Abstract
The centromere is the chromosomal region in which the kinetochore is assembled to orchestrate chromosome segregation. It is defined by the presence of a histone H3 variant called Centromere Protein A (CENP-A) or CenH3. Propagation of centromere identity entails deposition of new CENP-A upon exit from mitosis in vertebrate cells. A group of 16 proteins that co-immunoprecipitate with CENP-A, the Constitutive Centromere Associated Network or CCAN, contribute to kinetochore assembly and function. For most of them it is still unclear how and when they are recruited to centromeres and whether they have a role in CENP-A deposition. Taking advantage of the Xenopus egg cell-free system, we have addressed these issues for CCAN proteins CENP-C, CENP-T and CENP-W. CENP-C recruitment occurs as soon as sperm DNA, containing CENP-A, is added to the egg extract, and continues after de novo incorporation of CENP-A in early interphase. In contrast, centromeric recruitment of CENP-T occurs in late interphase and precedes that of CENP-W, which occurs in mitosis. Unlike CENP-C, CENP-T and CENP-W do not participate in CENP-A deposition. However, like CENP-C, they play a major role in kinetochore assembly. Depletion of CENP-C results in reduced amount of CENP-T at centromeres, an effect more prominent in mitosis than in interphase. In spite of this, kinetochores can still be assembled under this condition although the recruitment of Ndc80 and Mis12 is decreased. Our results support the existence of 2 pathways for kinetochore assembly directed by CENP-C and CENP-T/W, which can be reconstituted in Xenopus egg extracts.
Keywords: CCAN, CENP-A, CenH3, kinetochore
Abbreviations
- CCAN
Constitutive Centromere Associated Network
- CENP
centromere protein
Introduction
The kinetochore is an assembly of more than 100 proteins that provides the interface through which spindle microtubules interact with the sister chromatids to power their separation in mitosis and meiosis. Central to this interface is the KMN network, composed by 3 subcomplexes, KNL1/Blinkin/Spc105p, MIND/MIS12/Mtw1 and NDC80/Hec1.1,2 Kinetochores are assembled at a single locus in the chromosome, the centromere.3,4 In metazoans, this locus is epigenetically specified by the presence of nucleosomes containing a histone H3 variant known as centromere protein A (CENP-A), or cenH3, interspersed with canonical H3 nucleosomes.5-8 Centromere identity is maintained from one generation to the next by deposition of new CENP-A in every cell cycle. This deposition occurs upon mitotic exit in vertebrate cells 9 and requires a dedicated histone chaperone known as HJURP 10,11 and additional factors like the Mis18 complex.12,13 Kinetochore assembly downstream of CENP-A starts with the Constitutive Centromere Associated Network (CCAN), a group of 16 proteins that were identified based on their interaction with CENP-A and named from their continued presence at centromeres throughout the cell cycle.14,15 Importantly, while CENP-A nucleosomes are stably bound to centromeric chromatin,16 the binding of CCAN proteins appears to be dynamic.17-19 CENP-C and CENP-T serve as a platform for recruitment of the KMN network through pathways that are not yet well understood.20-29
From the CCAN components, CENP-N and CENP-C recognize CENP-A.30-32 Depletion of CENP-N or CENP-C in human cells by RNA interference leads to decreased incorporation of new CENP-A at centromeres.30,31 The contribution of CENP-N to the CENP-A deposition pathway is unknown whereas CENP-C has been proposed to drive HJURP targeting to centromeres through the Mis18 complex.33,34 CENP-T and CENP-W do not bind CENP-A but instead recognize H3 nucleosomes present in centromeric chromatin.35,36 These two proteins together with CENP-S and CENP-X, all 4 containing histone-fold domains, have been proposed to form a nucleosome-like particle capable of supercoiling DNA in vitro.37,38 How CENP-T/W/S/X are targeted to the centromere is currently unclear although it has been reported that their incorporation occurs in late S phase/G2 and requires CENP-A.18,35,36,39 It is also unclear whether they contribute to CENP-A deposition.
We employed the Xenopus egg cell-free extract system to investigate the recruitment of CENP-T and CENP-W and their role in de novo loading of CENP-A and kinetochore assembly. One advantage of this experimental system is the use of naïve templates in which centromeres are marked by the sole presence of CENP-A nucleosomes whereas all the rest of CCAN components must be recruited from the soluble egg extract to which the template DNA is added. Another advantage is that we analyze the defect caused by removing a protein in a single cell cycle without accumulation of errors from previous cycles in down regulation conditions (e.g., with siRNA). We previously developed an immunofluorescence-based assay to monitor de novo CENP-A incorporation.40 Using this assay we demonstrated that this incorporation requires exit from mitosis and the Xenopus homolog of HJURP.41 Here we show CENP-T and CENP-W are dispensable for CENP-A deposition whereas CENP-C is essential. CENP-T recruitment to centromeres occurs during DNA replication but is independent of this process. Incorporated CENP-T may be stabilized by CENP-C, particularly in mitosis. The amount of CENP-T at mitotic centromeres is drastically reduced in the absence of CENP-C, but it is nevertheless sufficient to recruit the outer kinetochore components Ndc80 and Mis12 to an extent similar to that observed in kinetochores assembled in the absence of CENP-T/W. These results suggest the existence of 2 parallel pathways of kinetochore assembly in the egg cell-free system.
Results
CENP-C, CENP-T and CENP-W are recruited to chromatin at different times
We developed specific antibodies against Xenopus CENP-C, CENP-T and CENP-W that recognize proteins in the soluble egg extract that migrate around 200 kDa, 120 kDa and 10 kDa, respectively (Fig. 1A). It has been previously shown that sperm chromatin contains CENP-A but no CENP-C.41,42 When this sperm chromatin is added to CSF extracts, prepared from unfertilized eggs arrested meiosis II, single-chromatid chromosomes are assembled. CENP-C co-localizes with CENP-A at the centromeres of these chromosomes (Fig. 1B, left). If calcium is added to the assembly mixture, a signaling cascade takes place that elicits exit from the metaphase II arrest into interphase. Chromosomes decondense, a nuclear membrane surrounds the chromatin and DNA replication ensues. CENP-C and CENP-A are still present at the centromeres in these interphase nuclei (Fig. 1B, middle). After 90 to 120 minutes, when replication is completed, CSF extract is added to the mixture providing enough cyclin B to drive CDK1 activation and entry into mitosis. The nuclear membrane breaks down and chromosomes condense. Now, CENP-C and CENP-A staining appear as double dots that correspond to the sister kinetochores of the replicated chromosomes (Fig. 1B, right).
Figure 1.
