Abstract
Short hairpin RNAs (shRNAs) are widely used for gene knockdown by inducing the RNA interference (RNAi) mechanism. The shRNA precursor is processed by Dicer into small interfering RNAs (siRNAs) and subsequently programs the RNAi-induced silencing complex (RISC) to find a complementary target mRNA (mRNA) for post-transcriptional gene silencing. Recent evidence indicates that shRNAs with a relatively short basepaired stem bypass Dicer to be processed directly by the Ago2 nuclease of the RISC complex. We named this design AgoshRNA as these molecules depend on Ago2 both for processing and subsequent silencing activity. This alternative AgoshRNA processing route yields only a single active RNA strand, an important feature to restrict off-target effects induced by the passenger strand of regular shRNAs. It is therefore important to understand this novel AgoshRNA processing route in mechanistic detail such that one can design the most effective and selective RNA reagents. We performed a systematic analysis of the optimal base pair (bp) composition at the top and bottom of AgoshRNA molecules. In this study, we document the importance of the 5′ end nucleotide (nt) and a bottom mismatch. The optimized AgoshRNA design exhibits improved RNAi activity across cell types. These results have important implications for the future design of more specific RNAi therapeutics.
Keywords: agoshRNA, argonaute2, dicer, RNA processing, shRNA
Introduction
The RNAi mechanism is widely conserved among vertebrates and invertebrates and uses microRNAs (miRNAs) to control cellular gene expression. Recent studies have reported an increasing number of miRNA classes that use alternative processing routes. The canonical route for miRNA processing employs the nuclear Drosha enzyme to convert the primary miRNA transcript into the precursor miRNA and subsequently the cytoplasmic Dicer endonuclease to create the mature miRNA duplex. One strand of this miRNA duplex is preferentially loaded into the Ago protein to form the RISC complex that targets partially complementary mRNAs for translational suppression and/or destruction. Additional layers of complexity were added to miRNA-regulated gene expression by the recent description of non-canonical RNAi pathways, most notable Drosha-independent RNA classes like “mirtrons” and tRNA precursors,1-3 and Dicer-independent miRNAs of which miR-451 is the prototype.4 We will discuss the latter miR-451 class in more detail as it relates to recent developments with man-made shRNA reagents.5
Although Dicer is essential for the processing of most miRNAs, some notable exceptions have recently been reported. The first indication came from studies showing that RISC does accommodate pre-miRNAs in the absence of Dicer.6,7 A direct involvement of the Ago2 protein in processing of Dicer-independent miRNAs was suggested by the specific disappearance of miR-451 in mice with a genetic Ago2 defect.8 This miRNA is well conserved among vertebrates and has some unusual features, most prominently the short stem of only 17 bp that seems too short to facilitate Dicer binding.9 It was suggested that pre-miR-451 is processed instead by Ago2, which could explain the unusual point of cleavage halfway the 3′ arm of the duplex. Subsequently, the miRNA has to unfold to allow binding to a complementary mRNA and Ago2-mediated silencing. This means that Ago2 plays a dual role in the processing and gene silencing activity of this miRNA class. Additional miRNA candidates that are processed by Ago2 were reported.10 For miR-451, the critical importance of the short stem length for routing toward Ago2 instead of Dicer was confirmed in a detailed mutational analysis.11 Additional characteristic features include a small hairpin loop of 4 nt and G-U as the top bp.12
ShRNA molecules can be synthesized in the cell from a transgene plasmid or vector and enter the RNAi pathway at the Dicer level. The first evidence for Dicer-independent shRNA processing came from studies on synthetic shRNA molecules that are transfected directly into cells. A new class of short shRNAs with a stem region of only 16–19 bp was described, but it remained unknown how and by whom these molecules are processed.13,14 We described transgenes encoding a specific shRNA design with a short stem length and small loop that triggered this alternative processing pathway.15 Processing was blocked in a cell line encoding a catalytically inactive Ago2 mutant and sequencing mapped the actual cleavage site halfway the 3′ arm between bp 10 and 11 of the shRNA duplex. All these characteristics are consistent with a role for Ago2 in processing of this special shRNA class that we therefore termed AgoshRNAs. Inspired by miR-451 findings, we subsequently described a modulating role for the top G-U bp in AgoshRNA activity.16 We now performed a more systematic analysis of the optimal nucleotide and bp composition at the top and bottom of AgoshRNA molecules.
Results
The experimental system
Figure 1 depicts regular Dicer and alternative Ago2-mediated processing for shRNA and AgoshRNA substrates, which results in the production of different short RNA molecules that trigger an RNAi response. In analogy to miR-451, AgoshRNA molecules are typically shorter (17–19 bp) than regular shRNAs (≥20 bp) and prefer a small loop of 3–6 nt.15 Regular Dicer-cleavage of both strands of the basepaired stem near the loop region generates the small interfering RNA (siRNA), consisting of 2 strands of approximately 21 nt, marked as black arrow for the guide strand and white arrow for the passenger strand (Fig. 1A). Activity of these strands can be scored by silencing of the matching Luc-sense or Luc-antisense reporters, respectively (Fig. 1B). Ago2-mediated cleavage on the 3′ side of the hairpin between bp 10 and 11 generates a single guide RNA molecule of approximately 33 nt (gray arrow) that exclusively targets the Luc-antisense reporter.
Figure 1.
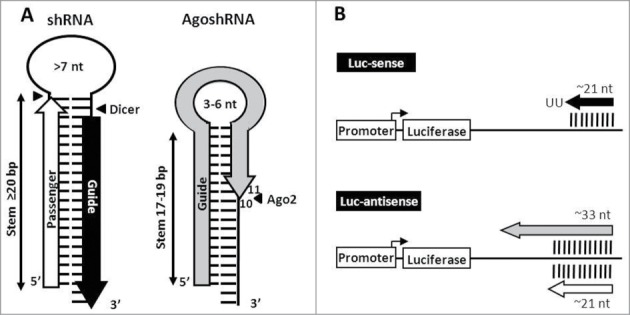
Characteristics of the traditional shRNA and novel AgoshRNA design. (A) Secondary structure of a shRNA and AgoshRNA molecule. The shRNA is processed by Dicer (▸◂) into an siRNA duplex of ≥20 bp with 3′ overhangs that is loaded into RISC. The passenger strand (white arrow) is cleaved and subsequently degraded, the guide strand (black arrow) is active in RNAi-silencing. The shorter AgoshRNA duplex (stem 17–19 bp) is not recognized by Dicer and consequently processed by Ago2. The AgoshRNA duplex is cleaved (◂) on the 3′ side between bp 10 and 11 to yield a single RNA molecule of ∼33 nt (gray arrow) that instructs Ago2 for RNAi-silencing. (B) Luc reporter constructs with sense and antisense target sequences. The Luc-sense reporter scores canonical shRNA guide activity, the Luc-antisense reporter scores both shRNA passenger and AgoshRNA activity.
