Abstract
mRNA localization ensures correct spatial and temporal control of protein synthesis in the cell. We show that an in vitro single molecule approach, using purified recombinant full-length proteins and synthesized mRNA, provides insight into the mechanism by which localizing mRNAs are carried to their destination. A messenger ribonucleoprotein (mRNP) complex was reconstituted from a budding yeast class V myosin motor complex (Myo4p–She3p), an mRNA-binding adaptor protein (She2p), and a localizing mRNA (ASH1). The motion of the mRNP was tracked with high spatial (∼10 nm) and temporal (70 ms) resolution. Using this “bottom-up” methodology, we show that mRNA triggers the assembly of a high affinity double-headed motor-mRNA complex that moves continuously for long distances on actin filaments at physiologic ionic strength. Without mRNA, the myosin is monomeric and unable to move continuously on actin. This finding reveals an elegant strategy to ensure that only cargo-bound motors are activated for transport. Increasing the number of localization elements (“zip codes”) in the mRNA enhanced both the frequency of motile events and their run length, features which likely enhance cellular localization. Future in vitro reconstitution of mRNPs with kinesin and dynein motors should similarly yield mechanistic insight into mRNA transport by microtubule-based motors.
Keywords: Myo4p, She3p, She2p, ASH1 mRNA, mRNA transport, mRNA localization, messenger ribonucleoprotein
Abbreviations
- mRNP
messenger ribonucleoprotein
Introduction
Localizing mRNAs often rely on molecular motors to reach their destination in the cell. Budding yeast provides an attractive model organism to study the spatial and temporal control of mRNA localization, because yeast exclusively use one class V myosin motor for this process. The mRNA binding protein She2p links the myosin motor to its mRNA cargo. The simplicity of this system led to a large number of cell biological experiments in budding yeast, primarily aimed at understanding the features required for correct localization of mRNA transcripts to the bud tip (Fig. 1A).1-7 One of the most widely-studied localizing mRNAs is ASH1, which codes for a cell fate determinant. ASH1 mRNA contains four cis-acting sequences called “zip codes” that bind She2p, which in turn recruit the myosin motors that move the mRNA. Many of the earlier cell biological studies focused on features of the mRNA that were required for correct localization,8 and less on the properties of the motor itself. The single molecule in vitro reconstitution approach described here investigates mRNA transport primarily from the motor's point of view.
Figure 1.
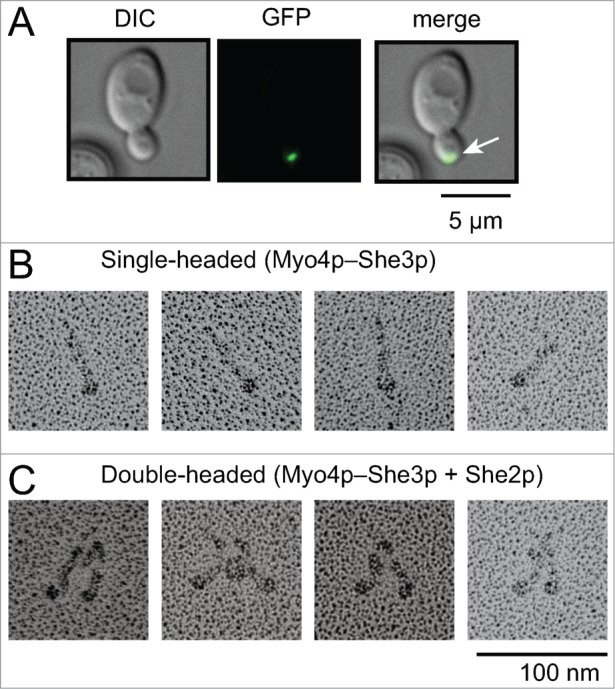
Myo4p–She3p bound to She2p forms a double-headed complex at low ionic strength. (A) Image of ASH1 mRNA localization to the bud tip (green dot, white arrow) in budding yeast. (B, C) Electron microscopy of metal-shadowed images showing (B) single-headed Myo4p–She3p complexes, and (C) double-headed Myo4p–She3p–She2p motor complexes at low ionic strength.