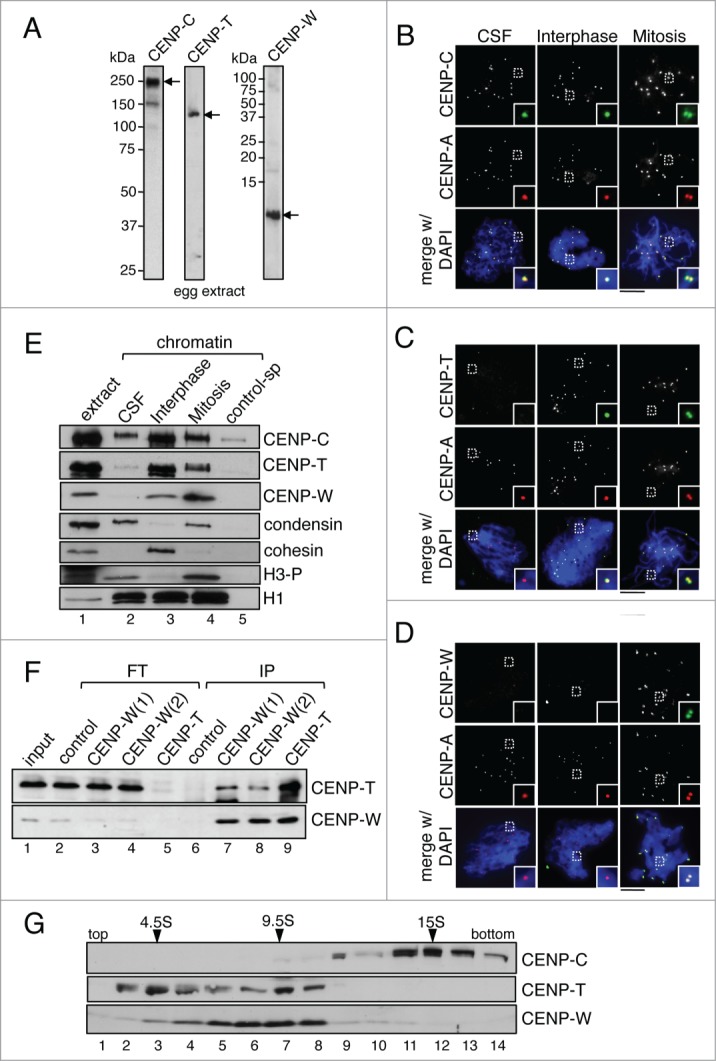
CENPC, CENP-T and CENP-W are recruited to chromatin at different times. (A) Immunoblot analysis of an egg extract with the indicated antibodies. For CENP-W, 2 different antibodies were raised, CENP-W(1) and CENP-W(2) (see Methods for description). While both worked for immunoprecipitation (see Fig. 1F), CENP-W(1) worked best for immunoblot and immunofluorescence. (B-D) Immunofluorescence analysis of CSF (unreplicated) chromosomes, interphase nuclei and mitotic (replicated) chromosomes with antibodies against CENP-C (B), CENP-T (C) and CENP-W (D), in green, and Cy3-labeled anti-CENP-A in red to label centromeres. DNA was counterstained with DAPI (blue). Colored images are shown for the insets and the merge with DAPI while single antibody stainings appear as gray scale images. Scale bar is 10μm. (E) Immunoblot analysis of chromatin assembled in CSF, interphase and mitotic extracts and purified by centrifugation through a sucrose cushion. Chromatin purified in the same way from a mock assembly reaction without sperm serves as control (control-sp, lane 5). Cohesin (Rad21) is a marker for interphase chromatin whereas condensin (CAP-G) and phospho H3 (H3-P) are present in CSF and mitotic chromatin. Histone H1 is used as loading control. (F) Immunoprecipitates (IP) obtained from soluble egg extracts were analyzed by immunoblot together with aliquots of the flow through (FT) from each reaction and of the input extract (1%). Immunoprecipitation with non-immune rabbit IgG was used as control. (G) A soluble egg extract was fractionated on a sucrose gradient (5-20% sucrose) and analyzed by immunoblot. The sedimentation coefficients of the major peaks of CENP-T, CENP-W and CENP-C are indicated.
In contrast to CENP-C, we do not detect CENP-T or CENP-W staining on the CSF chromosomes (Fig. 1C and D, left). For CENP-T, centromere signals can be observed in interphase nuclei and mitotic chromosomes whereas CENP-W antibodies only stain centromeres in mitosis (Fig. 1C and D, middle and right). To confirm these results, we isolated chromatin from the different assembly mixtures and analyzed it by immunoblot (Fig. 1E). Consistent with the immunofluorescence results, only CENP-C is present in CSF chromatin and CENP-T is recruited during interphase. Although some CENP-W can be detected in interphase chromatin, the bulk of CENP-W is recruited in mitosis. Thus, the 3 proteins appear to bind centromeric chromatin at different times. This result is most surprising in the case of CENP-T and CENP-W, since it has been reported that they assemble into a nucleosome-like particle together with CENP-S/X.37 For that reason, we asked whether CENP-T and CENP-W associate in the soluble egg extract and indeed, immunoprecipitation reactions show that CENP-T antibodies pull down CENP-W and vice versa (Fig. 1F). However, we also noticed that most of the CENP-T and CENP-W are removed from the extract after immunoprecipitation with the CENP-T antibody whereas immunoprecipitation with 2 distinct CENP-W antibodies is less efficient in removing CENP-W from the extracts and does not greatly effect CENP-T levels (compare lanes 3 and 4 with lane 5 in Fig. 1F). This suggests that there are 2 different populations of CENP-T in the extract, with and without CENP-W. Fractionation of an egg extract in a sucrose gradient confirms that CENP-T is present in 2 major peaks, one around 4.5S and other around 9.5S, and only the latter contains also CENP-W (Fig. 1G). It is therefore possible that CENP-T molecules that are not bound to CENP-W in the soluble extracts are the ones targeted to centromeric chromatin in interphase.
Centromeric localization of CENP-T in interphase does not require DNA replication
CENP-T recruitment to centromeres during interphase coincides with DNA replication, which can be followed by incorporation of a labeled nucleotide into replicated chromatin (Fig. 2A). To check if these 2 processes are actually coupled, we assembled interphase nuclei in the presence of the DNA polymerase inhibitor aphidicolin. Under this condition, no incorporation of biotin-dUTP can be detected, but CENP-T is properly localized at centromeres (Fig. 2B) and only a small decrease in the amount of centromeric CENP-T is observed (Fig. 2C). Next, we asked whether CENP-C must be present in addition to CENP-A in order for CENP-T to recognize centromeric chromatin. Extracts were depleted from CENP-C with a specific antibody. Immunoblot analysis shows that more than 90% of CENP-C is removed from the soluble extracts whereas the levels of CENP-T remain unchanged (Fig. 2E). CENP-T can still be found at centromeres of interphase nuclei assembled in these extracts (Fig. 2D), although signals are reduced in intensity by 40% (Fig. 2C). This partial effect is unlikely to be due to the presence of residual CENP-C at centromeres (Fig. S1). Thus, CENP-C may stabilize CENP-T after its incorporation at centromeres but its presence is not strictly required for this process, which is also independent of DNA replication.
Figure 2.
