Abstract
The existence of sense overlapping transcripts that share regulatory and coding information in the same genomic sequence shows an additional level of prokaryotic gene expression complexity. Here we report the discovery of ncRNAs associated with IS1341-type transposase (tnpB) genes, at the 3'-end of such elements, with examples in archaea and bacteria. Focusing on the model haloarchaeon Halobacterium salinarum NRC-1, we show the existence of sense overlapping transcripts (sotRNAs) for all its IS1341-type transposases. Publicly available transcriptome compendium show condition-dependent differential regulation between sotRNAs and their cognate genes. These sotRNAs allowed us to find a UUCA tetraloop motif that is present in other archaea (ncRNA family HgcC) and in a H. salinarum intergenic ncRNA derived from a palindrome associated transposable elements (PATE). Overexpression of one sotRNA and the PATE-derived RNA harboring the tetraloop motif improved H. salinarum growth, indicating that these ncRNAs are functional.
Keywords: sense overlaping transcripts, IS1341, HgcC RNA family, IS1341-type, RNA-seq, non-coding RNA, overlaping transcripts, transposons, IS200/605, tetraloop
Introduction
Overlapping genomic signals have been discovered across all domains of life, showing that DNA sequences are multifunctional, harboring regulatory and coding information simultaneously. Transcriptome complexity includes not only interweaving promoter elements and intergenic transcription but also a plethora of transcripts that overlap annotated genes.1,2 Sense and antisense non-coding (ncRNA) transcripts have been identified, with a clear predominance of antisense transcripts characterization. Antisense ncRNAs are ubiquitous and affect diverse cellular processes, from transcription initiation to translation efficiency3; they have also been shown to regulate transposition events.4 Even with the widespread use of strand-specific sequencing technologies, the identification and characterization of sense overlapping transcripts have expanded more modestly, since the signals for these molecules are convoluted with mRNA signals and thus, can be difficult to detect.
Recently, we have identified sense ncRNAs overlapping the 5′ end of coding sequences in archaea with focus in the haloarchaeon Halobacterium salinarum NRC-1.5 This extremophile thrives in environments with 4.5M NaCl concentration and has been used as a model organism for systems biology studies. As such, a massive amount of microarray and tiling array-based transcriptome data in diverse environmental conditions is currently available for this organism.6-11 In the present work, by revisiting the dataset from Koide et al.8, where transcriptome dynamics was assessed over a growth curve using high resolution tiling array data, we detected signals that could indicate the presence of sense overlapping transcripts inside IS1341-type transposase genes in H. salinarum.
IS1341-type transposases genes are also known in the literature as tnpB or orfB. They are part of the widespread IS200/605 family, the most ancient family in the archaeal domain of life.12 Based on gene composition, IS200/605 family can be classified into 3 distinct groups: (a) IS200 group, with only the tnpA gene (also known as the IS200 gene); (b) IS605 group, where tnpA and tnpB are found adjacent and (c) IS1341 group, where tnpB is found alone.13,14 Transposition of IS200/605 members requires the protein TnpA, which binds conserved palindromic sequences at the left end (LE) and right end (RE) of the insertion element.15,16 The mechanism of transposition is unique among IS elements since it requires single stranded DNA as substrates.15,17 The tnpB gene is not required for transposition14,18, however, it is known to play a regulatory role by inhibiting excision and insertion of ISDra2 in Deinococcus radiodurans.19 Mechanisms of TnpB action are currently unknown.
Sense overlapping transcripts in IS1341-type transposase genes have been detected in other archaea as part of studies for the identification of ncRNAs. In Sulfolobus solfataricus, 2 ncRNAs overlapping transposases in the same strand were identified and in Thermococcus kodakarensis, 3 RNAs transcribed from transposase loci were detected.20,21 In Pyrococcus abyssi, ncRNAs are transcribed from intergenic repetitive loci and have sequence similarity to the 3′ end of IS1341-type transposases.22 This apparent conservation of sense overlapping RNAs and similar sequences in intergenic ncRNAs prompted us to investigate not only the presence of these transcripts in H. salinarum NRC-1, but also to evaluate if they have a functional role. In the present work, we show that all IS1341-type transposases of H. salinarum possess sense overlapping transcripts, explore their expression profiles in diverse conditions, and show a conserved tetraloop motif that might be relevant for function of these ncRNAs.
Results and Discussion
H. salinarum has a large number of insertion sequences (114 according to ISfinder and ISbrowser23,24, https://www-is.biotoul.fr), which are believed to be responsible for the unusual genome plasticity observed in this organism.25 In H. salinarum NRC-1, reannotation based on R1 strain identified 10 IS1341-type transposases.26 Five of these IS1341-type transposases are located adjacent to tnpA, 1 in divergent and 4 in convergent orientation (Table 1).
Table 1.
