Abstract
Several proteins and RNAs expressed by mammalian viruses have been reported to interfere with RNA interference (RNAi) activity. We investigated the ability of the HIV-1-encoded RNA elements Trans-Activation Response (TAR) and Rev-Response Element (RRE) to alter RNAi. MicroRNA let7-based assays showed that RRE is a potent suppressor of RNAi activity, while TAR displayed moderate RNAi suppression. We demonstrate that RRE binds to TAR-RNA Binding Protein (TRBP), an essential component of the RNA Induced Silencing Complex (RISC). The binding of TAR and RRE to TRBP displaces small interfering (si)RNAs from binding to TRBP. Several stem-deleted RRE mutants lost their ability to suppress RNAi activity, which correlated with a reduced ability to compete with siRNA-TRBP binding. A lentiviral vector expressing TAR and RRE restricted RNAi, but RNAi was restored when Rev or GagPol were coexpressed. Adenoviruses are restricted by RNAi and encode their own suppressors of RNAi, the Virus-Associated (VA) RNA elements. RRE enhanced the replication of wild-type and VA-deficient adenovirus. Our work describes RRE as a novel suppressor of RNAi that acts by competing with siRNAs rather than by disrupting the RISC. This function is masked in lentiviral vectors co-expressed with viral proteins and thus will not affect their use in gene therapy. The potent RNAi suppressive effects of RRE identified in this study could be used to enhance the expression of RNAi restricted viruses used in oncolysis such as adenoviruses.
Keywords: adenovirus, HIV-1, lentiviral vectors, RNA interference, RNA silencing suppressor, Rev-Response Element RNA, Trans-Activation Response Element, TAR RNA Binding Protein (TRBP)
Abbreviations
- Ago2
Argonaute-2
- ds
double-stranded
- EGFP
enhanced green fluorescent protein
- EMSA
electrophoresis mobility shift assay
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HIV
human immunodeficiency virus
- FL
firefly luciferase
- IP
immunoprecipitation
- miRNA
micro RNA
- NC
nucleocapsid
- PAGE
polyacrylamide gel electrophoresis
- pre-miRNA
precursor miRNA
- RT
reverse transcription
- RISC
RNA-Induced Silencing Complex
- RL
Renilla luciferase
- RNAi
RNA interference
- RRE
Rev Response Element
- RSS
RNA silencing suppressor
- siRNA
small interfering RNA
- TAR
trans-activation responsive element
- TRBP
TAR RNA Binding Protein
- UTR
untranslated region
- VA
virus-associated
- WT
wild-type
Introduction
RNA interference (RNAi) is a highly conserved cellular process that regulates gene expression at the post-transcriptional level. Since their identification, micro (mi)RNAs encoded by the cell have been shown to regulate various cellular pathways, including development, immune responses, inflammation and cell growth.1 In mammals, miRNAs are transcribed in the nucleus as primary-miRNAs, which are processed by Drosha and DGCR8 into precursor (pre-) miRNAs.2 Pre-miRNAs are then exported into the cytoplasm via an exportin-5 mediated pathway, where they are bound by Dicer and the TAR-RNA-Binding Protein (TRBP).3-7 These 2 proteins process pre-miRNAs into mature double-stranded (ds) miRNAs. The active strand of this mature miRNA is retained in the Dicer-TRBP complex, which then recruits the endonuclease Argonaute-2 (Ago2).8 This loaded RNA-Induced Silencing Complex (RISC) targets mRNAs that are complementary to the miRNA.
In mammals, RNAi interacts with viruses but the outcome of this interaction is not uniformly anti-viral. Several studies have shown that RNAi can directly inhibit viral replication while others have demonstrated that viruses can and do utilize cellular and viral miRNAs and RNAi-associated proteins to enhance viral replication.9-15 For example, Human Immunodeficiency Virus-1 (HIV-1) is not restricted by RNAi,16 while adenovirus is severely restricted, which may limit its use as an oncolytic virus.17-19 The apparent absence of RNAi restriction of some viruses could be explained by the counteraction of encoded viral RNA silencing suppressors (RSSs). Several mammalian viruses encode RSSs,17,20-29 although the efficiency of a few of them has been debated.16,30-32 Proposed RSSs encoded by HIV-1 are the transactivator Tat20,27 and the Trans Activation Response (TAR) RNA.33 Tat highly upregulates the expression of cotransfected plasmids under certain promoters, which has called its ability to suppress RNAi into question.16, 32 TAR has been proposed to disrupt RNAi activity by binding TRBP, thereby sequestering it from the RISC.33 TRBP has been shown to be a crucial, non-redundant component of RNAi,4,34 so its removal from the RISC would be highly detrimental to RNAi function. However, knockdown of TRBP decreases HIV-1 replication, suggesting that either RNAi does not restrict HIV-1 RNA or that TRBP has other crucial functions for the virus independent of RNAi.35-37
HIV-1 has a (+) RNA genome that displays many regions of complex secondary structure. One structured RNA element is the Rev-Response Element (RRE). The RRE secondary structure consists of a circular “head” surrounded by 5 stems, called stems I-V. These structured dsRNA regions are essential for mediating binding of Rev to RRE. Stem II acts as an initial docking site for Rev, but during viral RNA export from the nucleus, the entire RRE molecule becomes coated with Rev.38-40 The RRE-Rev interaction is critical in enabling the export of unspliced and singly spliced RNAs from the nucleus through the interaction with proteins involved in nuclear export.41,42 Its complex secondary structure makes it probable that RRE may interact with other RNA-binding proteins. In addition, the RRE sequence is very well conserved across different lentiviruses.43,44 The TAR-TRBP interaction has been well characterized,45-48 and we hypothesized that other highly structured RNA regions in HIV-1 may also bind to TRBP. In addition, viral miRNAs derived from RRE have been described.49 We therefore examined the ability of RRE to suppress RNAi, we explored the mechanism of this function and its ability to affect or enhance the replication of a lentiviral vector or an adenovirus.
Results
RRE is a suppressor of miRNA-mediated RNAi activity
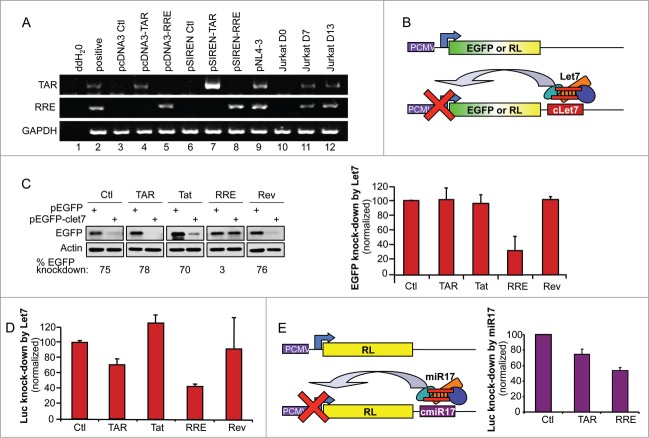
To investigate the RSS functions of different HIV-1 elements, we first expressed both TAR and RRE from 2 different expression vectors to achieve different levels of RNA expression. We cloned these 2 DNAs into the pcDNA3 (Invitrogen) and the pSIREN (Clontech) vectors, which use an RNA polymerase II and III promoter, respectively, to drive RNA expression. RT-PCR experiments with primers amplifying the complete RNA structure demonstrated that TAR (Fig. 1A, lanes 4, 7) and RRE (lanes 5, 8) are detectable at a higher level when cells are transfected with the pSIREN constructs. Because transfection rates are low in the lymphocytic Jurkat cells and the further assays based on let7 expression could not be done in these cells, NL4–3 infection was compared to HeLa cell transfection. When HeLa cells were transfected with either TAR or RRE vectors, we found that the expression levels obtained from pcDNA3 closely resemble expression levels in HIV-1 NL4–3 infection of Jurkat cells and therefore we used these constructs to assess their RSS activity (Fig. 1A).
Figure 1.
RRE inhibits miRNA-mediated RNAi activity in the cell. (A) Comparison of TAR and RRE expression in transfected HeLa cells and in HIV-infected Jurkat lymphocytes. HeLa cells (lanes 1–9) were transfected with 1 μg of pcDNA3, pcDNA3-TAR, pcDNA3-RRE, pSIREN, pSIREN-TAR, pSIREN-RRE or pNL4–3, as indicated. Jurkat cells (lanes 10–12) were infected with pNL4–3 and harvested at days 0, 7 and 13 of infection. Expression of TAR, RRE and GAPDH were detected by RT-PCR. (B) Schematic representation of the EGFP and Renilla Luciferase (RL) assays to measure the RNAi activity mediated by let7. (C) RRE prevents let7-mediated inhibition of EGFP expression. HeLa cells were cotransfected with 0.4 μg pEGFP or pEGFP-clet7 and with 1μg of pcDNA3, pcDNA3-TAR or pcDNA3-RRE, or 1 μg pCMV1-Tat or pCMV-Rev as indicated. Effects on knockdown of EGFP by miRNA let7 were observed by immunoblot (left), and quantified by densitometry analysis. Histogram (right) shows the average RNAi activity (EGFP knockdown) over 3 independent experiments ± SEM. (D) TAR and RRE prevent let7-mediated inhibition of RL expression. HeLa cells were transfected with 5 ng of pRL or pRL-clet7 and with 0.25 μg of pcDNA3, pcDNA3-TAR, or pcDNA3-RRE, or 0.25 μg pCMV1-Tat or pCMV-Rev as indicated. (E) TAR and RRE prevent miR17-mediated inhibition of RL expression. HEK 293T cells were transfected with 5 ng of pRL or pRL-cmiR17 and with 0.25 μg of pcDNA3, pcDNA3-TAR, or pcDNA3-RRE as indicated. (D and E) Luciferase signals were measured using a luminometer. Signals were normalized to the cotransfected pCMV-firefly luciferase (FL). RNAi activity (Luc knockdown) is calculated as the ratio of RL/FL to RL-let7/FL or to RL-miR17/FL, and displayed as per cent of RNAi activity with the empty plasmid. Histogram shows the average of 3 independent experiments ± SEM.
