Abstract
microRNA is necessary for osteoclast differentiation, function and survival. It has been reported that miR-199/214 cluster plays important roles in vertebrate skeletal development and miR-214 inhibits osteoblast function by targeting ATF4. Here, we show that miR-214 is up-regulated during osteoclastogenesis from bone marrow monocytes (BMMs) with macrophage colony stimulating factor (M-CSF) and receptor activator of nuclear factor-κB ligand (RANKL) induction, which indicates that miR-214 plays a critical role in osteoclast differentiation. Overexpression of miR-214 in BMMs promotes osteoclastogenesis, whereas inhibition of miR-214 attenuates it. We further find that miR-214 functions through PI3K/Akt pathway by targeting phosphatase and tensin homolog (Pten). In vivo, osteoclast specific miR-214 transgenic mice (OC-TG214) exhibit down-regulated Pten levels, increased osteoclast activity, and reduced bone mineral density. These results reveal a crucial role of miR-214 in the differentiation of osteoclasts, which will provide a potential therapeutic target for osteoporosis.
Keywords: miRNA, osteoclast, osteoporosis, PTEN, PI 3-kinase
Abbreviations
- BMMs
bone marrow monocytes
- M-CSF
macrophage colony stimulating factor
- RANKL
receptor activator of nuclear factor-κB ligand
- Pten
phosphatase and tensin homolog
- OC-TG214
osteoclast specific miR-214 transgenic mice
- NFATc1
nuclear factor of activated T-cells cytoplasmic
- TRAP
tartrate-resistant acid phosphatase
- Dnm3os
Dnm3 opposite strand
- WT
wild-type
- micro CT
Micro computed tomography
- BMD
bone mineral density
- BV/TV
ratio of bone volume to tissue volume
- Tb.Sp
trabecular spacing
- qRT-PCR
quantitative real-time PCR
Introduction
Osteoclasts are derived from monocyte-macrophage precursors that arise from multipotent haematopoietic stem cells. Many cytokines and exogenous hormones have been identified to be involved in osteoclastogenesis through transcription factors that positively or negatively modulate osteoclast proliferation, survival, differentiation and function.1-4 M-CSF and RANKL, both produced by osteoblast or activated T cells, are significant cytokines for osteoclastogenesis.5-7 RANKL can induce the expression of nuclear factor of activated T-cells cytoplasmic (NFATc1).8 Osteoclast-specific markers, such as tartrate-resistant acid phosphatase (TRAP) and cathepsin K, have multiple sites recognized by NFATc1,8-10 a key transcription factor for osteoclastogenesis.
microRNAs (miRNAs) are small non-coding RNAs of ~22 nucleotides known to play important roles in post-transcriptional regulation in various cellular processes.11 They contribute to every step of osteogenesis from embryonic bone development to maintenance of adult tissue, by regulating the growth, differentiation and functional activity of osteoblasts, osteoclasts and osteocytes.12 Bone remodeling is a continuous process requiring the physiological coupling of osteoclast with osteoblast activities.13 Various miRNAs have been reported to regulate osteoblastic differentiation, proliferation and bone formation.14-19 Recently, the function of miRNAs in osteoclastic differentiation and activity is beginning to be disclosed.
miR-214 is highly conserved across vertebrates and is encoded within a larger non-coding RNA, Dnm3 opposite strand (Dnm3os).20 It is transcribed together with miR-199a-2 from the opposite strand of the Dnm3 gene on mouse chromosome 1. The miR-199 and miR-214 genes cluster not only participates in skeleton formation, but maintains the skeleton in a healthy state as well. In osteoblasts, our previous results demonstrate that miR-214 targets ATF4 to inhibit bone formation.19 However, its expression and function in osteoclasts remain elusive.
In this paper, we find that the expression of miR-214 is upregulated in the process of M-CSF and RANKL induced osteoclastogenesis. Inhibition of miR-214 levels significantly reduces osteoclast activity. It functions by targeting Pten to activate PI3K/Akt/NFATc1 pathway. In vivo, osteoclast specific miR-214 transgenic mice (OC-TG214) exhibits downregulated Pten levels, increases osteoclast activity, and reduces bone mineral density.
Results
miR-214 is up-regulated in osteoclastogenesis induced by M-CSF and RANKL
To investigate the miRNAs involved in osteoclastogenesis, microarray assays were performed. RAW 264.7 osteoclast precursor cells were induced by treatment with RANKL. RAW 264.7 cells were cultured without RANKL as controls. Our results showed that 10 miRNAs are differently expressed during the differentiation of RAW 264.7 cells into osteoclasts (Fig. 1A). Six miRNAs (miR-92a, miR-20b, miR-5112, miR-1957a, miR-455 and miR-214) are significantly increased. Conversely, miR-3064, miR-5109, miR-155 and miR-511 are down-regulated. The expressions of 6 upregulated miRNAs are further confirmed by qRT-PCR, the results were consistent with that of miRNA microarray (Fig. 1B). Among them, miR-214 is the most obviously up-regulated miRNA. Its level is increased about 2.5 folds.
Figure 1.
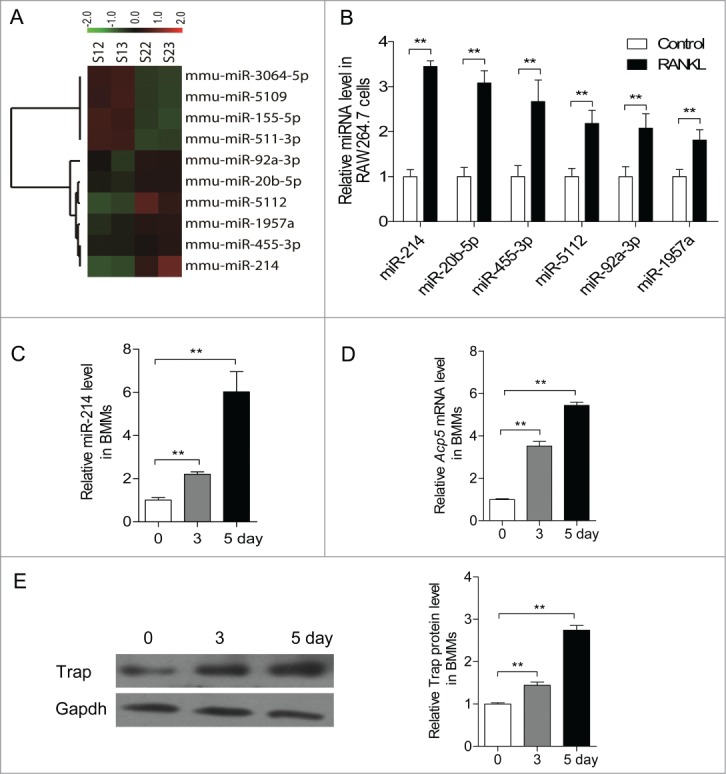
miR-214 was upregulated during osteoclastogenesis. (A) Microarray assays were performed in RAW 264.7 cells with or without RANKL induction for 2 d Red and green indicate high expression levels and low expression levels, respectively. Shown are the miRNAs that changed more than 1.fold5-. (B) Relative miRNA levels in RANKL-induced RAW 264.7 cells was analyzed by qRT-PCR, miRNA levels were normalized to U6. (C) BMMs were cultured with M-CSF (10 ng/ml) alone for 1 day, and followed with M-CSF (30 ng/ml) and RANKL (50 ng/ml) for indicated times. Relative miR-214 levels in M-CSF+RANKL-induced BMMs were analyzed by qRT-PCR, miR-214 levels were normalized to U6. (D) Relative Acp5 mRNA levels in M-CSF+RANKL-induced BMMs were analyzed by qRT-PCR and normalized to Gapdh. (E) Trap protein levels in BMMs with RANKL-induction for indicated times were determined by western blot analysis and normalized to Gapdh. The data represent the mean ± SEM of 3 experiments in triplicate. **P < 0.01.