A recent mutational analysis indicated that the length of the basepaired stem is a major determinant for shRNA activity via the regular Dicer route versus AgoshRNA activity via the non-canonical Ago2 route.15 However, other structural elements or sequence motifs may also influence the pathway selection. In this study, we zoom in on the identity of the top and bottom bp in AgoshRNAs as candidate determinants for Dicer-independent processing.
Toward the optimal top of an AgoshRNA molecule
We recently suggested a modulating role of a weak top G-U bp in a set of AgoshRNA mutants ranging from 15 to 23 bp, similar to recent miR-451 findings.12,16 Replacing the top Watson-Crick bp by a weak G-U bp seems to improve AgoshRNA activity. However, only a small AgoshRNA activity window was observed for molecules in the size range of 17–20 bp, and the most prominent knockdown was scored for AgoshRNA molecules of 19 bp. In this study, a more systematic analysis of the 2 top bp in AgoshRNA molecules of 19 bp was performed. Variants were made to test whether the G-U improvement holds true also for the reverse U-G pair and for the penultimate bp position. Thus, a total of 6 variants were made for 2 molecules, AgoshRT5 and AgoshPol47, both with the optimal stem/loop 19/5 configuration (Fig. 2). We introduced a top U-G (Mut1) or G-U (Mut4) or a double bp change (Mut2, Mut3, Mut5 and Mut6). Mut 2 and 3 were previously tested in a 21/5 setting (as Mut6 and Mut7, respectively).16 We used 2 luciferase reporters with complementary target sequences to measure shRNA and AgoshRNA silencing activity. Please note that the Luc-antisense reporter detects AgoshRNA activity but also potential passenger strand activity of the regular Dicer-route (Fig. 1B). To determine the knockdown abilities of the different AgoshRNA variants, we performed transfection series in HEK293T cells and the AgoshRNA molecule was titrated (1, 5 and 25 ng; Fig. 3). A fixed amount of renilla luciferase plasmid was included as control for the transfection efficiency. An unrelated shRNA (shNef) served as negative control for which the activity was set at 100%. Regular shRNA molecules with stem length of 21 bp and 5 nt loop (21/5) were included as positive controls. We will first describe the Dicer-route on the Luc-sense reporter and then the Ago2-route on the Luc-antisense reporter for the shRT5 inhibitor.
Figure 2.
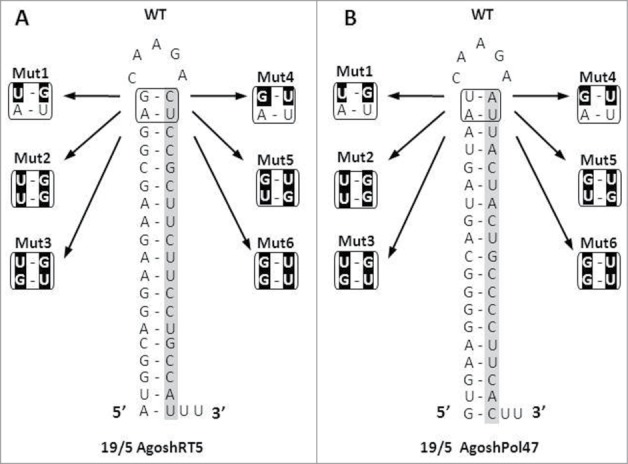
Design of AgoshRNAs with variation in the 2 top base pairs. AgoshRT5 and AgoshPol47 with 19 bp stem and 5 nt loop (19/5) were used as wild-type (WT) backbones. The encoded guide sequence is boxed. The top bp was modified (mutated bp in black).
Figure 3.
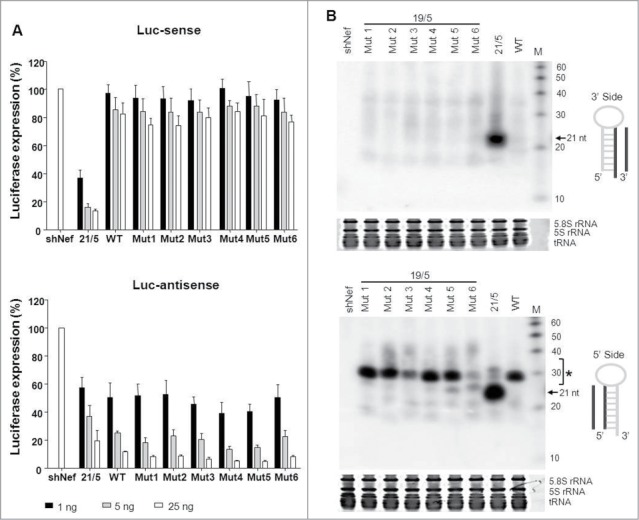
Knockdown activity and processing of AgoshRT5 variants. (A) The knockdown activity of the different AgoshRNA variants was determined by co-transfection with a luciferase reporter containing either the sense- or antisense-target sequence. HEK293T cells were co-transfected with 100 ng of the respective firefly luciferase reporter plasmid, 1 ng renilla luciferase plasmid as internal control and 1, 5 or 25 ng of the corresponding shRNA construct. An unrelated shRNA (shNef) served as negative control, this activity was set at 100% luciferase expression. We performed 3 independent transfections, each in duplicate, and standard deviations were calculated. (B) Processing of the 3′ strand (upper panel) and 5′ strand (lower panel) of the AgoshRT5 variants was analyzed by RNA gel blot. The AgoshRNAs varied in the 2 top bp. HEK293T cells were transfected with 5 μg of the indicated constructs. Size markers were included in the far right lane (length indicated in nt). An unrelated shRNA (shNef) was included as negative control. The regular shRNA ∼21 nt products are marked and * indicates the AgoshRNA ∼33 nt products.
The control 21/5 shRT5 construct exhibited good silencing activity on the Luc-sense reporter, with luciferase levels dropping to <20% with 25 ng of the inhibitor construct (Fig. 3A, upper part). The 19/5 AgoshRT5 wild-type (WT) exhibited much less Luc-sense silencing, which was expected because this design has poor shRNA activity. Mutating the top bp in G-U variants did not improve silencing activity (Mutants 1 to 6). Next, all constructs were tested in combination with the Luc-antisense reporter that scores both AgoshRNA activity and potential passenger strand activity of the regular Dicer-route. Some activity was apparent for the control 21/5, suggesting passenger strand activity as previously observed.15,16 WT exhibited enhanced knockdown activity, and mutants 1 to 6 marginally improved the AgoshRNA activity.
