
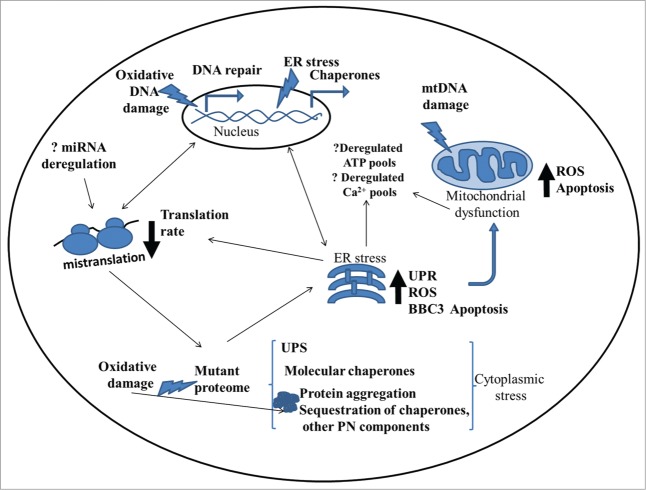
The role of mistranslation in human diseases has been overlooked and is poorly understood, but its strong effect on mitochondrial dysfunction, oxidative stress, mtDNA and gDNA damage and ER stress, uncovered in this study, suggest that it may have a strong effect on cell viability and degeneration (Fig. 8). Our data also raises the hypothesis that long-term therapies, environmental stressors, pathologies, mutations and other biological or environmental processes that interfere with translational fidelity will have direct impact on protein aggregation and consequently on proteotoxic stress, mutation rate, genome stability and aging.
Material and Methods
Maintenance of zebrafish (Danio rerio)
The AB zebrafish strain was maintained at 28°C on a 14 h-light/10 h-dark cycle. Stages of embryonic development were determined as described before.83 Animal husbandry and experimentation was authorized by the local animal welfare committee and followed the Portuguese law for animal experimentation (Regulatory Guideline nº 1005/92, October 23rd, 1992).
Mutant tRNA plasmid construction and microinjection
Serine tRNA genes with flanking regions were amplified from zebrafish genomic DNA and digested with ApaI and SacII restriction enzymes. The fragments were cloned into the pCS2+EGFP vector, producing the plasmid pCS2+EGFPtRNASer. Mutant tRNA constructs were produced using the plasmid pCS2+EGFPtRNASer by site directed mutagenesis of nucleotide positions corresponding to the anticodon of the mature Ser-tRNA (Table S1). Mutations were selected according to similarity of amino acid properties, blossom score, codon usage and total codon counts (Table S2).24,25,28,29,84 65 ng/μL of plasmid in phenol red/KCl were injected in one-cell stage embryos and embryos were observed under a stereo-microscope equipped for fluorescence measurements (Nikon).
Northern blot analysis
Total RNA of 24hpf embryos were resolved on 15% polyacrylamide gels and transferred to a nitrocellulose membrane (Hybond N, Amershan) that was cross linked using a UV Stratalinker-1800 from Stratagene. Probes for each mutant tRNA and lysine tRNA (loading control) were prepared by 5′ phosphorylation of 10pmol of short oligonucleotides with ɣ-32P-ATP. Membranes were hybridized overnight at the oligos Tm minus 5°C with rotation, then exposed to a K-screen and scanned using Molecular Imager FX (Bio-Rad).
