Abstract
U12-type introns are a rare class of nuclear introns that are removed by a dedicated U12-dependent spliceosome and are thought to regulate the expression of their target genes owing through their slower splicing reaction. Recent genome-wide studies on the splicing of U12-type introns are now providing new insights on the biological significance of this parallel splicing machinery. The new studies cover multiple different organisms and experimental systems, including human patient cells with mutations in the components of the minor spliceosome, zebrafish with similar mutations and various experimentally manipulated human cells and Arabidopsis plants. Here, we will discuss the potential implications of these studies on the understanding of the mechanism and regulation of the minor spliceosome, as well as their medical implications.
Introduction
U12-type introns, also called minor introns, are a distinct class of nuclear introns that contain highly conserved 5′ splice site (5′ss) and branch point sequences.1,2 Approximately 700–800 genes containing U12-type introns are known in the mammalian system,3,4 but while the numbers of U12-type introns vary in other organisms, they are mostly found from the same orthologous genes despite of large evolutionary distances.5-7 With few exceptions, most genes contain a single U12-type intron, surrounded by normal U2-type introns (also referred to as major introns). Minor introns are spliced by a divergent U12-dependent spliceosome, which contains 4 unique snRNA species and 7 unique protein components. Specifically, U11, U12, U4atac and U6atac snRNAs replace the respective U1, U2, U4 and U6 snRNAs found from the major spliceosome, while the U5 snRNA is shared between the 2 spliceosomes. All the unique proteins of the minor spliceosome are found in the U11/U12 di-snRNP, which carries out the initial intron recognition. In contrast, the U4atac/U6atac.U5 tri-snRNP which is needed for the final assembly of catalytically active spliceosome, is thought to have the same protein composition as the U4/U6.U5 tri-snRNP of the major spliceosome.8-10
One of the key functional differences between the 2 systems is that minor introns are only rarely involved in alternative splicing, most likely owing to their highly conserved sequence elements and a lack of usable alternative splice sites.11,12 Another difference is that minor introns have been reported to be spliced more slowly, both in vitro13,14 and also in vivo.15-18 The slower splicing in vivo has been inferred from the elevated levels of unspliced U12-type introns in the cellular steady-state RNA pool. On average, U12-type introns show approximately 2-fold higher retention levels compared to the U2-type introns in the same gene. The expected outcome of less efficient splicing is nuclear retention of mRNAs containing unspliced U12-type introns, possibly followed by nuclear decay. Accordingly, U12-type introns have been proposed to regulate the cellular levels of mature mRNAs.16,19 In support of this hypothesis, replacement of a minor intron by a major one within otherwise identical context leads to up-to 6–8 fold increase in protein yields16, and more recently, upregulation of U6atac snRNA levels have been shown to increase expression levels of genes containing minor introns.19 Conversely, a presence of a U12-type intron leads to reduced levels of protein production, as shown in microinjection experiments using Xenopus oocytes.15
Inefficient Versus Slow Splicing of Minor Introns