IS1341-type transposases (tnpB genes) analyzed in this work and their respective sense overlapping transcripts (sotRNAs)
| Gene | IS200/605 family group | chromosome | tnpB start codon | tnpB stop codon | strand | sotRNA transcript start | sotRNA transcript end | sotRNA size [nt] | R1 strain equivalent genes | |
|---|---|---|---|---|---|---|---|---|---|---|
| VNG0013C | IS1341 | chr | 12734 | 11478 | reverse | 11641 | 11428 | 214 | OE1019R | |
| VNG0026C | IS1341 | chr | 21789 | 20533 | reverse | 20701 | 20498 | 204 | OE1040R | |
| VNG0042G | IS1341 | chr | 35931 | 34651 | reverse | 34775 | 34570 | 206 | OE1070R | |
| VNG0044H | IS605 (VNG0043H – VNG0044H), convergent | chr | 36715 | 37971 | forward | 37813 | 38042 | 230 | OE1074F | |
| VNG0286C | IS605 (VNG0285C – VNG0286C), convergent | chr | 229443 | 230597 | forward | 230444 | 230629 | 186 | OE1440F | |
| VNG2652H | IS605 (VNG2653C – VNG2652H), divergent | chr | 1989188 | 1987884 | reverse | 1988005 | 1987819 | 187 | OE4727R | |
| VNG6181H | IS605 (VNG6182H – VNG6181H), convergent | pNRC200 | 155603 | 154368 | reverse | 154508 | 154259 | 250 | OE5062R | |
| VNG6221H | IS605 (VNG6222H – VNG6221H), convergent | pNRC200 | 183054 | 181795 | reverse | 182008 | 181724 | 285 | OE5102R | |
| VNG6361G | IS1341 | pNRC200 | 287504 | 288844 | forward | 288696 | 288915 | 220 | OE6034F | |
| VNG6406H | IS1341 | pNRC200 | 316931 | 315786 | reverse | 315833 | 315631 | 203 | OE6089R |
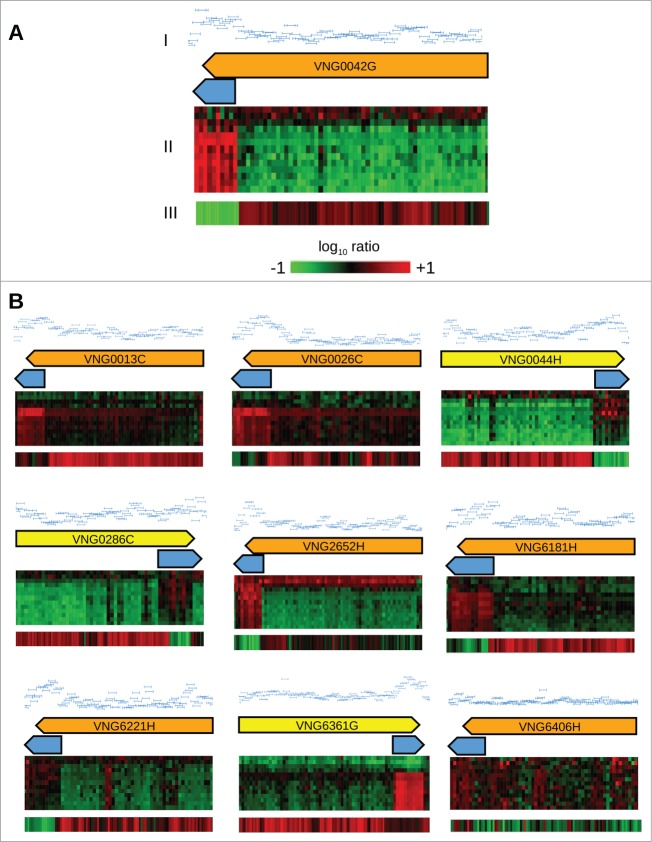
Analyzing publicly available tiling microarray data obtained in reference growth condition for H. salinarum NRC-18, we discovered a conserved transcriptional signature in all 10 IS1341-type transposases genes. This signature features a change in the expression signal inside the insertion sequence near the 3′ end, indicating the existence of possible sense overlapping transcripts (sotRNAs). Figure 1A-I illustrates this feature for VNG0042G. When the dynamics of these transcripts were analyzed over a growth curve, the putative sense overlapping transcripts showed an increase in relative expression levels, whereas the remaining of IS1341-type transposase gene showed decreasing levels (Fig. 1A–II). Tiling array data from TfbD transcription factor overexpression showed that sotRNAs presented decreasing expression levels, whereas IS1341-type genes presented increasing levels, stating an opposite regulation in varied conditions (Fig. 1A–III).8 Transcription signatures in the reference condition, together with the dynamical information from the differential expression profiles provided evidence for the existence of sense overlapping transcripts at the 3′ end of IS1341-type transposases genes in Halobacterium salinarum NRC-1 (Fig. 1A, B).
Figure 1.
Identification of sotRNAs in IS1341-type transposases using tiling array data. (A) (I) Tiling array signal in reference condition for VNG0042G and VNG_sot0042; (II) expression profiles over the growth curve8; (III) expression profiles during TfbD overexpression.8 Heatmaps are color-coded according to log10 expression ratios between each of the 13 time points relative to reference condition. (B) Tiling array signal in reference condition and expression profiles of the remaining 9 IS1341-type transposases (arrows in yellow for genes in forward strand and in orange for genes in reverse strand) and their sotRNAs (light blue arrows) over the growth curve and TfbD overexpression, as described in (A).
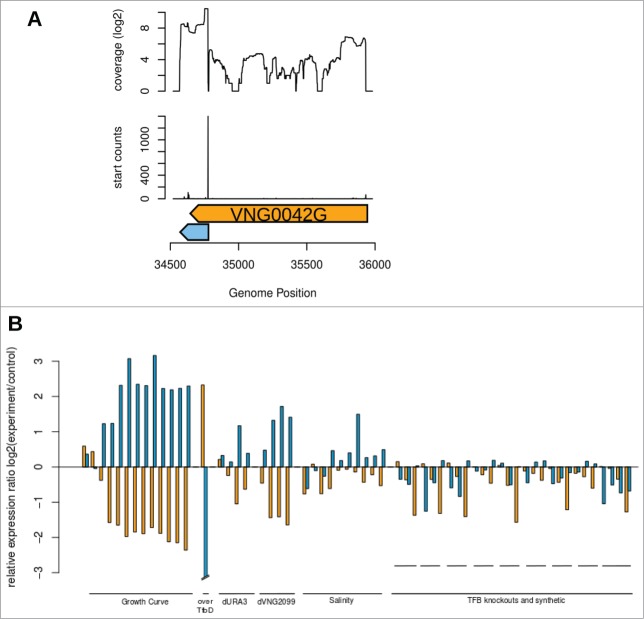
To validate their existence and define their boundaries with precision, we analyzed previously published small RNA-seq data (sRNA-seq).5 Figure 2A shows sRNA-seq based discovery of boundaries for VNG0042G associated sotRNA. An extended analysis (Fig. S1A) showed that all IS1341-type transposases in Halobacterium salinarum NRC-1 have an associated sotRNA. In addition, it was possible to map the 3′ and 5′ ends of all sotRNAs (Table 1) (Fig. S1A). We named these discovered features using “VNG_sot” as prefix and making reference to their cognate gene in the suffix. All sotRNAs mapped in this study start inside the coding sequence of IS1341-type transposase genes, at approximately 1100 nt from the start codon, with an average size of 218 nt and end, on average, 74 nt after gene's stop codon (Table 1).
Figure 2.