GFP fluorescence reporter
A green fluorescent protein (GFP) reporter was used to check the decoding activity of the mutant tRNAs. The GFP gene contains a Ser codon at position 65 which is essential for chromophore formation and mutations at this position result in significant alteration or even loss of GFP fluorescence. Various reporter constructs were built by site directed mutagenesis of codon 65 of the GFP gene cloned into plasmid pCS2+EGFP. The construct with a Ser codon at position 65 (S65) was used as the positive control, while the constructs bearing codon (A65 – GCG, G65 – GGA, L65 – CTG, V65 – GTG) were used as gain of function reporters. In these cases wild type levels of GFP fluorescence were dependent on the misincorporation of Ser by the mutant tRNAs at position 65. These reporters were cloned into the pCS2+EGFP plasmid yielding the gain/loss of function controls pCS2+EGFP(Ser65), pCS2+EGFP(Ala65), pCS2+EGFP(Gly65), pCS2+EGFP(Leu65), pCS2+EGFP(Val65) and the gain/loss of function test plasmids pCS2+EGFP(Ala65)Ser-tRNACGCAla, pCS2+EGFP(Gly65)Ser-tRNAUCCGly, pCS2+EGFP(Leu65)Ser-tRNACAGLeu and pCS2+EGFP(Val65)Ser-tRNACACVal (Table S3). Plasmids were prepared and microinjected as described previously. Embryos were observed under an epifluorescence microscope Imager.Z1 (Zeiss), a GFP filter, AxioCam HRm camera (Zeiss) and AxioVision software (Zeiss). The fluorescence levels were determined using ImageJ software; a minimum of 20 embryos were analyzed per assay. 24hpf control and test embryos were collected for western blot analysis. GFP protein expression determined by western blot was used to normalize fluorescence levels.
Western immunoblotting
For western blot analysis 24hpf embryos were dechorionated and deyolked. The whole embryo lysate was extracted using 1x SDS sample buffer at 95°C for 5 min. Approximately 50 μg/lane of protein were loaded on 12% or 15% SDS–PAGE minigels. After electrophoresis proteins were wet transferred to a nitrocellulose membrane. After blocking, membranes were incubated with primary antibodies: anti-GFP (Clontech), anti-β-tubulin (Invitrogen), anti-poliubiquitin (Covance), anti-HSP90 (StressMarq), anti-HSP70 (StressMarq), anti-HSP27 (Abcam), anti-eIF2α (Abcam) and eIF2α-P (Abcam) diluted in TBS-T with 5% LFM. Detection was performed using a secondary antibody labeled with an infrared dye (IRDye 800CW, Li-COR) at room temperature and in the dark. Membranes were scanned and analyzed using the Odyssey® IR scanner.
Mitochondrial and nuclear DNA damage
Quantitative PCR was performed according to.58 Briefly, 10 ng DNA (quantified with PicoGreen® dye, Invitrogen) was amplified with Gene Amp® XL PCR kit (Applied Biosystems) using the primers and conditions described for zebrafish. Small targets were amplified for normalization/verification of DNA concentration. PCR products were quantified using PicoGreen® dye, and DNA concentrations were converted to lesion frequencies per 10kB DNA.
Mitochondrial membrane potential
Mitochondria of 24hpf zebrafish embryos were isolated following the MITOISO1 isolation kit protocol (MITOISO1; Sigma). All steps were performed on ice or with icecold buffers. Mitochondrial membrane potential was determined with the JC-1 dye according to the manufacturer's instructions (MITOISO1, Sigma). Batches of 60 embryos were dechorionated and homogenized in 1X extraction buffer A, using a Teflon pestle. Embryo lysates were centrifuged at 600xg 5 min at 4°C; supernatants were centrifuged again at 11,000 × g 10 min at 4°C. The final pellets were resuspended in extraction buffer centrifuged again as described above. Pellets containing the mitochondrial fraction were resuspended in the supplied 1X storage buffer. For the detection of mitochondrial membrane potential, fractions were incubated with JC-1 dye for 7 min at room temperature in the dark. Finally, fluorescence was measured at490/590nm with a Perkin Elmer Fluorescence Spectrometer (LS 50B).