Budding yeast has only two class V myosins. One of them, Myo4p, is specialized to transport cortical ER, and over 20 different localizing mRNAs from the mother to the bud tip on actin cables.2-4,9-11 Imaging of tagged mRNA in living cells showed its continuous transport over several microns at a rate of ∼0.3 μm/sec,12 as expected for a motor-driven process. Surprisingly, when Myo4p was purified from yeast cells13 and characterized by ensemble in vitro motility assays, it appeared to have a low duty cycle, meaning that that the myosin head stays attached to actin for only a small fraction of its ATPase cycle. A cargo transporter needs to have a high duty cycle, (i.e., the myosin head needs to stay attached to actin for a large fraction of its ATPase cycle), which enables the motor to move micron-long distances along actin tracks as a single molecule, a feature known as processivity.
The discrepancy between the in vivo and in vitro observations needed to be reconciled. One possibility was that mRNA transport is accomplished by teams of non-processive motors that act together to move cargo continuously. Alternatively, other factors such as the adaptor protein She2p or the mRNA cargo itself, which were not present in the simplified in vitro assays, could affect the properties of Myo4p. If true, cargo transport could be regulated in the cell by accessory factors that turn the motor “on” or “off” for processive transport. To address these possibilities we used a single molecule approach to study the motor in vitro, but under conditions that more closely mimicked the cellular context. Full-length Myo4p and its tightly bound adaptor She3p were expressed and purified from Sf9 cells using the baculovirus/Sf9 cell expression system, while the mRNA binding protein She2p was expressed in bacteria. The purified proteins were reconstituted in vitro to form a double-headed motor/adaptor complex which was visualized moving processively on yeast actin–tropomyosin tracks using total internal reflection fluorescence (TIRF) microscopy.14 To address how mRNA cargo activates the complex for transport, we further increased the complexity of the single molecule assay by adding a full-length ASH1 mRNA transcript to form a fully reconstituted mRNP in vitro that moves processively on actin even at physiologic ionic strength.15 Depending on the goal of the experiment, labeling strategies to visualize the complex included binding a Quantum dot to the Myo4p motor, expressing She2p as a YFP fusion protein, or labeling the ASH1 transcript with an Alexa Fluor-UTP.
Two Motor Heads are Needed for Processivity
Myo4p has many of the attributes of a processive motor. Expressed chimeric constructs showed that its motor domain was capable of supporting processive motion as a single molecule when fused to the dimeric mammalian myosin Va lever arm and coiled-coil rod.16 Like myosin Va, Myo4p has a long extended lever arm that allows a head to take ∼72 nm steps on actin, and a cargo-binding globular tail. However, the number of heptad repeats (signature of an alpha-helical coiled coil) in the rod region of Myo4p is small (∼5) compared with the number found in vertebrate myosin Va (>20). The coiled coil is responsible for myosin being a double-headed motor. The most unusual feature of Myo4p is that its strong interaction with the adaptor protein She3p prevents dimerization with another Myo4p molecule.17 Thus, the simple reason why the Myo4p–She3p motor complex cannot “walk” processively on actin filaments like other class V myosins is because it is a single-headed motor.17,18
The stoichiometry of the Myo4p–She3p complex is 1:2.19,20 Structural insights from X-ray crystallography showed that an interaction between Myo4p and a truncated dimeric fragment of She3p is mediated through a hydrophobic patch in the globular tail domain of Myo4p.19 But we also know that a truncated version of Myo4p lacking the globular tail still binds She3p tightly,15,17 implying that there are likely to be additional interactions between the Myo4p rod and She3p that were not observed in the crystal structure. Future structural studies on additional domains of Myo4p–She3p will be needed to fully understand how the motor complex is stabilized.