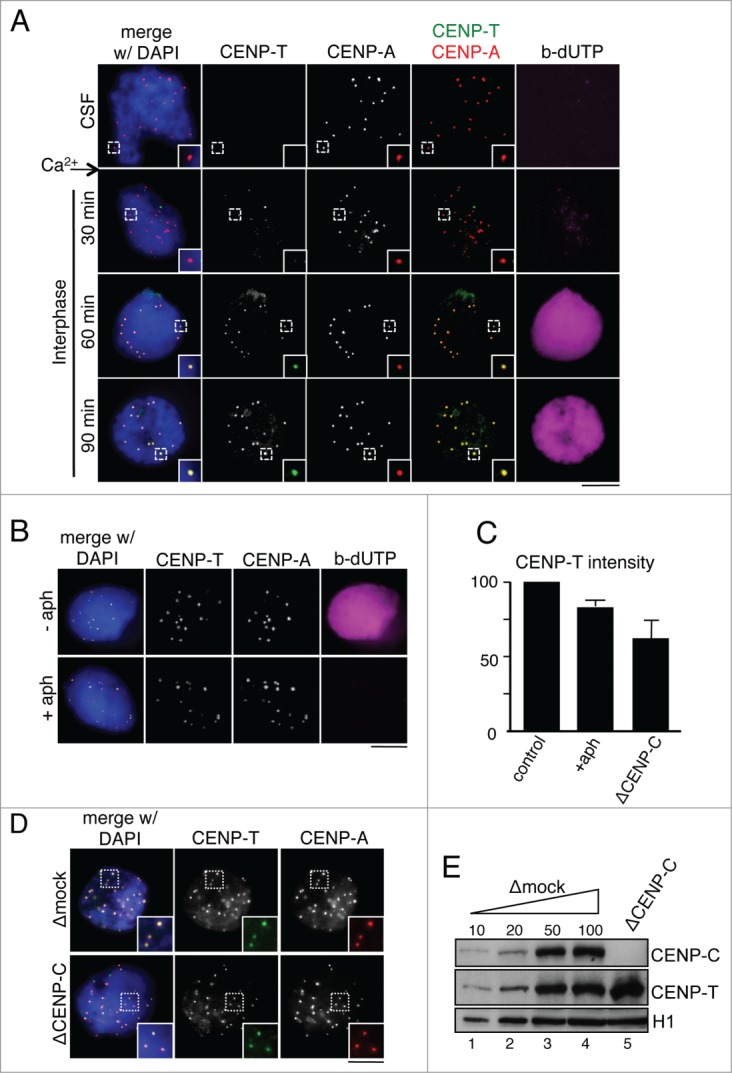
Requirements of CENP-T recruitment to interphase centromeres. (A) Time course analysis of CENP-T (green) recruitment to centromeres marked with Cy3-labeled anti-CENP-A (red). DNA replication is followed by incorporation of biotin-dUTP (magenta) and DNA is counterstained with DAPI (blue). The CENP-T signals at t = 30 min do not overlap with centromeres and are likely background. (B) Interphase nuclei assembled in the presence or absence of aphidicolin were analyzed with antibodies against CENP-T (green) and CENP-A (red). No incorporation of biotin-dUTP (magenta) can be detected in the presence of aphidicolin. (C) Intensity of CENP-T signals at centromeres of nuclei assembled under the indicated conditions. At least 200 centromeres were measured in 3 independent experiments per condition. Intensities were normalized to the mean value of the corresponding control experiment (no aphidicolin or mock-depletion). Bars represent mean± SEM. (D) Immunofluorescence analysis of CENP-T (green) localization in replicated interphase nuclei assembled in the presence (Δmock) or absence of CENP-C (ΔCENP-C). Centromeres are stained with Cy3-labeled anti-CENP-A (red). Scale bars, 10 μm. (E) Immunoblot analysis of CENP-C depleted extracts used in (D). Increasing amounts of mock-depleted CSF extract (Δmock), expressed as percentage, and extracts depleted from the indicated proteins were analyzed side by side by immunoblot. H1 was used as loading control.
Loading of new CENP-A in interphase is accompanied by loading of new CENP-C
It has been reported that CENP-C recognizes CENP-A present in centromeric chromatin.31,32 We asked whether the loading of new CENP-A upon exit from mitosis also provides new binding sites for CENP-C. To answer this question, we adapted the CENP-A loading assay to CENP-C. In this assay, CSF chromosomes and interphase nuclei are spun onto the same coverslip, stained and imaged together.40 CSF and interphase chromosomes can be distinguished by their distinct morphology and because only the latter incorporates UTP (Fig. 3A). On these images, we quantify signals at individual centromeres and calculate the average increase in staining for at least 15 pairs of CSF chromosomes/interphase nuclei. We observed a clear increase in CENP-C intensity in chromatin assembled in control extracts but not in extracts depleted of soluble CENP-A, in which CENP-A loading is impaired (Fig. 3B and C). Thus, CENP-A deposition is accompanied by loading of new CENP-C.
Figure 3.
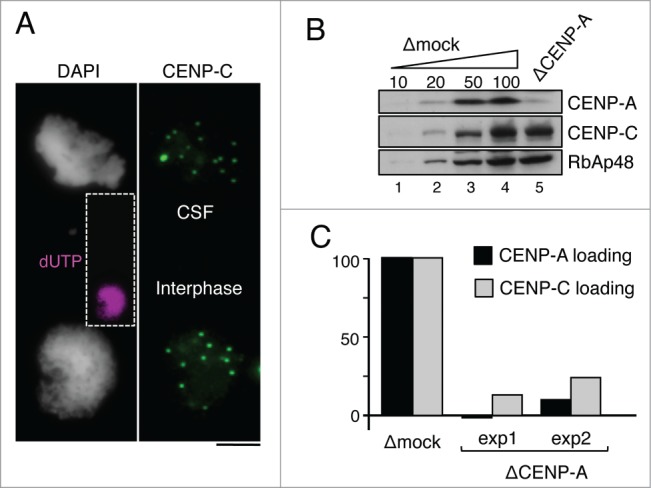
De novo loading of CENP-C following CENP-A incorporation in interphase. (A) Representative image of a pair used for analysis of CENP-C loading. CENP-C signals (green) from a mass of CSF chromosomes (top) are compared with those from an interphase nucleus (bottom), the latter already replicated as indicated by incorporation of dUTP (inset in magenta). Scale bar, 10 μm. (B) Increasing amounts of mock-depleted CSF extract (Δmock), expressed as percentage, and a CENP-A depleted extract (ΔCENP-A), were analyzed side by side to estimate the extent of depletion. RbAp48 is used as loading control. (C) Quantification of CENP-A loading (black bars) and CENP-C loading (gray bars) in CENP-A depleted extracts relative to the corresponding mock depleted extract using the loading assay described in the main text. At least 250 centromeres from more than 10 pairs as the one shown in (A) were measured for each condition in 2 independent experiments (exp1 and exp2).
CENP-T and CENP-W are not required for CENP-A deposition
Next, we asked if CENP-C, CENP-T and CENP-W participate in CENP-A deposition using the CENP-A loading assay. Depletion of CENP-C from the soluble extract severely impairs loading of new CENP-A (Fig. 4A and D). When mRNA of a myc-tagged version of CENP-C is added back to the depleted extract the corresponding mycCENP-C is synthesized (lane 6 in Fig. 4A) and CENP-A loading is restored to normal levels. Thus, CENP-C is essential for the assembly of new CENP-A, as previously suggested.33 In contrast, depletion of CENP-T to less than 10% of its normal levels in the extract does not have a major effect on CENP-A deposition (Fig. 4A and 4D). Depletion of CENP-W impairs CENP-A loading (Fig. 4C and 4F), but this is due to co-depletion of CENP-C (lane 5 in Fig. 4C). Addition of mycCENP-W to the CENP-W depleted extract does not rescue the CENP-A loading defect whereas addition of only mycCENP-C does. Moreover, depletion of CENP-W with another antibody that does not co-deplete CENP-C has no effect on CENP-A loading (Fig. S2). Thus, unlike CENP-C, neither CENP-T nor CENP-W is required for CENP-A deposition.
Figure 4.