Mitochondria staining
Live imaging of mitochondria in zebrafish was performed using MitoTracker® Red CM-H2XRos (Molecular Probes®). 24hpf embryos were dechorionated and incubated with MitoTracker 500nM for 30 min in the dark. For imaging, anesthetized zebrafish embryos were embedded in methyl cellulose and overlayed with vectashield. Mito Tracker fluorescence was documented by epifluorescence microscopy. Fluorescence images were obtained using an Imager.Z1 (Zeiss), a Rhod filter, AxioCam HRm camera (Zeiss) and AxioVision software (Zeiss).
Statistical analysis
Data was analyzed using GraphPad Prism. The differences between control and conditioned embryos were assessed using a Student's t test or ANOVA with post test Dunnett's Multiple Comparison Test. In all cases, p values < 0.05 were considered statistically significant.
Figure 8.
General overview of biological deregulation in the mistranslating metazoan cell. Misincorporation of amino acids during translation leads to the production of mutant proteins which aggregate with subsequent chaperone and accessory factors sequestration, disrupting the proteostais network which is crucial for cellular homeostasis. Aberrant proteins also activate ER stress and UPR response. ER stress and subsequent mitochondrial dysfunction are believed to produce increased levels of ROS that has the potential to damage nucleic acids, proteins and lipids amplifying the consequences of mistranslation. Deregulation of these organelles can influence calcium homeostasis and ATP production. However, several questions remain elusive, in particular how the apoptotic pathways are activated, what is the role of mistranslation in miRNA expression deregulation, and how is the native (WT) proteome affected and maintained in such stress conditions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
The authors are most grateful to the Portuguese Foundation for Science and Technology (FCT) for funding our work through project N° FCT-ANR/IMI-MIC/0041/2012, SFRH/BD/47868/2008 to M.R. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/ 10.1101/gad.1956510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell 2010; 37:668-78; PMID:20227371; http://dx.doi.org/ 10.1016/j.molcel.2010.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009; 185:35-42; PMID:19332886; http://dx.doi.org/ 10.1083/jcb.200811106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thompson DM, Parker R. Stressing out over tRNA cleavage. Cell 2009; 138:215-9; PMID:19632169; http://dx.doi.org/ 10.1016/j.cell.2009.07.001 [DOI] [PubMed] [Google Scholar]
- 5. Francklyn CS, Minajigi A. tRNA as an active chemical scaffold for diverse chemical transformations. FEBS Lett 2010; 584:366-75; PMID:19925795; http://dx.doi.org/ 10.1016/j.febslet.2009.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shah P, Gilchrist MA. Effect of correlated tRNA abundances on translation errors and evolution of codon usage bias. PLoS Genet 2010; 6:e1001128; PMID:20862306; http://dx.doi.org/ 10.1371/journal.pgen.1001128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yasukawa T, Suzuki T, Ueda T, Ohta S, Watanabe K. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAs(Leu)(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J Biol Chem 2000; 275:4251-7; PMID:10660592; http://dx.doi.org/ 10.1074/jbc.275.6.4251 [DOI] [PubMed] [Google Scholar]