The Myo4p–She3p complex is recruited to localizing transcripts through the mRNA-binding protein She2p.14,21 She2p is a tetramer,14,21,22 and the four monomers provide a potential mechanism to recruit multiple Myo4p–She3p motor complexes in close proximity. Electron microscopy of metal-shadowed images showed that in the absence of She2p, the Myo4p–She3p complex is single-headed (Fig. 1B). Addition of She2p to this complex revealed V-shapes structures that closely resemble double-headed vertebrate myosin Va (Fig. 1C). She2p is thus capable of recruiting two single-headed Myo4p–She3p motors, resulting in a stoichiometry of two Myo4p heavy chains: two She3p dimers: one She2p tetramer.19,20
Importantly, this two-headed motor complex, recruited via She2p, steps processively on actin filaments with a 72 nm step-size and hand-over-hand stepping pattern identical to that observed with myosin Va (Fig. 2A).23 The coordinated stepping of the two heads bound to She2p was unexpected because it is generally assumed that processive motion requires a strain-dependent communication between the heads, mediated through the coiled-coil rod of the motor. One intriguing possibility is that once She2p recruits two motor complexes, the Myo4p heavy chains can form a short coiled coil via the ∼5 heptads of predicted coiled-coil sequence in the rod. Cargo-mediated dimerization has been previously proposed for vertebrate myosin VI.24 Based on our studies with the Myo4p motor complex and She2p at low ionic strength (50 mM KCl), we proposed the dimerization scheme depicted in Figure 2B.
Figure 2.
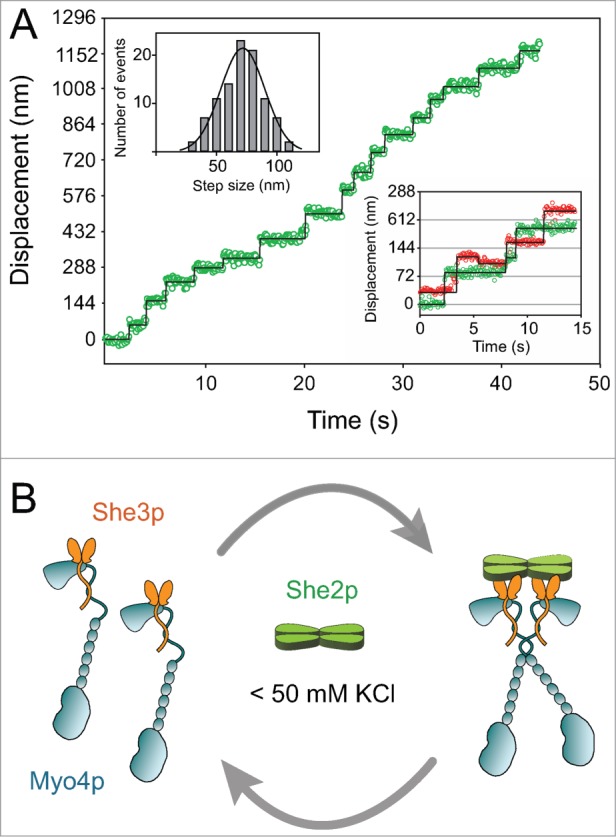
(A) Trace showing the displacement vs. time of the Myo4p–She3p–She2p motor complex on an actin filament at low ionic strength (50 mM KCl). The multiple steps without dissociation illustrate that the complex is processive. Myo4p–She3p is labeled with a Quantum dot (Qdot) on only one head of the two-headed motor complex. Using this labeling strategy, the head position of myosin can be tracked on actin with high time and spatial resolution by fitting the Qdot image of each frame to a two-dimensional Gaussian, which approximates its point spread function and allows its x and y positions to be determined with ∼6 nm resolution. The center of a distribution can be determined much more accurately than the width, which is limited by the ∼200 nm resolution of a light microscope. The top left inset shows the histogram of step sizes, that average 72 ± 18 nm. The bottom right inset shows a stepping trace of a two-headed complex in which each head is labeled with a different colored Quantum dot. These data illustrate the hand-over-hand stepping pattern of the two heads, which implies communication between the heads. (B) Diagram showing that She2p recruits two single-headed Myo4p–She3p complexes to form a processive double-headed motor complex at low ionic strength (< 50 mM KCl). She3p is shown as a dimer interacting with the Myo4p globular tail and rod.
mRNA is Necessary to Stabilize a Double-Headed Processive Motor Complex at Physiologic Ionic Strength
When reconstituting multi-component systems in vitro, it is necessary to consider cellular factors that may be important for understanding how the complex is regulated. Track modifications, protein concentration, molecular crowding, ionic strength and cargo are all important factors that may influence the assembly and biophysical properties of motor/adaptor complexes. In the case of Myo4p, it became apparent that ionic strength was an important factor to consider in the in vitro reconstitution.