Analysis of sRNA-seq and expression profiles for VNG0042G and VNG_sot0042. (A) Strand-specific sRNA-seq data5 was used to map the 5' end of sotRNAs in IS1341-type transposases of H. salinarum NRC-1. sRNA-seq data is visualized as log2 of total reads aligned in each genomic position for VNG0042G and VNG_sot0042. Enrichment of 5' ends of mapped reads are visualized as peaks right below small RNA-seq coverage. Light blue arrow: sotRNA. Dark orange arrows: genes annotated on reverse strand. (B) Expression profiles of VNG0042G (orange) and VNG_sot0042 (blue) in different environmental and genetic backgrounds. Each pair of orange/blue columns represent one published experimental condition. Data for the remaining genes and for each individual experiment is available in Supplementary Figure 1. Data for intergenic ncRNAs can be found in Figure 4 and Supplementary Figure 7.
Two targets were selected for further experimental validations: VNG_sot0042 and VNG_sot2652. C-RACE experiment for VNG_sot0042 confirmed its 5′ and 3′ ends (Fig. S2) and primer extension for VNG_sot2652 confirmed its 5′ end (Fig. S3).
To further investigate the expression profiles of sotRNAs and IS1341-type transposases, we considered all tiling microarray data sets available in public databases (see Materials and Methods) and focused on probes designed to measure expression levels in the same region defined by sRNA-seq analysis. All sotRNAs were found to be expressed with diverse patterns of modulation, as would be expected for a typical functional transcript (Fig. S1B). Most of the sotRNAs were differentially expressed relative to the remaining signal of IS1341-type transposase cognate gene in different environmental conditions, which could possibly indicate some kind of regulation between these elements. Figure 2B shows the data for VNG0042G and its sotRNA. There is one case where the sotRNA (VNG_sot0286) is located antisense to the probable 3′ UTR of VNG0287a (a putative RNA-binding protein). Analysis of their expression profiles over different conditions show that VNG_sot0286 and VNG0287a have anti-correlated expression profiles in some conditions, which could suggest a cis-antisense interaction (Fig. S4). None of the other 9 sotRNAs mapped in H. salinarum NRC-1 have this putative pattern for cis-antisense regulation.
The available data cannot conclusively differentiate between the 2 most probable sotRNA biogenesis hypotheses: primary transcription or processing products. When the upstream regions of the sotRNAs were analyzed, no classical BRE-TATA promoter sequence was found. Northern-blot experiments for 2 sotRNAs (VNG_sot0042 and VNG_sot2652, Fig. S5) might argue for processing, since the full-length transcript is not detected. Also, deep-sequencing data from primary transcript enriched libraries (dRNA-seq)5 only shows evidence of primary transcript enrichment for VNG_sot0026. However, it was previously reported that dRNA-seq data is prone to false negative results27 which reinforces the finding that VNG_sot0026 is a primary transcript but does not necessarily disprove the others. In addition, some sotRNAs were also shown as primary transcripts in T. kodakarensis.21 Chromatin immunoprecipitation data (ChIP-chip) points to transcription factor binding sites at upstream vicinity of some sotRNAs along with statistically derived H. salinarum gene regulatory elements (Table S1).6,7,41 Finally, the distinct expression patterns of sotRNAs and their cognate genes across several experiments are unlikely to be generated by processing a single precursor molecule.
The presence in H. salinarum NRC-1 of a probable sotRNA in a nonfunctional IS1341-type transposase gene (Fig. S1C), a truncated small 120 bp long pseudogene identical to OE5220R, suggests that sotRNA presence may somehow be the reason why these defective elements have not been lost from the genomes. Mining public available transcriptome data for other archaea and bacteria, we found that transcripts overlapping 3′ ends of tnpB are more prevalent than previously appreciated. Besides the previously described S. solfataricus P220 and T. kodakarensis KOD121 cases, we found examples in all archaea that contain IS1341 group genes and RNA-seq data available at NCBI's SRA database: Methanopyrus kandleri AV19, S. acidocaldarius DSM639 and P. furiosus DSM3638. Moreover, we found bacterial examples in Helicobacter pylori 26695 and Escherichia coli K12 data (Figure S6, File S1). These observations suggest a conserved role for sotRNAs in prokaryotes. Computational secondary structure analysis reveals a very distinctive feature among almost all sotRNAs in H. salinarum: the presence of a stable hairpin motif near the end of the sequence. Using five different secondary structure prediction algorithms on each RNA sequence, it was possible to identify: (i) a clear tetraloop motif comprising an 8 bp long stem with a UUCA loop (VNG_sot0042, VNG_sot0044, VNG_sot6181 and VNG_sot6221); (ii) single nucleotide variants of this tetraloop (VNG_sot0286 with UUUA, VNG_sot0013 with CUCA and VNG_sot0026 with GUCA) (Fig. 3); and (iii) stem-loop structures created by other sequences (VNG_sot6406 with a 3 nt long loop in a 6 bp stem, VNG_sot2652 with a 6 nt long loop in a 10 bp stem, and VNG_sot6361 with a 3 nt long loop in a 4 bp stem) (File S2). While the 5 algorithms are considerably different in their approaches and underlying assumptions to predict secondary structures28, they all made consistent predictions of tetraloops and collectively added to the significance of our discovery of this feature.
Figure 3.
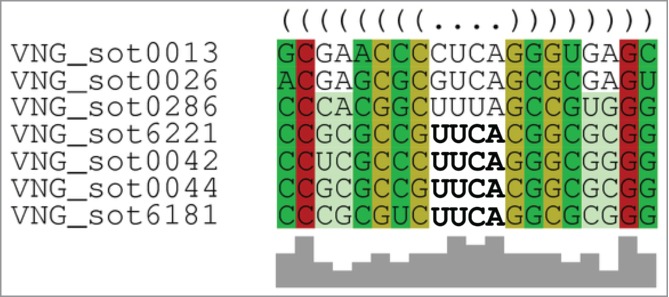
Tetraloop motif found in most sotRNAs in H. salinarum NRC-1. Colors represent base pairing groups and color saturation corresponds to conservation, parenthesis correspond to pairing and dots to unpaired regions. Gray blocks below the sequences represent a histogram of the most conserved nucleotide. Raw data is available at Supplementary File 2.