- 8. Bykhovskaya Y, Casas K, Mengesha E, Inbal A, Fischel-Ghodsian N. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet 2004; 74:1303-8; PMID:15108122; http://dx.doi.org/ 10.1086/421530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zheng J, Ji Y, Guan MX. Mitochondrial tRNA mutations associated with deafness. Mitochondrion 2012; 12:406-13; PMID:22538251; http://dx.doi.org/ 10.1016/j.mito.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 10. Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J 2001; 20:4794-802; PMID:11532943; http://dx.doi.org/ 10.1093/emboj/20.17.4794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li X, Fischel-Ghodsian N, Schwartz F, Yan Q, Friedman RA, Guan MX. Biochemical characterization of the mitochondrial tRNASer(UCN) T7511C mutation associated with nonsyndromic deafness. Nucleic Acids Res 2004; 32:867-77; PMID:14960712; http://dx.doi.org/ 10.1093/nar/gkh226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci U S A 2008; 105:11043-9; PMID:18682559; http://dx.doi.org/ 10.1073/pnas.0802862105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Latour P, Thauvin-Robinet C, Baudelet-Méry C, Soichot P, Cusin V, Faivre L, Locatelli MC, Mayençon M, Sarcey A, Broussolle E, et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet 2010; 86:77-82; PMID:20045102; http://dx.doi.org/ 10.1016/j.ajhg.2009.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stum M, McLaughlin HM, Kleinbrink EL, Miers KE, Ackerman SL, Seburn KL, Antonellis A, Burgess RW. An assessment of mechanisms underlying peripheral axonal degeneration caused by aminoacyl-tRNA synthetase mutations. Mol Cell Neurosci 2011; 46:432-43; PMID:21115117; http://dx.doi.org/ 10.1016/j.mcn.2010.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, Davisson MT, Sundberg JP, Schimmel P, Ackerman SL. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 2006; 443:50-5; PMID:16906134; http://dx.doi.org/ 10.1038/nature05096 [DOI] [PubMed] [Google Scholar]
- 16. Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med 2014; 20:306-14; PMID:24581449; http://dx.doi.org/ 10.1016/j.molmed.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez V, Chen Y, Elkahloun A, Dutra A, Pak E, Chandrasekharappa S. Chromosome 8 BAC array comparative genomic hybridization and expression analysis identify amplification and overexpression of TRMT12 in breast cancer. Genes Chromosomes Cancer 2007; 46:694-707; PMID:17440925; http://dx.doi.org/ 10.1002/gcc.20454 [DOI] [PubMed] [Google Scholar]
- 18. Freed EF, Bleichert F, Dutca LM, Baserga SJ. When ribosomes go bad: diseases of ribosome biogenesis. Mol Biosyst 2010; 6:481-93; PMID:20174677; http://dx.doi.org/ 10.1039/b919670f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koga H, Kaushik S, Cuervo AM. Protein homeostasis and aging: The importance of exquisite quality control. Ageing Res Rev 2011; 10:205-15; PMID:20152936; http://dx.doi.org/ 10.1016/j.arr.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol 2007; 18:716-31; PMID:18023214; http://dx.doi.org/ 10.1016/j.semcdb.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen B, Retzlaff M, Roos T, Frydman J. Cellular strategies of protein quality control. Cold Spring Harb Perspect Biol 2011; 3:a004374; PMID:21746797; http://dx.doi.org/ 10.1101/cshperspect.a004374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lenhard B, Orellana O, Ibba M, Weygand-Durasević I. tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res 1999; 27:721-9; PMID:9889265; http://dx.doi.org/ 10.1093/nar/27.3.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cusack S, Yaremchuk A, Tukalo M. The crystal structure of the ternary complex of T.thermophilus seryl-tRNA synthetase with tRNA(Ser) and a seryl-adenylate analogue reveals a conformational switch in the active site. EMBO J 1996; 15:2834-42; PMID:8654381 [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor WR. The classification of amino acid conservation. J Theor Biol 1986; 119:205-18; PMID:3461222; http://dx.doi.org/ 10.1016/S0022-5193(86)80075-3 [DOI] [PubMed] [Google Scholar]