Even though increased retention levels for minor introns have been reported in multiple systems, neither the generality of this observation, i.e. whether all or a subset of U12-type introns show increased retention, nor the subsequent fate of transcripts containing unspliced U12-type introns had been addressed until recently. Using cellular fractionation and RNAseq, we investigated global intron retention in mammalian cells and found that the majority of minor introns are indeed retained on average at 2-fold higher levels compared to the neighboring U2-type introns in the same transcript.20 Furthermore, we showed that unspliced U12-type introns are further stabilized upon knockdown of the nuclear exosome. This indicates that transcripts containing unspliced U12-type introns are actively degraded by the nuclear quality control machinery. Together, these 2 observations provide strong support to the rate-limiting control hypothesis which states that U12-type introns limit the expression levels of their host genes. Most likely the degradation activity is linked to nuclear retention whereby a delay in processing increases the probability of mRNA degradation, as suggested by the kinetic surveillance hypothesis of nascent pre-mRNAs.21,22 Alternatively, the surveillance mechanisms could be actively recruited to transcripts containing unspliced introns through interactions between the components of stalled spliceosome and the nuclear quality control machinery as shown recently.23
However, a central but yet unanswered question relates to the mechanistic interpretation of increased retention of U12-type introns. The elevated U12-type intron retention levels on steady-state mRNA pool have been thought to indicate a slower splicing rate for U12-type introns as originally proposed both theoretically and experimentally by Patel et al.16 The key underlying assumption of this model is that splicing rates for the individual pre-mRNAs within a pre-mRNA population are the same. However, by allowing heterogeneity in splicing rates it is possible to come up with an alternative hypothesis, namely inefficient instead of slow splicing to explain the observed overrepresentation of unspliced U12-type introns. For example, by assuming 2 subpopulations of U12-type intron-containing transcripts, with one splicing at the same rate as U2-type introns and the other one failing splicing entirely would result in a pre-mRNA profile in RT-PCR analysis that is indistinguishable from the one resulting from kinetically slow splicing. Such inefficient splicing could arise if one of the specific components of the minor spliceosome would be limiting the formation of catalytically active spliceosomes. An alternative scenario could be a strict dependency on co-transcriptionality in the splicing of minor introns. Co-transcriptional splicing of minor introns itself is supported by qRT-PCR analyses of U12-type intron splicing on long pre-mRNAs24 and by detection of U6atac snRNA in chromatin fractions in RNAseq analyses25, but at the moment it is not known if these introns can also be spliced in a post-transcriptional manner. Nevertheless, it is tempting to speculate that such a co-transcriptional splicing requirement would provide only a narrow window of opportunity for splicing to succeed, which would in turn lead to elevated levels of aborted or halted splicing complexes that are targeted by the nuclear quality control mechanism as described earlier with defective U2-type introns (Fig. 1).26
Figure 1.
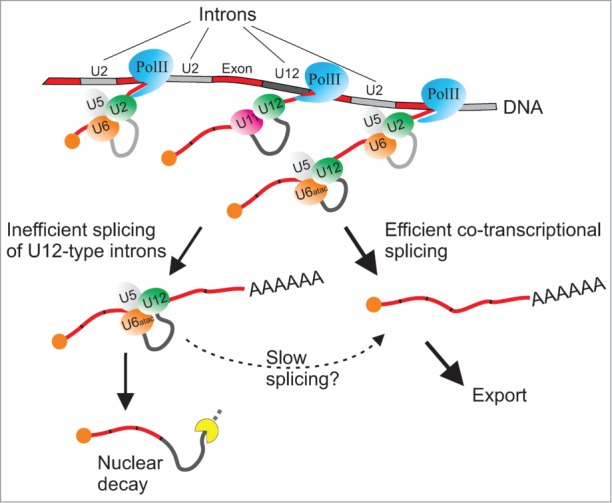
A revised model for rate-limiting regulation of gene expression by U12-type introns. U12-type introns are spliced in a co-transcriptional manner similarly to the U2-type introns to produce mature mRNA molecules. A majority or all U12-type introns are correctly recognized by the U11/U12 di-snRNP, and most likely can also assemble later spliceosomal complexes. However, a subset of them fails to carry out the splicing reaction and is targeted by the nuclear quality control mechanisms. Alternatively, it is possible that this subset can be spliced more slowly post-transcriptionally. Exons are indicated with a red color, U2-type introns are with light gray, and U12-type introns are with dark gray.