We next evaluated the ability of HIV-1-encoded elements expressed at the most physiologic level to affect miRNA-mediated RNAi activity. By transfecting a plasmid encoding a reporter gene [(either enhanced green fluorescent protein (EGFP) or Renilla luciferase (RL)] with the complementary sequence to cellular miRNA let7 inserted into its 3′ untranslated region (UTR)50 we assessed the interfering activity of let7 expressed in HeLa cells (Fig. 1B). Tat was previously found to be an RSS,20,27 but this was contradicted by others,32 so we also evaluated its potential in these assays along with Rev, TAR and RRE. Using the EGFP-reporter, we found that the previously described RSSs Tat and TAR were unable to decrease RNAi activity. In contrast, RRE was a potent suppressor of let7-mediated RNAi activity with an average of 67% inhibition (Fig. 1C). When we used the luciferase reporter, RRE suppressed let7-mediated RNAi activity by 57%. In this assay, TAR also had a suppressive effect, with a 29% inhibition (Fig. 1D). In both assays, neither Tat nor Rev had an RSS activity (Fig. 1B and C), emphasizing the role of viral dsRNA elements in RNAi inhibition. Our assays rely on the expression of let7, which is present in large amounts in HeLa cells. To determine if the same RSS activity could be observed in other cells expressing miRNAs in different proportions, we also tested TAR and RRE in HEK 293T cells using an assay based on miR17 instead of let7 (Fig. 1E). We found that both TAR and RRE had similar RSS activity in these cells.
RRE binds to TRBP
TRBP was originally isolated through its binding to TAR RNA, and this interaction has been extensively studied.4 TAR has been suggested to suppress RNAi by sequestering TRBP, which may prevent its interaction with Dicer.33 To evaluate whether RRE may act in a similar manner, we first examined whether RRE was also able to bind to TRBP. Although TRBP has previously been isolated in a screen with RRE RNA, their direct interaction has not been shown.51 In contrast, the protein has been shown to directly bind small interfering (si)RNAs.52-54 By electrophoresis mobility shift assay (EMSA), using a radiolabelled control siRNA and recombinant MBP or MBP-TRBP, we observed several low mobility bands, which indicates that TRBP binds to the siRNA either as a monomer or as a multimer (Fig. 2A, left). In a similar EMSA experiment using radiolabelled RRE, we also observed the formation of multiple TRBP-RRE complexes in a dose-dependent manner (Fig. 2A, center left) similar to those observed with TAR RNA.45 In contrast, incubation of a control 20 (Ctl1) or 165 (Ctl2) nt-long RNA with TRBP produced no or weak monomeric band shifts (Fig. 2A, right panels), indicating that the TRBP-RRE and TRBP-siRNA complexes are specific for binding and forming multiple complexes.
Figure 2.
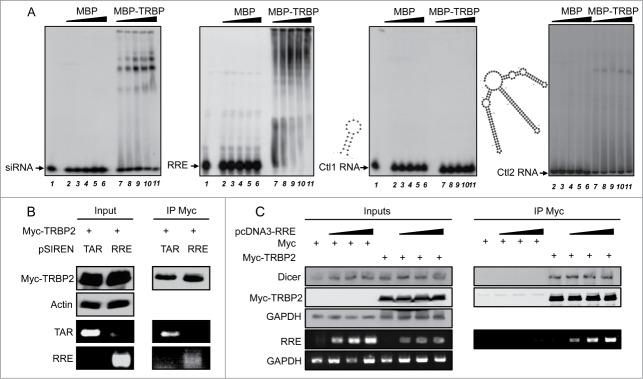
RRE RNA binds to TRBP. (A) EMSAs of siRNA, RRE RNA with TRBP or control (ctl) RNAs. Radiolabelled siRNA (left panel), RRE (center left), Ctl1 (center right) or Ctl2 RNA (right) were incubated with 0 (lane 1), 0.15 (lanes 2, 7), 0.3 (lanes 3, 8), 0.6 (lanes 4, 9), 1.2 (lanes 5, 10) or 2.4 (lanes 6, 11) μM of purified MBP or MBP-TRBP as indicated. Samples were separated on a 4% (Ctl1 and siRNA) or 3.5% (Ctl2 and RRE) polyacrylamide gel. Complexes are visualized by an upward shift in the radioactive signal. The structures and stabilities of Ctl1 (20nt, ΔG = −6.10) and Ctl2 (165 nt, ΔG = −73.60) were predicted using the M-fold software and are shown to the left of the corresponding blot. (B) RNA-IP experiments between Myc-TRBP2 and TAR or RRE RNAs. Cells were cotransfected with 7.5 μg of pCMV-MycTRBP2 and 7.5 μg of either pSIREN-TAR or pSIREN-RRE as indicated. Lysates were immunoprecipitated using an anti-Myc antibody. Proteins in the input (10%) and in the immunoprecipitate were analyzed by immunoblot with anti-Myc and anti-actin antibodies (top). RNAs were recovered and analyzed by RT-PCR, with primers specific for TAR and RRE (bottom). (C) RNA-IP experiments with increasing amounts of RRE. As in (B), except that cells were transfected with 0, 5, 10 or 15 μg of pcDNA3-RRE and topped up to 15 μg with pcDNA3 empty. Cells were cotransfected with pCMV-Myc or pCMV-MycTRBP2, as indicated.
We next assessed whether RRE is able to bind to TRBP in cells. Cells were cotransfected with the highly expressed pSIREN-TAR or pSIREN-RRE vectors, and a plasmid expressing Myc-tagged TRBP. RNA-IP experiments using an anti-Myc antibody followed by RT-PCR showed that we recovered both TAR and RRE as co-immunoprecipitates with TRBP (Fig. 2B). This indicates that in cells, TRBP is able to bind to both of these structured RNAs and that RRE may suppress RNAi by using this interaction. We further evaluated the ability of RRE to bind to TRBP at a broader range of concentrations and examined the impact on the expression of RISC proteins. Cells were transfected with increasing amounts of pcDNA3-RRE and co-transfected with Myc or Myc-TRBP (Fig. 2C). RRE specifically bound to Myc-TRBP even at the lowest concentration used. Expression of RRE in the cell did not appear to affect the expression of Dicer to any significant degree and did not disrupt the binding of Dicer to Myc-TRBP.
HIV-1 TAR and RRE RNAs do not affect the stability of the RISC but displace an siRNA bound to TRBP
Previous reports have indicated that TAR disrupts the TRBP-Dicer complex formed by the overexpressed proteins, but the stability of the endogenous RISC was not determined.33 We first evaluated if RRE expression was affecting the amount of the RISC proteins in the cell over time. We found that Dicer, Ago2 and TRBP were equally expressed up to 72 h whether pcDNA3 or pcDNA3-RRE was transfected, suggesting that the RNA affects neither their expression nor their stability (Fig. 3A). We next evaluated the ability of the TAR and RRE RNA elements to disrupt the endogenous TRBP-Dicer-Ago2 complex. We first established that an anti-Dicer antibody was able to immunoprecipitate Dicer, Ago2 and TRBP whereas a control antibody was not (Fig. 3B, top). Cells were then transfected with constructs expressing TAR or RRE, and lysates were immunoprecipitated using the same anti-Dicer antibody (Fig. 3B, bottom). The presence of both proteins in the IP showed that neither TAR nor RRE were able to impede TRBP-Dicer interaction even with the pSIREN vector, which allows high expression of either TAR or RRE. In addition, recruitment of Ago2 to the RISC was maintained, suggesting that these suppressors do not affect the assembly of the endogenous RISC.
Figure 3.
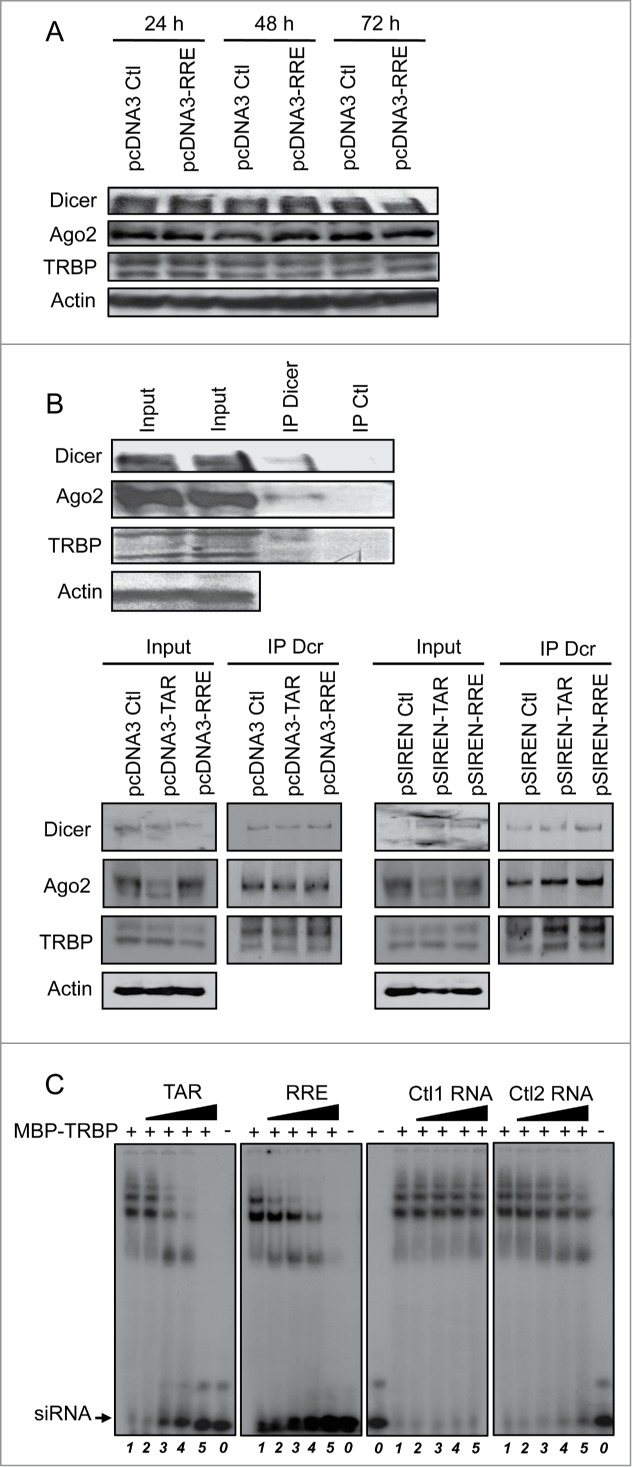
TAR and RRE compete with siRNA for binding to TRBP. (A) Expression and stability of Dicer, Ago2 and TRBP remain unchanged in the presence of RRE. HeLa cells were transfected with 7.5 μg of pcDNA3 or pcDNA3-RRE as indicated. Cell lysates were analyzed by immunoblot with anti-Dicer, anti-TRBP, anti-Ago2 or anti-actin after 24 h, 48 h or 72 h post transfection as indicated. (B) TAR and RRE do not displace endogenous TRBP-Dicer-Ago2 complex. HeLa cells were transfected with none (top), 7.5 μg of pcDNA3, pcDNA3-RRE, pcDNA3-TAR, pSIREN, pSIREN-RRE or pSIREN-TAR (bottom) as indicated. Lysates were immunoprecipitated with an anti-Dicer or an anti-EGFP (Ctl) antibody. Proteins in the input (10%) and in the immunoprecipitate were analyzed by immunoblot with anti-Dicer, anti-TRBP, anti-Ago2 or anti-actin as indicated. (C) TAR and RRE displace siRNA in a TRBP complex. Radiolabelled siRNA was incubated alone (lanes 0) or with 2.4 μM of MBP-TRBP (lanes 1–5) prior to the addition of 0 (lanes 1), 3.75 (lanes 2), 15 (lanes 3), 37.5 (lanes 4) or 150 (lanes 5) pmoles of cold TAR (left panel), RRE (center left), Ctl1 (center right) or Ctl2 (right) RNA. Samples were separated on a 4% polyacrylamide gel.