- 25. Betts MJ, Russell RB. Amino acid properties and consequences of subsitutions. John Wiley & Sons, Ltd, 2003. [Google Scholar]
- 26. Haig D, Hurst LD. A quantitative measure of error minimization in the genetic code. J Mol Evol 1991; 33:412-7; PMID:1960738; http://dx.doi.org/ 10.1007/BF02103132 [DOI] [PubMed] [Google Scholar]
- 27. White SH. Amino acid preferences of small proteins. Implications for protein stability and evolution. J Mol Biol 1992; 227:991-5; PMID:1433304; http://dx.doi.org/ 10.1016/0022-2836(92)90515-L [DOI] [PubMed] [Google Scholar]
- 28. Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A 1992; 89:10915-9; PMID:1438297; http://dx.doi.org/ 10.1073/pnas.89.22.10915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Berleant D, White M, Pierce E, Tudoreanu E, Boeszoermenyi A, Shtridelman Y, Macosko JC. The genetic code–more than just a table. Cell Biochem Biophys 2009; 55:107-16; PMID:19639425; http://dx.doi.org/ 10.1007/s12013-009-9060-9 [DOI] [PubMed] [Google Scholar]
- 30. Reid BG, Flynn GC. Chromophore formation in green fluorescent protein. Biochemistry 1997; 36:6786-91; PMID:9184161; http://dx.doi.org/ 10.1021/bi970281w [DOI] [PubMed] [Google Scholar]
- 31. Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry 1993; 32:1212-8; PMID:8448132; http://dx.doi.org/ 10.1021/bi00056a003 [DOI] [PubMed] [Google Scholar]
- 32. Geslain R, Cubells L, Bori-Sanz T, Alvarez-Medina R, Rossell D, Martí E, Ribas de Pouplana L. Chimeric tRNAs as tools to induce proteome damage and identify components of stress responses. Nucleic Acids Res 2010; 38:e30; PMID:20007146; http://dx.doi.org/ 10.1093/nar/gkp1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ojha J, Masilamoni G, Dunlap D, Udoff RA, Cashikar AG. Sequestration of toxic oligomers by HspB1 as a cytoprotective mechanism. Mol Cell Biol 2011; 31:3146-57; PMID:21670152; http://dx.doi.org/ 10.1128/MCB.01187-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bryantsev AL, Kurchashova SY, Golyshev SA, Polyakov VY, Wunderink HF, Kanon B, Budagova KR, Kabakov AE, Kampinga HH. Regulation of stress-induced intracellular sorting and chaperone function of Hsp27 (HspB1) in mammalian cells. Biochem J 2007; 407:407-17; PMID:17650072; http://dx.doi.org/ 10.1042/BJ20070195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang Y, Frejtag W, Dai R, Mivechi NF. Heat shock factor-4 (HSF-4a) is a repressor of HSF-1 mediated transcription. J Cell Biochem 2001; 82:692-703; PMID:11500947; http://dx.doi.org/ 10.1002/jcb.1191 [DOI] [PubMed] [Google Scholar]
- 36. Marzec M, Eletto D, Argon Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim Biophys Acta 2012; 1823:774-87; PMID:22079671; http://dx.doi.org/ 10.1016/j.bbamcr.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gething MJ. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol 1999; 10:465-72; PMID:10597629; http://dx.doi.org/ 10.1006/scdb.1999.0318 [DOI] [PubMed] [Google Scholar]
- 38. Hu MC, Gong HY, Lin GH, Hu SY, Chen MH, Huang SJ, Liao CF, Wu JL. XBP-1, a key regulator of unfolded protein response, activates transcription of IGF1 and Akt phosphorylation in zebrafish embryonic cell line. Biochem Biophys Res Commun 2007; 359:778-83; PMID:17560942; http://dx.doi.org/ 10.1016/j.bbrc.2007.05.183 [DOI] [PubMed] [Google Scholar]