One of the key characteristics of IS200/605 family is the presence of conserved imperfect palindromic sequences that form a hairpin-like structure at DNA level at both left and right ends of the insertion elements, sometimes just upstream of CDS stop codons.15,16,19 Therefore, IS1341-type transposase genes should harbor the right end (RE) of the IS element, which may explain the sotRNA motifs found as a retained structure formed at DNA level, also stable at RNA level. We named the motif found, the RE-like tetraloop.
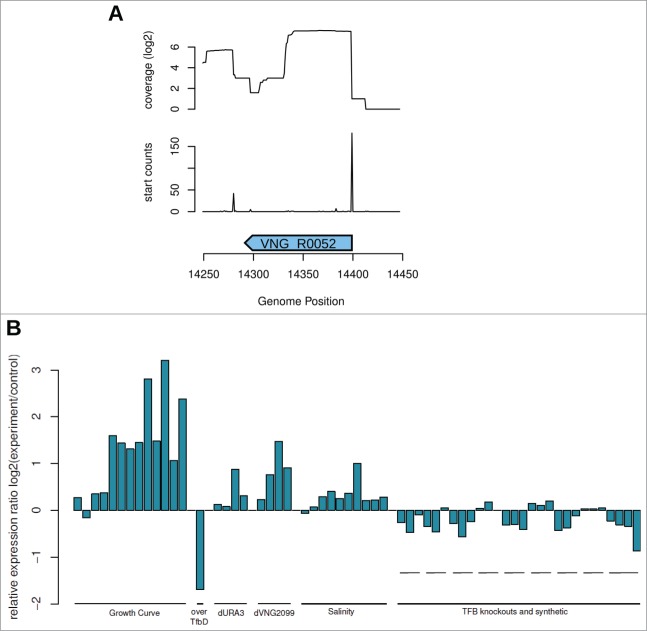
Similarly, in P. abyssi, sequence alignments of intergenic ncRNAs sRK48 and sRK52 with other thermococcal genomes showed similar conserved structures and the most conserved one (P1 loop)22 corresponds to the RE-like tetraloop, reinforcing the probable functionality of this region in archaea. A sequence similarity search for the RE-like tetraloops in H. salinarum NRC-1 genome showed that, besides all sotRNAs regions, 3 intergenic regions also have this secondary structure: 14335 to 14359 on main chromosome, 268252 to 268281 on pNRC200 plasmid, and 19532 to 19556 on main chromosome (File S3). From these 3, only the first 2 had sufficient sRNA-seq read coverage signal: VNG_R0052 and VNG_R6334 (Table 2). We determined the 5′ and 3′ end of these transcripts using RNA-seq data (Fig. 4A and Fig. S7A) and verified the expression profiles over diverse environmental and genetic perturbations. Intergenic ncRNAs showed differential expression, with trends similar to sotRNAs (Fig. 4B and Fig. S7B).
Table 2.
Intergenic RNAs harboring the RE-like tetraloop motif
| ncRNA | VNG_R0052 | VNG_R6334 |
|---|---|---|
| Start | 14397 | 268030 |
| End | 14299 | 268304 |
| Length | 99 | 275 |
| Chr | chr | pNRC200 |
| Strand | reverse | forward |
| RE-like Start | 14359 | 268256 |
| RE-like End | 14340 | 268277 |
Figure 4.
(A) Strand-specific sRNA-seq data5 was used to map 5' ends of intergenic RNAs in H. salinarum NRC-1. sRNA-seq data is visualized as log2 of total reads aligned in each genomic position for VNG_R0052. Enrichment of 5' ends of mapped reads are visualized as peaks right below small RNA-seq coverage profile. (B) Expression profiles of VNG_R0052 in different environmental and genetic backgrounds.
When we analyze the sequence of the intergenic ncRNA VNG_R0052, we can find not only the stem loop corresponding to the RE of the IS element, but also, fused to it at the 5′ end, we can find part of the left end (LE) sequence of IS200/605 family (File S2). This phenomena has been previously observed in Haloquadratum walsbyi as a result of transposase deletion, generating elements that are denominated PATEs (palindrome associated transposable elements).29 We show that a PATE element is indeed expressed at the RNA level and similarity search in the NCBI database shows that VNG_R0052 is not an exception: fused LE and RE are common among haloarchaea (File S4). We predict that, as in VNG_R0052 case, PATEs might also be functionally transcribed in other organisms. Moreover, since PATEs have counterparts in bacteria (REPINS, found for example in Pseudomonas fluorescens30), we extend this prediction to bacteria.
Interestingly, the RE-like tetraloop is a previously unannotated structural motif in the HgcC ncRNA family (RFAM v12 accession code RF00062) (Fig. S8). HgcC family was first discovered in the genome of Methanococcus jannaschii by an in silico analysis designed for the identification of high guanine-cytosine content regions.31 Yet, functions of this ncRNA family are still unknown, thus the presence of the RE-like tetraloop inside RNAs from this family might help further studies focusing on elucidating their function.
To test if RNAs harboring this conserved structural motif may have some functional role, we overexpressed one sotRNA (VNG_sot0042) and one intergenic RNA (VNG_R0052), and analyzed the consequences on growth dynamics in standard batch culture. Remarkably, both strains harboring the RE-like tetraloops had improved growth relative to the control strain providing preliminary evidence that this conserved motif is somehow functional in H. salinarum NRC-1 (Fig. 5). In the future, the specific mechanism by which the tetraloop acts can potentially be investigated by analyzing the effect of systematically disrupting its stem and loop structures on this growth phenotype.
Figure 5.