We next investigated the possibility that TAR and RRE may be competing with siRNAs for binding to TRBP and inclusion in the endogenous RISC. We assessed the ability of TAR or RRE to compete with siRNAs for binding to TRBP by EMSA (Fig. 3C). In this assay, TAR and RRE, but not the control RNAs, displaced the radiolabelled siRNA from TRBP, suggesting that these structured RNAs compete with siRNAs for incorporation into the RISC.
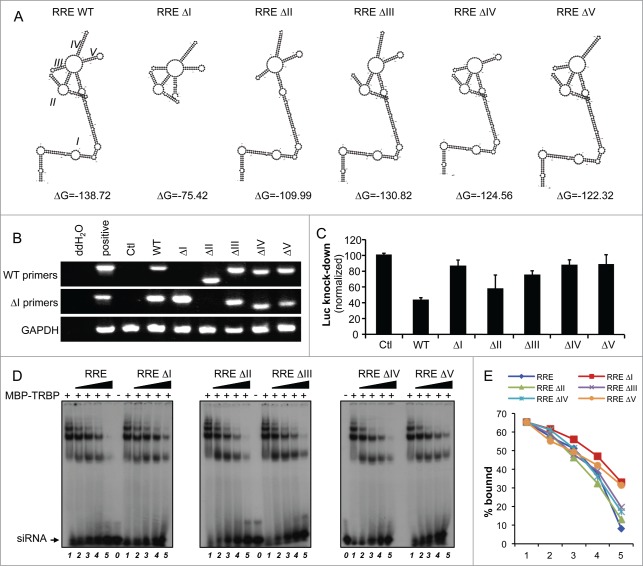
Mutants of RRE have a decreased ability to suppress RNAi activity and to compete with siRNA-TRBP binding
To determine if a specific structure was responsible for the RRE RSS activity, we next generated a series of mutants in which one stem-loop of RRE was deleted at a time. The sequence for each of these mutants was folded in silico using the M-fold software.55 This analysis suggested that each deletion affects only the target stem, and not the overall structure of the molecule (Fig. 4A). The expression of each mutant evaluated by RT-PCR was similar to levels of the WT RRE construct (Fig. 4B). We next assessed the suppression of RNAi mediated by each mutant. Using the let7-luc reporter, we observed a complete loss of suppression of RNAi for mutants ΔI, ΔIV and ΔV, and a partial loss of function for mutant ΔIII, whereas mutant ΔII retained RSS activity close to WT levels (Fig. 4C). To determine if the RSS activity correlates with siRNA-TRBP binding, we evaluated the ability of each mutant to compete for this interaction. Using the same molar ratio of RRE or of each mutant, RRE mutants ΔI and ΔV were the weakest competitors (Fig. 4D, E). Mutant ΔII retained near-wild-type competitive ability, whereas mutants ΔIII and ΔIV had an intermediate competitive ability. This showed a close correlation between the competition and the RSS activity.
Figure 4.
Mutants of RRE display decreased RNAi suppression. (A) Secondary structure of mutant RRE RNAs. Mutants of RRE were generated by deleting one dsRNA stem at a time. Secondary structures of WT RRE and each of the mutants were predicted using the M-fold software. (B) Expression of mutant RRE RNAs. HeLa cells were transfected with 1 μg of each WT or mutant plasmid. RNA expression was visualized by RT-PCR using either the WT or the ΔI primers for RRE RNAs, as indicated, and compared to GAPDH expression. (C) Activity of RRE mutants on the inhibition mediated by let7 on RL expression. HeLa cells were cotransfected with 5 ng of pRL or pRL-let7 and with 0.25 μg of pcDNA3, pcDNA3-RRE or pcDNA3-RRE mutants as indicated. RNAi activity (Luc knockdown) was calculated as in Fig. 1D and the histogram shows the average of 3 independent experiments ± SEM. (D) Displacement of siRNA-TRBP complex by RRE mutants. Binding of TRBP to radiolabelled siRNA and competition was performed as in Fig. 3B with the indicated mutants. (E) Quantitation of siRNA-TRBP complex displacement by RRE mutants. The signal for the siRNA-TRBP complex was quantified using ImageJ 1.47 software.
A lentiviral vector expressing RRE inhibits RNAi activity, only in the absence of Rev or GagPol
Lentiviruses used as vectors generally express both TAR and RRE and have the potential to suppress RNAi.56 In contrast, full-length HIV-1 does not suppress RNAi activity.16 We therefore evaluated whether a lentiviral vector derived from HIV-1, from which viral proteins are not expressed, could inhibit RNAi activity. Using the pHIV7-tdTomato vector, derived from pHIV7-CGFP harbouring both TAR and RRE,57 we assessed RNAi activity using the same let7-luc reporter in comparison to HIV-1 NL4–3 (Fig. 5A). The expression of HIV7-tdTomato reduced RNAi activity by 35%, whereas NL4–3 did not. This shows that the lentivirus acts, at least partially, as an RSS. This result suggests that the difference between a lentiviral vector and HIV-1 to suppress RNAi activity may be related to the presence of viral proteins that could bind to TAR and RRE and mask their function as RSSs. To test this idea, we used the let7-EGFP reporter with overexpressed Rev, which binds RRE, or GagPol, which binds HIV-1 RNA through the nucleocapsid (NC) in the cytoplasm58,59 to determine if they prevent the RSS activity of pHIV7-tdTomato (Fig. 5B). Indeed, the RSS activity of HIV7-tdTomato was similar to that observed in the let7-luc reporter, with 24% inhibition of RNAi activity, whereas no inhibition could be observed when co-expressed with Rev or GagPol. Because the RSS activity of the HIV7-tdTomato was not as high as RRE, we evaluated if it could be due to its lower expression in the cells at the amounts we used in the assay. We measured TAR and RRE RNA expression from HIV7-tdTomato and found that they were less expressed than TAR or RRE independently (Fig. 5C). This result indicates that the lower RSS activity of the lentiviral vector compared to TAR or RRE is likely due to its low expression in cells.
Figure 5.
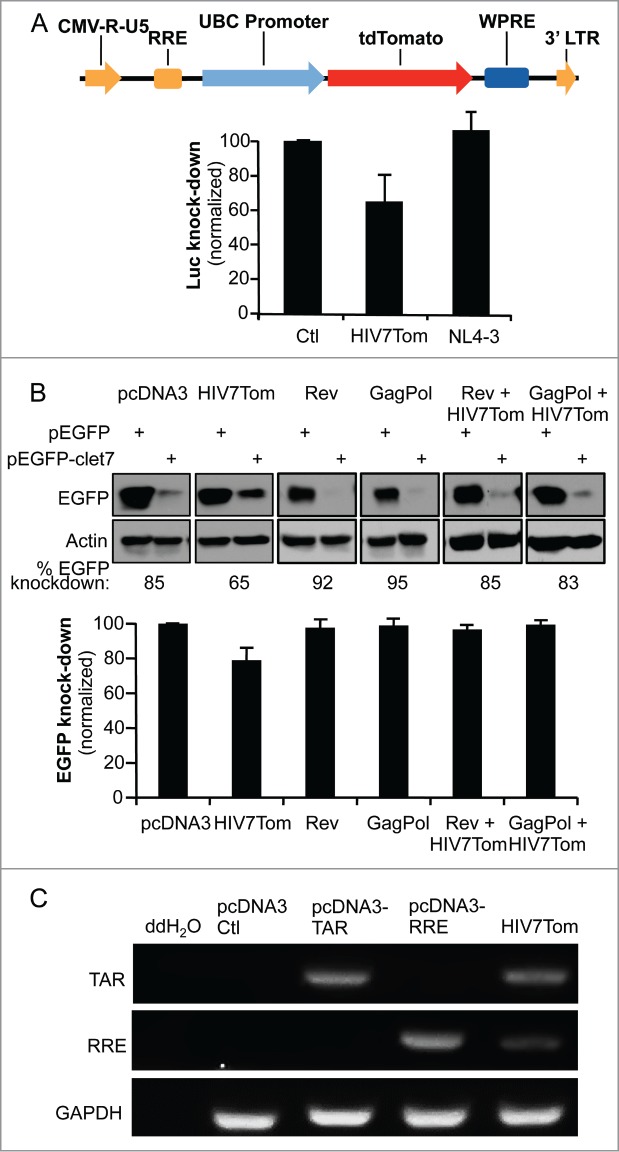
A lentiviral vector expressing RRE decreases miRNA-mediated RNAi activity in the absence of Rev or GagPol. (A) pHIV7-tdTomato lentiviral vector expressing RRE partially prevents let7-mediated inhibition of RL. (top) Schematic representation of pHIV7-tdTomato vector. Relevant genes and regulatory elements are shown. (bottom) HeLa cells were transfected with 0.25μg of pcDNA3, pHIV7-tdTomato or pNL4–3 as indicated and the assay was carried out as in Fig. 1D. (B) Rev and GagPol restore RNAi suppressed by HIV7-tdTomato lentiviral vector. HeLa cells were cotransfected with 0.4 μg pEGFP or pEGFP-clet7 and with 1μg of pcDNA3, pHIV7-tdTomato, pCMV-Rev, pCHGP expressing GagPol or pHIV7-tdTomato with pCMV-Rev, pCHGP as indicated. The assay was carried out as in Fig. 1C. (C) Expression of TAR and RRE RNAs from HIV7-tdTomato. HeLa cells were mock-transfected or transfected with 1 μg of pcDNA3, pcDNA3-TAR, pcDNA3-RRE or pHIV7-tdTomato as indicated. RNAs were recovered and analyzed by RT-PCR, with primers specific for TAR, RRE and GAPDH.