- 39. Schäfer A, Wolf DH. Sec61p is part of the endoplasmic reticulum-associated degradation machinery. EMBO J 2009; 28:2874-84; PMID:19696741; http://dx.doi.org/ 10.1038/emboj.2009.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lang S, Erdmann F, Jung M, Wagner R, Cavalie A, Zimmermann R. Sec61 complexes form ubiquitous ER Ca2+ leak channels. Channels (Austin) 2011; 5:228-35; PMID:21406962; http://dx.doi.org/ 10.4161/chan.5.3.15314 [DOI] [PubMed] [Google Scholar]
- 41. Oda Y, Okada T, Yoshida H, Kaufman RJ, Nagata K, Mori K. Derlin-2 and Derlin-3 are regulated by the mammalian unfolded protein response and are required for ER-associated degradation. J Cell Biol 2006; 172:383-93; PMID:16449189; http://dx.doi.org/ 10.1083/jcb.200507057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim TY, Kim E, Yoon SK, Yoon JB. Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem Biophys Res Commun 2008; 369:741-6; PMID:18307982; http://dx.doi.org/ 10.1016/j.bbrc.2008.02.086 [DOI] [PubMed] [Google Scholar]
- 43. Wakana Y, Takai S, Nakajima K, Tani K, Yamamoto A, Watson P, Stephens DJ, Hauri HP, Tagaya M. Bap31 is an itinerant protein that moves between the peripheral endoplasmic reticulum (ER) and a juxtanuclear compartment related to ER-associated Degradation. Mol Biol Cell 2008; 19:1825-36; PMID:18287538; http://dx.doi.org/ 10.1091/mbc.E07-08-0781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Rutkowski DT, Kang SW, Goodman AG, Garrison JL, Taunton J, Katze MG, Kaufman RJ, Hegde RS. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol Biol Cell 2007; 18:3681-91; PMID:17567950; http://dx.doi.org/ 10.1091/mbc.E07-03-0272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chuang JZ, Zhou H, Zhu M, Li SH, Li XJ, Sung CH. Characterization of a brain-enriched chaperone, MRJ, that inhibits Huntingtin aggregation and toxicity independently. J Biol Chem 2002; 277:19831-8; PMID:11896048; http://dx.doi.org/ 10.1074/jbc.M109613200 [DOI] [PubMed] [Google Scholar]
- 46. Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci 2006; 63:2560-70; PMID:16952052; http://dx.doi.org/ 10.1007/s00018-006-6192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Braun JE, Wilbanks SM, Scheller RH. The cysteine string secretory vesicle protein activates Hsc70 ATPase. J Biol Chem 1996; 271:25989-93; PMID:8824236; http://dx.doi.org/ 10.1074/jbc.271.42.25989 [DOI] [PubMed] [Google Scholar]
- 48. Fortier S, Yang X, Wang Y, Bennett RA, Strauss PR. Base excision repair in early zebrafish development: evidence for DNA polymerase switching and standby AP endonuclease activity. Biochemistry 2009; 48:5396-404; PMID:19374445; http://dx.doi.org/ 10.1021/bi900253d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bladen CL, Lam WK, Dynan WS, Kozlowski DJ. DNA damage response and Ku80 function in the vertebrate embryo. Nucleic Acids Res 2005; 33:3002-10; PMID:15914672; http://dx.doi.org/ 10.1093/nar/gki613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ringvoll J, Moen MN, Nordstrand LM, Meira LB, Pang B, Bekkelund A, Dedon PC, Bjelland S, Samson LD, Falnes PO, et al. AlkB homologue 2-mediated repair of ethenoadenine lesions in mammalian DNA. Cancer Res 2008; 68:4142-9; PMID:18519673; http://dx.doi.org/ 10.1158/0008-5472.CAN-08-0796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kawahara A, Che YS, Hanaoka R, Takeda H, Dawid IB. Zebrafish GADD45beta genes are involved in somite segmentation. Proc Natl Acad Sci U S A 2005; 102:361-6; PMID:15623554; http://dx.doi.org/ 10.1073/pnas.0408726102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schmidt EK, Clavarino G, Ceppi M, Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 2009; 6:275-7; PMID:19305406; http://dx.doi.org/ 10.1038/nmeth.1314 [DOI] [PubMed] [Google Scholar]