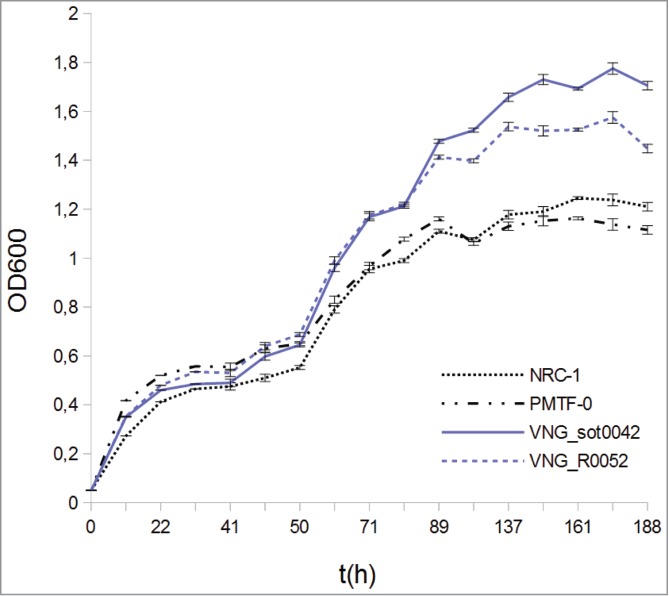
Growth curve of 4 different strains of H. salinarum NRC-1 under reference conditions. Strains overexpressing RNAs (VNG_R0052 and VNG_sot0042) that harbors the RE-like tetraloop present higher growth when compared to control strains (NRC-1, wild type and PMTF-0, which is the wild type strain with an empty overexpression vector).
The exact role of IS1341-type transposases is still an open problem but recent reports indicate that they have an inhibitory effect on IS excision and insertion.19 Given that all transcripts analyzed in this work (sotRNAs and a PATE-derived RNA) harbor similar motifs, we might speculate whether these molecules are related to this mechanism. A possibility is that this motif could work as a competitor for TnpA binding, and thus, making it less available for binding at single stranded DNA regions. PATE-derived RNAs could also present similar functionality. Thus, IS1341 group of transposases could act as reservoirs of stem loops, as previously hypothesized.32 Further experimental work would be required to test this hypothesis, nevertheless, our data point to the functionality of ncRNAs derived from IS1341-type transposases that harbor the conserved RE-like tetraloop motif in their sequences.
Conclusions
In the present work, we report the discovery of sense overlapping transcripts in IS1341-type transposases in H. salinarum NRC-1 and show that they are generally differentially expressed relative to their cognate transposase gene in diverse environmental conditions. In addition, we detected the presence of a conserved tetraloop motif in several sotRNAs and in intergenic ncRNAs that correspond to palindrome associated transposable elements (PATEs). We provide evidence that transcripts harboring this conserved tetraloop motif, which are very similar to right end (RE) of the insertion sequence, are probably functional since their overexpression improved H. salinarum NRC-1 growth.
Materials and Methods
H. salinarum NRC-1 transcriptome analysis
Publicly available RNA-seq data was used to locate ncRNA transcripts with high spatial resolution. Publicly available tiling microarray data was used to interrogate expression dynamics over time or combinatorial experimental conditions for the ncRNAs found in this work. The RNA-seq data is composed by deep-sequencing of replicated sRNA enriched libraries (<200 bp) obtained from standard H. salinarum NRC-1 growth conditions5 available at NCBI's SRA database (SRX433542). Tiling microarray data is a compendium of all publicly available datasets (Aug/2014) obtained by different platforms (NCBI's GEO database accession numbers: GPL17005, GPL13426, GPL7369, GPL7255 and GPL8468). The data set used were: 13 points of a standard growth-curve (GSE12923); overexpression of the TfbD transcription factor (GSE15788)8; knockouts of transcription factor Tfb and overexpression of synthetically enhanced versions of it (GSE31308)9; knockout of ribonuclease VNG2099C (GSE45988)10; variation of extracellular salinity concentrations for 3 different salts (GSE53544).11 Normalized expression ratios appropriate for each study were obtained directly from GEO records as published. Growth curve dynamics and TfbD overexpression tiling array data were visualized in detail at probe level using Gaggle Genome Browser.33
Sequence similarity search in public databases
Sense overlapping transcripts for specific tnpB genes were found in different organism using standard BLASTn-SRA suite sequence similarity search. The RNA-seq datasets considered comprised archaeal and bacterial data: S. acidocaldarius DSM639 (SRA accession SRX327494), Methanopyrus kandleri AV19 (accession SRP019041), H. pylori 26695 (accessions SRX014018, SRX014030 to SRX014033 and SRX014050) and E. coli K12 (accession SRX254762). H. salinarum NRC-1 intergenic ncRNA containing RE-like tetraloop motifs where found through standard BLASTn searches using all 10 sotRNAs sequences as queries.
RNA secondary structure computational analysis
RNA sequences of all 10 sotRNAs and 2 intergenic ncRNAs were used for structural predictions using 5 different methods: RNAfold-MFE, which predicts the Minimum Free Energy (MFE) structure; RNAfold using thermodynamic partition function centroids algorithm34; ContextFold, which uses a very large number of parameters in a machine learning approach35; Vsfold5, which allow the prediction of pseudoknots by using sequential (5′ to 3′) folding and thermodynamically most-probable folding pathways36 and Cylofold, which simulates the 3D folding process in a coarse-grain model.37 All predictions are shown in extended dot-bracket format to include pseudoknots. All 5 methods were run using their respective web-server versions and default parameters.
Strains and culturing
H. salinarum NRC-1 was grown in enriched complex media (CM) consisting of 25% NaCl, 2% MgSO4.7H2O, 0.2% KCl, 0.3% Na-citrate and 1% peptone, at 37°C with constant light and agitation of 225 rpm. When required, the media was supplemented with 20 μg/mL of mevinolin (A.G. Scientific, San Diego, CA). E. coli strain DH5α was grown at 37°C with air shaking at 225 rpm in Luria-Bertani media and supplemented with 50 μg/mL of carbenicilin (Sigma) when necessary.38
RNA preparation
Total RNA was prepared using the MirVana RNA extraction kit (Ambion) and treated with Turbo DNAse I (Fermentas). Samples were analyzed by PCR to rule out DNA contamination.
Primer extension
10 μg of RNA was hybridized with 1pmol of 5′ fluorophore (VIC or NED) labeled oligonucleotide and 1 μl of 10 mM dNTPs in a total volume of 10μl (Temperature cycle for annealing: 95°C 5′, 62°C 15′, 59°C 15′, 53°C 15′, 50°C 15′). After annealing, a Synthesis Mix 2 μl 10x RT Buffer (200 mM Tris-HCl (pH 8.4), 500 mM KCl); 4 μl MgCl2(25 mM); 2 ul 0.1 M DTT; 1 μl RNAse Out; 1 μl SuperScript III (Invitrogen) was added and the reaction was incubated at 50°C for 50′. The cDNA was purified and analyzed in an ABI 3500XL (Applied Biosystems). Data processing was made using Gene Mapper V5.0 (Life Technologies).