Myo4p–She3p assembles into a processive dimer when bound to She2p at low ionic strength (50 mM KCl), but this complex dissociates when the ionic strength is raised to near physiological (140 mM KCl). Salt concentrations ≤50 mM KCl are typically used in in vitro motility assays to enhance interactions, but these conditions may lead to conclusions that are not relevant in the cell. In this case, our observation that the complex dissociates at 140 mM KCl implies that to function as a cargo transporter in the cell, other cellular factors are required to stabilize the two-headed motor complex. An obvious candidate was the mRNA cargo itself.
We therefore reconstituted a messenger ribonucleoprotein (mRNP) by adding the budding yeast localizing mRNA transcript ASH1, to the Myo4p–She3p–She2p complex. The mRNA was synthesized with a fluorescently labeled UTP to visualize mRNPs moving on actin filaments with high spatial (∼10 nm) and temporal (70 ms) resolution. At ionic strength approximating physiologic (140 mM KCl), ASH1 mRNA was required to stabilize the double-headed motor complex, which moved for long distances on actin (Fig. 3A). In the presence of sufficient motor, the mRNP with a full-length ASH1 transcript, bound as many as eight motor heads (Fig. 3B). The spacing between the motor heads varied during a run, highlighting the dynamic nature of the transport complex.15 Cargo is usually thought to be a passive player during transport, but in this case, mRNA participates in the assembly of the mRNP by providing additional interactions that stabilize the complex.
Figure 3.
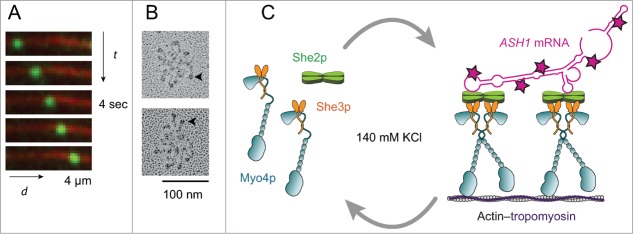
Single molecule reconstitution of a motile ASH1 mRNP. (A) mRNPs (green) are visualized moving on actin filaments (red) at near physiological ionic strength (140 mM KCl) using total internal reflection fluorescence microscopy. mRNPs are assembled with the native full-length ASH1 mRNA, which was synthesized with an Alexa 488–labeled UTP for visualization. Images were acquired at 67 ms/frame at 1 mM MgATP. (B) Electron microscopy of metal-shadowed images showing fully reconstituted mRNPs. The mRNP, assembled with the native four zip code ASH1 mRNA transcript, binds up to eight motor heads (arrowheads). (C) Diagram showing that budding yeast ASH1 mRNA and She2p synergize the assembly of motile mRNPs near physiological ionic strength. The ASH1 transcript contains multiple zip codes, each capable of recruiting a motor complex.
The More Zip Codes the Better
mRNA is an ideal cargo for studying the single molecule properties of motor/cargo complexes because it is easy to synthesize and modify. The ASH1 mRNA zip code sequences are modular, making it easy to alter motor recruitment to the transcript. Like many other localizing mRNA transcripts, the ASH1 transcript contains multiple zip code elements, each capable of recruiting a motor complex for transport. By altering the zip code number of the ASH1 transcript, we find that transcripts containing more zip codes are transported for longer distances. This is expected from a team of motors, because the probability of having at least one head bound to the track increases. This observation provides a molecular understanding of why localizing mRNAs tend to have multiple localization elements. Multiple zip codes also enhance the initial engagement of a transcript with the actin track, simply by increasing the probability that at least one motor complex is bound to the mRNA.15
In fact, any change to our in vitro assay that more closely mimicked cellular conditions enhanced the characteristic run length of the mRNP. This included adding budding yeast tropomyosin (Tpm1p) to the actin track, and assembling single actin filaments into bundles, to more closely mimic the actin cables found in vivo. The ability to move cargo for long distances most likely enhances mRNA localization in the cell.
Interestingly, one of the four zip codes in the budding yeast ASH1 mRNA has a higher affinity than the other three for the Myo4p motor complex.15 The reason for this difference is not clear, but raises the possibility that zip codes can fine tune the recruitment of motors through adaptor binding affinity. In higher eukaryotes this property may be important for co-ordinating the activities of multiple and/or different motors on mRNPs.