Northern blot analysis was performed to analyze processing of the different 19/5 AgoshRT5 variants (Fig. 3B). We used LNA oligonucleotides to detect products from the 3′ side (upper panel) and 5′ side (lower panel). An unrelated shRNA (shNef) was used as negative control. Consistent with the luciferase knockdown data, we did not observe 3′ strand siRNA production for the 19/5 variants. A prominent ∼21 nt RNA fragment was only observed for the positive control 21/5. A clear product, with the size of a typical AgoshRNA (∼33 nt), is visible when the 5′ strand of the 19/5 constructs was analyzed on a northern blot. This indicates that these variants bypass Dicer to be processed by Ago2. The RNA gel blot revealed some differences in the intensity of the ∼33 nt product among mutants 1 to 6. In particular, mutant 3 and 6 produced less ∼33 nt fragment than the other mutants and WT, but this did not result in reduced knockdown activity. We currently do not fully understand this difference, but one should realize that the guide strands will differ in 1 or 2 nt positions from those of the WT hairpins, which may affect their mRNA-annealing capacity. Please also note that the probe-annealing capacity also differs for the WT vs. mutant guides, such that one should not use the northern blot data in a quantitative manner.
To study whether this top bp effect is a general phenomenon that also applies to other AgoshRNAs with a completely different sequence, we tested the same G-U combinations in the context of the 19/5 AgoshPol47 construct. The control 21/5 shRNA showed good silencing activity on the Luc-sense reporter and exhibited very little activity on Luc-antisense (Fig. 4A), indicating that the 3′ strand is indeed selected as guide by Ago2. The WT 19/5 construct exhibited moderate shRNA activity on the Luc-sense reporter despite the suboptimal stem length of 19 bp. Mutants 1 to 6 lost most shRNA activity (Fig. 4A, upper panel). Possibly, the weak top G-U bp may open transiently to present a stem of 18 or only 17 bp, thus frustrating Dicer recognition. However, this did not coincide with a gain of AgoshRNA activity for mutants 1 to 6 (Fig. 4A, lower panel). Thus, the G-U effects seem to vary per inhibitory molecule.
Figure 4.
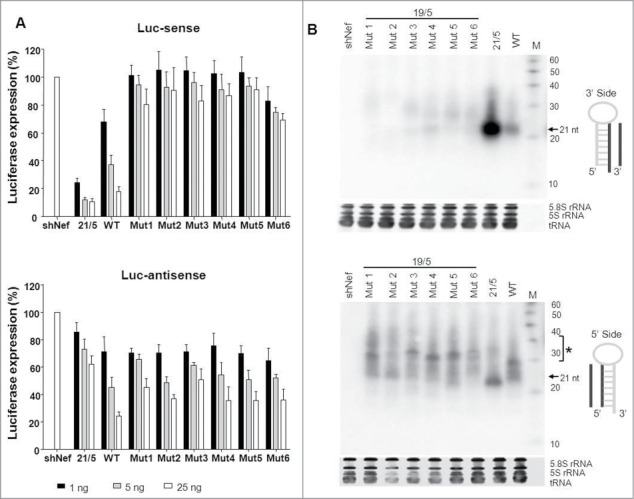
Knockdown activity and processing of AgoshPol47 variants. (A) The knockdown activity of the guide strand on Luc-sense (upper panel) and passenger strand on Luc-antisense (lower panel) of the AgoshPol47 was determined by co-transfection of a luciferase reporter encoding the sense and antisense target sequence, respectively, in HEK293T cells. We performed 3 independent transfections, each in duplicate, and standard deviations were calculated. (B) Total RNA was analyzed by northern blot for processing products derived from the 3′ strand (upper panel) and 5′ strand (lower panel). Size markers are indicated on the right. The regular shRNA ∼21 nt products are marked and * indicates the AgoshRNA ∼30 nt products. See Figure 3 for more details.
We next investigated processing of these AgoshPol47 variants by RNA gel blot analysis. Consistent with the luciferase knockdown data, we observed the expected ∼21 nt fragment for the positive control 21/5 with the 3′ side probe (Fig. 4B, upper panel). A less prominent fragment was apparent for WT 19/5, whereas lack of 3′ strand siRNA production was observed for mutants 1 to 6. We observed modest ∼33 nt RNA fragments with the 5′ side probe (Fig. 4B, lower panel). Some differences in size and amount of the ∼33 nt product were apparent for the mutants. Mutant 2 and 6 produced less prominent ∼33 nt fragments, although no significant impact on the knockdown activity was measured. We currently do not understand the subtle size differences. These combined results indicate that a top G-U bp can influence AgoshRNA processing and activity, but the effect varies per shRNA molecule and is modest.
Toward the optimal bottom of an AgoshRNA molecule
There is accumulating evidence that the AgoshRNA pathway is very similar or even identical to that of miR-451. One of the hallmarks of miR-451 is that it starts with 5′ A instead of 5′ U that is more common in other miRNAs. In fact, this A remains unpaired at the bottom of the stem (A·C mismatch below a bottom A-U bp). Inspired by recent miR-451 findings we transplanted both the A·C and U·C mismatches that proved optimal for miR-451 activity onto AgoshRNA molecules.11 To study the general value of such manipulations, we again tested 2 inhibitors (Fig. 5, RT5 and Pol47) in both the shRNA (21/5) and AgoshRNA (19/5) context. The bottom bp (A-U for RT5, G-C for Pol47) was replaced with these mismatches.
Figure 5.