Northern blot
For Northern blot analyzes, 50 μg of total RNA treated with RNase-free DNAseI (Fermentas) was separated on polyacrylamide gel (8% acrylamide:bisacrylamide [29:1], 8M urea, 1xTris–borate–EDTA buffer). RNAs were transferred to Hybond-N+ membranes (GE Healthcare) and hybridized with 32P-labeled oligonucleotides (5′-GACGTAGCCTGTTCTTGAGGTGCGAACGCACCAGTTTCAT-3′ for VNG0042G and VNG_sot0042; 5′-CCCTGTTCCGCGAGGGAACGGGATGAGGTTGGGCGGCTTA-3′ for VNG2652H and VNG_sot2652) using Rapid-hyb buffer (GE Healthcare). Signals were detected by autoradiography using a M35A X-Omat Processor (Kodak).
Circularization and RACE (Rapid Amplification of cDNA Ends)
5′ and 3′ ends of RNAs were determined by RNA circularization.39 Briefly, RNA free of DNA contamination was treated with Tobacco Acid Phosphatase (Epicentre) following manufacturer's instructions and treated with T4 RNA ligase (Thermo Scientific) for intramolecular ligation of 5′ and 3′ ends. cDNA was synthesized using SuperScript III First Strand Synthesis System (Life Technologies) using random hexamers. Divergent PCR (CircVNG0042-F —- 5′-ATGGTTTACGACGTAGCC-TGTTC −3′, CircVNG0042-R —- 5′-GTTCCTGTTTGACAATGAAACTG -3′) using circular cDNA was made using Kapa Taq ReadyMix (Kapa Biosystems). Amplification products were purified from agarose gels and cloned into pGEM T-easy Vector (PROMEGA) for sequencing. RACE assays were performed as previously described40 using 5′/3′ RACE Kit 2nd Generation (Roche) for VNG_R0052 using the following primer sequences: RSP1 — 5′ - GAAGGGCGAGGCTTTCTTCTC –- 3′, RSP2 –- 5′ – CGAGGCTTTCTTCTCGATTC –- 3′, RSP3 –- 5′ - CTGTTACGGAGAATCGAGAAG –- 3′, RSP4 –- 5′ GAATCGAGAAGAAAGCCTCG — 3′.
General cloning procedures
VNG_R0052 and VNG_sot0042 were cloned into pMTF overexpression vector for H. salinarum NRC-1.41 DNA fragments were amplified by PCR using 50 ng of H. salinarum NRC-1 genomic DNA, 10pmol of specific oligos, 6.25 μl of 2x KAPA2G Fast ReadyMix (with loading dye) (Product Number: KK5102), 0.25 μl of MgCl2 (25 mM), 3.875 μl of DMSO and DEPC treated water to a final volume of 12.5μl. The oligos used for VNG_R0052 were 5′-GACATATGCGGAGAATCGAGAAGA AAGCC-3′ and 5′-GGGGATCCTCAGTCTAAACT-TCTGGGGTGG-3′; and for VNG_sot0042 were 5′-GCCGGGCATATGACGTATCTCCGAGTCCCGTCA-3′ and 5′-GCGGGGGGATCCACAGGATAGCTGAGACTGCAGCAT-3′. The selected oligos added a BamHI site located at the 5′ region and a NdeI site located at 3′ region. PCR products were purified from 1% agarose gel by illustra GFX PCR DNA and Gel Extraction Kit (GE Healthcare). All fragments were cloned as BamHI/NdeI fragments into pMTF vector and transformed in chemically competent E. coli DH5α. Plasmidial DNA was extracted using Wizard Plus SV Miniprep Kits (Promega) and sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). The confirmed plasmids were used to transform H. salinarum NRC-1. Transformant selection was obtained by plating the cells into solid CM supplemented with mevinolin (20 μg/ml).
Growth curves
For phenotypical characterization, isolated colonies of each H. salinarum strain were grown in liquid CM, supplemented with 20μg/ml of mevinolin when needed, until they reached OD600 = 0.5. Each culture was diluted to OD600 = 0.05 and grown under standard conditions for 8 days, all growth curves were made in 2 biological and technical replicates.42
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Silvia Epifânio for technical support. We thank reviewers for constructive criticisms that improved our work.
Funding
This work was supported by Projeto Jovem Pesquisador em Centros Emergentes da Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP, http://fapespbr/en/) [09/09532–0 to TK]; Edital Universal do Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) [473660/2013–0 to TK, 470120/2009–6 to TK, 476724/2013–9 to RZNV]; Fundação de Apoio ao Ensino, Pesquisa e Assistência do Hospital das Clínicas da Faculdade de Medicina de Ribeirão Preto da Universidade de São Paulo (FAEPA) [1640/2009 to TK]; Núcleo de Pesquisa em Ciência Genômica (NAP-CG) da Universidade de São Paulo; and fellowships FAPESP [11/07487–7 to LSZ, 2013/21522–5 to JVGF, 10/10195–5 to VCSI].