Conflicting Viewpoints
Subsequent to our publication,15 a single molecule study by Heym et al.20 drew several conclusions contradictory to ours. They concluded that She2p alone is necessary for Myo4p to undergo processive motion, and that neither mRNA nor zip codes are necessary. Unfortunately, these broad conclusions are based solely on data obtained at low ionic strength (50 mM KCl), which we believe gives rise to the conflicting opinions. Their low ionic strength data recapitulate our earlier findings at 50 mM KCl which showed that She2p alone can recruit a pair of single-headed Myo4p–She3p motor complexes that move processively on actin in a hand-over-hand fashion, without mRNA.14 We also showed that upon addition of mRNA at low ionic strength, processive movement is independent of both zip codes and She2p,15 implying that motors are being recruited via non-specific interactions between Myo4p–She3p and the highly charged mRNA.
Only by working near physiologic ionic strength (140 mM KCl) does mRNA transport require both zip codes and She2p for transport,15 consistent with cell biological data.25 A stable Myo4p–She3p–She2p complex does not form at 140 mM KCl in vitro in the absence of mRNA, and thus Myo4p would be non-motile in the cell without cargo. Only in the presence of mRNA does She2p form a stable complex with Myo4p–She3p under physiologic ionic strength, a regulatory checkpoint that ensures that only cargo-bound motors can move processively.
Heym et al.20 suggested several reasons other than ionic strength for these differing conclusions, but we do not believe that these other factors are responsible. They suggested that mRNP run length increases with zip code number because we used high stoichiometric ratios of myosin over mRNA. Excess motor was used in our experiments to ensure that all zip codes had the potential to bind a pair of motor heads, with excess non-processive single-headed motors remaining in solution. While this may not mimic in vivo concentrations, the cell has macromolecular crowding which facilitates biological interactions. Increasing the number of zip codes increased run length near physiologic ionic strength, as expected when a larger team of motors works together.26 In addition, metal-shadowed electron microscopic images obtained with equimolar concentrations of mRNA, Myo4p and She2p, showed that zip codes are required for recruitment of Myo4p to localizing transcripts, and that up to eight motor heads can bind to the native four zip code ASH1 mRNA.15 In agreement, Heym et al.20 also conclude that the vast majority of zip code elements in the cell are bound to motor complexes. Multiple zip codes are not essential for cellular mRNA localization, but their presence offers advantages by enhancing both the percentage of cells with correct mRNA localization, and the quality of localization at the bud tip.27 This might be due to the ability of multi-zip code transcripts to recruit multiple motors which enhance engagement with the actin track, facilitate navigation of obstacles encountered on the track, and allow movement even under load. We show that altering the track has a large impact on Myo4p run length,15 and that the closer the track resembled the actin cables found in budding yeast (i.e bundled yeast actin decorated with yeast tropomyosin Tpm1p), the better the movement. The study by Heym et al.20 drew conclusions based on movement on single skeletal actin filaments.
They also speculated that our use of She2p containing four Cys to Ser point mutations was responsible for the differing conclusions. These mutations were previously made to prevent intermolecular disulfide formation and maintain She2p in a homogeneous state necessary for crystallization,28 features that are equally important to interpret the single molecule assays. We showed that She2p(Ser to Cys) was indistinguishable from wild type She2p in an in vivo mRNA localization assay,14 which is generally considered good evidence that the mutant protein functions like wild type.
Alternative Single Molecule Approaches to Understand mRNA Transport
Advances in optical and single molecule techniques have given researchers the tools to take a number of different single molecule approaches to study the mechanisms of mRNA transport. Only through a synthesis of results obtained from multiple approaches will a thorough understanding be obtained. Here we used a “bottom-up” approach, with purified recombinant proteins and synthesized mRNA transcripts, allowing full control over the reconstitution. This approach is particularly useful when all of the components in the complex have been identified. Bullock and colleagues used a complementary “top-down” approach, isolating motor/mRNA complexes from Drosophila embryonic cell extracts.29,30 The top-down approach is less controlled than a complete in vitro reconstitution, but has the advantage of bringing along potentially important, but less well-characterized binding partners that can affect motor activity. Their work shows the importance of both dynein–dynactin copy number29 as well as regulation of dynein–dynactin activity.30 Future work identifying additional components of isolated mRNPs will help to fully understand how multiple copies of a single type of motor, as well as oppositely directed motors, coordinate their activities on localizing transcripts to determine assembly and directionality.