At present, there is little information to conclusively rule out either one of the possibilities as an explanation for the observed elevated intron retention levels. Tentative support for slow splicing of U12-type introns comes from in vivo measurements of transcription and splicing rates by Singh and Padgett.24 By synchronizing transcription initiation with DRB, a CDK7 inhibitor that prevents reinitiation of transcription but allows elongation to proceed, followed by qPCR identification of specific exon-exon junctions they reported approximately 2-fold slower appearance of spliced exon junctions with U12-type introns compared to U2-type junctions. While this agrees well with the earlier steady-state measurements which generally show a 2-fold higher intron retention levels for U12-type introns, the observed splicing rates for U2- and U12-type introns are 5 and 10 minutes, respectively. However, the observed 5 min difference is close to the sampling frequency and therefore the resolution limit of the experimental setup. In contrast, our recent analysis of the decay rates of unspliced U12-type intron containing pre-mRNA molecules provides a provisional support for the inefficient splicing scenario. We found that the decay of the unspliced U12-type intron signal was not only particularly slow in exosome knockdown cells, but more importantly, the levels of unspliced introns were nearly invariant up to 2 hours after inhibition of transcription initiation. Such a pattern is more consistent with inefficient splicing (where a subset of introns fail to splice) than with slow splicing (where one would expect a gradual loss of U12-type intron pre-mRNA signal). However, either piece of evidence is not conclusive as such. To settle this issue it will be necessary to use other methods such as in vivo single molecule microscopy to accurately estimate splicing rates of U12-type introns as has been done with U2-type introns.27
Regardless of whether the U12-type introns are spliced slowly or inefficiently, the molecular events responsible for the elevated retention levels of U12-type introns appear to take place after the intron recognition step. Inhibition of intron recognition step either by human disease-causing mutations28 or by knocking down specific protein components in the U11/U12 di-snRNP responsible for the initial intron recognition29,30 all lead to, in addition to a defect in the splicing of U12-type introns, activation of cryptic U2-type splice sites in the vicinity of a subset of U12-type introns. Importantly, this has been observed not only in human or mammalian cells, but also in Arabidopsis.29 In contrast, under normal conditions the unspliced U12-type introns accumulate but there is no indication of concomitant activation of nearby cryptic splice sites,19,20,28 arguing that U12-type introns are in fact recognized with similar efficiency as U2-type introns, but one or more subsequent steps in the spliceosome assembly or catalytic activation of the spliceosome are either slow or fail altogether, thus resulting in the elevated intron retention signal seen with the RT-PCR and RNAseq analyses (Fig. 1). Because inhibition of U6atac snRNA with a morpholino oligonucleotides in human cells also result in activation of cryptic U2-type splice sites,19 it is possible that a stable recognition of U12-type introns and suppression of cryptic U2-type splice sites requires an exchange of U11 to U6atac snRNA at the 5′ss and possibly formation of the mature spliceosomal complexes (see Fig. 1).
Implications for Human Diseases
The outcome of splicing decisions between the U12- and U2-type introns, that is, whether to retain an unspliced U12-type intron, or to activate nearby cryptic U2-type splice sites may also have important clinical implications. Specifically, 2 congenital human diseases affecting specific components of the minor spliceosome have been described to date. MOPD1/TALS is a disease caused by mutations in the U4atac snRNA component of the minor spliceosome.31,32 In this case, the mutations disable the formation of U4atac/U6atac.U5 tri-snRNP and cause a relatively mild splicing defect with a small increase in the retention levels of U12-type introns in the endogenous genes.31,33 Nevertheless, the resulting disease is very severe with multiple developmental defects and eventual death typically within the first 3 y of life. In contrast, mutations in the 65K gene lead to rather dramatic activation of cryptic U2-type splice sites, but a very mild disease (IGHD1) phenotype with pituitary hypoplasia resulting in postnatal dwarfism, but apparently no other major symptoms.28 While detailed transcriptome analysis of the MOPD1/TALS patients has not yet been described, the results seem somewhat counterintuitive, with more severe splicing defects leading to a milder disease. One possible explanation is that that perhaps the activation of cryptic U2-type splice sites seen with IGHD1 are better tolerated than the intron retention events reported with MOPD1/TALS. This could be because because many of the mRNAs resulting from cryptic splicing events may be eliminated via NMD pathway, while excessive accumulation of unspliced U12-type introns may interfere with the normal functioning and regulation of the U12-dependent spliceosome. In support of this hypothesis, another 65K mutation in zebrafish, which also seems to affect later stages of spliceosome assembly and does not lead to activation of cryptic splice sites also displays a more severe phenotype with multiple developmental defects.34
A key question related to the regulatory potential of the U12-dependent spliceosome is whether its activity can be regulated via intracellular or extracellular signals. To date, 2 regulatory pathways have been described that affect minor spliceosome components and/or splicing activity. One of them is a negative feedback regulation that targets 2 key proteins, 48K and 65K, that are both components of the U11/U12 intron recognition complex.35,36 The other one is the p38MAPK signaling pathway that controls the U6atac snRNA levels, which in turn regulate the overall efficiency of U12-type intron splicing.19 It is likely that these 2 regulatory pathways have necessary but separate roles for the splicing of U12-type introns. Consequently, the negative feedback regulation of 48K and 65K proteins is most likely maintaining the cellular homeostasis of the U11/U12 di-snRNP (the intron recognition complex) levels, which enables accurate recognition of U12-type introns. In contrast, the p38MAPK regulation of U6atac snRNA levels could determine the overall splicing efficiency of the minor spliceosome, with a direct impact on expression levels with genes containing U12-type introns.19 Regulation of minor spliceosome activity through the p38MAPK pathway may also explain the tissue specificity seen with the human diseases caused by mutations in the minor spliceosome.