RRE alleviates RNAi mediated restriction of adenovirus
To determine whether RNAi suppression by RRE could have implications in the context of other viral infections, we examined the impact of coexpressing RRE with an adenovirus. Adenoviruses are known to be restricted by RNAi and to encode their own RNA-based RSSs, the VA RNAs I and II.17,19 We therefore evaluated the ability of RRE to restore replication levels in VA-deficient constructs of adenovirus, previously designed to enhance their oncolytic activity.18 We first verified the amount of DNA in the cells 1 h (input), 24 h, 48 h or 72 h after transfection of HeLa cells and observed that the replicating DNA is 2.9 and 2.35 fold increased for pAdWT or pAdΔVA respectively, over the input DNA after 48 h (Fig. 6A). HeLa cells were then cotransfected with RRE or the empty vector and adenovirus constructs pAdWT or pAdΔVA. Viral expression was assessed by immunoblotting against early adenovirus protein E1A. While AdΔVA displayed a reduced expression level compared to AdWT (Fig. 6B, lane 3 compared to lane 5) its expression was increased in the presence of RRE to reach the WT level (Fig. 6B, lane 4). In contrast, RRE did not increase E1A expression from AdWT (Fig. 6B, lanes 5–6). To accurately quantify the increase in replication, qPCR experiments detecting the adenovirus genomes were also carried out (Fig. 6C). In the presence of RRE, DNA levels of the adenovirus ΔVA mutant approximated those of WT adenovirus without RRE. RRE also increased the expression of WT adenovirus over twofold, suggesting that the WT virus is still restricted by RNAi to some degree. Overall, RRE alleviates the natural RNAi restriction of adenoviruses and can replace the RSS activity of the VA RNAs.
Figure 6.
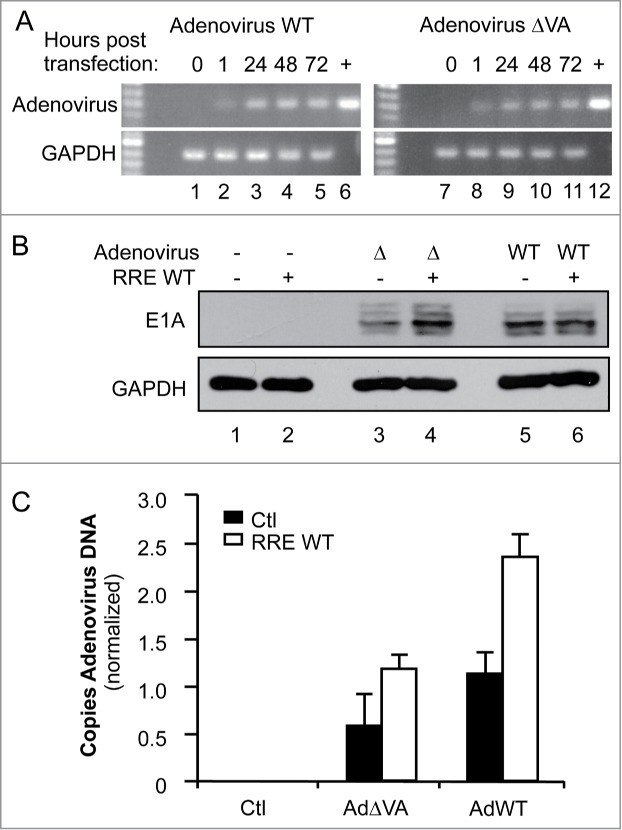
RRE restores the replication of VA-defective adenovirus. (A) Amplification of adenovirus DNA. HeLa cells were transfected with 1 μg of pcDNA3 (lanes 1, 7), pAd5 WT (lanes 2–5), or pAd5ΔVA (lanes 8–11). DNA was collected at 1 h (lanes 2, 8), 24 h (lanes 3, 9), 48 h (lanes 1, 4, 7, 10) or 72 h (lanes 5, 11) post transfection. (+) represents amplified plasmid DNA. Viral and GAPDH DNAs were amplified by 25 and 30 cycles of PCR, respectively. (B and C) HeLa cells were cotransfected with 1 μg of pcDNA3 (lanes 1–2), pAd5ΔVA (lanes 3–4), or pAd5 WT (lanes 5–6), and with 1μg of pcDNA3 (lanes 1, 3, 5) or pcDNA3-RRE (lanes 2, 4, 6). (B) Expression of adenoviral proteins. Expression of adenoviral proteins was assessed by immunoblot with anti-E1A protein compared to the total proteins represented by GAPDH expression. (C) Quantitation of adenoviral DNA. Viral DNA was quantified by real-time qPCR using a FAM-TAMRA probe. The histogram represents the average of 3 independent experiments ± SEM.
Discussion
Since the discovery of RNAi as a major mechanism of gene regulation, it has been demonstrated to be a strategy of innate immunity against viruses in plants and lower eukaryotes. No consensus has been reached concerning its antiviral effect in mammals and the effect may be dependent on the virus and the cells used.15,60-62 RNAi inactivation results in increased viral replication when cells from different species than the natural host are infected, in latently infected cells or in embryonic stem cells,12,13,25,63 but has the opposite effect in cells that allow high levels of viral replication.35,64,65 Viral si- (vsi)RNAs are highly abundant in many, if not all, herpesviridae species, where they regulate the viral life cycle. They are present in various amounts in a large number of viruses and they impact cellular functions.11,66,67 Many described proteins and RNAs acting as RSSs have mostly been shown to increase viral replication in cell lines from different origin than the natural host, although some have been shown in natural host cells.68 In addition, the induction of RISC proteins upon virus entry has not been shown, pointing to an intrinsic mechanism rather than an inducible pathway. Among the RNAs shown to have RSS activity, adenovirus VA RNAs I and II and HIV-1 TAR have been characterized.17,19,33 We have identified HIV-1 RRE RNA as a new RSS, which inhibits RNAi mediated by the endogenous let7 or miR17 miRNAs, and found that its activity as an RSS is stronger than that of the previously described TAR (Fig. 1).
The characterization of RRE as a potent RSS when expressed alone but not when expressed within the entire HIV-116 raises the question of its mechanism of action, its role in lentiviral expression and its potential to increase the replication of RNAi-restricted viruses. We have demonstrated that RRE binds to TRBP both in vitro and in a cellular context (Fig. 2). Therefore, the mechanism of RNAi restriction could occur through sequestration of TRBP away from the RISC by preventing its binding to Dicer. However, when we assessed the interactions between RISC proteins, neither TAR nor RRE disrupted the endogenous RISC (Fig. 3A). Our results are in contrast to the TAR-mediated disruption of the TRBP-Dicer interaction observed in the context of overexpressed TRBP, Dicer and TAR.33 In our assays, interactions between the key endogenous proteins of the RISC, TRBP, Dicer and Ago23,5,69,70 were maintained despite high expression levels of either TAR or RRE RNAs (Fig. 3A). This result shows that the endogenous RISC is much stronger than the TRBP-Dicer dimer, which may be due to a stabilization of the complex by additional proteins. In contrast, we show that both TAR and RRE are able to displace siRNAs from TRBP, which may reflect what happens in the cell with miRNAs and the entire RISC (Fig. 3B). The mutants of RRE all displayed decreased RNAi suppression as compared to WT RRE. Mutant ΔI completely lost its ability to suppress RNAi, which is consistent with its decreased stability (ΔG = −75.42 vs. ΔG = −138.72 for WT, Fig. 4A), which would affect the molecule's secondary structure. Mutant ΔII was the only mutant that retained significant suppressive activity, indicating that RNAi suppression by RRE does not rely on the Rev docking stem (stem II) but requires the distal region of RRE, specifically stems IV and V (Fig. 4C). These two stems most resemble miRNA structure, which could explain why they compete with siRNA for binding to the RISC. The weaker competition of RRE ΔI and ΔV mutants for siRNA-TRBP interaction shows the importance of stem-loop V within the overall RRE structure for the RSS activity (Fig. 4D). In addition, the correlation between the competition assays and the RSS activity of the RRE mutants strongly indicates that TRBP-siRNA interaction is a key target of RSSs. Because our assays require functional endogenous let7 and miR17 miRNAs, we cannot exclude that RRE might also function at the level of pre-miRNA export. Testing RRE activity in other miRNA-based assays and on TRBP-Dicer-Ago2 complex binding to other si/miRNAs will provide a more global view of its RSS activity in the future.
HIV-1 has been successfully targeted by si and short hairpin RNAs suggesting that neither Tat, TAR nor RRE act as RSSs during HIV-1 replication.56 However, this raises the question of their activity in a lentiviral vector used in gene therapy. We therefore assessed the ability of a clinically relevant lentiviral vector, pHIV7-tdTomato, to suppress RNAi activity. This vector encodes TAR and RRE but no viral protein. Our findings show that in the absence of viral protein, RNAi activity is decreased (Fig. 5A and B), indicating that lentiviral vectors expressing TAR and RRE restrict RNAi function of transfected cells. The weaker RSS activity of tdTomato compared to RRE could be explained by a lower expression of the RRE RNA in the context of this vector (Fig. 5C). Because expressing Rev or GagPol restores RNAi activity (Fig. 5B), this effect is not detectable in packaging cells expressing viral proteins. The effect of Rev suppressing the RSS activity is likely linked to its binding to RRE. The effect of GagPol likely occurs through binding to HIV-1 RNA by the NC part. Indeed, NC is able to cover all HIV-1 RNA in the virion and this may occur in the cell when NC is included in Gag or GagPol or when it is partially cleaved by the protease.71 These results also explain why, despite containing potent RSSs such as TAR and RRE described in this study, HIV-1 has not been observed to suppress RNAi.16 Furthermore, when integrated, a self-inactivating lentivirus with no LTR will not express TAR or RRE, therefore confirming the use of RNAi as a valid therapeutic option to target HIV-1 in gene therapy.
Our results raise the question of whether RRE is able to act as an RSS during HIV-1 replication, and if so, when. Reports have indicated that RRE may serve as a substrate to the RISC, as viral miRNAs derived from stem I of RRE have been described.49 This suggests that at some point during HIV-1 replication, RRE is accessible to RISC proteins and may compete with endogenous miRNAs for loading into the RISC. The low amount of TAR and RRE vsiRNAs identified by various groups may prove to be of importance during a relatively short time frame at the beginning of HIV-1 RNA expression.49,72-75 The displacement of endogenous miRNAs into the RISC mediated by TAR or RRE competition may contribute to mechanisms leading to changes in gene expression and associated pathogenicity in patients.