We also found that the expression of miR-214 is increased progressively during the process of RANKL and M-CSF induced osteoclastogenesis from BMMs (Fig. 1C). In detail, miR-214 starts to be increased progressively on day 3 after induction. On day 5, the expression of miR-214 reaches 6 folds of that on day 1. The change of miR-214 level concurs with the up-regulation of expression of osteoclast effector gene Acp5 (Fig. 1D, E). The results suggested that miR-214 plays an important role in osteoclastogenesis.
miR-214 is essential to osteoclastogenesis
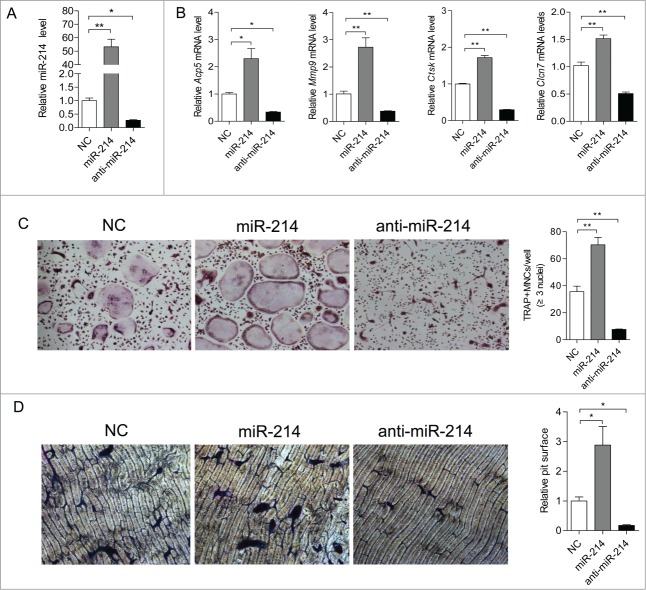
To assess the significance of miR-214 for osteoclast differentiation, we treated BMMs with miR-214 mimics or anti-miR-214 in the course of osteoclastogenesis. The results showed that cellular miR-214 levels are substantially upregulated by miR-214 mimics treatment and markedly downregulated by anti-miR-214 treatment (Fig. 2A). TRAP, MMP9, CLC7 and Cathepsin K are important markers for the differentiation and activity of osteoclast.21-25 After 5 days of induction, the expression of Acp5, Clcn7, Mmp9 and Ctsk are significantly up-regulated by miR-214 mimics and down-regulated by anti-miR-214 compared to treatment with their negative control, respectively (Fig. 2B). Functionally, the number of TRAP-positive, multinucleated osteoclasts per well are substantially increased in the miR-214 mimics treatment group and markedly reduced in the anti-miR-214 treatment group compared to the negative control group (Fig. 2C). In addition, the relative surface of osteoclast bone-resorbing pits is also reduced due to downregulated osteoclast activity in the anti-miR-214 treatment group (Fig. 2D). These results showed that miR-214 is essential for osteoclastogenesis.
Figure 2.
miR-214 regulates osteoclastogenesis from BMMs. (A) BMMs were induced with M-CSF and RANKL for 5 d Relative miR-214 levels were determined by qRT-PCR after treatment with miR-214 mimics or anti-miR-214 or negative control (NC) and miR-214 levels were normalized to U6. (B) Relative Acp5, Mmp9, Ctsk and Clcn7 mRNA levels were determined by qRT-PCR and normalized to Gapdh. (C) Representative photographs of OC formation (original magnification 200ċ) and the number of TRAP-positive multinucleated cells (MNCs) are shown. (D) Bone resorption areas on bovine bone slices were measured by image analysis. Representative images of resorbed bone slices (original magnification 4ċ). The data represent the mean ± SEM of 3 experiments in triplicate. *P < 0.05, **P < 0.01.
miR-214 directly targets Pten
Bioinformatics analysis suggests that the 3′UTR of Pten contains a miR-214 targeting site. It targets Pten in T cells, human ovarian cancer cells and gastric cancer cells to increase their proliferation ability.26-28 Using two computer-aided algorithms, miRecords, and CHIP Base, we obtained a list of predicted target genes that are potentially regulated by miR-214. One of the target genes, Pten, which was shown to play a critical role in RANKL-induced osteoclast differentiation,29 was selected for experimental verification. We found that 3′-UTR of mouse Pten contains a putative region (nucleotides 3102–3109, NM_008960) that is complementary with the seed sequence of mmu-miR-214 (Fig. 3A). To test whether miR-214 targets Pten, we examined that the effect of miR-214, anti-miR-214 and NC on luciferase activity in RAW 264.7 cells transfected with the Pten 3′ UTR reporter. We found that miR-214 substantially inhibits the luciferase reporter activity of the Pten 3′ UTR. Moreover, the luciferase reporter activity is significantly increased after reducing the endogenous levels of miR-214 by treating with anti-miR-214 (Fig. 3B). We then examined the effect of miR-214 and anti-miR-214 on the Pten mRNA and protein levels in RANKL-induced RAW 264.7 osteoclast precursors. The results demonstrated that miR-214 leads to a decrease in Pten protein level in RANKL-induced RAW 264.7 cells, and anti-miR-214 leads to an increase in Pten protein level compared with cells transfected with miR-214 negative control (Fig. 3C). However, there is no detectable change in Pten mRNA level (Fig. 3D). ATF4 is another important transcription factor that plays a direct role in regulating osteoclast differentiation, and it is a target of miR-214 in osteoblasts.19 However, there are no changes found in Atf4 protein levels after transfection with miR-214 mimics in RANKL-induced RAW 264.7 cells (Fig. 3E). These results showed that Pten is the target of miR-214 in osteoclast differentiation
Figure 3.