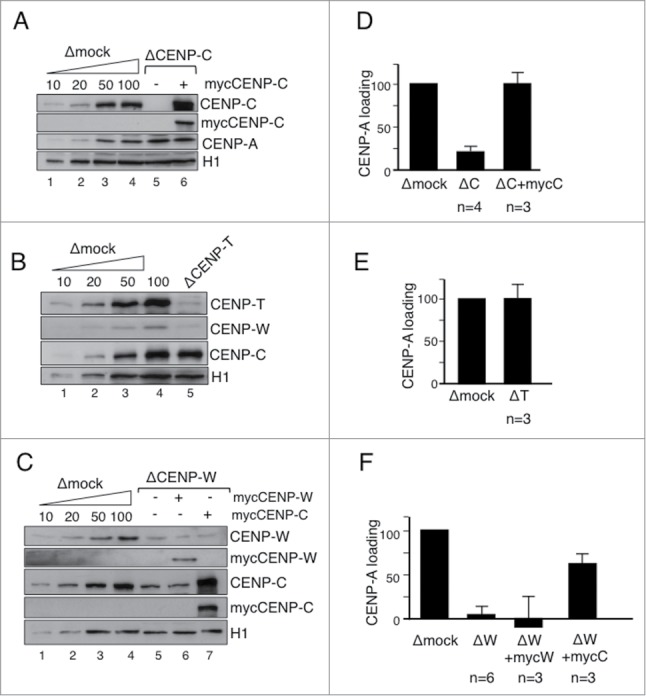
CENP-T and CENP-W are not required for CENP-A loading. (A-C) Immunoblot analyses of extracts used in the CENP-A loading assays shown in (D-F). Increasing amounts of mock depleted CSF extract (Δmock) and extracts depleted from the indicated proteins with or without add-back of myc-tagged proteins were analyzed side by side. H1 is used as a loading control. In (C), the CENP-W(2) antibody was used for depletion. (D-F) Bar graphs representing CENP-A loading efficiency in nuclei assembled in the indicated extracts. More than 250 centromeres were quantitated per condition and experiment. The number of experiments (n) is indicated below each bar. Bars represent mean± SEM.
We confirmed the interaction between CENP-C and CENP-W by immunoprecipitation from soluble egg extracts (Fig. S3A). The CENP-W(1) antibody pulls down both CENP-C and CENP-T in addition to CENP-W whereas the CENP-T antibody pulls down CENP-T and CENP-W, and the CENP-C antibody pulls down only CENP-C. These results, together with the results in Figure 1F and G, indicate that only a minor fraction of CENP-W is stored in the oocyte cytoplasm associated to CENP-C, independently of CENP-T, while most CENP-W is in association with CENP-T. In contrast, most CENP-C is present in the CSF extract in a complex with CENP-W, since depletion of CENP-W with the CENP-W(1) antibody co-depletes CENP-C to similar extent (around 80%, lane 5 in Fig. 4C). Although the functional significance of this association is currently unclear, it is interesting to note that it is cell cycle specific, as it is not detected in interphase (Fig. S3B).
CENP-C and CENP-T are required for kinetochore assembly in mitosis
We have previously shown that CENP-C depletion reduces by 40% the amount of centromeric CENP-T in interphase nuclei (Fig. 2C and D). When these nuclei are driven to mitosis the amount of CENP-T further decreases to around 20% of the amount present in chromosomes assembled in mock depleted extracts (Fig. 5A). CENP-W cannot be detected at mitotic centromeres by immunofluorescence in the absence of CENP-C (Fig. 5B), although immunoblot analysis of chromatin fractions reveals a small population remaining in these chromosomes (compare lanes 4 and 7 in Fig. 5C). On the other hand, CENP-T depletion does not affect CENP-C localization (Fig. 5D).
Figure 5.
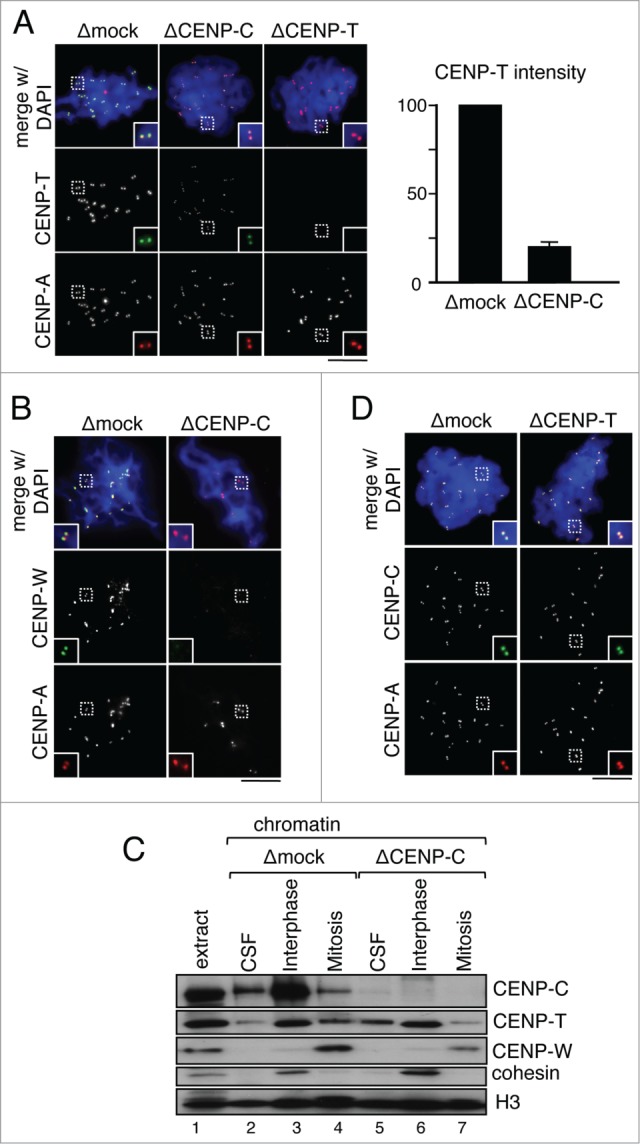
Maintenance of CENP-T/W at mitotic centromeres depends on CENP-C. (A) Immunofluorescence analysis of CENP-T (green) localization at mitotic centromeres (labeled by CENP-A, red) assembled in extracts that had been mock-depleted or depleted of CENP-C. The graph on the right shows the centromeric fluorescent intensity for CENP-T as percentage of the mean value obtained in the corresponding control experiment. At least 970 centromeres were measured for each condition in 5 independent experiments. (B) CENP-W localization in chromosomes assembled as in (A). (C) Immunoblot analysis of chromatin fractions assembled in CSF, interphase and mitotic extracts following depletion of CENP-C or mock depletion as control. Cohesin (Rad21) is a marker for interphase. Histone H3 is used as loading control. (D) Immunofluorescence analysis of CENP-C (green) localization at mitotic centromeres (labeled by CENP-A, red) in chromosomes from mock depleted or CENP-T depleted extracts. Scale bars, 10 μm.
The CCAN protein network connects centromeric chromatin to the outer kinetochore in mitosis. Some experimental evidences suggest a certain redundancy of 2 parallel pathways directed by CENP-C and CENP-T/W, the former being more important for Mis12 complex (Mis12C) recruitment and the latter for recruitment of Ndc80 complex (Ndc80C).23,24,28 Other evidences, however, are more consistent with the existence of a single pathway with CENP-C at the top.29 To investigate this issue in our experimental system, we performed immunofluorescent staining of mitotic chromosomes assembled in egg extracts lacking CENP-C, CENP-T or both with antibodies against Mis12 and Ndc80. Importantly, the levels of these KMN components in the soluble extracts remain unchanged after the depletions (Fig. 6A). We observe a partial reduction of Mis12C (Fig. 6B) and Ndc80C (Fig. 6C) signals in the singly depleted extracts while neither protein can be detected when CENP-T and CENP-C are eliminated simultaneously. Thus, despite the fact that depletion of CENP-C reduces significantly the amount of CENP-T/W at mitotic centromeres (Fig. 5A–C), the remaining CENP-T/W can recruit both Mis12C and Ndc80C. We conclude that 2 parallel pathways driven by CENP-C and CENP-T/W promote kinetochore assembly and can be reconstituted in Xenopus egg extracts.