The ability of exogenous RRE to act as an RSS to increase HIV-1 replication could not be tested because RRE acts as an RNA decoy for Rev and inhibits viral replication.76 We therefore assessed if, in the context of a virus restricted by RNAi, RRE would be able to alleviate RNAi-associated restriction of viral replication. Adenovirus VA RNA releases viral restriction by RNAi and ΔVA adenovirus replicates less efficiently than WT virus.17,18 When RRE was expressed in trans, it was able to increase the expression of the early protein E1A and to restore the replication of ΔVA adenovirus to WT levels suggesting that the RNA coding for the early protein is largely restricted by RNAi (Fig. 6). In addition, RRE increased the replication of both the ΔVA and the WT adenovirus twofold. These trans-complementation assays confirm that RRE can act as an RSS in the context of a viral infection, similarly to what has been shown for other RSSs.22,23,27 Because E1A is synthesized in part from the incoming DNA,77 the increase of E1A expression from ΔVA adenovirus but not from WT adenovirus suggests that RRE mainly compensates for the lack of VA when the RNA is transcribed from the incoming DNA. The RRE-mediated increase in DNA replication observed on both ΔVA and WT adenovirus suggests that RRE further impacts the RNA transcribed from the replicating DNA whether it is mutated in VA or not. Suppression of RNAi may be of particular interest in specific situations. In light of our results, expression of RRE or its ΔII mutant could increase the expression and stability of a target gene on adenovirus and other viral vectors as observed with other RSSs.21 It could also increase the activity of oncolytic adenoviruses, which would widen their applications.18 In certain viral infections11,14 and cancers,78 RNAi activity is detrimental to the host. In these cases, a global suppression of RNAi may also be beneficial.
Materials and Methods
Plasmids
pCMV-Myc-TRBP2,36 pCMV-RL, pCMV-RL-clet7 and pCMV-FL,50 pCMV-RL-cmiR17,5 pHIV7-CGFP,57 pCHGP,79 pCMV1-Tat48 and pCMV-Rev80 have been described. pcDNA3-TAR and pcDNA3-RRE were cloned from pNL4–3 and inserted into pcDNA3 using the HindIII and XhoI restriction sites for TAR and EcoRI and XhoI for RRE. Mutant ΔI of RRE was cloned by PCR from pcDNA3-RRE and inserted into pcDNA3 using the BamHI and NotI.
RREΔI-F-5′ATGTAGGATCCGGGAGCAGCAGGAAGCACT3′
RREΔI-R-5′ATATGCGGCCGCCCACAGCCAGGATTCTTG3′.
Mutants ΔII-ΔV were cloned in a 2-step process. Fragment A was cloned using the generic forward primer F-5′ATAGAGGATCCTAGCACCCAACGCAAAGAG3′, and a specific reverse primer for each mutant, and fragment B was cloned using a specific forward primer for each and a generic reverse primer, R-5′ATATGCGGCCGCTAGCATTCCAAGGCATAG3′. Mutant-specific primers are as follows:
pcDNA3-RREΔII (A)
R-5′GCGGCCGCGAATTCTTCCTGCTGCTGCTCCCAAGAACCCAA3′
pcDNA3-RREΔII (B) F-5′ATAGAGAATTCGCAGCAGAACAATTTGCTGAGGGC3′
pcDNA3-RREΔIII (A) R-5′GCGGCCGCGAATTCCTGCACTATATCAGACAATAATTG3′
pcDNA3-RREΔIII (B) F-5′ATAGAGAATTCAGGGCTATTGAGGCGCAACAGCAT3′
pcDNA3-RREΔIV (A) R-5′GCGGCCGCGAATTCTAGCCCTCAGCAAATTGTTCTGCT3′
pcDNA3-RREΔIV (B) F-5′ATAGAGAATTCCAGTCTGGGGCATCAAACAGCTCC3′
pcDNA3-RREΔV (A) R-5′GCGGCCGCGAATTCCTGTGAGTTGCAACAGATGCTGTT3′
pcDNA3-RREΔV (B) F-5′ATAGAGAATTCAAGAATCCTGGCTGTGGAAAGATA3′
pSIREN-RRE was cloned by amplifying RRE from the HIV-1 provirus pNL4–3 using the following primers:
RRE F-5′ATATTGGATCCTAGCACCCACCAAGGCAAAGAGAA3′
RRE R-5′GCTATGAATTCTAGCATTCCAA GGCACAGCAGTGG3′
The resulting PCR product was digested by EcoRI and BamHI, and ligated into pSIREN-DNR (Clontech). pSIREN-TAR was cloned using the same approach.
TAR F-5′AATTAGGATCCAAAATTTTAAAATTTTGGGTCTCTCTGGTTAGACCAGATC3′
TAR R-5′AATTAAGAATTCATATATATATATATATTGGGTTCCCTAGTTAGCCACAG3′
The plasmid expressing the Enhanced Green Fluorescence Protein (EGFP), pEGFP-C1, was obtained from Clontech. pEGFP-clet7 was constructed by cloning a perfectly complementary sequence to let7 into the 3′ region of EGFP. The let7 sequence published in50 was used to design oligonucleotides with XhoI and SalI restriction enzyme sites. The annealed oligonucleotides were ligated into XhoI- and SalI- digested pEGFP-C1.
Let7 F-5′TCGAGACTACCTGCACTGTAAGCACTTT G3′
Let7 R-5′TCGACAAAGTGCTTACAGTGCAGGTAGT C3′
pHIV7-tdTomato was derived from pHIV7-CGFP81 by removing a Not I/Kpn I fragment containing the CMV promoter, eGFP, WPRE and HIV 5′LTR. To introduce an EagI site at the 5′ end of the WPRE, the WPRE and the HIV 5′LTR were then amplified by PCR with primers JRO 8 and JRO 9 and cloned back into the HIV7 backbone. To make the UBC::tdTomato fusion, the tdTomato coding region was first released by BamHI and EcoRI from pRSET-B-tdTomato (gift from Dr. Roger Tsien) and cloned into pcDNA3.1(+)Neo (Invitrogen), resulting in pCMV-tdTomato. The CMV promoter was then removed by cutting with Bgl II and BamHI and replaced by the Ubiquitin C (UBC) promoter82 that had been amplified from Jurkat genomic DNA with primer JRO128 and JRO129, resulting in pUBC-tdTomato. JRO128 contains NotI, XbaI and XmaI restriction sites for additional cloning. The entire UBC::tdTomato fragment was then cut by EagI and cloned back into the empty HIV7 vector to generate pHIV7-tdTomato.
JRO 8 F-5′TAACGGCCGGCTTATCGATAATCAACCTCTGG3′
JRO 9 R-5′GCGAATTGGGTACCGGGC3′
JRO128 F-5′aaagatctattaatgcggccgctctagacccgGGCCTCCGCGCCGGGTTTTGG3′
JRO129 R-5′aaggatccTGTCTAACAAAAAAGCCAAAAACGGCCAG3′
Cell culture and cell infection of Jurkat with HIV-1
HeLa, HEK 293T and Jurkat cells were cultured as previously.34,83,84 For virus production, HEK 293T cells were transfected with HIV-1 provirus pNL4–3 and supernatants were harvested 48 h post-transfection. Viral content of supernatant was evaluated by performing a reverse-transcriptase assay as described.84 For the infection, volumes of supernatant corresponding to 1 or 2 million cpms of reverse transcriptase activity were applied to Jurkat cells for 3 h before washing.
RT-PCR
For reverse transcription-PCR (RT-PCR), total RNA was extracted 48 h post-transfection with Trizol isolation reagent (Invitrogen) and treated by DNase I (Promega). Total cDNAs were reverse transcribed from 5 μg of total RNA using 150 ng of random primers (Invitrogen) in a 25-μl reaction mixture containing 40 U of RNaseOUT (Invitrogen), 1 mM of each of the deoxynucleoside triphosphates, 10 mM dithiothreitol, and 300 U of Superscript II (Invitrogen). Incubation was performed at 42°C for 90 min, and 5 μl of the resulting reaction mixture containing the single-strand cDNA template was used for PCR amplification. TAR, RRE and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were amplified by PCR in a 100-μl reaction mixture containing 0.5 mM of each forward and reverse primer, 2.5 U of Taq DNA polymerase (Invitrogen), 2.5 mM MgCl2, 0.2 mM of each of the deoxynucleoside triphosphates, and 1X Taq buffer (Invitrogen). The PCR reaction was carried out for 30 cycles, each consisting of 1 min at 94°C, 1 min at 63°C for TAR, 66°C for RRE and 55°C for GAPDH, 1 min at 72°C. The number of cycles was chosen to be in the linear range of the reaction. The products were fractionated on a 1% agarose gel.
GAPDH F-5′CCTTCATTGACCTCAACTACAT3′
GAPDH R-5′CCAAAGTTGTCATGGATGACC3′
PCR
For PCR on cells transfected with the adenovirus vectors, cells were digested in 0.3 mg/ml proteinase K at 37°C for 18 h and DNA was isolated by salting out.85 Viral and cellular DNA was verified by conducting PCR at different times post-transfection. For adenovirus DNA, the PCR reaction was carried out as follows: 5 min 94oC, [1 min at 94°C, 1 min at 56°C and 1 min at 72°C] × 25 cycles, 5 min 72oC. For GAPDH, the PCR reaction was: 5 min 94oC, [1 min at 94°C, 1 min at 60°C and 1 min at 72°C] × 30 cycles, 5 min 72oC. The number of cycles was chosen to be in the linear range of the reaction. The products were fractionated on a 1% agarose gel.