- 53. Malhotra JD, Miao H, Zhang K, Wolfson A, Pennathur S, Pipe SW, Kaufman RJ. Antioxidants reduce endoplasmic reticulum stress and improve protein secretion. Proc Natl Acad Sci U S A 2008; 105:18525-30; PMID:19011102; http://dx.doi.org/ 10.1073/pnas.0809677105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, Paules R, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol Cell 2003; 11:619-33; PMID:12667446; http://dx.doi.org/ 10.1016/S1097-2765(03)00105-9 [DOI] [PubMed] [Google Scholar]
- 55. Haynes CM, Titus EA, Cooper AA. Degradation of misfolded proteins prevents ER-derived oxidative stress and cell death. Mol Cell 2004; 15:767-76; PMID:15350220; http://dx.doi.org/ 10.1016/j.molcel.2004.08.025 [DOI] [PubMed] [Google Scholar]
- 56. Chance B, Sies H, Boveris A. Hydroperoxide metabolism in mammalian organs. Physiol Rev 1979; 59:527-605; PMID:37532 [DOI] [PubMed] [Google Scholar]
- 57. Cui H, Kong Y, Zhang H. Oxidative stress, mitochondrial dysfunction, and aging. J Signal Transduct 2012; 2012:646354; PMID:21977319; http://dx.doi.org/ 10.1155/2012/646354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Hunter SE, Jung D, Di Giulio RT, Meyer JN. The QPCR assay for analysis of mitochondrial DNA damage, repair, and relative copy number. Methods 2010; 51:444-51; PMID:20123023; http://dx.doi.org/ 10.1016/j.ymeth.2010.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Santos JH, Mandavilli BS, Van Houten B. Measuring oxidative mtDNA damage and repair using quantitative PCR. Methods Mol Biol 2002; 197:159-76; PMID:12013794 [DOI] [PubMed] [Google Scholar]
- 60. Van Houten B, Woshner V, Santos JH. Role of mitochondrial DNA in toxic responses to oxidative stress. DNA Repair (Amst) 2006; 5:145-52; PMID:15878696; http://dx.doi.org/ 10.1016/j.dnarep.2005.03.002 [DOI] [PubMed] [Google Scholar]
- 61. Cline SD. Mitochondrial DNA damage and its consequences for mitochondrial gene expression. Biochim Biophys Acta 2012; 1819:979-91; PMID:22728831; http://dx.doi.org/ 10.1016/j.bbagrm.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Springer W, Kahle PJ. Regulation of PINK1-Parkin-mediated mitophagy. Autophagy 2011; 7:266-78; PMID:21187721; http://dx.doi.org/ 10.4161/auto.7.3.14348 [DOI] [PubMed] [Google Scholar]
- 63. Ved R, Saha S, Westlund B, Perier C, Burnam L, Sluder A, Hoener M, Rodrigues CM, Alfonso A, Steer C, et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J Biol Chem 2005; 280:42655-68; PMID:16239214; http://dx.doi.org/ 10.1074/jbc.M505910200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Giaime E, Yamaguchi H, Gautier CA, Kitada T, Shen J. Loss of DJ-1 does not affect mitochondrial respiration but increases ROS production and mitochondrial permeability transition pore opening. PLoS One 2012; 7:e40501; PMID:22792356; http://dx.doi.org/ 10.1371/journal.pone.0040501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Saotome M, Safiulina D, Szabadkai G, Das S, Fransson A, Aspenstrom P, Rizzuto R, Hajnóczky G. Bidirectional Ca2+-dependent control of mitochondrial dynamics by the Miro GTPase. Proc Natl Acad Sci U S A 2008; 105:20728-33; PMID:19098100; http://dx.doi.org/ 10.1073/pnas.0808953105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tondera D, Czauderna F, Paulick K, Schwarzer R, Kaufmann J, Santel A. The mitochondrial protein MTP18 contributes to mitochondrial fission in mammalian cells. J Cell Sci 2005; 118:3049-59; PMID:15985469; http://dx.doi.org/ 10.1242/jcs.02415 [DOI] [PubMed] [Google Scholar]
- 67. Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 2008; 19:2402-12; PMID:18353969; http://dx.doi.org/ 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Inohara N, Nuñez G. Genes with homology to mammalian apoptosis regulators identified in zebrafish. Cell Death Differ 2000; 7:509-10; PMID:10917738; http://dx.doi.org/ 10.1038/sj.cdd.4400679 [DOI] [PubMed] [Google Scholar]