Figure 6.
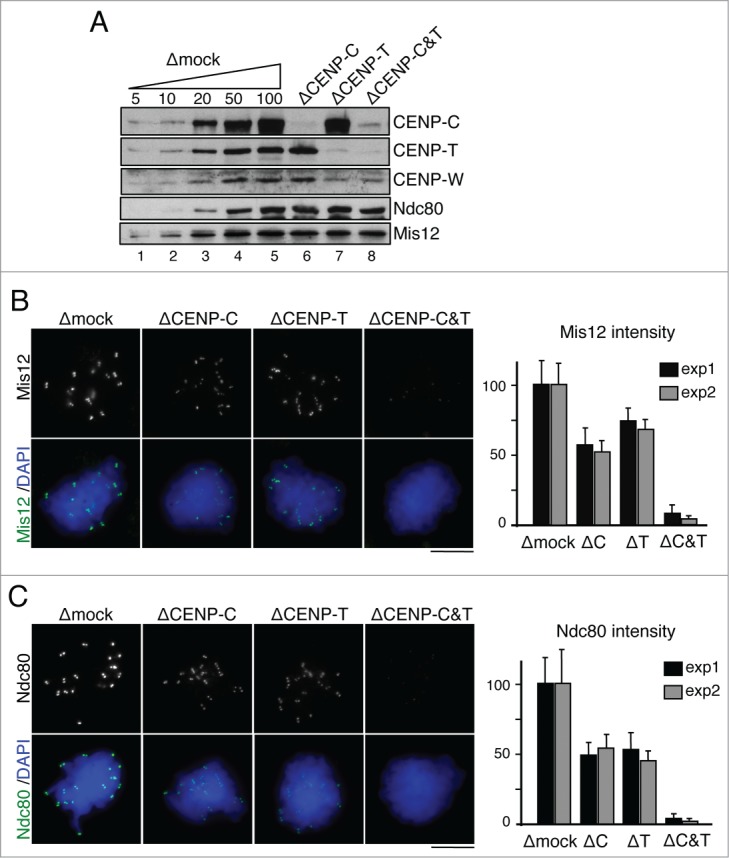
CENP-C and CENP-T/W contribute to kinetochore assembly. (A) Immunoblot analysis of the extracts used to assemble the chromosomes shown in (B) and (C). (B-C) Representative examples of chromosomes assembled in mock depleted extracts and extracts lacking CENP-C, CENP-T or both, and stained with antibodies against Mis12 (B) and Ndc80 (C). Quantification of total fluorescence in dots per chromosome mass is shown in the graphs on the right. Bars represent mean± SD. Between 10 and 20 nuclei per condition were measured in 2 independent experiments (exp1 and exp2).
Discussion
Here we have addressed the role of the CCAN components CENP-C and CENP-T/W in the 2 major aspects of centromere function, self-propagation and kinetochore assembly, using Xenopus egg extracts. We confirm the importance of CENP-C in the CENP-A deposition pathway, previously observed by Moree et al. 33 with a different assay that measures incorporation of exogenously added CENP-A. Importantly, we show that CENP-T and CENP-W are not required in this pathway. This is consistent with a recent study showing that down regulation of CENP-C, but not of CENP-T, reduces Mis18 complex localization to G1 centromeres in human cells.43 Another interesting finding is the sequential recruitment of CENP-C, CENP-T and CENP-W to a naïve chromatin template whose centromeres contain CENP-A nucleosomes but no CCAN proteins (Fig. 7). CENP-C present in the soluble egg extract is capable of recognizing and binding CENP-A as soon as the sperm chromatin is added to CSF extract.42 Moreover, when additional CENP-A is incorporated in the DNA upon exit from mitosis, new CENP-C is loaded as well. However, we find little CENP-T in CSF chromatin and, instead, its recruitment occurs in late interphase. This recruitment could depend on a factor that is only activated at this stage and/or could require a modification of the chromatin template that results from some interphase event.44 In the Xenopus egg cell-free system there is little transcription and the main process happening in interphase chromatin is DNA replication. We show here that CENP-T assembles onto centromeres even if DNA replication is inhibited by addition of aphidicolin. CENP-C is not required for CENP-T or its putative loader to recognize centromeric chromatin. However, it may stabilize the incorporated CENP-T either directly or through the HIKM complex.29
Figure 7.
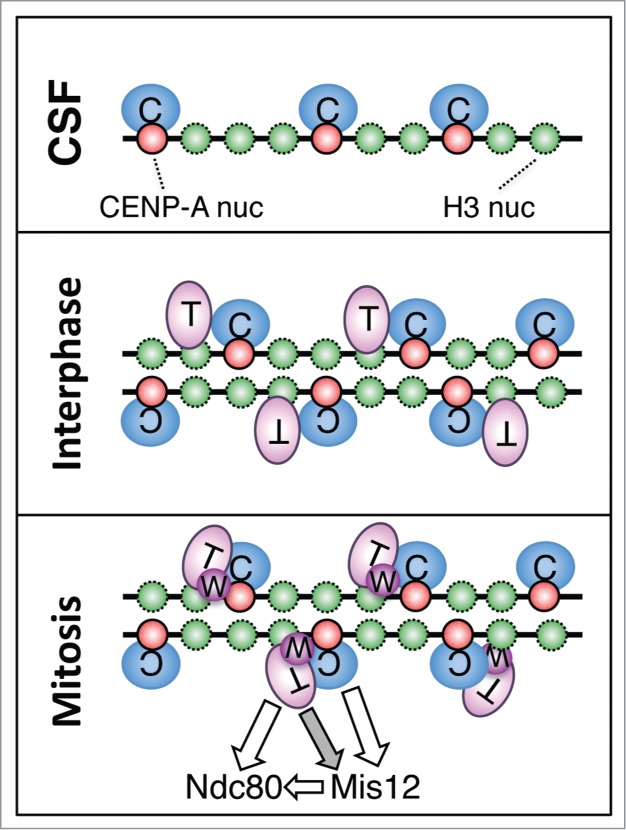
Sequential recruitment of CENP-C, CENP-T and CENP-W to centromeres in Xenopus egg extracts. CENP-C recognizes CENP-A and binds to centromeric chromatin when sperm is added to CSF extracts. When these extracts are driven to interphase, de novo loading of CENP-A occurs, which requires CENP-C but no CENP-T or CENP-W. The new CENP-A also recruits more CENP-C. Soon after DNA replication, CENP-T binds to centromeres. CENP-C is not strictly required for this process, but its presence may stabilize the incorporated CENP-T. Upon entry in mitosis, CENP-W can be detected at centromeres. CENP-C becomes more important for maintenance of CENP-T/W at this stage, most likely by providing stability to centromeric chromatin. Both CENP-C and CENP-T/W direct kinetochore assembly by recruiting Mis12C and Ndc80C, respectively. It is currently unclear how CENP-T affects Mis12C recruitment independently of Ndc80C (gray arrow), although a weak interaction between them has been detected in vitro.24
Unexpectedly, we observe little CENP-W in interphase chromatin. While we cannot discard the possibility that the antibody that we use is unable detect CENP-W at interphase centromeres by immunofluorescence, immunoblot analyses show a clear increase of CENP-W on chromatin in mitosis with respect to interphase. Thus, even if a small population of CENP-W is recruited to interphase centromeres along with CENP-T, we find that bulk recruitment of CENP-W occurs in mitosis. Myc-tagged CENP-W added to the extracts cannot localize to centromeres at any cell cycle stage, maybe because it cannot compete with endogenous CENP-W already stored in a complex with CENP-T in the egg cytoplasm. Our findings of a different recruitment time for CENP-T and CENP-W could be consistent with the differences in dynamics reported for the 2 proteins in FRAP studies in human cells.18 In the same study, the fluorescence staining of centromere-associated CENP-T increases by 5-fold from early S-phase to G2 but only 2-fold for CENP-W. Thus, CENP-T could be assembled first onto replicated chromatin and later on recruit CENP-W/S/X. In vitro assays suggest the existence of multiple DNA binding sites in CENP-T and CENP-W.28,38 While in human cells the assembly of the CENP-T/W/S/X complex on chromatin occurs in late S/G2 phase cells,18,36 it may take place in mitosis in the egg extracts. Xenopus homologues of CENP-S and CENP-X are present in the database but no antibodies against them are available yet to test when they are recruited to centromeres.