Direct real-time observation of single mRNPs in the cell has also been critical for our understanding of how mRNAs localize.31 One of the first pieces of evidence that mRNA transport in budding yeast was powered by a myosin was the observation that mRNA movement was directed.12 Recently, single particle tracking in cultured neurons was used to show that mRNPs containing multiple mRNA transcripts disassemble into multiple mRNPs upon KCl-depolarization,32 highlighting the highly dynamic nature of mRNPs during transport. The ability to reconstitute these systems will be important for determining the molecular basis for this and other dynamic properties of mRNPs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Susan Lowey and members from the lab for helpful comments on the manuscript.
Funding
This work was funded by NIH grant GM078097 to K.M.T.
References
- 1.Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell 1996; 84:687-97; PMID:8625407; http://dx.doi.org/ 10.1016/S0092-8674(00)81047-8 [DOI] [PubMed] [Google Scholar]
- 2.Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell 1996; 84:699-709; PMID:8625408; http://dx.doi.org/ 10.1016/S0092-8674(00)81048-X [DOI] [PubMed] [Google Scholar]
- 3.Long RM, Singer RH, Meng X, Gonzalez I, Nasmyth K, Jansen RP. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science 1997; 277:383-7; PMID:9219698; http://dx.doi.org/ 10.1126/science.277.5324.383 [DOI] [PubMed] [Google Scholar]
- 4.Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature 1997; 389:90-3; PMID:9288973; http://dx.doi.org/ 10.1038/38015 [DOI] [PubMed] [Google Scholar]
- 5.Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 1999; 9:333-6; PMID:10209102; http://dx.doi.org/ 10.1016/S0960-9822(99)80144-4 [DOI] [PubMed] [Google Scholar]
- 6.Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J 2000; 19:5514-24; PMID:11032818; http://dx.doi.org/ 10.1093/emboj/19.20.5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci U S A 2000; 97:5273-8; PMID:10792032; http://dx.doi.org/ 10.1073/pnas.080585897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jambhekar A, Derisi JL. Cis-acting determinants of asymmetric, cytoplasmic RNA transport. RNA 2007; 13:625-42; PMID:17449729; http://dx.doi.org/ 10.1261/rna.262607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Estrada P, Kim J, Coleman J, Walker L, Dunn B, Takizawa P, Novick P, Ferro-Novick S. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J Cell Biol 2003; 163:1255-66; PMID:14691136; http://dx.doi.org/ 10.1083/jcb.200304030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci U S A 2003; 100:11429-34; PMID:13679573; http://dx.doi.org/ 10.1073/pnas.2033246100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2005; 102:18005-10; PMID:16326802; http://dx.doi.org/ 10.1073/pnas.0509229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell 1998; 2:437-45; PMID:9809065; http://dx.doi.org/ 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- 13.Reck-Peterson SL, Tyska MJ, Novick PJ, Mooseker MS. The yeast class V myosins, Myo2p and Myo4p, are nonprocessive actin-based motors. J Cell Biol 2001; 153:1121-6; PMID:11381095; http://dx.doi.org/ 10.1083/jcb.153.5.1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krementsova EB, Hodges AR, Bookwalter CS, Sladewski TE, Travaglia M, Sweeney HL, Trybus KM. Two single-headed myosin V motors bound to a tetrameric adapter protein form a processive complex. J Cell Biol 2011; 195:631-41; PMID:22084309; http://dx.doi.org/ 10.1083/jcb.201106146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sladewski TE, Bookwalter CS, Hong MS, Trybus KM. Single-molecule reconstitution of mRNA transport by a class V myosin. Nat Struct Mol Biol 2013; 20:952-7; PMID:23812374; http://dx.doi.org/ 10.1038/nsmb.2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krementsova EB, Hodges AR, Lu H, Trybus KM. Processivity of chimeric class V myosins. J Biol Chem 2006; 281:6079-86; PMID:16377634; http://dx.doi.org/ 10.1074/jbc.M510041200 [DOI] [PubMed] [Google Scholar]
- 17.Hodges AR, Krementsova EB, Trybus KM. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J Biol Chem 2008; 283:6906-14; PMID:18175803; http://dx.doi.org/ 10.1074/jbc.M708865200 [DOI] [PubMed] [Google Scholar]