Primers used to amplify adenoviral DNA:
F-5′TATGTTTTGTTTGAAGTCTTTGACG3′
R-5′ GATGTTGCTTGCTTCTTTATGTTGT3′
Primers used to amplify GAPDH DNA:
F-5′ATGGGGAAGGTGAAGGTC
R-5′ GGTGCCATGGAATTTGCC
Immunoblotting
Cells were washed twice with phosphate buffered saline (PBS) and lysed in cold lysis buffer [50mM Tris-HCl (pH 7.4), 150 mM NaCl, 5 mM EDTA (pH 8), 10% Glycerol, 1% Nonidet (N) P-40] with the protease inhibitor cocktail (Roche) and with the phosphatase inhibitor PhosSTOP (Roche). Cell lysates were prepared, separated and transferred for immunoblotting as previously described.34,83,84 Membranes were blocked for 1 h in 5% nonfat milk and 0.05% Tris-buffered saline (TBS)-Tween and incubated overnight at 4°C with anti-EGFP (Santa Cruz) antibody at a 1/1000 dilution, anti-Myc (Santa Cruz) antibody at a 1/1000 dilution, anti-actin (Chemicon) antibody at a 1/10,000 dilution, anti-Dicer3495 or anti-Dicer (Medimabs, Montréal, QC, CA) at a 1/5000 dilution, anti-TRBPjbx36 at a 1/500 dilution, anti-Ago2 7C6 (from Dr. T. Hobman) at a 1/5000 dilution, anti-HIV-1 p24 183-H12–5C83,84 at a 1/1000 dilution, anti-Ad2/5 E1A (Santa Cruz) at a 1/1000 dilution, or anti-GAPDH (Santa Cruz) at a 1/1000 dilution in 5% milk/TBST. After 5 washes in TBST, membranes were incubated with peroxidase-conjugated secondary donkey anti-rabbit antibody (Amersham) for TRBP, Ago2, Dicer and E1A, and sheep anti-mouse (Amersham) for EGFP, Myc, Actin, p24 and GAPDH at a 1/5000 dilution. The bands were visualized as described86 and quantified by densitometry analysis, using Adobe Photoshop CS3 10.0.1 software.
Dual Luciferase assays
HeLa or HEK 293T cells were cotransfected in 24-well plates with 5 ng of pCMV-RL, pCMV-RL-clet7 or pCMV-RL-cmiR17 and 20 ng pCMV-FL using Polyethylenimine (PEI) (Polysciences) at a 1:3 DNA:PEI ratio. Cells were lysed 48 h after transfection and luciferase activities were measured using the Dual-Luciferase Reporter System (Promega). Renilla luciferase (RL) expression was normalized to Firefly luciferase (FL) expression to correct for transfection efficiency (RL/FL). Percentage RNAi activity was obtained by calculating RL/RL-clet750 or RL/RL-cmiR17.5
Electrophoresis Mobility Shift Assay (EMSA)
MBP and MBP-TRBP were produced in vitro from pMal-MBP and pMal-MBP-TRBP plasmids.45,47 RNAs were in vitro transcribed from linearized plasmids using the Riboprobe® System – T7 (Promega), using α32P-UTP when radiolabelled. TAR (80 nt) and Ctl1 (20 nt) RNAs were produced from pUC18-T7TAR-CAT48 cut by HindIII or BglII, respectively. Ctl2 RNA (165 nt) was produced from pcDNA3 plasmid linearized by Bcl1. RRE and RRE mutants were produced from their pcDNA3-derived plasmids linearized by Xho1. siRNA (Control non-silencing siRNA, Qiagen, 1022076) was radiolabelled using T4 polynucleotide kinase and γ32P-ATP. EMSAs were carried out as described.45 A concentration of 0.15–2.4 μM MBP or MBP-TRBP was incubated with labeled siRNA (2 × 103 cpm) or RRE RNA (12 × 103 cpm) for 20 min at 20°C. The samples were then separated by electrophoresis in a non-denaturing polyacrylamide gel (19:1, W/W, acrylamide/bisacrylamide) at 200V for 4 (siRNA) or 14 (RRE) h. Gels were fixed in 10% acetic acid, dried and autoradiographed for 64 or 16 h, respectively. In competition experiments, labeled siRNA (4 × 103 cpm) was incubated with MBP-TRBP 20 min prior to the addition of increasing amounts of cold RNA (TAR, RRE, mutants derived from RRE, Ctl1 or Ctl2 RNAs), and samples were separated by electrophoresis in a 4% polyacrylamide gel. Gels were fixed, dried, and autoradiographed for 64 h.
Immunoprecipitation (IP) and RNA-IP
IPs were carried out as described34 using anti-Dicer mAbDcr73,5 polyclonal anti-Dicer (Medimabs) or polyclonal anti-EGFP (control) antibodies. For RNA-IPs, HeLa cells or HEK 293T cells were transfected with pCMV-Myc or pCMV-Myc-TRBP2 and cotransfected with pSIREN-TAR, pSIREN-RRE or pcDNA3-RRE. Cells were lysed and extracts were immunoprecipitated in RNase-free conditions as described34 using anti-Myc (Santa Cruz) antibodies. One quarter of the immunoprecipitate was used to recover bound proteins by boiling the beads for 10 min and fractionated by 7.5% SDS-PAGE. The immunoprecipitates were analyzed by immunoblot by using anti-Dicer, anti-Myc and anti-actin or anti-GAPDH antibodies. The remainder of the immunoprecipitate was used to recover RNA by resuspension in 1 ml Trizol reagent (Invitrogen) and RNA extraction. RT-PCRs were performed as described above.
M-fold RNA modeling
M-fold software (http://mfold.rna.albany.edu/?q=mfold/RNA-Folding-Form55) was used to predict the secondary structures of WT RRE and mutants derived from RRE.
Adenovirus expression and detection
pAdWT (Type 5) and pAdΔVA were obtained from Dr. R. Alemany.18 Plasmids were transfected into HeLa cells. Viral and cellular DNA was verified by PCR at different times post-transfection and quantified by real-time qPCR at 48 h post infection. Viral expression was measured after 48 h of viral growth by detection of E1A early stage protein by Western Blotting.
Real-time qPCR
Cells were digested in 0.3 mg/ml proteinase K at 37°C for 18 h and DNA was isolated by salting out.85 Quality of total DNA was verified by conducting PCR against GAPDH before running qPCR. qPCR was run on Applied Biosystems Fast 7500 series thermocycler using a FAM-TAMRA probe and primers targeting the adenoviral genome:
Probe-5′6FAM-ATCGAAACCGTGTACCTGCGCAC-TAMRA3′
F-5′TATGTTTTGTTTGAAGTCTTTGACG3′
R-5′ GATGTTGCTTGCTTCTTTATGTTGT3′
Results were analyzed using the Applied Biosystems SDS 7500 v2.0.4 software. DNA expression levels in the linear range were calculated by the standard curve method using the pAdWT plasmid.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed
Acknowledgments
We thank Drs. L. Kleiman and E.A. Cohen for helpful suggestions, W. Filipowicz (F. Miescher Inst., Basel, Switzerland) for pRL, pRL-let7 and pRL-miR17 constructs and for Dicer antibodies, T. Hobman (U. of Alberta, Edmonton, Canada) for the Ago2 antibody, R. Tsien (U. California, San Diego) for the pRSET-B-tdTomato and R. Alemany (Institut Català d'Oncologia, Barcelona, Spain) for the adenovirus vectors. We thank Drs. D. Rekosh and M.L. Hammarskjöld for the pCMV-Rev through the NIH AIDS Reagent Program.
Funding
This work has been supported by Canadian Institutes of Health Research grants [HOP93434 and DCB120266 to AG], by a Vanier Canada Graduate Scholarship [CGV-204699 to SMD] and by a Banting and Best Canada Graduate Scholarship [CGD-96479 to RJS].
References
- 1. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev 2011; 91:827-87; PMID:21742789; http://dx.doi.org/ 10.1152/physrev.00006.2010 [DOI] [PubMed] [Google Scholar]
- 2. Lim LP, Glasner ME, Yekta S, Burge CB, Bartel DP. Vertebrate microRNA genes. Science 2003; 299:1540; PMID:12624257; http://dx.doi.org/ 10.1126/science.1080372 [DOI] [PubMed] [Google Scholar]
- 3. Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the dicer complex to ago2 for microRNA processing and gene silencing. Nature 2005; 436:740-4; PMID:15973356; http://dx.doi.org/ 10.1038/nature03868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Daniels SM, Gatignol A. The multiple functions of TRBP, at the hub of cell responses to viruses, stress, and cancer. Microbiol Mol Biol Rev 2012; 76:652-66; PMID:22933564; http://dx.doi.org/ 10.1128/MMBR.00012-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Haase AD, Jaskiewicz L, Zhang H, Lainé S, Sack R, Gatignol A, Filipowicz W. TRBP, a regulator of cellular PKR and HIV-1 virus expression, interacts with dicer and functions in RNA silencing. EMBO Rep 2005; 6:961-7; PMID:16142218; http://dx.doi.org/ 10.1038/sj.embor.7400509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme dicer in the maturation of the let-7 small temporal RNA. Science 2001; 293:834-8; PMID:11452083; http://dx.doi.org/ 10.1126/science.1062961 [DOI] [PubMed] [Google Scholar]
- 7. Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 2003; 17:3011-6; PMID:14681208; http://dx.doi.org/ 10.1101/gad.1158803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004; 305:1437-41; PMID:15284456; http://dx.doi.org/ 10.1126/science.1102513 [DOI] [PubMed] [Google Scholar]
- 9. Corbeau P. Interfering RNA and HIV: reciprocal interferences. PLoS Pathog 2008; 4:e1000162; PMID:18818734; http://dx.doi.org/ 10.1371/journal.ppat.1000162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gatignol A, Lainé S, Clerzius G. Dual role of TRBP in HIV replication and RNA interference: viral diversion of a cellular pathway or evasion from antiviral immunity? Retrovirol 2005; 2:65; PMID:16253139; http://dx.doi.org/ 10.1186/1742-4690-2-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virol 2011; 411:325-43; PMID:21277611; http://dx.doi.org/ 10.1016/j.virol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y, Lu J, Han Y, Fan X, Ding SW. RNA interference functions as an antiviral immunity mechanism in mammals. Science 2013; 342:231-4; PMID:24115437; http://dx.doi.org/ 10.1126/science.1241911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science 2013; 342:235-8; PMID:24115438; http://dx.doi.org/ 10.1126/science.1241930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ouellet DL, Provost P. Current knowledge of MicroRNAs and noncoding RNAs in virus-infected cells. Method Mol Biol 2010; 623:35-65; PMID:20217543; http://dx.doi.org/ 10.1007/978-1-60761-588-0_3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sagan SM, Sarnow P. Molecular biology. RNAi, antiviral after all. Science 2013; 342:207-8; PMID:24115433; http://dx.doi.org/ 10.1126/science.1245475 [DOI] [PubMed] [Google Scholar]