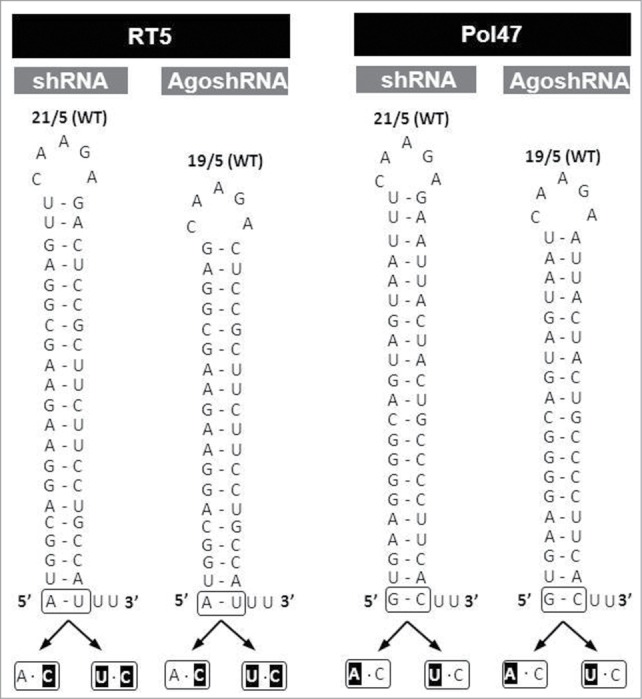
Design of shRNA and AgoshRNA mutants with variation in the bottom bp. shRNAs (21/5) and AgoshRNAs (19/5) based on the RT5 and Pol47 inhibitors were used as WT backbones. The bottom bp was substituted by the mismatches A·C and U·C (mutated nt in black).
The activity of the regular shRNA guide on the Luc-sense reporter was measured (Fig. 6, upper panel). Activity scored in the presence of the unrelated shNef was set at 100%. The 21/5 and 19/5 WT molecules were included as controls. Both 21/5 WT shRNA constructs (RT5 and Pol47) demonstrated good shRNA activity, with luciferase levels dropping to <20% with 25 ng of the inhibitory construct. A minor loss of activity was observed for the A·C variants, but a pronounced decrease in activity was measured for the U·C variants. The 19/5 WT RT5 exhibited no Luc-sense silencing activity, consistent with the AgoshRNA profile, and no effect was apparent for the A·C and U·C mutants. The 19/5 WT Pol47 showed good Luc-sense silencing activity, but this shRNA activity was lost for the A·C and U·C variants. The length of the basepaired stem is a major determinant for shRNA versus AgoshRNA activity and a mismatch (A·C and U·C) in place of the bottom bp shortens the molecule by 1 bp. Reduced stem length may thus explain the loss of shRNA activity observed for 21/5 and 19/5 on Luc-sense as the shRNA may be more prone to Ago2-cleavage. However, it does not explain the differential loss of shRNA activity for the A·C vs. the U·C mutant in the 21/5 context.
Figure 6.
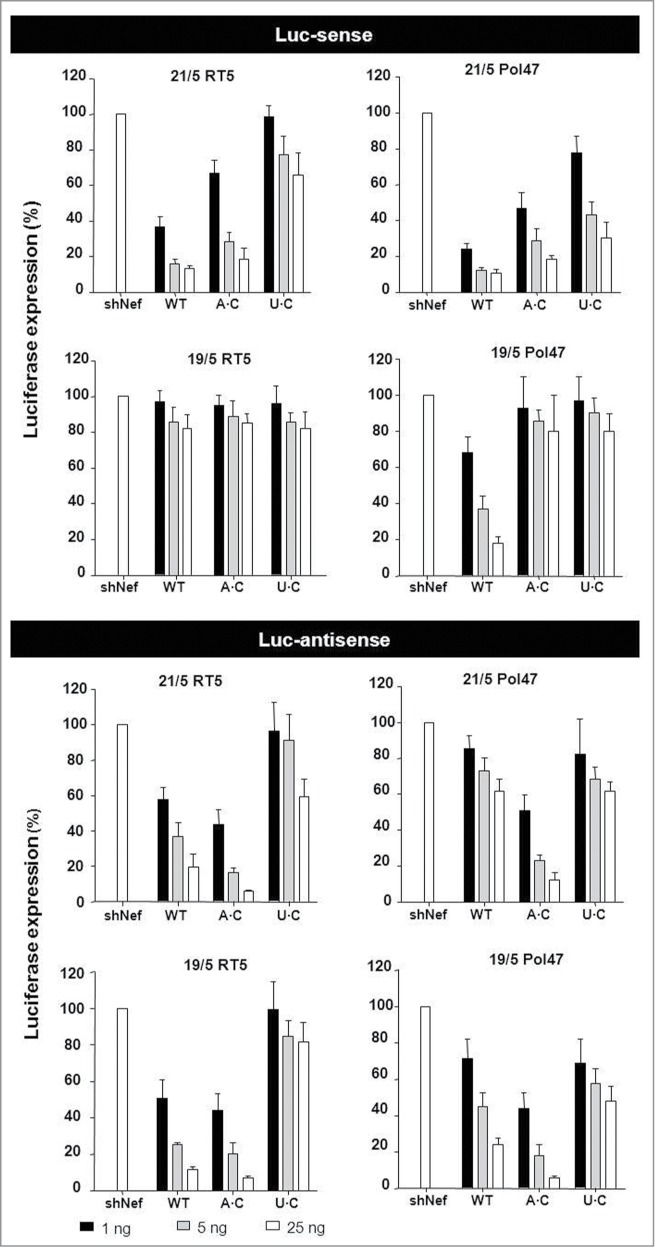
Knockdown activity of the A·(C) and U·(C) AgoshRNA variants. The knockdown activity of the guide strand on Luc-sense (upper panel) and the passenger strand on Luc-antisense (lower panel) was determined by co-transfection of a luciferase reporter encoding the sense (top) and antisense (bottom) target sequence in HEK293T cells. We performed 3 independent transfections, each in duplicate, and standard deviations were calculated. See Figure 3 for additional details.
We used Luc-antisense reporters to score non-canonical AgoshRNA activity (Fig. 6, lower panel). The A·C mutants showed a remarkable increase in AgoshRNA activity compared to the WT shRNAs. This effect was observed for both molecules (RT5 and Pol47) in both contexts (21/5 and 19/5). Thus, the A·C configuration causes a loss of shRNA activity (Fig. 6 upper panel, except for the inactive 19/5 RT5) and a concomitant gain of AgoshRNA activity (Fig. 6, lower panel). In contrast, the U·C variants did not improve silencing activity compared to WT in the AgoshRNA test (Pol47 hairpins 21/5 and 19/5) or lost most activity (RT5 hairpins 21/5 and 19/5). Thus, opening the bottom bp can have profound effects, but only the A·C variants do consistently yield increased AgoshRNA activity.