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 2007; 8:413-23; PMID:17486121; http://dx.doi.org/ 10.1038/nrg2083 [DOI] [PubMed] [Google Scholar]
- 2.Sorek R, Cossart P. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 2010; 11:9-16; PMID:19935729; http://dx.doi.org/ 10.1038/nrg2695 [DOI] [PubMed] [Google Scholar]
- 3.Pelechano V, Steinmetz LM. Gene regulation by antisense transcription. Nat Rev Genet [Internet] 2013. [cited 2014 Jul 14]; 14:880-93. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24217315; http://dx.doi.org/ 10.1038/nrg3594 [DOI] [PubMed] [Google Scholar]
- 4.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol 2007; 10:102-9; PMID:17387036; http://dx.doi.org/ 10.1016/j.mib.2007.03.012 [DOI] [PubMed] [Google Scholar]
- 5.Zaramela LS, Vencio RZN, ten-Caten F, Baliga NS, Koide T. Transcription start site associated RNAs (TSSaRNAs) are ubiquitous in all domains of life. PLoS One 2014; 9:e107680; PMID:25238539; http://dx.doi.org/ 10.1371/journal.pone.0107680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonneau R, Facciotti MT, Reiss DJ, Schmid AK, Pan M, Kaur A, Thorsson V, Shannon P, Johnson MH, Bare JC, et al.. A predictive model for transcriptional control of physiology in a free living cell. Cell [Internet] 2007. [cited 2011 Jul 22]; 131:1354-65. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18160043; PMID:18160043; http://dx.doi.org/ 10.1016/j.cell.2007.10.053 [DOI] [PubMed] [Google Scholar]
- 7.Brooks AN, Reiss DJ, Allard A, Wu W-J, Salvanha DM, Plaisier CL, Chandrasekaran S, Pan M, Kaur A, Baliga NS. A system-level model for the microbial regulatory genome. Mol Syst Biol 2014; 10:740; PMID:25028489; http://dx.doi.org/ 10.15252/msb.20145160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koide T, Reiss DJ, Bare JC, Pang WL, Facciotti MT, Schmid AK, Pan M, Marzolf B, Van PT, Lo F-Y, et al.. Prevalence of transcription promoters within archaeal operons and coding sequences. Mol Syst Biol [Internet] 2009. [cited 2011 Aug 12]; 5:285. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2710873&tool=pmcentrez&rendertype=abstract; PMID:19536208; http://dx.doi.org/ 10.1038/msb.2009.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turkarslan S, Reiss DJ, Gibbins G, Su WL, Pan M, Bare JC, Plaisier CL, Baliga NS. Niche adaptation by expansion and reprogramming of general transcription factors. Mol Syst Biol 2011; 7:554; PMID:22108796; http://dx.doi.org/ 10.1038/msb.2011.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wurtmann EJ, Ratushny AV, Pan M, Beer KD, Aitchison JD, Baliga NS. An evolutionarily conserved RNase-based mechanism for repression of transcriptional positive autoregulation. Mol Microbiol 2014; 92:369-82; PMID:24612392; http://dx.doi.org/ 10.1111/mmi.12564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beer KD, Wurtmann EJ, Pinel NN, Baliga NS. Model organisms retain an “ecological memory” of complex ecologically relevant environmental variation. Appl Environ Microbiol [Internet] 2014. [cited 2014 Jul 31]; 80:1821-31. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24413600; PMID:24413600; http://dx.doi.org/ 10.1128/AEM.03280-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Filee J, Siguier P, Chandler M, Filée J, Siguier P, Chandler M. Insertion sequence diversity in archaea. Microbiol Mol Biol Rev [Internet] 2007. [cited 2013 Mar 31]; 71:121-57. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1847376&tool=pmcentrez&rendertype=abstract; PMID:17347521; http://dx.doi.org/ 10.1128/MMBR.00031-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev [Internet] 2014. [cited 2014 Jul 15]; 38(5):865-91. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24499397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ton-Hoang B, Guynet C, Ronning DR, Cointin-Marty B, Dyda F, Chandler M. Transposition of ISHp608, member of an unusual family of bacterial insertion sequences. EMBO J 2005; 24:3325-38; PMID:16163392; http://dx.doi.org/ 10.1038/sj.emboj.7600787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barabas O, Ronning DR, Guynet C, Hickman AB, Ton-Hoang B, Chandler M, Dyda F. Mechanism of IS200/IS605 family DNA transposases: activation and transposon-directed target site selection. Cell 2008; 132:208-20; PMID:18243097; http://dx.doi.org/ 10.1016/j.cell.2007.12.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hickman AB, James JA, Barabas O, Pasternak C, Ton-Hoang B, Chandler M, Sommer S, Dyda F. DNA recognition and the precleavage state during single-stranded DNA transposition in D. radiodurans. EMBO J 2010; 29:3840-52; PMID:20890269; http://dx.doi.org/ 10.1038/emboj.2010.241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pasternak C, Ton-Hoang B, Coste G, Bailone A, Chandler M, Sommer S. Irradiation-Induced Deinococcus radioduransGenome Fragmentation Triggers Transposition of a Single Resident Insertion Sequence. PLoS Genet [Internet] 2010; 6:e1000799. Available from: http://dx.doi.org/10.1371/journal.pgen.1000799; PMID:20090938; http://dx.doi.org/ 10.1371/journal.pgen.1000799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kersulyte D, Kalia A, Zhang M, Lee H-K, Subramaniam D, Kiuduliene L, Chalkauskas H, Berg DE. Sequence Organization and Insertion Specificity of the Novel Chimeric ISHp609 Transposable Element of Helicobacter pylori. J Bacteriol [Internet] 2004; 186:7521-8. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC524915/; PMID:15516563; http://dx.doi.org/ 10.1128/JB.186.22.7521-7528.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasternak C, Dulermo R, Ton-Hoang B, Debuchy R, Siguier P, Coste G, Chandler M, Sommer S. ISDra2 transposition in Deinococcus radiodurans is downregulated by TnpB. Mol Microbiol 2013; 88:443-55; PMID:23461641; http://dx.doi.org/ 10.1111/mmi.12194 [DOI] [PubMed] [Google Scholar]
- 20.Zago MA, Dennis PP, Omer AD. The expanding world of small RNAs in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Microbiol [Internet] 2005. [cited 2013 May 27]; 55:1812-28. Available from: http://www.ncbi.nlm.nih.gov/pubmed/15752202; PMID:15752202; http://dx.doi.org/ 10.1111/j.1365-2958.2005.04505.x [DOI] [PubMed] [Google Scholar]