- 69. Chinnadurai G, Vijayalingam S, Gibson SB. BNIP3 subfamily BH3-only proteins: mitochondrial stress sensors in normal and pathological functions. Oncogene 2008; 27(Suppl 1):S114-27; PMID:19641497; http://dx.doi.org/ 10.1038/onc.2009.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. He C, Bartholomew CR, Zhou W, Klionsky DJ. Assaying autophagic activity in transgenic GFP-Lc3 and GFP-Gabarap zebrafish embryos. Autophagy 2009; 5:520-6; PMID:19221467; http://dx.doi.org/ 10.4161/auto.5.4.7768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet 2009; 10:715-24; PMID:19763154; http://dx.doi.org/ 10.1038/nrg2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature 2008; 454:1088-95; PMID:18756251; http://dx.doi.org/ 10.1038/nature07195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang S, Kaufman RJ. The impact of the unfolded protein response on human disease. J Cell Biol 2012; 197:857-67; PMID:22733998; http://dx.doi.org/ 10.1083/jcb.201110131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cinaroglu A, Gao C, Imrie D, Sadler KC. Activating transcription factor 6 plays protective and pathological roles in steatosis due to endoplasmic reticulum stress in zebrafish. Hepatology 2011; 54:495-508; PMID:21538441; http://dx.doi.org/ 10.1002/hep.24396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Proud CG. eIF2 and the control of cell physiology. Semin Cell Dev Biol 2005; 16:3-12; PMID:15659334; http://dx.doi.org/ 10.1016/j.semcdb.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 76. Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, et al. Sustained translational repression by eIF2α-P mediates prion neurodegeneration. Nature 2012; 485:507-11; PMID:22622579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem 2005; 280:26185-92; PMID:15899901 [DOI] [PubMed] [Google Scholar]
- 78. Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol 2003; 160:189-200; PMID:12527753; http://dx.doi.org/ 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Suen DF, Norris KL, Youle RJ. Mitochondrial dynamics and apoptosis. Genes Dev 2008; 22:1577-90; PMID:18559474; http://dx.doi.org/ 10.1101/gad.1658508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Detmer SA, Chan DC. Functions and dysfunctions of mitochondrial dynamics. Nat Rev Mol Cell Biol 2007; 8:870-9; PMID:17928812; http://dx.doi.org/ 10.1038/nrm2275 [DOI] [PubMed] [Google Scholar]
- 81. Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: The mitochondria-associated membrane (MAM). Biochim Biophys Acta 2013; 1833:213-24; PMID:22575682 [DOI] [PubMed] [Google Scholar]
- 82. Csordás G, Hajnóczky G. SR/ER-mitochondrial local communication: calcium and ROS. Biochim Biophys Acta 2009; 1787:1352-62; PMID:19527680; http://dx.doi.org/ 10.1016/j.bbabio.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn 1995; 203:253-310; PMID:8589427; http://dx.doi.org/ 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- 84. Moura G, Pinheiro M, Silva R, Miranda I, Afreixo V, Dias G, Freitas A, Oliveira JL, Santos MA. Comparative context analysis of codon pairs on an ORFeome scale. Genome Biol 2005; 6:R28; PMID:15774029; http://dx.doi.org/ 10.1186/gb-2005-6-3-r28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, Li J, Thiagarajan M, White JA, Quackenbush J. TM4 microarray software suite. Methods Enzymol 2006; 411:134-93; PMID:16939790; http://dx.doi.org/ 10.1016/S0076-6879(06)11009-5 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.