Perspectives
The hypothesis that U12-type introns are providing rate-limiting regulation for their host genes was proposed some 10 y ago based on the analysis of a subset of U12-type introns which showed elevated levels of U12-type intron retention in RT-PCR analysis. Recent genome-wide analyses have now confirmed that inefficient splicing is indeed a general feature of the majority of U12-type introns, but have also shown that transcripts containing U12-type introns are degraded by the nuclear quality control mechanisms. While the distinction of whether the U12-type introns are spliced inefficiently or slowly will need to wait for future experiments, possibly single-molecule microscopy, there is also now strong evidence that the activity of the minor spliceosome can be regulated via cellular signaling pathways. Future research building on earlier data and the most recent genome-wide results will reveal the impact of the U12-type intron splicing on cellular pathways and also as a possible modulator of human disease phenotypes.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of Frilander lab for critical reading and suggestions to the manuscript.
Funding
This work has been supported by Academy of Finland (grant 140087 to M.J.F.).
References
- 1. Patel AA, Steitz JA. Splicing double: Insights from the second spliceosome. Nat Rev Mol Cell Biol 2003; 4:960-70; PMID:14685174; http://dx.doi.org/ 10.1038/nrm1259 [DOI] [PubMed] [Google Scholar]
- 2. Turunen JJ, Niemelä EH, Verma B, Frilander MJ. The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA 2013; 4:61-76; PMID:23074130; http://dx.doi.org/ 10.1002/wrna.1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res 2006; 34:3955-67; PMID:16914448; http://dx.doi.org/ 10.1093/nar/gkl556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res 2007; 35:D110-5; PMID:17082203; http://dx.doi.org/ 10.1093/nar/gkl796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Basu MK, Makalowski W, Rogozin IB, Koonin EV. U12 intron positions are more strongly conserved between animals and plants than U2 intron positions. Biol Direct 2008; 3:19; PMID:18479526; http://dx.doi.org/ 10.1186/1745-6150-3-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin C-F, Mount S, Jarmołowski A, Makałowski W. Evolutionary dynamics of U12-type spliceosomal introns. BMC Evol Biol 2010; 10:47; PMID:20163699; http://dx.doi.org/ 10.1186/1471-2148-10-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhu W, Brendel V. Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res 2003; 31:4561-72; PMID:12888517; http://dx.doi.org/ 10.1093/nar/gkg492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schneider C, Will CL, Makarova OV, Makarov EM, Lührmann R. Human U4/U6.U5 and U4atac/U6atac.U5 tri-snRNPs exhibit similar protein compositions. Mol Cell Biol 2002; 22:3219-29; PMID:11971955; http://dx.doi.org/ 10.1128/MCB.22.10.3219-3229.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Will CL, Schneider C, Hossbach M, Urlaub H, Rauhut R, Elbashir S, Tuschl T, Lührmann R. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 2004; 10:929-41; PMID:15146077; http://dx.doi.org/ 10.1261/rna.7320604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Will CL, Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 2005; 386:713-24; PMID:16201866; http://dx.doi.org/ 10.1515/BC.2005.084 [DOI] [PubMed] [Google Scholar]