Our results also show that assembly and/or maintenance of CENP-T/W at mitotic centromeres requires CENP-C. Previous results in human and chicken somatic cells using siRNA and genetic conditional deletion, respectively, suggest a more independent behavior of CENP-C and CENP-T/W in some studies 24,35 but not in other.29 It is important to note that in our experimental system the mitotic centromeric chromatin assembled in CENP-C depleted extracts does not contain any CENP-C while in other systems complete removal of chromatin-bound CENP-C is difficult to achieve.8 As shown in other organisms,22,23 CENP-C likely interacts with Mis12C and this, in turn, can recruit Ndc80C to centromeres even in the absence of CENP-T/W.28,45 A previous study also showed that Ndc80 and Mis12 co-immunoprecipitate from Xenopus egg extracts.46 When CENP-C is missing, centromeric chromatin may be destabilized and only a small fraction of CENP-T/W remains at centromeres. Recent measurements of the relative positions of kinetochore components by super resolution microscopy suggest that only a fraction of CENP-C is actually involved in interactions with Mis12C 47 whereas the rest could be performing a scaffolding role by interacting directly with centromeric chromatin.48 Importantly, the fraction of CENP-T/W that remains at mitotic centromeres lacking CENP-C is still able to recruit both Mis12C and Ndc80C. The interaction between Ndc80C components Spc24/25 and the N-terminal region of CENP-T has been described and shown to be mutually exclusive with the interaction between Ndc80C and Mis12C.28,45 Thus, another link must exist between CENP-T and Mis12C (Fig. 7, gray arrow).
In summary, our results support the existence of 2 pathways for kinetochore assembly directed by CENP-C and CENP-T/W,21,24,28,29,45 which can be reconstituted in the Xenopus cell-free system. Moreover, kinetochores can be assembled onto sperm either directly in CSF extracts or by allowing sperm to replicate and then reenter mitosis. Our studies demonstrate that these 2 experimental approaches build very different kinetochores. Kinetochores assembled in CSF extracts lack the entire CENP-T/W and presumably S/X branch of assembly and appear to utilize only the CENP-C branch. In contrast, kinetochores assembled after passage through interphase have both branches. This distinction provides an experimental opportunity to separate these 2 branches of kinetochore assembly. Moreover, some previous results using CSF extracts to test kinetochore function may require reinterpretation in light of this distinction, which must be taken into account in future experiments using this system.
Materials and Methods
Antibody preparation
Full-length cDNAs of Xenopus tropicalis CENP-T and Xenopus laevis CENP-W were transferred from IMAGE clones 7793391 and 6323080, respectively, to pENTR/D-TOPO first and then to pDEST-17 (Invitrogen) to produce a His-tagged polypeptides in E. coli that were purified and injected in rabbits for antibody production (Innovagen AB, Sweden). Rabbit polyclonal antibodies were also raised against an N-terminal fragment of Xenopus laevis CENP-C (amino acids 207-296, a gift from A. F. Straight) and a C-terminal peptide of CENP-W (CIKAVAKTALKKSKG) coupled to KLH. The antibody raised against full-length CENP-W, CENP-W(1), was used for immunoblot and immunofluorescence in all the Figures. The antibody against the C-terminal peptide, CENP-W(2) was used for immunoprecipitation and depletion as indicated. Other antibodies used in this study have been described before. Embryonic histone H1, CENP-T N-terminal peptide, RbAp48 41; Xenopus CENP-A 49; cohesin Rad21 50; condensin CAP-G 51; phospho H352; Ndc80 and Mis12 46.
Preparation and use of Xenopus egg extracts
Cytostatic factor (CSF)-arrested low speed supernatants of Xenopus eggs were prepared in XBE2 buffer (10 mM K-Hepes [pH 7.7], 0.1 M KCl, 2 mM MgCl2, 0.1 mM CaCl2, 5 mM EGTA and 50 mM sucrose). To drive CSF extracts to interphase 100 μg/ml cycloheximide and 0.5 mM CaCl2 were added. When required, aphidicolin (Sigma) was added at 50 μg/ml and biotin-16-dUTP (Roche) at 10 μM. Chromosome assembly reactions containing 800-1000 sperm nuclei/μl were processed for chromatin fractionation and immunoblot or for immunofluorescence as described.49 Immunoprecipitation reactions were carried out on 10 μl of protein A agarose beads using 100 μl of extract and 2-5 μg of antibody. Fractionation of a CSF extract in a 5-20% sucrose gradient was performed as described.51
Immunodepletion and add back
For immunodepletions antibodies were bound to Affiprep Protein A support (Bio-Rad Laboratories). Depletion of 100 μl of extract required one (CENP-C) or 2 rounds (CENP-A, CENP-T, CENP-W) of incubation with 50 μl of beads bound to 30 μg of antibody. For add back experiments, the full-length cDNA of CENP-W was cloned into pCS2+myc vector and the corresponding myc-tagged proteins was produced with TNT Quick Coupled Transcription/Translation system (Promega). The reticulocyte lysate containing the protein was added to the depleted extract (up to 10% of extract volume) before addition of the sperm. Full length CENP-C cloned in pCS2+myc was a generous gift of A.F. Straight.33 In this case, the vector was linearized with SalI and used for the production of mRNA using mMESSAGE mMACHINE SP6 transcription kit (Ambion). Purified mRNAs were added to CSF extracts at 0.05-0.1 mg/ml and the extracts were incubated for 2 h at 22°C before starting the assembly reaction to allow protein translation before sperm addition.
Immunofluorescence
Chromosomes and nuclei assembled in Xenopus extract were processed for immunofluorescence by fixation with 10 volumes of 2% paraformaldehyde in XBE2 containing 0.5% of Triton X-100. After 10 min, the fixed assembly mixtures were spun down on coverslips through a 5 ml-cushion of 30% glycerol in XBE2 at 6500×g for 15 min at 4°C. Coverslips were blocked for 1 h in 3% BSA in TBS-0.1% Triton X-100 and incubated for 1-2 hours with primary antibodies diluted in blocking solution at 2-5μg/ml followed by 1 hour incubation in 1:200 donkey anti-Rabbit FITC (Vector Labs). Next, coverslips were incubated with 1 mg/ml non-immune rabbit IgG to saturate open IgG binding sites in the secondary antibody and then incubated with Dylight 594-labeled CENP-A or CENP-C antibodies for 1 hour. After washing, coverslips were stained with DAPI and mounted with Mowiol. Samples were analyzed with a Leica DM6000 microscope. Black and white images were taken with a CCD camera and later processed with Photoshop. The same corrections in intensity and contrast were applied for all the images corresponding to the same staining in different conditions for a given experiment. Only in the case of fluorescently-labeled CENP-A or CENP-C, used to mark the position of centromeres but never to score differences in intensity, the images may have been processed differently. Quantification of fluorescence intensity was conducted on unprocessed images using Image J (National Institutes of Health) except for Figure 6, in which we used Definiens Developer XD v2 software. The CENP-A loading assay has been described before 40,41.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to acknowledge Diego Megías (Head of the Confocal Microscopy Unit at CNIO) for help with image quantification. We are grateful to members of the Losada and Méndez labs as well as to colleagues in the Marie Curie ITN “Nucleosome 4D” network, in particular G. Almouzni, for helpful discussions. We also thank A.F. Straight for providing the xCENP-C plasmids.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This research has been supported by MINECO (CSD2007-00015 INESGEN, BFU2013-48481-R to A.L. and FPI fellowship to P.S.). I.K. was a fellow of the Marie Curie ITN “Nucleosome 4D” (EU-FP7) and S.W. is supported by La Caixa International PhD Program.