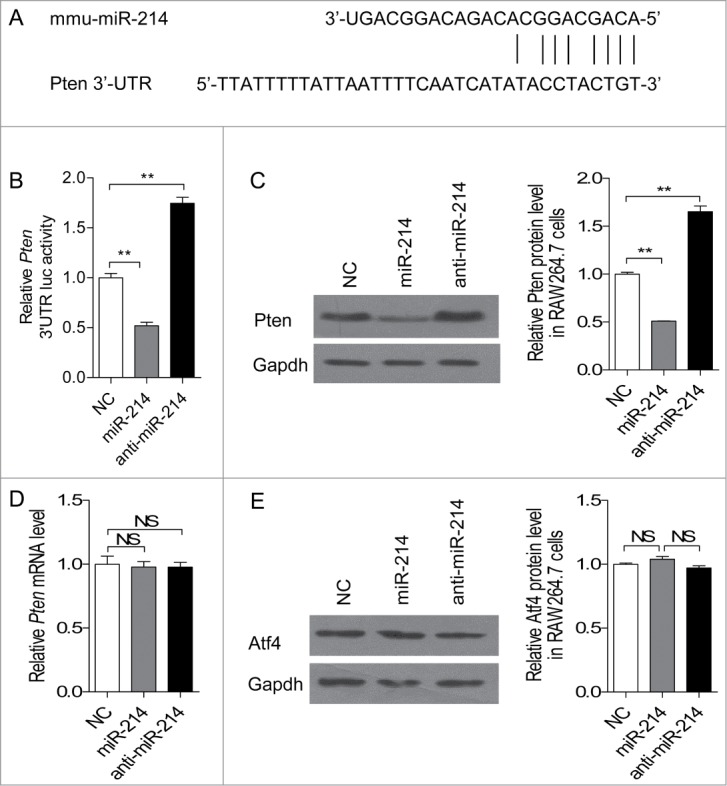
miR-214 directly targets Pten in osteoclasts. (A) Sequence alignment of mouse miR-214 with 3'-UTR of Pten. The seed sequences of miR-214 (up) and 3'-UTR of Pten (down) are complementary. (B) The effect of miR-214, anti-miR-214 and NC on luciferase activity in RAW 264.7 cells transfected with the Pten 3' UTR reporter. (C) Effect of miR-214, anti-miR-214 and NC on Pten protein level by protein gel blot analysis. miR-214 mimics, anti-miR-214 or NC were transfected into RANKL-induced RAW 264.7 cells. The protein levels were normalized to Gapdh. (D) Pten mRNA level was determined by qRT-PCR. The level of Pten were normalized to Gapdh. (E) Atf4 protein level was determined by western blot analysis and normalized to Gapdh. The data represent the mean ± SEM of 3 experiments in triplicate. *P < 0.05, **P < 0.01, NS, not significant.
miR-214 promotes osteoclast differentiation through Pten involved pathway
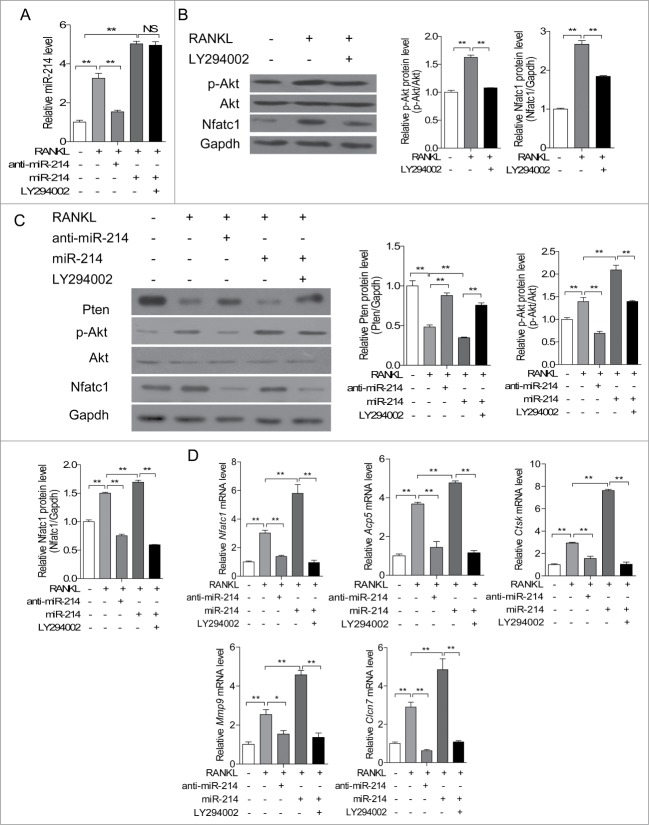
It has been demonstrated that Pten regulates RANKL-induced osteoclast differentiation from RAW 264.7 osteoclast precursors through PI3K/Akt pathway.29 To test the effect of miR-214 on the Pten/Akt signaling pathway during RANKL induced osteoclast differentiation, RAW 264.7 cells were treated with RANKL, RANKL with anti-miR-214, miR-214 mimics, and miR-214 mimics with PI3K inhibitor LY294002, respectively. Intracellular miR-214 levels are able to be upregulated by RANKL induction and reduced by anti-miR-214 treatment. LY294002 has no effect on intracellular miR-214 level (Fig. 4A). We also verified that PI3K inhibitor LY294002 can inhibit phosphorylated Akt and NFATc1 in RAW 264.7 cells (Fig. 4B). Therefore, we further detected the protein levels of Pten, Akt, phosphorylated Akt and NFATc1 in RAW 264.7 cells in Figure 4A. The results demonstrated that Pten protein level is inhibited, phosphorylated Akt and NFATc1 are up-regulated after RANKL induction. Furthermore, anti-miR-214 is able to inhibit Akt phosphorylation and NFATc1 levels through up-regulating Pten levels in this process. Under miR-214 treatment, Pten protein level is reduced in RAW 264.7 cells. Accordingly, the phosphorylation of Akt and NFATc1 levels are increased. PI3K inhibitor LY294002 strongly down-regulates miR-214 induced Akt activation (Fig. 4C). Moreover, the increased mRNA levels of Nfatc1, Acp5, Clcn7, Mmp9 and Ctsk after induction with RANKL or treatment with miR-214 are partly blocked by anti-miR-214 and PI3K inhibitor LY294002 (Fig. 4D). These data suggested that miR-214 regulates RANKL-activated PI3K/Akt signaling pathway for osteoclast differentiation by targeting Pten.