- 11. Levine A, Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res 2001; 29:4006-13; PMID:11574683; http://dx.doi.org/ 10.1093/nar/29.1.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang W-C, Chen Y-C, Lee K-M, Tarn W-Y. Alternative splicing and bioinformatic analysis of human U12-type introns. Nucleic Acids Res 2007; 35:1833-41; PMID:17332017; http://dx.doi.org/ 10.1093/nar/gkm026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Frilander MJ, Steitz JA. Initial recognition of U12-dependent introns requires both U11/5' splice-site and U12/branchpoint interactions. Genes Dev 1999; 13:851-63; PMID:10197985; http://dx.doi.org/ 10.1101/gad.13.7.851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tarn W-Y, Steitz JA. A novel spliceosome containing U11, U12 and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 1996; 84:801-11; PMID:8625417; http://dx.doi.org/ 10.1016/S0092-8674(00)81057-0 [DOI] [PubMed] [Google Scholar]
- 15. Bozzoni I, Fragapane P, Annesi F, Pierandrei-Amaldi P, Amaldi F, Beccari E. Expression of two Xenopus laevis ribosomal protein genes in injected frog oocytes. A specific splicing block interferes with the L1 RNA maturation. J Mol Biol 1984; 180:987-1005; PMID:6084725; http://dx.doi.org/ 10.1016/0022-2836(84)90267-5 [DOI] [PubMed] [Google Scholar]
- 16. Patel AA, McCarthy M, Steitz JA. The splicing of U12-type introns can be a rate-limiting step in gene expression. EMBO J 2002; 21:3804-15; PMID:12110592; http://dx.doi.org/ 10.1093/emboj/cdf297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pessa H, Ruokolainen A, Frilander MJ. The abundance of the spliceosomal snRNPs is not limiting the splicing of U12-type introns. RNA 2006; 12:1883-92; PMID:16957280; http://dx.doi.org/ 10.1261/rna.213906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Santoro B, De Gregorio E, Caffarelli E, Bozzoni I. RNA-protein interactions in the nuclei of Xenopus oocytes: complex formation and processing activity on the regulatory intron of ribosomal protein gene L1. Mol Cell Biol 1994; 14:6975-82; PMID:7935414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G. Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. eLife 2013; 2:e00780; PMID:23908766; http://dx.doi.org/ 10.7554/eLife.00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Niemelä EH, Oghabian A, Staals RHJ, Pruijn GJM, Frilander MJ. Global analysis of the nuclear processing of unspliced U12-type introns by the exosome. Nucleic Acids Res 2014; 42:7358-69; PMID:24848017; http://dx.doi.org/ 10.1093/nar/gku391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burgess SM, Guthrie C. Beat the clock: paradigms for NTPases in the maintenance of biological fidelity. Trends Biochem Sci 1993; 18:381-4; PMID:8256287; http://dx.doi.org/ 10.1016/0968-0004(93)90094-4 [DOI] [PubMed] [Google Scholar]
- 22. Doma MK, Parker R. RNA Quality Control in Eukaryotes. Cell 2007; 131:660-8; PMID:18022361; http://dx.doi.org/ 10.1016/j.cell.2007.10.041 [DOI] [PubMed] [Google Scholar]
- 23. Nag A, Steitz JA. Tri-snRNP-associated proteins interact with subunits of the TRAMP and nuclear exosome complexes, linking RNA decay and pre-mRNA splicing. RNA Biol 2012; 9:334-42; PMID:22336707; http://dx.doi.org/ 10.4161/rna.19431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singh J, Padgett RA. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 2009; 16:1128-33; PMID:19820712; http://dx.doi.org/ 10.1038/nsmb.1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tilgner H, Knowles DG, Johnson R, Davis CA, Chakrabortty S, Djebali S, Curado J, Snyder M, Gingeras TR, Guigó R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 2012; 22:1616-25; PMID:22955974; http://dx.doi.org/ 10.1101/gr.134445.