References
- 1. Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol 2008; 9:33-46; PMID:18097444; http://dx.doi.org/ 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- 2. Santaguida S, Musacchio A. The life and miracles of kinetochores. Embo J 2009; 28:2511-31; PMID:19629042; http://dx.doi.org/ 10.1038/emboj.2009.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cleveland DW, Mao Y, Sullivan KF. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 2003; 112:407-21; PMID:12600307; http://dx.doi.org/ 10.1016/S0092-8674(03)00115-6 [DOI] [PubMed] [Google Scholar]
- 4. Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet 2008; 9:923-37; PMID:19002142; http://dx.doi.org/ 10.1038/nrg2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blower MD, Sullivan BA, Karpen GH. Conserved organization of centromeric chromatin in flies and humans. Dev Cell 2002; 2:319-30; PMID:11879637; http://dx.doi.org/ 10.1016/S1534-5807(02)00135-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell 2007; 25:309-22; PMID:17244537; http://dx.doi.org/ 10.1016/j.molcel.2006.12.018 [DOI] [PubMed] [Google Scholar]
- 7. Dunleavy EM, Almouzni G, Karpen GH. H3.3 is deposited at centromeres in S phase as a placeholder for newly assembled CENP-A in G(1) phase. Nucleus 2011; 2:146-57; PMID:21738837; http://dx.doi.org/ 10.4161/nucl.2.2.15211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fachinetti D, Diego Folco H, Nechemia-Arbely Y, Valente LP, Nguyen K, Wong AJ, Zhu Q, Holland AJ, Desai A, Jansen LE, et al. A two-step mechanism for epigenetic specification of centromere identity and function. Nat Cell Biol 2013; 15:1056-66; PMID:23873148; http://dx.doi.org/ 10.1038/ncb2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jansen LE, Black BE, Foltz DR, Cleveland DW. Propagation of centromeric chromatin requires exit from mitosis. J Cell Biol 2007; 176:795-805; PMID:17339380; http://dx.doi.org/ 10.1083/jcb.200701066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dunleavy EM, Roche D, Tagami H, Lacoste N, Ray-Gallet D, Nakamura Y, Daigo Y, Nakatani Y, Almouzni-Pettinotti G. HJURP is a cell-cycle-dependent maintenance and deposition factor of CENP-A at centromeres. Cell 2009; 137:485-97; PMID:19410545; http://dx.doi.org/ 10.1016/j.cell.2009.02.040 [DOI] [PubMed] [Google Scholar]
- 11. Foltz DR, Jansen LE, Bailey AO, Yates JR, Bassett EA, Wood S, Black BE, Cleveland DW. Centromere-specific assembly of CENP-a nucleosomes is mediated by HJURP. Cell 2009; 137:472-84; PMID:19410544; http://dx.doi.org/ 10.1016/j.cell.2009.02.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fujita Y, Hayashi T, Kiyomitsu T, Toyoda Y, Kokubu A, Obuse C, Yanagida M. Priming of centromere for CENP-A recruitment by human hMis18alpha, hMis18beta, and M18BP1. Dev Cell 2007; 12:17-30; PMID:17199038; http://dx.doi.org/ 10.1016/j.devcel.2006.11.002 [DOI] [PubMed] [Google Scholar]
- 13. Kim IS, Lee M, Park KC, Jeon Y, Park JH, Hwang EJ, Jeon TI, Ko S, Lee H, Baek SH, et al. Roles of Mis18alpha in epigenetic regulation of centromeric chromatin and CENP-A loading. Mol Cell 2012; 46:260-73; PMID:22516971; http://dx.doi.org/ 10.1016/j.molcel.2012.03.021 [DOI] [PubMed] [Google Scholar]
- 14. Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol 2006; 8:458-69; PMID:16622419; http://dx.doi.org/ 10.1038/ncb1397 [DOI] [PubMed] [Google Scholar]
- 15. Okada M, Cheeseman IM, Hori T, Okawa K, McLeod IX, Yates JR, Desai A, Fukagawa T. The CENP-H-I complex is required for the efficient incorporation of newly synthesized CENP-A into centromeres. Nat Cell Biol 2006; 8:446-57; PMID:16622420; http://dx.doi.org/ 10.1038/ncb1396 [DOI] [PubMed] [Google Scholar]
- 16. Bodor DL, Valente LP, Mata JF, Black BE, Jansen LE. Assembly in G1 phase and long-term stability are unique intrinsic features of CENP-A nucleosomes. Mol Biol Cell 2013; 24:923-32; PMID:23363600; http://dx.doi.org/ 10.1091/mbc.E13-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hemmerich P, Weidtkamp-Peters S, Hoischen C, Schmiedeberg L, Erliandri I, Diekmann S. Dynamics of inner kinetochore assembly and maintenance in living cells. J Cell Biol 2008; 180:1101-14; PMID:18347072; http://dx.doi.org/ 10.1083/jcb.200710052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Prendergast L, van Vuuren C, Kaczmarczyk A, Doering V, Hellwig D, Quinn N, Hoischen C, Diekmann S, Sullivan KF. Premitotic assembly of human CENPs -T and -W switches centromeric chromatin to a mitotic state. PLoS Biol 2011; 9:e1001082; PMID:21695110; http://dx.doi.org/ 10.1371/journal.pbio.1001082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hellwig D, Emmerth S, Ulbricht T, Doring V, Hoischen C, Martin R, Samora CP, McAinsh AD, Carroll CW, Straight AF, et al. Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci 2011; 124:3871-83; PMID:22100916; http://dx.doi.org/ 10.1242/jcs.088625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol 2006; 175:41-53; PMID:17030981; http://dx.doi.org/ 10.1083/jcb.200606020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 2006; 127:983-97; PMID:17129783; http://dx.doi.org/ 10.1016/j.cell.2006.09.039 [DOI] [PubMed] [Google Scholar]
- 22. Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C Is a Structural Platform for Kinetochore Assembly. Curr Biol 2011; 21:399-405; PMID:21353555; http://dx.doi.org/ 10.1016/j.cub.2011.02.005 [DOI] [PubMed] [Google Scholar]
- 23. Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of cenp-C to the mis12 complex joins the inner and outer kinetochore. Curr Biol 2011; 21:391-8; PMID:21353556; http://dx.doi.org/ 10.1016/j.cub.2010.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gascoigne KE, Takeuchi K, Suzuki A, Hori T, Fukagawa T, Cheeseman IM. Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 2011; 145:410-22; PMID:21529714; http://dx.doi.org/ 10.1016/j.cell.2011.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schleiffer A, Maier M, Litos G, Lampert F, Hornung P, Mechtler K, Westermann S. CENP-T proteins are conserved centromere receptors of the Ndc80 complex. Nat Cell Biol 2012; 14:604-13; PMID:22561346; http://dx.doi.org/ 10.1038/ncb2493 [DOI] [PubMed] [Google Scholar]