- 21.Jager D, Forstner KU, Sharma CM, Santangelo TJ, Reeve JN. Primary transcriptome map of the hyperthermophilic archaeon Thermococcus kodakarensis. BMC Genomics 2014; 15:684; PMID:25127548; http://dx.doi.org/ 10.1186/1471-2164-15-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phok KK, Moisan A, Rinaldi D, Brucato N, Carpousis AJ, Gaspin C, Clouet-d'Orval BB. Identification of CRISPR and riboswitch related RNAs among novel noncoding RNAs of the euryarchaeon Pyrococcus abyssi. BMC Genomics [Internet] 2011. [cited 2011 Aug 8]; 12:312. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3124441&tool=pmcentrez&rendertype=abstract; PMID:21668986; http://dx.doi.org/ 10.1186/1471-2164-12-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 2006; 34:D32-6; PMID:16381877; http://dx.doi.org/ 10.1093/nar/gkj014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandler M, Kichenaradja P, Siguier P, Pe J, Pérochon J, Chandler M. ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res [Internet] 2010. [cited 2014 Jul 29]; 38:D62-8. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2808865&tool=pmcentrez&rendertype=abstract; PMID:19906702; http://dx.doi.org/ 10.1093/nar/gkp947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng WV, Kennedy SP, Mahairas GG, Berquist B, Pan M, Shukla HD, Lasky SR, Baliga NS, Thorsson V, Sbrogna J, et al.. Genome sequence of Halobacterium species NRC-1. Proc Natl Acad Sci U S A [Internet] 2000; 97:12176-81. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=311145&tool=pmcentrez&rendertype=abstract; PMID:11016950; http://dx.doi.org/ 10.1073/pnas.190337797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfeiffer F, Schuster SC, Broicher A, Falb M, Palm P, Rodewald K, Ruepp A, Soppa J, Tittor J, Oesterhelt D. Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics 2008; 91:335-46; PMID:18313895; http://dx.doi.org/ 10.1016/j.ygeno.2008.01.001 [DOI] [PubMed] [Google Scholar]
- 27.Shao W, Price MN, Deutschbauer AM, Romine MF, Arkin AP. Conservation of transcription start sites within genes across a bacterial genus. MBio 2014; 5:e01398-14; PMID:24987095; http://dx.doi.org/ 10.1128/mBio.01398-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puton T, Kozlowski LP, Rother KM, Bujnicki JM. CompaRNA: a server for continuous benchmarking of automated methods for RNA secondary structure prediction. Nucleic Acids Res 2013; 41:4307-23; PMID:23435231; http://dx.doi.org/ 10.1093/nar/gkt101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dyall-Smith ML, Pfeiffer F, Klee K, Palm P, Gross K, Schuster SC, Rampp M, Oesterhelt D. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One [Internet] 2011. [cited 2013 Apr 30]; 6:e20968. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3119063&tool=pmcentrez&rendertype=abstract; PMID:21701686; http://dx.doi.org/ 10.1371/journal.pone.0020968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bertels F, Rainey PB. Curiosities of REPINs and RAYTs. Mob Genet Elements [Internet] 2011; 1:262-8. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3337134/; PMID:22545236; http://dx.doi.org/ 10.4161/mge.18610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klein RJ, Misulovin Z, Eddy SR. Noncoding RNA genes identified in AT-rich hyperthermophiles. Proc Natl Acad Sci U S A [Internet] 2002; 99:7542-7. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=124278&tool=pmcentrez&rendertype=abstract; PMID:12032319; http://dx.doi.org/ 10.1073/pnas.112063799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Delihas N. Stem loop sequences specific to transposable element IS605 are found linked to lipoprotein genes in Borrelia plasmids. PLoS One [Internet] 2009. [cited 2014 Jul 28]; 4:e7941. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2775950&tool=pmcentrez&rendertype=abstract; PMID:19936201; http://dx.doi.org/ 10.1371/journal.pone.0007941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bare JC, Koide T, Reiss DJ, Tenenbaum D, Baliga NS. Integration and visualization of systems biology data in context of the genome. BMC Bioinformatics 2010; 11:382; PMID:20642854; http://dx.doi.org/ 10.1186/1471-2105-11-382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The Vienna RNA Websuite. Nucleic Acids Res [Internet] 2008; 36:W70-4. Available from: http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2447809/; PMID:18424795; http://dx.doi.org/ 10.1093/nar/gkn188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zakov S, Goldberg Y, Elhadad M, Ziv-Ukelson M. Rich parameterization improves RNA structure prediction. J Comput Biol 2011; 18:1525-42; PMID:22035327; http://dx.doi.org/ 10.1089/cmb.2011.0184 [DOI] [PubMed] [Google Scholar]
- 36.Dawson WK, Fujiwara K, Kawai G. Prediction of RNA Pseudoknots Using Heuristic Modeling with Mapping and Sequential Folding. PLoS One [Internet] 2007; 2:e905. Available from: http://dx.plos.org/10.1371/journal.pone.0000905; PMID:17878940; http://dx.doi.org/ 10.1371/journal.pone.0000905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bindewald E, Kluth T, Shapiro BA. CyloFold: secondary structure prediction including pseudoknots. Nucleic Acids Res 2010; 38:W368-72; PMID:20501603; http://dx.doi.org/ 10.1093/nar/gkq432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gerhardt P. Methods for General and Molecular Bacteriology [Internet]. American Society for Microbiology; 1994. Available from: http://books.google.co.in/books?id=fGYXAQAAIAAJ [Google Scholar]
- 39.Abdelrahman YM, Rose LA, Belland RJ. Developmental expression of non-coding RNAs in Chlamydia trachomatis during normal and persistent growth. Nucleic Acids Res 2011; 39:1843-54; PMID:21051342; http://dx.doi.org/ 10.1093/nar/gkq1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schaefer BC. Revolutions in rapid amplification of cDNA ends: new strategies for polymerase chain reaction cloning of full-length cDNA ends. Anal Biochem 1995; 227:255-73; PMID:7573945; http://dx.doi.org/ 10.1006/abio.1995.1279 [DOI] [PubMed] [Google Scholar]
- 41.Facciotti MT, Reiss DJ, Pan M, Kaur A, Vuthoori M, Bonneau R, Shannon P, Srivastava A, Donohoe SM, Hood LE, et al.. General transcription factor specified global gene regulation in archaea. Proc Natl Acad Sci U S A [Internet] 2007; 104:4630-5. Available from: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1838652&tool=pmcentrez&rendertype=abstract; PMID:17360575; http://dx.doi.org/ 10.1073/pnas.0611663104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robb FT, DasSarma S, Fleischmann EM. Archaea: a laboratory manual. Halophiles [Internet]. Cold Spring Harbor Laboratory; 1995. Available from: http://books.google.com.br/books?id=sYvjSAAACAAJ [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.