Figure 4.
miR-214 promotes osteoclast differentiation through Pten involved pathway. (A) miR-214 levels were determined by qRT-PCR in RAW 264.7 cells after treatment with RANKL, miR-214 mimics, anti-miR-214 and PI3K inhibitor LY294002. miR-214 levels were normalized to U6. (B) Effect of RANKL and PI3K inhibitor LY294002 on the protein levels of Nfatc1 and phosphorylated Akt in RAW 264.7 cells. Nfatc1 and phosphorylated Akt levels were analyzed by protein gel blot, and Gapdh was used as the internal control. (C) Effect of miR-214, anti-miR-214 and PI3K inhibitor LY294002 on the protein levels of Pten, Nfatc1 and phosphorylated Akt in RAW 264.7 cells. Pten, Nfatc1 and phosphorylated Akt levels were analyzed by western blot, and Gapdh was used as the internal control. (D) Effect of miR-214, anti-miR-214 and PI3K inhibitor LY294002 on NFATc1, Acp5, Ctsk, Mmp9, Clcn7 mRNA levels in RAW 264.7 cells. The mRNA levels were analyzed by qRT-PCR, and products were normalized to Gapdh. The data represent the mean ± SEM of 3 independent experiments in triplicate. *P < 0.05, **P < 0.01, NS, not significant.
Promotion of osteoclast activity by miR-214 in vivo
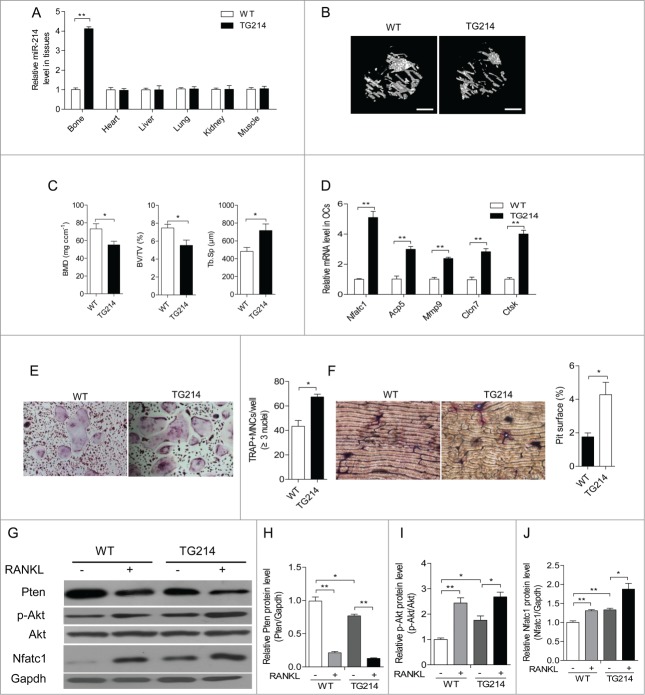
To investigate the role of miR-214 in vivo, we developed a construct that expresses miR-214 under the control of the Acp5 promoter, which has been shown to be specially activated in osteoclasts.30 Subsequently, we established Acp5-miR-214 transgenic (OC-TG214) mice. We selected one OC-TG214 mouse line for further experiments. Compared to littermate wild-type (WT) mice, OC-TG214 mouse line showed more than 4 times higher miR-214 expression in bone tissue at 8 weeks of age (Fig. 5A). Micro CT (Micro computed tomography) analysis revealed that the bone mineral density (BMD) and ratio of bone volume to tissue volume (BV/TV) in OC-TG214 mice are markedly lower than that in WT mice. We also found that the trabecular spacing (Tb.Sp) in OC-TG214 mice are notably increased compared to WT mice (Fig. 5B, C). Compared to WT mice, Nfatc1, Acp5, Clcn7, Mmp9 and Ctsk mRNA levels are significantly increased in osteoclasts sorting from OC-TG214 mice (Fig. 5D). Moreover, TRAP-positive osteoclast formation are promoted in RANKL induced BMMs from OC-TG214 mice (Fig. 5E). In addition, osteoclast bone-resorbing activity is also increased because of upregulation of osteoclast formation (Fig. 5F). Furthermore, we isolated BMMs from WT and OC-TG214 mice and observed the changes of Pten protein levels after RANKL-induction. We found that Pten protein levels are much lower in the BMMs from OC-TG214 mice than that from WT mice and rapidly reduced with RANKL treatment. The reduction is more obvious in cells from OC-TG214 mice. Consistently, the levels of phosphorylation of Akt and NFATc1 are higher in BMMs from OC-TG214 mice than that from WT mice. After treatment with RANKL, the up-regulation of these proteins is much higher in RANKL induced OC-TG214 mice derived BMMs (Fig. 5G, H, I, J). These results revealed that miR-214 overexpression in osteoclasts leads to activated osteoclast activity and bone loss.
Figure 5.
Promotion of osteoclast activity by miR-214 in vivo. (A) qRT-PCR analysis of miR-214 levels in bone and other tissues (normalized to those in WT mice) from WT and Acp5-miR-214 transgenic (OC-TG214) mouse lines. miR-214 levels were normalized to U6. (B) MicroCT examination and representative images showing The 3-dimensional trabecular architecture in OC-TG214 and WT mice. Scale bars, 1 mm. (C) Micro CT measurements for BMD, BV/TV and Tb.Sp in the proximal tibia of WT and OC-TG214 mice. n=5. (D) Nfatc1, Acp5, Mmp9, Clcn7 and Ctsk mRNA levels in Oscar+ separated by cell sorting with flow cytometry were analyzed by qRT-PCR. The expression levels were normalized to Gapdh. (E) TRAP staining (original magnification 200ċ) and the number of TRAP-positive MNCs of RANKL-induced BMMs isolated from bone marrow of WT and OC-TG214 mice. (F) Bone resorption areas on bovine bone slices were measured by image analysis. Representative images of resorbed bone slices (original magnification 4ċ). (G, H, I, J) Pten, Nfatc1 and phosphorylated Akt levels in M-CSF and RANKL induced BMMs from WT and OC-TG214 mice for 4 d were measured by protein gel blot and expressed as the ratios of densitometry to Gapdh. Gapdh was used as the internal control. The data represent the mean ± SEM of 3 experiments in triplicate. *P < 0.05, **P < 0.01.