Northern blot analysis was performed to analyze RNA processing (Fig. 7). Please note that the actual lengths of the predicted Dicer- and Ago-cleavage products vary according to the actual stem length. Thus, 21/5 variants yield RNA fragments of 21 (Dicer) or 37 nt (Ago2) and 19/5 variants yield fragments of 19 (Dicer) and 33 nt (Ago2). First, the RT5 results obtained with the 3′ and 5′ probes will be discussed (Fig. 7A and 7B, respectively). The siRNAs derived from the 3′ side of the shRNA were observed (∼21 nt) for WT 21/5 shRT5 but a less prominent band is visible for the A·C variant and especially the U·C variant (Fig. 7A, compare lanes 2, 4 and 5). This pattern correlates with the reduced luciferase knockdown activity. The 19/5 AgoshRT5 showed the expected loss of the 3′ side shRNA fragment and appearance of the ∼33 nt 5′ side fragment (Fig. 7A and 7B, respectively, lanes 3, 6 and 7). Most importantly, we observed the typical Ago2 products (∼37 nt) for the 21/5 A·C variant, but not the U·C variant (Fig. 7B, lanes 4 and 5). This result seems to confirm that the increased Luc-antisense knockdown activity scored for the 21/5 A·C variant is due to the shift from Dicer to Ago2 processing. The prominent ∼33 nt AgoshRNA of 19/5 was increased for the A·C variant and reduced for the U·C variant (Fig. 7B, lanes 6 and 7), fully consistent with the luciferase silencing data.
Figure 7.
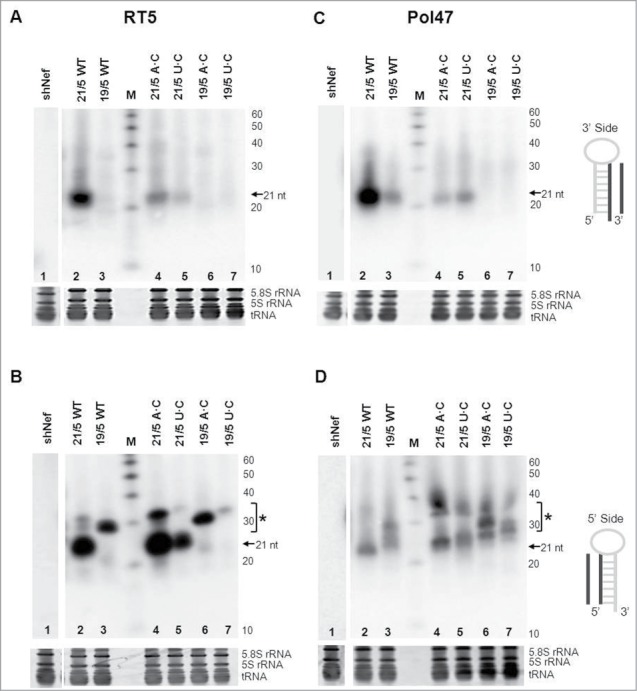
Processing of shRNAs and AgoshRNAs is influenced by the bottom base pair. (A, B) RT5 and (C, D) Pol47 variants were analyzed by RNA gel blot. HEK293T cells were transfected with 5 μg of the corresponding RNA constructs. The shRNAs and AgoshRNAs (21/5 and 19/5) varied in bottom bp (WT is A-U for RT5 and G-C for Pol47, and mutants A·C or U·C). Total RNA was isolated and analyzed by northern blot using an LNA probe to detect processing products derived from the 3′ side (A, C) and the 5′ side (B, D). Size markers were included (length indicated in nt). An unrelated shRNA (shNef) was included as negative control. The regular shRNA ∼21 nt products are marked and * indicates the AgoshRNA ∼30 nt products.
A globally similar pattern was observed for the 3′ side of Pol47 (Fig. 7C). The abundant siRNAs (∼21 nt) made by 21/5 WT were much reduced for the A·C and U·C variants (compare lanes 2, 4 and 5). The AgoshRNA design 19/5 produces much less siRNAs from the 3′ side (lane 3), but this signal remains absent for the 2 mismatch variants (lanes 6 and 7). This complete loss of the 3′ signal correlates with the nearly complete loss of knockdown activity on the Luc-sense reporter (Fig. 6). A low level of ∼21 nt RNA fragment was detected for 21/5 WT with the 5′ side probe, reflecting the passenger strand (Fig. 7D, lane 2). Interestingly, a new and abundant AgoshRNA fragment of ∼37 nt was observed for the A·C mutant, although the ∼21 nt shRNA band was still present (Fig. 7D, lane 4). The AgoshRNA fragments (∼37 nt) made by the 21/5 U·C variant were reduced compared to the A·C mutant. The 19/5 variants produce smaller RNA bands that may represent 3′-trimmed products as described for miR-451.17 A deep sequencing analysis may be needed to describe this phenomenon in more detail. Again, the A·C mutant generates more product than the U·C variant (Fig. 7D, lanes 6 and 7). Overall, the A·C variation seems to provide a significant and general improvement of the AgoshRNA design. Because only 2 AgoshRNA templates were tested in this study, future experimentation should reveal the general value of this new design rule.
General AgoshRNA improvement in a cell type-independent manner
To investigate whether the A·C optimization of the AgoshRNA design is apparent in other cell types, we tested the set of Pol47 inhibitors in C33A and Vero cells. C33A is a human cervical cancer cell line and Vero is an African green monkey kidney-derived cell line. These cell lines are not related to the human embryonic kidney 293T cells (HEK293T) used in our initial tests. Cells were co-transfected with the appropriate reporter plasmid and the shRNA/AgoshRNA constructs were titrated (Fig. 8). Similar luciferase knockdown profiles were scored in the C33A and Vero cells when compared to HEK293T cells. In other words, a decrease in activity was apparent for the A·C and U·C variants on the Luc-sense reporter (Fig. 8, upper panel) and the highest knockdown activity on the Luc-antisense reporter was observed for the A·C variants (Fig. 8, lower panel). The activity of the A·C variant was improved from around 50% to more than 90% knockdown efficiency for 21/5 shRNAs and from around 70% to more than 90% inhibition for 19/5 AgoshRNAs (both measured with 25 ng input shRNA). The U·C variant generally showed reduced knockdown activity compared to WT in all cell types. These results confirm the unique properties of the A·C variant and demonstrate a general improvement of AgoshRNA activity.
Figure 8.