- 16. Sanghvi VR, Steel LF. A re-examination of global suppression of RNA interference by HIV-1. PloS one 2011; 6:e17246; PMID:21386885; http://dx.doi.org/ 10.1371/journal.pone.0017246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Andersson MG, Haasnoot PC, Xu N, Berenjian S, Berkhout B, Akusjärvi G. Suppression of RNA interference by adenovirus virus-associated RNA. J Virol 2005; 79:9556-65; PMID:16014917; http://dx.doi.org/ 10.1128/JVI.79.15.9556-9565.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cascallo M, Gros A, Bayo N, Serrano T, Capella G, Alemany R. Deletion of VAI and VAII RNA genes in the design of oncolytic adenoviruses. Hum Gene Ther 2006; 17:929-40; PMID:16972761; http://dx.doi.org/ 10.1089/hum.2006.17.929 [DOI] [PubMed] [Google Scholar]
- 19. Lu S, Cullen BR. Adenovirus VA1 noncoding RNA can inhibit small interfering RNA and MicroRNA biogenesis. J Virol 2004; 78:12868-76; PMID:15542639; http://dx.doi.org/ 10.1128/JVI.78.23.12868-12876.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immun 2005; 22:607-19; PMID:15894278; http://dx.doi.org/ 10.1016/j.immuni.2005.03.010 [DOI] [PubMed] [Google Scholar]
- 21. de Vries W, Berkhout B. RNAi suppressors encoded by pathogenic human viruses. Int J Biochem Cell Biol 2008; 40:2007-12; PMID:18571459; http://dx.doi.org/ 10.1016/j.biocel.2008.04.015 [DOI] [PubMed] [Google Scholar]
- 22. Fabozzi G, Nabel CS, Dolan MA, Sullivan NJ. Ebolavirus proteins suppress the effects of small interfering RNA by direct interaction with the mammalian RNA interference pathway. J Virol 2011; 85:2512-23; PMID:21228243; http://dx.doi.org/ 10.1128/JVI.01160-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog 2007; 3:e86; PMID:17590081; http://dx.doi.org/ 10.1371/journal.ppat.0030086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kakumani PK, Ponia SS, Rajgokul KS, Sood V, Chinnappan M, Banerjea AC, Medigeshi GR, Malhotra P, Mukherjee SK, Bhatnagar RK. Role of RNA interference (RNAi) in dengue virus replication and identification of NS4B as an RNAi suppressor. J Virol 2013; 87:8870-83; PMID:23741001; http://dx.doi.org/ 10.1128/JVI.02774-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saïb A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science 2005; 308:557-60; PMID:15845854; http://dx.doi.org/ 10.1126/science.1108784 [DOI] [PubMed] [Google Scholar]
- 26. Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA., et al. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci U S A 2004; 101:1350-5; PMID:14745017; http://dx.doi.org/ 10.1073/pnas.0308308100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qian S, Zhong X, Yu L, Ding B, de Haan P, Boris-Lawrie K. HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc Natl Acad Sci U S A 2009; 106:605-10; PMID:19122141; http://dx.doi.org/ 10.1073/pnas.0806822106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xu N, Segerman B, Zhou X, Akusjärvi G. Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the RNA-induced silencing complex and associate with polyribosomes. J Virol 2007; 81:10540-9; PMID:17652395; http://dx.doi.org/ 10.1128/JVI.00885-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhu Y, Cherukuri NC, Jackel JN, Wu Z, Crary M, Buckley KJ, Bisaro DM, Parris DS. Characterization of the RNA silencing suppression activity of the Ebola virus VP35 protein in plants and mammalian cells. J Virol 2012; 86:3038-49; PMID:22238300; http://dx.doi.org/ 10.1128/JVI.05741-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kok KH, Jin DY. Influenza A virus NS1 protein does not suppress RNA interference in mammalian cells. J Gen Virol 2006; 87:2639-44; PMID:16894203; http://dx.doi.org/ 10.1099/vir.0.81764-0 [DOI] [PubMed] [Google Scholar]
- 31. Lantermann M, Schwantes A, Sliva K, Sutter G, Schnierle BS. Vaccinia virus double-stranded RNA-binding protein E3 does not interfere with siRNA-mediated gene silencing in mammalian cells. Virus Res 2007; 126:1-8; PMID:17306404; http://dx.doi.org/ 10.1016/j.virusres.2007.01.009 [DOI] [PubMed] [Google Scholar]
- 32. Lin J, Cullen BR. Analysis of the interaction of primate retroviruses with the human RNA interference machinery. J Virol 2007; 81:12218-26; PMID:17855543; http://dx.doi.org/ 10.1128/JVI.01390-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bennasser Y, Yeung ML, Jeang KT. HIV-1 TAR RNA subverts RNA interference in transfected cells through sequestration of TAR RNA-binding protein, TRBP. J Biol Chem 2006; 281:27674-8; PMID:16887810; http://dx.doi.org/ 10.1074/jbc.C600072200 [DOI] [PubMed] [Google Scholar]
- 34. Daniels SM, Melendez-Peña CE, Scarborough RJ, Daher A, Christensen HS, El Far M, Purcell DF, Lainé S, Gatignol A. Characterization of the TRBP domain required for dicer interaction and function in RNA interference. BMC Mol Biol 2009; 10:38; PMID:19422693; http://dx.doi.org/ 10.1186/1471-2199-10-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Christensen HS, Daher A, Soye KJ, Frankel LB, Alexander MR, Lainé S, Bannwarth S, Ong CL, Chung SW, Campbell SM., et al. Small interfering RNAs against the TAR RNA binding protein, TRBP, a dicer cofactor, inhibit human immunodeficiency virus type 1 long terminal repeat expression and viral production. J Virol 2007; 81:5121-31; PMID:17360756; http://dx.doi.org/ 10.1128/JVI.01511-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Daher A, Laraki G, Singh M, Melendez-Peña CE, Bannwarth S, Peters AH, Meurs EF, Braun RE, Patel RC, Gatignol A. TRBP control of PACT-induced phosphorylation of protein kinase R is reversed by stress. Mol Cell Biol 2009; 29:254-65; PMID:18936160; http://dx.doi.org/ 10.1128/MCB.01030-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sanghvi VR, Steel LF. The cellular TAR RNA binding protein, TRBP, promotes HIV-1 replication primarily by inhibiting the activation of double-stranded RNA-dependent kinase PKR. J Virol 2011; 85:12614-21; PMID:21937648; http://dx.doi.org/ 10.1128/JVI.05240-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heaphy S, Dingwall C, Ernberg I, Gait MJ, Green SM, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 regulator of virion expression (rev) protein binds to an RNA stem-loop structure located within the rev response element region. Cell 1990; 60:685-93; PMID:1689218; http://dx.doi.org/ 10.1016/0092-8674(90)90671-Z [DOI] [PubMed] [Google Scholar]
- 39. Cook KS, Fisk GJ, Hauber J, Usman N, Daly TJ, Rusche JR. Characterization of HIV-1 REV protein: binding stoichiometry and minimal RNA substrate. Nucleic Acid Res 1991; 19:1577-83; PMID:2027765; http://dx.doi.org/ 10.1093/nar/19.7.1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Karn J, Dingwall C, Finch JT, Heaphy S, Gait MJ. RNA binding by the tat and rev proteins of HIV-1. Biochimie 1991; 73:9-16; PMID:1903308; http://dx.doi.org/ 10.1016/0300-9084(91)90068-C [DOI] [PubMed] [Google Scholar]
- 41. Suhasini M, Reddy TR. Cellular proteins and HIV-1 rev function. Curr HIV Res 2009; 7:91-100; PMID:19149558; http://dx.doi.org/ 10.2174/157016209787048474 [DOI] [PubMed] [Google Scholar]
- 42. McLaren M, Marsh K, Cochrane A. Modulating HIV-1 RNA processing and utilization. Front Biosci 2008; 13:5693-707; PMID:18508616; http://dx.doi.org/ 10.2741/3110 [DOI] [PubMed] [Google Scholar]
- 43. Fernandes J, Jayaraman B, Frankel A. The HIV-1 rev response element: an RNA scaffold that directs the cooperative assembly of a homo-oligomeric ribonucleoprotein complex. RNA Biol 2012; 9:6-11; PMID:22258145; http://dx.doi.org/ 10.4161/rna.9.1.18178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jeang KT, Gatignol A. Comparison of regulatory features among primate lentiviruses. Curr Top Microbiol Immunol 1994; 188:123-44; PMID:7924423 [DOI] [PubMed] [Google Scholar]
- 45. Daviet L, Erard M, Dorin D, Duarte M, Vaquero C, Gatignol A. Analysis of a binding difference between the two dsRNA-binding domains in TRBP reveals the modular function of a KR-helix motif. Eur J Biochem 2000; 267:2419-31; PMID:10759868; http://dx.doi.org/ 10.1046/j.1432-1327.2000.01256.x [DOI] [PubMed] [Google Scholar]
- 46. Erard M, Barker DG, Amalric F, Jeang KT, Gatignol A. An Arg/Lys-rich core peptide mimics TRBP binding to the HIV-1 TAR RNA upper-stem/loop. J Mol Biol 1998; 279:1085-99; PMID:9642086; http://dx.doi.org/ 10.1006/jmbi.1998.1831 [DOI] [PubMed] [Google Scholar]
- 47. Gatignol A, Buckler C, Jeang KT. Relatedness of an RNA-binding motif in human immunodeficiency virus type 1 TAR RNA-binding protein TRBP to human P1/dsI kinase and drosophila staufen. Mol Cell Biol 1993; 13:2193-202; PMID:8455607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gatignol A, Buckler-White A, Berkhout B, Jeang KT. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science 1991; 251:1597-600; PMID:2011739; http://dx.doi.org/ 10.1126/science.2011739 [DOI] [PubMed] [Google Scholar]