Discussion
In present study, we found that miR-214 is upregulated during M-CSF and RANKL induced osteoclastogenesis from BMMs. In vitro, miR-214 can promote osteoclastogenesis by targeting Pten and activating PI3 kinase/Akt/NFATc1 pathway (Fig. 6). In vivo, osteoclast specific overexpression of miR-214 reduces bone mineral density and bone volume by increasing osteoclast activity. Under the treatment of RANKL, BMMs from OC-TG214 mice have stronger activity to differentiate into osteoclasts. In these cells, Pten levels are reduced, and the phosphorylation of Akt is up-regulated, and the expression of Nfatc1, the master regulator for the transcription of osteoclast-specific marker genes, is obviously increased. Accordingly, the activity of osteoclast is increased. To date, several miRNAs have been identified to be involved in the regulation of osteoclast differentiation.31-36 Among them, only miR-21 and miR-148a have been identified to promote osteoclastogenesis by targeting PDCD4 and MAFB, respectively. miR-214 is highly expressed in the osteoporosis bone tissues19 and in the differentiation of PBMCs into osteoclasts by microarray assay.37 The expression study showed predominant expression patterns for miR-199–214 cluster in tissues surrounding developing craniofacial skeletal elements and indicated a conserved role of miR-199 and miR-214 in vertebrate skeletogenesis.38 miR-199–214 is essential to skeletogenesis because miR-199–214 cluster knockout mice display skeletal abnormalities.20 In our study, we found that miR-214 plays an important role in osteoclastogenesis and affects osteoclast activity through regulating the expression of osteoclast functional genes.
Figure 6.
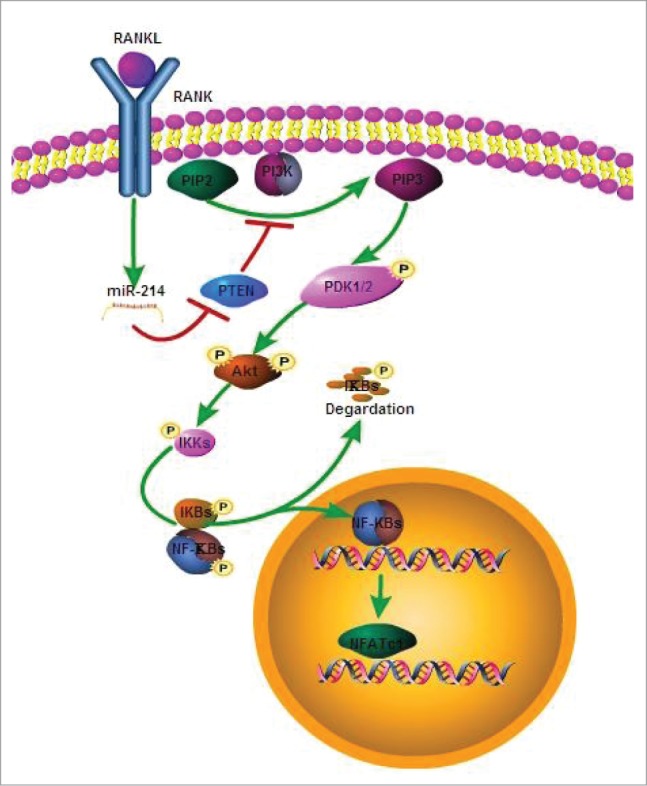
The model for effect of miR-214 on osteoclastogenesis. MiR-214 regulates RANKL-activated PI3K/Akt signaling pathway for osteoclast differentiation through targeting Pten.
miR-214 is widely expressed in different tissues and cells. It targets different genes to regulate the occurrence of cancer, immune, differentiation of skeletal muscle cells, protection of heart from ischemic injury, inhibition of angiogenesis. Among them, Pten is one of its main targets. In breast cancer, ovarian cancer, hepatic cancer and gastric cancer, miR-214 targets Pten to regulate the proliferation, migration and invasion of these cancer cells.39 In immune system, miR-214 targets Pten to increase monocyte survival and enhances T cell activity.40 In bone tissue, miR-214 has been identified to target ATF4 in osteoblasts.19 In this study, our results demonstrated that miR-214 promotes osteoclastogenesis by targeting Pten, but not Atf4 in RANKL and M-CSF induced RAW 264.7 cells, although both of them are essential for the function of osteoclast function. This reflects the importance of miR-214 in the fine-tuning of gene expression related to bone remodeling.
Osteoclasts and osteoblasts lineages communicate with each other through cell-cell contact, diffusible paracrine factors and cell-bone matrix interaction, which is essential for bone remodeling and mineral homeostasis.41,42 MiRNAs represent a sophisticated level of gene regulation that coordinates a broad spectrum of biological processes in the regulation of bone remodeling. However, the control of miRNAs in the crosstalk between the 2 metabolically opposing cell types, osteoblasts and osteoclasts, remains to be addressed. The role of miR-214 in the regulation of both osteoblasts and osteoclasts implicates that miR-214 could be such a mediator. In endothelial cells, miR-214 plays a dominant role in exosome-mediated signaling between endothelial cells.43 Exosomes from hepatic stellate cells are a conduit for uptake of miR-214 by primary mouse hepatocytes.44 These results suggested that there is a potential mechanism that miR-214 mediated crosstalk between osteoclasts and osteoblasts is involved in the capacity of exosome to shuttle miRNAs to target cells.
In summary, this study demonstrates that miR-214 regulates osteoclast differentiation by targeting Pten, which provides a new insight into the roles of miR-214 in osteoclastogenesis. Our previous study demonstrated that miR-214 is present in osteoblasts and acts to inhibit bone formation under various pathophysiological and physiological scenarios.19 In this research, we highlight the importance of miR-214 in the regulation of bone loss by directly demonstrating its effect on osteoclast. Based on this evidence, it can be concluded that miR-214 antagonist could promote bone formation through inhibiting osteoclast and promoting osteoblast at the same time. A therapeutic blockage of miR-214 could be helpful in the control of osteoporosis in post-menopausal women and in elderly patients of both sexes. Further investigation in our lab are underway to clarify the mechanisms in contributing to the upregulation of miR-214 in osteoclasts and miR-214 mediated crosstalk between osteoblasts and osteoclasts.