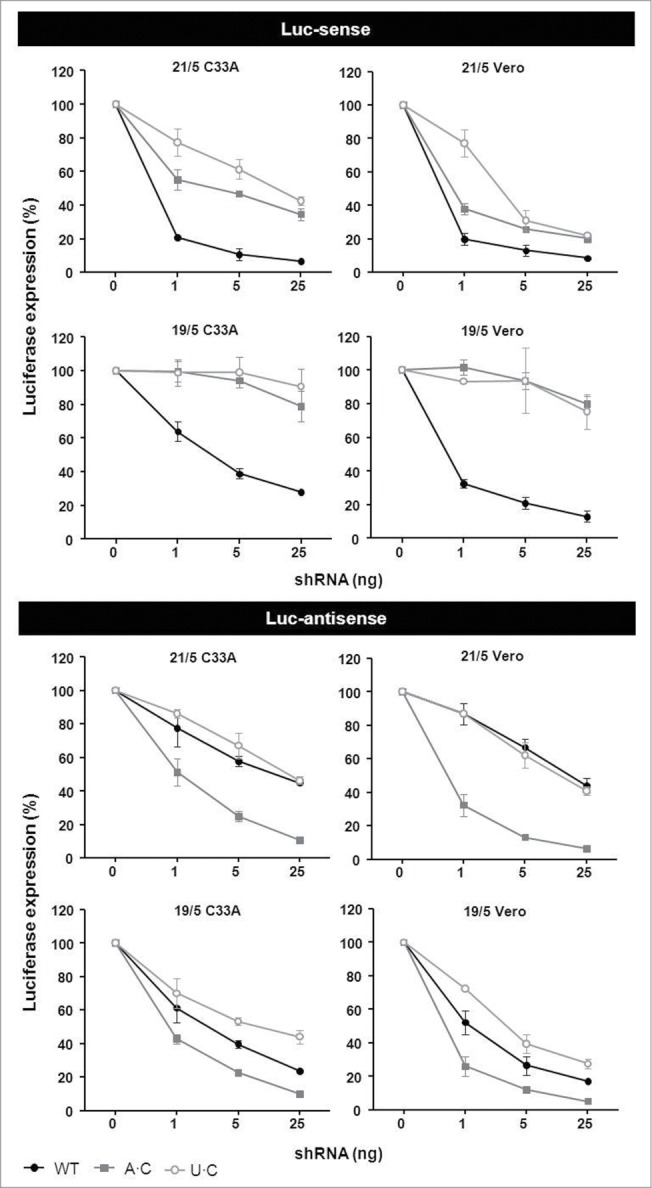
Improved AgoshRNA activity with a bottom A·(C) mismatch. C33A and Vero cells were co-transfected with 100 ng of the respective firefly luciferase reporter plasmid, 1 ng renilla luciferase plasmid as internal control and 1, 5 or 25 ng of the AgoshPol47 variants (WT with G-C and mutants A·C or U·C). An unrelated shRNA (shNef) served as negative control for which the activity obtained was set at 100% luciferase expression. We performed 3 independent transfections, each in duplicate, and standard deviations were calculated.
Discussion
Inspired by the natural miR-451 molecule that - like AgoshRNAs - is processed in a Dicer-independent manner, we studied the contribution of the 5′ nt and the bottom bp for AgoshRNA inhibitors. The most significant finding relates to the importance of the identity of the unpaired 5′ end nt for AgoshRNA activity, with the A·C bottom mismatch being most efficient. The results indicate that regular shRNAs (21/5) may be redirected from Dicer to Ago2 with the introduction of the A·C mismatch at the bottom of the hairpin. Nevertheless, such variants demonstrate hybrid processing with short Dicer and long Ago2 products. To avoid regular shRNA processing, shorter AgoshRNAs (19/5) with the A·C mismatch were tested. Analysis of the RNA products and activity measurements indicated that this novel AgoshRNA design bypasses Dicer effectively. Only a single RNAi-active guide strand is produced, which is an important property to restrict off target RNAi effects caused by the passenger strand.18 Most importantly, the optimized AgoshRNAs exhibited improved RNAi activity across different cell types.
AgoshRNA optimization by the A·C bottom mismatch may have multiple mechanistic implications. A bottom mismatch has an immediate impact on the hairpin stem length, which is an important determinant of AgoshRNA activity.14,16,19,20 Starting with the 19/5 AgoshRNA design, the bottom mismatch generates structures of only 18 bp that are too short for Dicer binding and consequently end up in Ago2 for alternative processing. The stem length parameter may explain the gain of AgoshRNA activity for the A·C variants, but it does not explain the loss of AgoshRNA activity for the U·C variants.
There may be additional unrelated effects of changing the +1 nt position. An “early” effect is possible at the transcriptional level if the H1 RNA polymerase III promoter prefers the use of a particular starting nucleotide. Several studies stated that the H1 promoter prefers +1 A as is the case in the human genome, but no experimental tests were performed.21,22 In fact, comparison of H1 promoter sequences among mammals indicated much sequence variation at the +1 position (unpublished results). Thus, the H1 promoter may be flexible with regard to +1 nt usage, although much remains currently unknown.23 A “late” effect of +1 variation can be considered when the bottom part of the AgoshRNA interacts with the Mid domain of Ago2. Crystal structures of Ago2 in complex with nucleoside monophosphates (AMP, CMP, GMP, and UMP), mimicking the 5′ end of miRNAs, indicate that the Mid domain makes specific contacts with AMP and UMP with a 30-fold higher affinity than GMP and CMP.24 However, we described a gain of AgoshRNA activity with 5′ A, but a loss of activity for 5′ U, which argues against this scenario. Thus, other sequences or structural elements within the AgoshRNAs may also determine the processing efficiency and subsequently the silencing activity. For instance, the duplex thermodynamics may affect the relative AgoshRNA stability and guide strand selection efficiency. We recently obtained evidence for the modulating role of a terminal G-U or U-G bp at the top of the basepaired stem, which also reflects miR‑451 architecture.16 We studied whether this observation may be applied as a general rule for AgoshRNA design. Some improved RNAi activity was observed for the AgoshRT5 G-U variants, independent on whether a top G-U or U-G bp was used. We did not measure a further improvement with a double G-U or U-G at the top of the hairpin. However, no such improvement was scored when tested in AgoshPol47, indicating that the G-U concept does not provide a general AgoshRNA improvement.