- 49. Yeung ML, Bennasser Y, Watashi K, Le SY, Houzet L, Jeang KT. Pyrosequencing of small non-coding RNAs in HIV-1 infected cells: evidence for the processing of a viral-cellular double-stranded RNA hybrid. Nucleic Acid Res 2009; 37:6575-86; PMID:19729508; http://dx.doi.org/ 10.1093/nar/gkp707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science 2005; 309:1573-6; PMID:16081698; http://dx.doi.org/ 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- 51. Park H, Davies MV, Langland JO, Chang HW, Nam YS, Tartaglia J, Paoletti E, Jacobs BL, Kaufman RJ, Venkatesan S. TAR RNA-binding protein is an inhibitor of the interferon-induced protein kinase PKR. Proc Natl Acad Sci U S A 1994; 91:4713-7; PMID:7515177; http://dx.doi.org/ 10.1073/pnas.91.11.4713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gredell JA, Dittmer MJ, Wu M, Chan C, Walton SP. Recognition of siRNA asymmetry by TAR RNA binding protein. Biochemistry 2010; 49:3148-55; PMID:20184375; http://dx.doi.org/ 10.1021/bi902189s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sakurai K, Amarzguioui M, Kim DH, Alluin J, Heale B, Song MS, Gatignol A, Behlke MA, Rossi JJ. A role for human dicer in pre-RISC loading of siRNAs. Nucleic Acid Res 2010; 39:1510-25; PMID:20972213; http://dx.doi.org/ 10.1093/nar/gkq846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Takahashi T, Miyakawa T, Zenno S, Nishi K, Tanokura M, Ui-Tei K. Distinguishable in vitro binding mode of monomeric TRBP and dimeric PACT with siRNA. PloS one 2013; 8:e63434; PMID:23658827; http://dx.doi.org/ 10.1371/journal.pone.0063434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mathews DH, Turner DH, Zuker M. RNA secondary structure prediction. Curr Protoc Nucleic Acid Chem 2007; 11:11 2; PMID:18428968; http://dx.doi.org/ 10.1002/0471142700.nc1102s28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chung J, Zhang J, Li H, Ouellet DL, DiGiusto DL, Rossi JJ. Endogenous MCM7 microRNA cluster as a novel platform to multiplex small interfering and nucleolar RNAs for combinational HIV-1 gene therapy. Hum Gene Ther 2012; 23:1200-8; PMID:22834872; http://dx.doi.org/ 10.1089/hum.2012.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yam PY, Li S, Wu J, Hu J, Zaia JA, Yee JK. Design of HIV vectors for efficient gene delivery into human hematopoietic cells. Mol Ther 2002; 5:479-84; PMID:11945076; http://dx.doi.org/ 10.1006/mthe.2002.0558 [DOI] [PubMed] [Google Scholar]
- 58. Lehmann M, Milev MP, Abrahamyan L, Yao XJ, Pante N, Mouland AJ. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J Biol Chem 2009; 284:14572-85; PMID:19286658; http://dx.doi.org/ 10.1074/jbc.M808531200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Molle D, Segura-Morales C, Camus G, Berlioz-Torrent C, Kjems J, Basyuk E, Bertrand E. Endosomal trafficking of HIV-1 gag and genomic RNAs regulates viral egress. J Biol Chem 2009; 284:19727-43; PMID:19451649; http://dx.doi.org/ 10.1074/jbc.M109.019844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pfeffer S. RNA silencing as a natural antiviral defense system in mammals: where are we now? Mol Ther 2010; 18:871-2; PMID:20436494; http://dx.doi.org/ 10.1038/mt.2010.68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev 2009; 23:1151-64; PMID:19451215; http://dx.doi.org/ 10.1101/gad.1793309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Whisnant AW, Bogerd HP, Flores O, Ho P, Powers JG, Sharova N, Stevenson M, Chen CH, Cullen BR. In-depth analysis of the interaction of HIV-1 with cellular microRNA biogenesis and effector mechanisms. mBio 2013; 4:e000193; PMID:23592263; http://dx.doi.org/ 10.1128/mBio.00193-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Triboulet R, Mari B, Lin YL, Chable-Bessia C, Bennasser Y, Lebrigand K, Cardinaud B, Maurin T, Barbry P, Baillat V., et al.. Suppression of microRNA-silencing pathway by HIV-1 during virus replication. Science 2007; 315:1579-82; PMID:17322031; http://dx.doi.org/ 10.1126/science.1136319 [DOI] [PubMed] [Google Scholar]
- 64. Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science 2005; 309:1577-81; PMID:16141076; http://dx.doi.org/ 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 65. Zhang C, Huys A, Thibault PA, Wilson JA. Requirements for human dicer and TRBP in microRNA-122 regulation of HCV translation and RNA abundance. Virol 2012; 433:479-88; PMID:22999255; http://dx.doi.org/ 10.1016/j.virol.2012.08.039 [DOI] [PubMed] [Google Scholar]
- 66. Narayanan A, Kehn-Hall K, Bailey C, Kashanchi F. Analysis of the roles of HIV-derived microRNAs. Expert Opin Biol Ther 2011; 11:17-29; PMID:21133815; http://dx.doi.org/ 10.1517/14712598.2011.540564 [DOI] [PubMed] [Google Scholar]
- 67. Parameswaran P, Sklan E, Wilkins C, Burgon T, Samuel MA, Lu R, Ansel KM, Heissmeyer V, Einav S, Jackson W., et al. Six RNA viruses and forty-one hosts: viral small RNAs and modulation of small RNA repertoires in vertebrate and invertebrate systems. PLoS Pathog 2010; 6:e1000764; PMID:20169186; http://dx.doi.org/ 10.1371/journal.ppat.1000764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bivalkar-Mehla S, Vakharia J, Mehla R, Abreha M, Kanwar JR, Tikoo A, Chauhan A. Viral RNA silencing suppressors (RSS): novel strategy of viruses to ablate the host RNA interference (RNAi) defense system. Virus Res 2010; 155:1-9; PMID:20951748; http://dx.doi.org/ 10.1016/j.virusres.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A 2008; 105:512-7; PMID:18178619; http://dx.doi.org/ 10.1073/pnas.0710869105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang HW, Noland C, Siridechadilok B, Taylor DW, Ma E, Felderer K, Doudna JA, Nogales E. Structural insights into RNA processing by the human RISC-loading complex. Nat Struct Mol Biol 2009; 16:1148-53; PMID:19820710; http://dx.doi.org/ 10.1038/nsmb.1673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Darlix JL, de Rocquigny H, Mauffret O, Mely Y. Retrospective on the all-in-one retroviral nucleocapsid protein. Virus Res 2014; 193:2-15; PMID:24907482; http://dx.doi.org/ 10.1016/j.virusres.2014.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Klase Z, Kale P, Winograd R, Gupta MV, Heydarian M, Berro R, McCaffrey T, Kashanchi F. HIV-1 TAR element is processed by dicer to yield a viral micro-RNA involved in chromatin remodeling of the viral LTR. BMC Mol Biol 2007; 8:63; PMID:17663774; http://dx.doi.org/ 10.1186/1471-2199-8-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Klase Z, Winograd R, Davis J, Carpio L, Hildreth R, Heydarian M, Fu S, McCaffrey T, Meiri E, Ayash-Rashkovsky M., et al. HIV-1 TAR miRNA protects against apoptosis by altering cellular gene expression. Retrovirol 2009; 6:18; PMID:19220914; http://dx.doi.org/ 10.1186/1742-4690-6-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ouellet DL, Plante I, Landry P, Barat C, Janelle ME, Flamand L, Tremblay MJ, Provost P. Identification of functional microRNAs released through asymmetrical processing of HIV-1 TAR element. Nucleic Acids Res 2008; 36:2353-65; PMID:18299284; http://dx.doi.org/ 10.1093/nar/gkn076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Schopman NC, Willemsen M, Liu YP, Bradley T, van Kampen A, Baas F, Berkhout B, Haasnoot J. Deep sequencing of virus-infected cells reveals HIV-encoded small RNAs. Nucleic Acids Res 2012; 40:414-27; PMID:21911362; http://dx.doi.org/ 10.1093/nar/gkr719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lee TC, Sullenger BA, Gallardo HF, Ungers GE, Gilboa E. Overexpression of RRE-derived sequences inhibits HIV-1 replication in CEM cells. New Biol 1992; 4:66-74; PMID:1536832 [PubMed] [Google Scholar]
- 77. Flint SJ, Enquist LW, Racaniello VR, Skalka AM. Principles of Virology. Washington, DC: ASM Press, 2009 [Google Scholar]
- 78. Esquela-Kerscher A, Slack FJ. Oncomirs – microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259-69; PMID:16557279; http://dx.doi.org/ 10.1038/nrc1840 [DOI] [PubMed] [Google Scholar]
- 79. Li MJ, Bauer G, Michienzi A, Yee JK, Lee NS, Kim J, Li S, Castanotto D, Zaia J, Rossi JJ. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol Ther 2003; 8:196-206; PMID:12907142; http://dx.doi.org/ 10.1016/S1525-0016(03)00165-5 [DOI] [PubMed] [Google Scholar]
- 80. Lewis N, Williams J, Rekosh D, Hammarskjöld ML. Identification of a cis-acting element in human immunodeficiency virus type 2 (HIV-2) that is responsive to the HIV-1 rev and human T-cell leukemia virus types I and II rex proteins. J Virol 1990; 64:1690-7; PMID:2157051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Unwalla HJ, Li HT, Bahner I, Li MJ, Kohn D, Rossi JJ. Novel Pol II fusion promoter directs human immunodeficiency virus type 1-inducible coexpression of a short hairpin RNA and protein. J Virol 2006; 80:1863-73; PMID:16439542; http://dx.doi.org/ 10.1128/JVI.80.4.1863-1873.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Schorpp M, Jager R, Schellander K, Schenkel J, Wagner EF, Weiher H, Angel P. The human ubiquitin C promoter directs high ubiquitous expression of transgenes in mice. Nucleic Acids Res 1996; 24:1787-8; PMID:8650001; http://dx.doi.org/ 10.1093/nar/24.9.1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Clerzius G, Shaw E, Daher A, Burugu S, Gélinas JF, Ear T, Sinck L, Routy JP, Mouland AJ, Patel RC, Gatignol A. The PKR activator, PACT, becomes a PKR inhibitor during HIV-1 replication. Retrovirol 2013; 10:96; PMID:24020926; http://dx.doi.org/ 10.1186/1742-4690-10-96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Scarborough RJ, Lévesque MV, Boudrias-Dalle E, Chute IC, Daniels SM, Ouellette RJ, Perreault JP, Gatignol A. A conserved Target Site in HIV-1 gag RNA is accessible to Inhibition by both an HDV ribozyme and a short hairpin RNA. Mol Ther Nucleic acids 2014; 3:e178; PMID:25072692; http://dx.doi.org/ 10.1038/mtna.2014.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988; 16:1215; PMID:3344216; http://dx.doi.org/ 10.1093/nar/16.3.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lainé S, Scarborough RJ, Lévesque D, Didierlaurent L, Soye KJ, Mougel M, Perreault JP, Gatignol A. In vitro and in vivo cleavage of HIV-1 RNA by new SOFA-HDV ribozymes and their potential to inhibit viral replication. RNA Biol 2011; 8:343-53; PMID:21422817; http://dx.doi.org/ 10.4161/rna.8.2.15200 [DOI] [PubMed] [Google Scholar]