Materials and Methods
Cell culture
Osteoclasts were induced with M-CSF (R&D Systems Inc., 416-ML/CF) and RANKL (R&D Systems Inc.., 462-TEC/CF) from BMMs. Bone marrow cells were cultured in 10 cm2 plate containing Alpha Modified Eagle's Medium (α-MEM, Hyclone, No.SH30265) supplemented with 10% FBS (Gibco Laboratories, No.16000–044), 1% penicillin and streptomycin and M-CSF (10 ng/ml) for 1 day. Non-adherent cells were considered as BMMs, and cultured in α-MEM containing 10% FBS, M-CSF (30 ng/ml) and RANKL (50 ng/ml). The RAW 264.7 cell line was maintained in Dulbecco's modified Eagle's medium (DMEM, Hyclone, No.SH30022) with 10% FBS and 1% penicillin and streptomycin. RAW 264.7 cells were induced with RANKL (50 ng/ml) for osteoclastogenesis. The cells were maintained under standard cell culture conditions of 5% CO2 and 95% humidity. The culture medium was replaced for fresh medium every 2 d BMMs or RAW 264.7 cells were transfected with 20 nM miR-214 mimics, anti-miR-214, and NC (negative control) (GenePharma) using Lipofectamine 2000 (Invitrogen, No.11668–019) for 5 d or 2 d respectively. For PI3K inhibition, RAW 264.7 cells were treated with 40μM LY294002 (MERKMILLIPORE, 440202) for 24 hours.
miRNA microarray assay
The RNA of RAW 264.7 cells induced with or without RANKL for 2 d were extracted. miRNAs were isolated and labeled either with Cy3 or Cy5. miRNA microarray assay was completed by Phalanx Biotech Group. The probe content was in accordance with Version 19.0 of the Sanger miRBase database. The signal intensity of each spot was process by R program (2.12.1). The median value of repeating spots were selected into analysis. We filtered out spots which flag <0 within all arrays. Spots that passed the criteria were normalized by invariant set normalization method. Normalized spot intensities were transformed to gene expression log2 ratios between the control and treatment groups by pair-wise t test. The spots with |log2 ratio| ≥ 0.585 and p values < 0.05 are selected for further analysis.
TRAP staining
The procedure was according to the protocol of Acid Phosphatase kit (Sigma Aldrich, No.387). Briefly, after 3 d of culture, RANKL-induced cells were fixed by immersing in Fixative Solution for 30 seconds in room temperature (18–26°C), then rinsed thoroughly in deionized water. TRAP staining fluid was added, and the plates were incubated at 37°C protected from light for 1 hour. After removal of the TRAP solution, the plates were washed 3 times with distilled water. The TRAP positive multinuclear cells were recorded using inverted microscope (Nikon).
Determination of bone resorption pits
For pit assay, BMMs (5ċ10 5 cells/well) were seeded on bovine bone slices in 24-well plates in proliferation medium for 1 day and switched to differentiation medium for 3 d Bovine bone slices were ultrasonicated in 1 mol/L NH4OH to remove adherent cells and stained with 0.1% toluidine blue solution. Pit area versus total bone area of each slice was measured using Image Pro Plus 6.2 software (Media Cybernetics Inc..).
miRNA extraction
The total RNA from the collected cells was extracted using the miRNeasy Mini Kit (QIAGEN, No. 217004). Briefly, the cells were collected in a reaction tube, lysed with 700 µl QIAzol and mixed with 140 µl chloroform. After being centrifuged at 12,000 g for 15 min at 4°C, the upper aqueous phase was transferred to an RNeasy Mini spin column in a 2 ml collection tube and mixed with 100% ethanol. After being washed with 700 µl Buffer RWT and 500 µl Buffer RPE, the total RNA was collected for real-time PCR analysis.
Reverse transcription and quantitative real-time PCR (qRT-PCR)
Total RNA from bone tissues or cells was extracted with TRIzol Reagent (Invitrogen, No.15596–026) as the manufacturer's instructions. RNA (0.5 μg) was reversely transcribed with PrimeScript RT reagent Kit (TaKaRa, No.037A) according to the manufacturer's instructions. Stem-loop RT-PCR was used for the quantification of miR-214. Two microliters of cDNA were used for detecting mRNA and miRNA expression by quantitative PCR using SYBR→ Premix Ex TaqTMII Kit (TaKaRa, No.820A). Gapdh was used as a normalization control for mRNA and U6 was used as a normalization control in all miRNA measurements. Primers used were synthesized by Beijing Sunbiotech Co., Ltd. and their sequence were as follows: Gapdh (Forward: 5′-AACATCAAATGGGGTGAGGCC-3′ and Reverse: 5′-GTTGTCATGGATGACCTTGGC-3′),
Pten (Forward: 5′-AATTCCCAGTCAGAGGCGCTATGT-3′ and Reverse: 5′-GATTGCAAGTTCCGCCACTGAACA-3′), Nfatc1 (Forward: 5′-ACGCTACAGCTGTTCATTGG-3′ and Reverse: 5′-CTTTGGTGTTGGACAGGATG-3′), Acp5 (Forward: 5′-GCGACCATTGTTAGCCACATACG-3′ and Reverse: 5′-CGTTGATGTCGCACAGAGGGAT-3′), Clcn7 (Forward: 5′-GTCCTTCAGCCTCAGTCG-3′ and Reverse: 5′-ACACAGCGTCTAATCACAAC-3′), Mmp9 (Forward: 5′-GCGGCCCTCAAAGATGAACGG-3′ and Reverse: 5′-GCTGACTACGATAAGGACGGCA-3′), Ctsk (Forward: 5′-GCGTTGTTCTTATTCCGAGC-3′ and Reverse: 5′-CAGCAGAGGTGTGTACTATG-3′), miR-214 (Forward: 5′-GACAGCAGGCACAGACA-3′), miR-20b-5p (Forward: 5′-CAAAGTGCTCATAGTGCAGGTAG-3′), miR-455–3p (Forward: 5′-GCAGTCCACGGGCATATACAC-3′), miR-5112 (Forward: 5′-TAGCTCAGCGGGAGAGCA-3′), miR-92a-3p (Forward: 5′-TATTGCACTTGTCCCGGCCT-3′), miR-1957a (Forward: 5′-CGCAGTGGTAGAGCATATGAC-3′), Regular miRNA Reverse: 5′-GTGCAGGGTCCGAG-3′.
Luciferase reporter assay
RAW 264.7 cells were placed on 6-well plates at a density of 1 ċ 106 cells per well. Cells were transfected with pGL3-PTEN 3′UTR (Addgene, 28104) and pRL-TK (Promega, USA) Renilla luciferase plasmid with miR-214, anti-miR-214 or NC following the manufacturer's instruction (Lipofectamine RNAiMAX, Invitrogen, USA). Luciferase assays were performed with the dual-luciferase reporter assay system (Promega, E1910) according to the manufacturer's instructions. Luminescent signals were quantified by luminometer (Glomax, Promega), and each value from the firefly luciferase construct was normalized by Renilla luciferase assay.