These combined results have important practical implications for the design of therapeutic AgoshRNA reagents, e.g. antivirals and anti-HIV molecules. Optimized AgoshRNA therapeutics with increased activity and reduced off-target effects may allow one to reduce the RNA dosage, thus reducing the risk of adverse effects, e.g., due to saturation of components of the RNAi pathway.25 We previously listed other putative advantages of the AgoshRNA design.5,15 The shorter duplex of AgoshRNAs may exhibit an improved safety profile because innate immunity sensor like interferon will be triggered less likely by shorter RNA duplexes.26 This advantage will be even greater for A·C mismatch variants. Ago2-mediated processing may also yield more precise RNA molecules than Dicer cleavage, which is known to create imprecise ends.27 AgoshRNAs may possibly also mimic miR‑451 in causing exclusive loading into Ago2, thus avoiding off targeting via Ago1, 3 and 4.12 Another potential advantage is that AgoshRNAs are fully active in Dicer-deficient cells, e.g. monocytes that lack Dicer expression or cells that lack the RNAi machinery.15,16 Overall, our current results combined with those of other laboratories suggest that there is a future for potent and specific AgoshRNA reagents in basic biology research and therapeutic applications.13,25,28 The AgoshRNA design may constitute a new platform for gene silencing that outperforms current miRNA and shRNA technology.
Material and Methods
DNA constructs
The shRNA expression plasmids were made by annealing complementary oligonucleotides (containing BamHI and HindIII sites) and inserting them into the BglII and HindIII sites of the pSUPER vector, as previously described.29–31 The RNA secondary structure of the shRNA transcript was predicted by the Mfold web server.32 Firefly luciferase reporter plasmids were constructed by insertion in the EcoRI and PstI sites of the pGL3 plasmid33 of a 50–70 nt HIV-1 sequence, with the 19 nt target region in the center. The luciferase reporters with the sense and antisense target sequences were described previously.15 All DNA constructs were sequence verified using the BigDye Terminator Cycle Sequencing kit (ABI, Foster City, CA, USA). Hairpin RNA constructs were sequenced using a sample denaturation temperature of 98˚C and upon addition of 1M Betaine.
Cell culture and DNA transfection
Human embryonic kidney (HEK) 293T, C33A and Vero cells were grown as monolayer in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal calf serum (FCS) (Hybond), penicillin (100 U/ml), streptomycin (100 μg/ml) and minimal essential medium non-essential amino acids (DMEM/10% FCS) at 37˚C and 5% CO2. For luciferase assays, all cell lines were plated one day before transfection in 24-well plates at a density of 1.4 × 105 cells per well in 0.5 ml DMEM/10%FCS without antibiotics. Cells were transfected with 100 ng of the firefly luciferase expression plasmid, 1 ng of Renilla luciferase expression plasmid (pRL) and 1, 5 or 25 ng of RNA vector using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Cells were lysed 48 h post transfection to measure firefly and renilla luciferase activities using the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). An unrelated shRNA (shNef) served as negative control, which was set at 100% luciferase expression. We performed 3 independent transfections, each in duplicate. The ratio between firefly and renilla luciferase activity was used for normalization of experimental variations such as differences in transfection efficiencies. The luciferase data were subsequently corrected for between session variation as described previously.34 The resulting 6 values were used to calculate the standard deviation shown as error bar.
siRNA detection by northern blotting
Northern blot experiments were performed as previously described.15,35 Briefly, HEK293T cells were transfected with 5 μg of shRNA constructs using Lipofectamine 2000. Total cellular RNA was isolated after 48 h with the mirVana miRNA isolation kit (Ambion) according to the manufacturer's protocol. The RNA concentration was measured using the Nanodrop 1000 (Thermo Fisher Scientific). Isolated RNA was analyzed by denaturing 15%-polyacrylamide gel electrophoresis (precast Novex TBU gel, Invitrogen) using a 32P-labeled Decade Marker (Ambion) for size estimation. To check for equal sample loading, the gel was stained with 2 μg/ml ethidium bromide for 20 min. The rRNA (5S rRNA) and tRNA bands were visualized with UV light. The RNA was electro-transferred to a positively charged nylon membrane (Boehringer Mannheim, GmbH, Mannheim, Germany) and cross linked to the membrane using UV (254 nm, 0.12 J). LNA oligonucleotide probes were 5′ end labeled with the kinaseMax kit (Ambion) in the presence of 1 μl of [γ-32P]ATP (0.37 MBq/μl Perkin Elmer). We used the following oligonucleotides probes (LNA positions underlined) to detect the 5′ and 3′ strand of the siRNA, respectively 5′-CTCCGCTTCTTCCTGCCAT-3′ and 5′-ATGGCAGGAAGAAGCGGAG-3′ for RT5 and 5′-ATTACTACTGCCCCTTCAC-3′ and 5′-GTGAAGGGGCAGTAGTAAT-3′ for Pol47. To remove unincorporated nucleotides, the probes were purified on Sephadex G-25 spin column (Amersham Biosciences). The blot was incubated in 10 ml ULTRAhyb hybridization buffer (Ambion) at 42°C for 30 min. After addition of the labeled LNA oligonucleotide, hybridization was performed at 42°C for 16 h. The blot was washed twice for 5 min at 42°C in 2 x SSC/0.1% SDS and twice for 15 min at 42°C in 0.1 × SSC/0.1% SDS and subsequently analyzed using a PhosphorImager (Amersham Biosciences) and the ImageQuant (v5.1) software package.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek - Chemische Wetenschappen (NWO-CW, Top Grant) and Zorg Onderzoek Nederland - Medische Wetenschappen (ZonMw, Translational Gene Therapy Grant).