- 18.Dunn BD, Sakamoto T, Hong MS, Sellers JR, Takizawa PA. Myo4p is a monomeric myosin with motility uniquely adapted to transport mRNA. J Cell Biol 2007; 178:1193-206; PMID:17893244; http://dx.doi.org/ 10.1083/jcb.200707080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H, Singh N, Esselborn F, Blobel G. Structure of a myosinbulletadaptor complex and pairing by cargo. Proc Natl Acad Sci USA 2014; 111:E1082-90; http://dx.doi.org/ 10.1073/pnas.1401428111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heym RG, Zimmermann D, Edelmann FT, Israel L, Ökten Z, Kovar DR, Niessing D. In vitro reconstitution of an mRNA-transport complex reveals mechanisms of assembly and motor activation. J Cell Biol 2013; 203:971-84; PMID:24368805; http://dx.doi.org/ 10.1083/jcb.201302095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller M, Richter K, Heuck A, Kremmer E, Buchner J, Jansen RP, Niessing D. Formation of She2p tetramers is required for mRNA binding, mRNP assembly, and localization. RNA 2009; 15:2002-12; PMID:19710186; http://dx.doi.org/ 10.1261/rna.1753309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chung S, Takizawa PA. Multiple Myo4 motors enhance ASH1 mRNA transport in Saccharomyces cerevisiae. J Cell Biol 2010; 189:755-67; PMID:20457760; http://dx.doi.org/ 10.1083/jcb.200912011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Warshaw DM, Kennedy GG, Work SS, Krementsova EB, Beck S, Trybus KM. Differential labeling of myosin V heads with quantum dots allows direct visualization of hand-over-hand processivity. Biophys J 2005; 88:L30-2; PMID:15764654; http://dx.doi.org/ 10.1529/biophysj.105.061903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phichith D, Travaglia M, Yang Z, Liu X, Zong AB, Safer D, Sweeney HL. Cargo binding induces dimerization of myosin VI. Proc Natl Acad Sci U S A 2009; 106:17320-4; PMID:19805065; http://dx.doi.org/ 10.1073/pnas.0909748106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Müller M, Heym RG, Mayer A, Kramer K, Schmid M, Cramer P, Urlaub H, Jansen RP, Niessing D. A cytoplasmic complex mediates specific mRNA recognition and localization in yeast. PLoS Biol 2011; 9:e1000611; PMID:21526221; http://dx.doi.org/ 10.1371/journal.pbio.1000611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klumpp S, Lipowsky R. Cooperative cargo transport by several molecular motors. Proc Natl Acad Sci U S A 2005; 102:17284-9; PMID:16287974; http://dx.doi.org/ 10.1073/pnas.0507363102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chartrand P, Meng XH, Huttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell 2002; 10:1319-30; PMID:12504008; http://dx.doi.org/ 10.1016/S1097-2765(02)00694-9 [DOI] [PubMed] [Google Scholar]
- 28.Niessing D, Hüttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA binding protein with a basic helical hairpin motif. Cell 2004; 119:491-502; PMID:15537539; http://dx.doi.org/ 10.1016/j.cell.2004.10.018 [DOI] [PubMed] [Google Scholar]
- 29.Amrute-Nayak M, Bullock SL. Single-molecule assays reveal that RNA localization signals regulate dynein-dynactin copy number on individual transcript cargoes. Nat Cell Biol 2012; 14:416-23; PMID:22366687; http://dx.doi.org/ 10.1038/ncb2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soundararajan HC, Bullock SL. The influence of dynein processivity control, MAPs, and microtubule ends on directional movement of a localising mRNA. Elife (Cambridge) 2014; 3:e01596; PMID:24737859; http://dx.doi.org/ 10.7554/eLife.01596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park HY, Buxbaum AR, Singer RH. Single mRNA tracking in live cells. Methods Enzymol 2010; 472:387-406; PMID:20580973; http://dx.doi.org/ 10.1016/S0076-6879(10)72003-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park HY, Lim H, Yoon YJ, Follenzi A, Nwokafor C, Lopez-Jones M, Meng X, Singer RH. Visualization of dynamics of single endogenous mRNA labeled in live mouse. Science 2014; 343:422-4; PMID:24458643; http://dx.doi.org/ 10.1126/science.1239200 [DOI] [PMC free article] [PubMed] [Google Scholar]