Western blot analysis
Cells were lysed in lysis buffer (50 mM Tris, pH 7.5, 250 mM NaCl, 0.1% SDS, 2 mM dithiothreitol (DTT), 0.5% NP-40, 1 mM PMSF and protease inhibitor cocktail on ice for 30 min. Protein fractions were collected by centrifugation at 15,000 g at 4°C for 30 min and then subjected to SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% BSA and incubated with specific antibodies overnight. A horseradish peroxidase–labeled secondary antibody was added and visualized using an chemiluminescence kit (Thermo Pierce, No.32109). We used antibodies recognizing mouse Pten (Santa Cruz Biotechnology, #9559), Akt (Cell Signaling Technology, #9272), P-Akt (Cell Signaling Technology, # 9271), NFATc1 (Cell Signaling Technology, #8032), ATF4 (Abcam, No.ab1371), and GAPDH (Santa Cruz Biotechnology, #25778) to detect protein levels, respectively. The ratios of the protein band intensities relative to that of GAPDH were calculated for each sample using Image J.
Generation of osteoclast-specific miR-214 transgenic mice
The plasmid (gift of Dr. Sakamuri V. Reddy), which contains the Acp5 promoter to drive osteoclast-specific gene expression, was used to generate OC-TG214 mice. We subcloned mouse pre-miR-214 cDNA into a vector and confirmed the expression of miR-214 in RAW 264.7 cells by PCR. Acp5-pre-miR-214 plasmid was constructed by inserting the Kpn□/Xba□ fragment of pre-miR-214 into the corresponding sites of downstream of Acp5 promoter. The fragments of the Acp5-pre-miR-214 were purified and microinjected into C57BL/6J mouse oocytes and the oocytes were then surgically transferred into pseudopregnant C57BL/6J dams at the Xiehe Medical University Animal Model Center. Transgenic founder mice were bred for 6 generations to obtain a defined genetic background. All the experimental procedures were approved by the Committees of Animal Ethics and Experimental Safety of China Astronaut Research and Training Center.
Cell sorting with flow cytometry
The bone marrow cells were collected from the WT and OC-TG214 mice femur and tibia. After washed by PBS/1%BSA, the cells were directly stained with antibody to mouse Oscar (1:40, Santa Cruz Biotechnology, sc-34237) and then stained with donkey anti-goat IgG-PE (1:100, R&D systems, F0107). After that, stained cell population was used for FACS. The obtained selected Oscar+ cell population was used for total RNA extraction and qRT-PCR.
Micro CT analysis
For the proximal tibia, the whole secondary spongiosa at the left distal femur from each mouse was scanned ex vivo using a micro CT system (Inveon Mm Gantry LG CT Camera). Briefly, 150 slices with a voxel size of 10 µm were scanned at the region of the proximal tibia beginning at the growth plate and extending distally along the tibia diaphysis. Eighty continuous slices beginning at 0.1 mm from the most proximal aspect of the growth plate in which both condyles were no longer visible were selected for analysis. All trabecular bone from each selected slice was segmented for 3-dimensional reconstruction to calculate the following parameters: BMD, BV/TV, Tb.Sp and structure model index.45
Statistical analysis
Data are presented as mean ± s.e.m per experimental condition unless noted otherwise. Considering the presence of unequal variance for the data, we firstly test the equality of variances across groups. If it shows the variances are unequal, we then use mixed model with heterogeneous variances for 2-way analysis or Welch's t test for 1-way analysis. Otherwise, we use the regular linear model or Student's t-test. A p value less than 0.05 was considered statistically significant. p value less than 0.01 was considered as very significant. Bonferroni adjustment was used for multiple comparisons. All the statistical tests are analyzed by Prism software (Graphpad prism for windows, version 5.01) and SPSS (Version 14.0).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Mr. Xiaogang Wang (Jinan University) for valuable advice and Dr. Sakamuri V. Reddy (Medical University of South Carolina) for kindly providing the Acp5 promoter vector.
Funding
This work was supported by National Natural Science Foundation of China Project (31325012, 31170811, 31271225, 31340064, and 81370971), National Basic Research 973 Program of China (2011CB711003) and State Key Lab of Space Medicine Fundamentals and Application grant (SMFA13A02).
References
- 1.Del Fattore A, Teti A, Rucci N. Osteoclast receptors and signaling. Arch Biochem Biophys 2008; 473:147–60; http://dx.doi.org/ 10.1016/j.abb.2008.01.011. [DOI] [PubMed] [Google Scholar]
- 2.Ross FP, Teitelbaum SL. alphavbeta3 and macrophage colony-stimulating factor: partners in osteoclast biology. Immunol Rev 2005; 208:88–105; PMID:16313343; http://dx.doi.org/ 10.1111/j.0105-2896.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- 3.Miyazaki T, Tanaka S, Sanjay A, Baron R. The role of c-Src kinase in the regulation of osteoclast function. Mod Rheumatol 2006; 16: 68–74; PMID:16633924; http://dx.doi.org/ 10.3109/s10165-006-0460-z. [DOI] [PubMed] [Google Scholar]
- 4.Edwards CM, Mundy GR. Eph receptors and ephrin signaling pathways: a role in bone homeostasis. Int J Med Sci 2008; 5:263–72; PMID:18797510; http://dx.doi.org/ 10.7150/ijms.5.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hattersley G, Owens J, Flanagan AM, Chambers TJ. Macrophage colony stimulating factor (M-CSF) is essential for osteoclast formation in vitro. Biochem Biophys Res Commun 1991; 177:526–31; PMID:2043138; http://dx.doi.org/ 10.1016/0006-291X(91)92015-C. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi N, Udagawa N, Suda T. A new member of tumor necrosis factor ligand family, ODF/OPGL/TRANCE/RANKL, regulates osteoclast differentiation and function. Biochem Biophys Res Commun 1999; 256:449–55; PMID:10080918; http://dx.doi.org/ 10.1006/bbrc.1999.0252. [DOI] [PubMed] [Google Scholar]