- 26. Bock LJ, Pagliuca C, Kobayashi N, Grove RA, Oku Y, Shrestha K, Alfieri C, Golfieri C, Oldani A, Dal Maschio M, et al. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat Cell Biol 2012; 14:614-24; PMID:22561345; http://dx.doi.org/ 10.1038/ncb2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hori T, Shang WH, Takeuchi K, Fukagawa T. The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 2013; 200:45-60; PMID:23277427; http://dx.doi.org/ 10.1083/jcb.201210106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nishino T, Rago F, Hori T, Tomii K, Cheeseman IM, Fukagawa T. CENP-T provides a structural platform for outer kinetochore assembly. EMBO J 2013; 32:424-36; PMID:23334297; http://dx.doi.org/ 10.1038/emboj.2012.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Basilico F, Maffini S, Weir JR, Prumbaum D, Rojas AM, Zimniak T, De Antoni A, Jeganathan S, Voss B, van Gerwen S, et al. The pseudo GTPase CENP-M drives human kinetochore assembly. 2014; 3:e02978; PMID:25006165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol 2009; 11:896-902; PMID:19543270; http://dx.doi.org/ 10.1038/ncb1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 2010; 189:1143-55; PMID:20566683; http://dx.doi.org/ 10.1083/jcb.201001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kato H, Jiang J, Zhou BR, Rozendaal M, Feng H, Ghirlando R, Xiao TS, Straight AF, Bai Y. A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 2013; 340:1110-3; PMID:23723239; http://dx.doi.org/ 10.1126/science.1235532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moree B, Meyer CB, Fuller CJ, Straight AF. CENP-C recruits M18BP1 to centromeres to promote CENP-A chromatin assembly. J Cell Biol 2011; 194:855-71; PMID:21911481; http://dx.doi.org/ 10.1083/jcb.201106079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dambacher S, Deng W, Hahn M, Sadic D, Frohlich J, Nuber A, Hoischen C, Diekmann S, Leonhardt H, Schotta G. CENP-C facilitates the recruitment of M18BP1 to centromeric chromatin. Nucleus 2012; 3:101-10; PMID:22540025; http://dx.doi.org/ 10.4161/nucl.18955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 2008; 135:1039-52; PMID:19070575; http://dx.doi.org/ 10.1016/j.cell.2008.10.019 [DOI] [PubMed] [Google Scholar]
- 36. Dornblut C, Quinn N, Monajambashi S, Prendergast L, van Vuuren C, Munch S, Deng W, Leonhardt H, Cardoso MC, Hoischen C, et al. A CENP-S/X complex assembles at the centromere in S and G2 phases of the human cell cycle. Open Biol 2014; 4:130229; PMID:24522885; http://dx.doi.org/ 10.1098/rsob.130229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishino T, Takeuchi K, Gascoigne KE, Suzuki A, Hori T, Oyama T, Morikawa K, Cheeseman IM, Fukagawa T. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 2012; 148:487-501; PMID:22304917; http://dx.doi.org/ 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Takeuchi K, Nishino T, Mayanagi K, Horikoshi N, Osakabe A, Tachiwana H, Hori T, Kurumizaka H, Fukagawa T. The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res 2014; 42:1644-55; PMID:24234442; http://dx.doi.org/ 10.1093/nar/gkt1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hellwig D, Munch S, Orthaus S, Hoischen C, Hemmerich P, Diekmann S. Live-cell imaging reveals sustained centromere binding of CENP-T via CENP-A and CENP-B. J Biophotonics 2008; 1:245-54; PMID:19412974; http://dx.doi.org/ 10.1002/jbio.200810014 [DOI] [PubMed] [Google Scholar]
- 40. Sanchez P, Losada A. New clues to understand how CENP-A maintains centromere identity. Cell Div 2011; 6:11; PMID:21554702; http://dx.doi.org/ 10.1186/1747-1028-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bernad R, Sanchez P, Rivera T, Rodriguez-Corsino M, Boyarchuk E, Vassias I, Ray-Gallet D, Arnaoutov A, Dasso M, Almouzni G, et al. Xenopus HJURP and condensin II are required for CENP-A assembly. J Cell Biol 2011; 192:569-82; PMID:21321101; http://dx.doi.org/ 10.1083/jcb.201005136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell 2009; 20:4246-55; PMID:19641019; http://dx.doi.org/ 10.1091/mbc.E09-05-0378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McKinley KL, Cheeseman IM. Polo-like kinase 1 licenses CENP-A deposition at centromeres. Cell 2014; 158:397-411; PMID:25036634; http://dx.doi.org/ 10.1016/j.cell.2014.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hori T, Shang WH, Toyoda A, Misu S, Monma N, Ikeo K, Molina O, Vargiu G, Fujiyama A, Kimura H, et al. Histone H4 lys 20 monomethylation of the CENP-A nucleosome is essential for kinetochore assembly. Dev Cell 2014; 29:740-9; PMID:24960696; http://dx.doi.org/ 10.1016/j.devcel.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malvezzi F, Litos G, Schleiffer A, Heuck A, Mechtler K, Clausen T, Westermann S. A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J 2013; 32:409-23; PMID:23334295; http://dx.doi.org/ 10.1038/emboj.2012.356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Emanuele MJ, McCleland ML, Satinover DL, Stukenberg PT. Measuring the stoichiometry and physical interactions between components elucidates the architecture of the vertebrate kinetochore. Mol Biol Cell 2005; 16:4882-92; PMID:16079178; http://dx.doi.org/ 10.1091/mbc.E05-03-0239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Suzuki A, Badger BL, Wan X, DeLuca JG, Salmon ED. The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev Cell 2014; 30:717-30; PMID:25268173; http://dx.doi.org/ 10.1016/j.devcel.2014.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ribeiro SA, Vagnarelli P, Dong Y, Hori T, McEwen BF, Fukagawa T, Flors C, Earnshaw WC. A super-resolution map of the vertebrate kinetochore. Proc Natl Acad Sci U S A 2010; 107:10484-9; PMID:20483991; http://dx.doi.org/ 10.1073/pnas.1002325107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rivera T, Losada A. Shugoshin regulates cohesion by driving relocalization of PP2A in Xenopus extracts. Chromosoma 2009; 118:223-33; PMID:18987869; http://dx.doi.org/ 10.1007/s00412-008-0190-4 [DOI] [PubMed] [Google Scholar]
- 50. Losada A, Hirano M, Hirano T. Identification of Xenopus SMC protein complexes required for sister chromatid cohesion. Genes Dev 1998; 12:1986-97; PMID:9649503; http://dx.doi.org/ 10.1101/gad.12.13.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hirano T, Kobayashi R, Hirano M. Condensins, chromosome condensation protein complexes containing XCAP-C, XCAP-E and a Xenopus homolog of the Drosophila Barren protein. Cell 1997; 89:511-21; PMID:9160743; http://dx.doi.org/ 10.1016/S0092-8674(00)80233-0 [DOI] [PubMed] [Google Scholar]
- 52. Kimura K, Hirano T. Dual roles of the 11S regulatory subcomplex in condensin functions. Proc Natl Acad Sci U S A 2000; 97:11972-7; PMID:11027308; http://dx.doi.org/ 10.1073/pnas.220326097 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