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eberle AB, Hessle V, Helbig R, Dantoft W, Gimber N, Visa N. Splice-site mutations cause Rrp6-mediated nuclear retention of the unspliced RNAs and transcriptional down-regulation of the splicing-defective genes. PLoS One 2010; 5:e11540; PMID:20634951; http://dx.doi.org/ 10.1371/journal.pone.0011540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin Robert M, Rino J, Carvalho C, Kirchhausen T, Carmo-Fonseca M. Live-Cell Visualization of Pre-mRNA Splicing with Single-Molecule Sensitivity. Cell Reports 2013; 4:1144-55; PMID:24035393; http://dx.doi.org/ 10.1016/j.celrep.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Argente J, Flores R, Gutiérrez-Arumí A, Verma B, Martos-Moreno GA, Cuscó I, Oghabian A, Chowen JA, Frilander MJ, Pérez-Jurado LA. Defective minor spliceosome mRNA processing results in isolated familial growth hormone deficiency. EMBO Mol Med 2014; 6:299-306; PMID:24480542; http://dx.doi.org/ 10.1002/emmm.201303573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jung HJ, Kang H. The Arabidopsis U11/U12-65K is an indispensible component of minor spliceosome and plays a crucial role in U12 intron splicing and plant development. Plant J 2014; 78:799-810; PMID:24606192; http://dx.doi.org/ 10.1111/tpj.12498 [DOI] [PubMed] [Google Scholar]
- 30. Turunen JJ, Will CL, Grote M, Lührmann R, Frilander MJ. The U11-48K protein contacts the 5' splice site of U12-type introns and the U11-59K protein. Mol Cell Biol 2008; 28:3548-60; PMID:18347052; http://dx.doi.org/ 10.1128/MCB.01928-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Edery P, Marcaillou C, Sahbatou M, Labalme A, Chastang J, Touraine R, Tubacher E, Senni F, Bober MB, Nampoothiri S, et al. Association of TALS Developmental Disorder with Defect in Minor Splicing Component U4atac snRNA. Science 2011; 332:240-3; PMID:21474761; http://dx.doi.org/ 10.1126/science.1202205 [DOI] [PubMed] [Google Scholar]
- 32. He H, Liyanarachchi S, Akagi K, Nagy R, Li J, Dietrich RC, Li W, Sebastian N, Wen B, Xin B, et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 2011; 332:238-40; PMID:21474760; http://dx.doi.org/ 10.1126/science.1200587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jafarifar F, Dietrich RC, Hiznay JM, Padgett RA. Biochemical defects in minor spliceosome function in the developmental disorder MOPD I. RNA 2014; 20:1078-89; PMID:24865609; http://dx.doi.org/ 10.1261/rna.045187.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Markmiller S, Cloonan N, Lardelli RM, Doggett K, Keightley M-C, Boglev Y, Trotter AJ, Ng AY, Wilkins SJ, Verkade H, et al. Minor class splicing shapes the zebrafish transcriptome during development. Proc Natl Acad Sci USA 2014; 111:3062-7; PMID:24516132; http://dx.doi.org/ 10.1073/pnas.1305536111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Verbeeren J, Niemelä EH, Turunen JJ, Will CL, Ravantti JJ, Lührmann R, Frilander MJ. An ancient mechanism for splicing control: U11 snRNP as an activator of alternative splicing. Mol Cell 2010; 37:821-33; PMID:20347424; http://dx.doi.org/ 10.1016/j.molcel.2010.02.014 [DOI] [PubMed] [Google Scholar]
- 36. Turunen JJ, Verma B, Nyman TA, Frilander MJ. HnRNPH1/H2, U1 snRNP and U11 snRNP co-operate to regulate the stability of the U11-48K pre-mRNA. RNA 2013; 19:380-9; PMID:23335637; http://dx.doi.org/ 10.1261/rna.036715.112 [DOI] [PMC free article] [PubMed] [Google Scholar]