References
- 1. Havens MA, Reich AA, Duelli DM, Hastings ML. Biogenesis of mammalian microRNAs by a non-canonical processing pathway. Nucleic Acids Res 2012; 40:4626-40; PMID:22270084; http://dx.doi.org/ 10.1093/nar/gks026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol 1998; 72:8150-7; PMID:9733856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 2011; 43:892-903; PMID:21925378; http://dx.doi.org/ 10.1016/j.molcel.2011.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires ago catalysis. Nature 2010; 465:584-9; PMID:20424607; http://dx.doi.org/ 10.1038/nature09092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Berkhout B, Liu YP. Towards improved shRNA and miRNA reagents as inhibitors of HIV-1 replication. Future Microbiol 2014; 9:561-71; PMID:24810353; http://dx.doi.org/ 10.2217/fmb.14.5 [DOI] [PubMed] [Google Scholar]
- 6. Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol 2005; 6:376-85; PMID:15852042; http://dx.doi.org/ 10.1038/nrm1644 [DOI] [PubMed] [Google Scholar]
- 7. Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007; 131: 1097-108; PMID:18083100; http://dx.doi.org/ 10.1016/j.cell.2007.10.032 [DOI] [PubMed] [Google Scholar]
- 8. Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 2008; 453:534-8; PMID:18404147; http://dx.doi.org/ 10.1038/nature06904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol 2005; 23:227-31; PMID:15619616; http://dx.doi.org/ 10.1038/nbt1052 [DOI] [PubMed] [Google Scholar]
- 10. Li N, You X, Chen T, Mackowiak SD, Friedländer MR, Weigt M, Du H, Gogol-Döring A, Chang Z, Dieterich C, et al. Global profiling of miRNAs and the hairpin precursors: insights into miRNA processing and novel miRNA discovery. Nucleic Acids Res 2013; 41:3619-34; PMID:23396444; http://dx.doi.org/ 10.1093/nar/gkt072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang JS, Maurin T, Lai EC. Functional parameters of dicer-independent microRNA biogenesis. RNA 2012; 18:945-57; PMID:22461413; http://dx.doi.org/ 10.1261/rna.032938.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dueck A, Ziegler C, Eichner A, Berezikov E, Meister G. microRNAs associated with the different human argonaute proteins. Nucleic Acids Res 2012; 40:9850-62; PMID:22844086; http://dx.doi.org/ 10.1093/nar/gks705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dallas A, Ilves H, Ge Q, Kumar P, Shorenstein J, Kazakov SA, Cuellar TL, McManus MT, Behlke MA, Johnston BH. Right- and left-loop short shRNAs have distinct and unusual mechanisms of gene silencing. Nucleic Acids Res 2012; 40:9255-71; PMID:22810205; http://dx.doi.org/ 10.1093/nar/gks662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge Q, Ilves H, Dallas A, Kumar P, Shorenstein J, Kazakov SA, Johnston BH. Minimal-length short hairpin RNAs: the relationship of structure and RNAi activity. RNA 2010; 16:106-17; PMID:19952116; http://dx.doi.org/ 10.1261/rna.1894510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu YP, Schopman NC, Berkhout B. Dicer-independent processing of short hairpin RNAs. Nucleic Acids Res 2013; 41:3723-33; PMID:23376931; http://dx.doi.org/ 10.1093/nar/gkt036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Herrera-Carrillo E, Harwig A, Liu YP, Berkhout B. Probing the shRNA characteristics that hinder dicer recognition and consequently allow ago-mediated processing and agoshRNA activity. RNA 2014; 20: 1410-18; PMID:25035295; http://dx.doi.org/ 10.1261/rna.043950.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yoda M, Cifuentes D, Izumi N, Sakaguchi Y, Suzuki T, Giraldez AJ, Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of argonaute2-cleaved precursor microRNAs. Cell Rep. 2013. Nov 14;5(3):715-26; PMID:24209750; http://dx.doi.org/ 10.1016/j.celrep.2013.09.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA "off-target" transcript silencing mediated by seed region sequence complementarity. RNA 2006; 12:1179-87; PMID:16682560; http://dx.doi.org/ 10.1261/rna.25706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu YP, von Eije KJ, Schopman NC, Westerink JT, ter Brake O, Haasnoot J, Berkhout B. Combinatorial RNAi against HIV-1 using extended short hairpin RNAs. Mol Ther 2009; 17:1712-23; PMID:19672247; http://dx.doi.org/ 10.1038/mt.2009.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McIntyre GJ, Yu YH, Lomas M, Fanning GC. The effects of stem length and core placement on shRNA activity. BMC Mol Biol 2011; 12:34; PMID:21819628; http://dx.doi.org/ 10.1186/1471-2199-12-34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA 2007; 13:1765-74; PMID:17698642; http://dx.doi.org/ 10.1261/rna.599107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sano M, Kato Y, Akashi H, Miyagishi M, Taira K. Novel methods for expressing RNA interference in human cells. Methods Enzymol 2005; 392:97-112; PMID:15644177; http://dx.doi.org/ 10.1016/S0076-6879(04)92006-X [DOI] [PubMed] [Google Scholar]
- 23. Tuschl T. Expanding small RNA interference. Nat Biotechnol 2002; 20:446-8; PMID:11981553; http://dx.doi.org/ 10.1038/nbt0502-446 [DOI] [PubMed] [Google Scholar]
- 24. Frank F, Sonenberg N, Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010; 465:818-22; PMID:20505670; http://dx.doi.org/ 10.1038/nature09039 [DOI] [PubMed] [Google Scholar]
- 25. Ma H, Zhang J, Wu H. Designing ago2-specific siRNA/shRNA to avoid competition with endogenous miRNAs. Mol Ther Nucleic Acids 2014; 3:e176; PMID:25025466; http://dx.doi.org/ 10.1038/mtna.2014.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bridge AJ, Pebernard S, Ducraux A, Nicoulaz AL, Iggo R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat Genet 2003; 34:263-4; PMID:12796781; http://dx.doi.org/ 10.1038/ng1173 [DOI] [PubMed] [Google Scholar]
- 27. Gu S, Jin L, Zhang Y, Huang Y, Zhang F, Valdmanis PN, Kay MA. The loop position of shRNAs and pre-miRNAs is critical for the accuracy of dicer processing in vivo. Cell 2012; 151:900-11; PMID:23141545; http://dx.doi.org/ 10.1016/j.cell.2012.09.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ma H, Dallas A, Ilves H, Shorenstein J, MacLachlan I, Klumpp K, Johnston BH. Formulated minimal-length synthetic small hairpin RNAs are potent inhibitors of hepatitis C virus in mice with humanized livers. Gastroenterology. 2014. Jan;146(1):63-6.e5; PMID:24076507; http://dx.doi.org/ 10.1053/j.gastro.2013.09.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002; 296:550-3; PMID:11910072; http://dx.doi.org/ 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- 30. Schopman NC, Liu YP, Konstantinova P, Ter Brake O, Berkhout B. Optimization of shRNA inhibitors by variation of the terminal loop sequence. Antiviral Res 2010; 86:204-11; PMID:20188764; http://dx.doi.org/ 10.1016/j.antiviral.2010.02.320 [DOI] [PubMed] [Google Scholar]
- 31. Ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther 2006; 14: 883-92; PMID:16959541; http://dx.doi.org/ 10.1016/j.ymthe.2006.07.007 [DOI] [PubMed] [Google Scholar]
- 32. Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/ 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res 2005; 33: 796-804; PMID:15687388; http://dx.doi.org/ 10.1093/nar/gki220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B, Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology 2006; 3:2; PMID:16398936; http://dx.doi.org/ 10.1186/1742-4690-3-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu YP, Haasnoot J, Ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res 2008; 36:2811-24; PMID:18346971; http://dx.doi.org/ 10.1093/nar/gkn109 [DOI] [PMC free article] [PubMed] [Google Scholar]