- 7.Yasuda H, Shima N, Nakagawa N, Yamaguchi K, Kinosaki M, Mochizuki S, Tomoyasu A, Yano K, Goto M, Murakami A, et al.. Osteoclast differentiation factor is a ligand for osteoprotegerin/osteoclastogenesis-inhibitory factor and is identical to TRANCE/RANKL. Proc Natl Acad Sci U S A 1998; 95:3597–602; PMID:9520411; http://dx.doi.org/ 10.1073/pnas.95.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al.. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 2002; 3:889–901; PMID:12479813; http://dx.doi.org/ 10.1016/S1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 9.Matsumoto M, Kogawa M, Wada S, Takayanagi H, Tsujimoto M, Katayama S, Hisatake K, Nogi Y. Essential role of p38 mitogen-activated protein kinase in cathepsin K gene expression during osteoclastogenesis through association of NFATc1 and PU.1. J Biol Chem 2004; 279:45969–79; PMID:15304486; http://dx.doi.org/ 10.1074/jbc.M408795200. [DOI] [PubMed] [Google Scholar]
- 10.Kim Y, Sato K, Asagiri M, Morita I, Soma K, Takayanagi H. Contribution of nuclear factor of activated T cells c1 to the transcriptional control of immunoreceptor osteoclast-associated receptor but not triggering receptor expressed by myeloid cells-2 during osteoclastogenesis. J Biol Chem 2005; 280:32905–13; PMID:16046394; http://dx.doi.org/ 10.1074/jbc.M505820200. [DOI] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–97; PMID:14744438; http://dx.doi.org/ 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 12.Kapinas K, Delany AM. MicroRNA biogenesis and regulation of bone remodeling. Arthritis Res Ther 2011; 13:220; PMID:21635717; http://dx.doi.org/ 10.1186/ar3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao H, Ito Y, Chappel J, Andrews NW, Teitelbaum SL, Ross FP. Synaptotagmin VII regulates bone remodeling by modulating osteoclast and osteoblast secretion. Dev Cell 2008; 14:914–25; PMID:18539119; http://dx.doi.org/ 10.1016/j.devcel.2008.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Guan X, Guo F, Zhou J, Chang A, Sun B, Cai Y, Ma Z, Dai C, Li X, et al.. miR-30e reciprocally regulates the differentiation of adipocytes and osteoblasts by directly targeting low-density lipoprotein receptor-related protein 6. Cell Death Dis 2013; 4:e845; PMID:24113179; http://dx.doi.org/ 10.1038/cddis.2013.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassan MQ, Maeda Y, Taipaleenmaki H, Zhang W, Jafferji M, Gordon JA, Li Z, Croce CM, van Wijnen AJ, Stein JL, et al.. miR-218 directs a Wnt signaling circuit to promote differentiation of osteoblasts and osteomimicry of metastatic cancer cells. J Biol Chem 2012; 287:42084–92; PMID:23060446; http://dx.doi.org/ 10.1074/jbc.M112.377515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang JF, Fu WM, He ML, Wang H, Wang WM, Yu SC, Bian XW, Zhou J, Lin MC, Lu G, et al.. MiR-637 maintains the balance between adipocytes and osteoblasts by directly targeting Osterix. Mol Biol Cell 2011; 22:3955–61; PMID:21880893; http://dx.doi.org/ 10.1091/mbc.E11-04-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kapinas K, Kessler C, Ricks T, Gronowicz G, Delany AM. miR-29 modulates Wnt signaling in human osteoblasts through a positive feedback loop. J Biol Chem 2010; 285:25221–31; PMID:20551325; http://dx.doi.org/ 10.1074/jbc.M110.116137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kapinas K, Kessler CB, Delany AM. miR-29 suppression of osteonectin in osteoblasts: regulation during differentiation and by canonical Wnt signaling. J Cell Biochem 2009; 108:216–24; PMID:19565563; http://dx.doi.org/ 10.1002/jcb.22243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, Guo B, Li Q, Peng J, Yang Z, Wang A, Li D, Hou Z, Lv K, Kan G, et al.. miR-214 targets ATF4 to inhibit bone formation. Nat Med 2013; 19:93–100; PMID:23223004; http://dx.doi.org/ 10.1038/nm.3026. [DOI] [PubMed] [Google Scholar]
- 20.Watanabe T, Sato T, Amano T, Kawamura Y, Kawamura N, Kawaguchi H, Yamashita N, Kurihara H, Nakaoka T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev Dyn 2008; 237:3738–48; PMID:18985749; http://dx.doi.org/ 10.1002/dvdy.21787. [DOI] [PubMed] [Google Scholar]
- 21.Ishibashi O, Niwa S, Kadoyama K, Inui T. MMP-9 antisense oligodeoxynucleotide exerts an inhibitory effect on osteoclastic bone resorption by suppressing cell migration. Life Sci 2006; 79:1657–60; PMID:16806280; http://dx.doi.org/ 10.1016/j.lfs.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 22.Kornak U, Kasper D, Bosl MR, Kaiser E, Schweizer M, Schulz A, Friedrich W, Delling G, Jentsch TJ. Loss of the ClC-7 chloride channel leads to osteopetrosis in mice and man. Cell 2001; 104:205–15; PMID:11207362; http://dx.doi.org/ 10.1016/S0092-8674(01)00206-9. [DOI] [PubMed] [Google Scholar]
- 23.Gowen M, Lazner F, Dodds R, Kapadia R, Feild J, Tavaria M, Bertoncello I, Drake F, Zavarselk S, Tellis I, et al.. Cathepsin K knockout mice develop osteopetrosis due to a deficit in matrix degradation but not demineralization. J Bone Miner Res 1999; 14:1654–63; PMID:10491212; http://dx.doi.org/ 10.1359/jbmr.1999.14.10.1654. [DOI] [PubMed] [Google Scholar]
- 24.Kiviranta R, Morko J, Alatalo SL, NicAmhlaoibh R, Risteli J, Laitala-Leinonen T, Vuorio E. Impaired bone resorption in cathepsin K-deficient mice is partially compensated for by enhanced osteoclastogenesis and increased expression of other proteases via an increased RANKL/OPG ratio. Bone 2005; 36:159–72; PMID:15664014; http://dx.doi.org/ 10.1016/j.bone.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 25.Saftig P, Hunziker E, Wehmeyer O, Jones S, Boyde A, Rommerskirch W, Moritz JD, Schu P, von Figura K. Impaired osteoclastic bone resorption leads to osteopetrosis in cathepsin-K-deficient mice. Proc Natl Acad Sci U S A 1998; 95:13453–8; PMID:9811821; http://dx.doi.org/ 10.1073/pnas.95.23.13453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li LM, Hou DX, Guo YL, Yang JW, Liu Y, Zhang CY, Zen K. Role of microRNA-214-targeting phosphatase and tensin homolog in advanced glycation end product-induced apoptosis delay in monocytes. J Immunol 2011; 186:2552–2560; PMID:21228352; http://dx.doi.org/ 10.4049/jimmunol.1001633. [DOI] [PubMed] [Google Scholar]
- 27.Jindra PT, Bagley J, Godwin JG, Iacomini J. Costimulation-dependent expression of microRNA-214 increases the ability of T cells to proliferate by targeting pten. J Immunol 2010; 185:990–997; PMID:20548023; http://dx.doi.org/ 10.4049/jimmunol.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al.. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 2008; 68:425–433; PMID:18199536; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 29.Sugatani T. PTEN regulates RANKL- and osteopontin-stimulated signal transduction during osteoclast differentiation and cell motility. J Biol Chem 2002; 278:5001–5008; PMID:12460992; http://dx.doi.org/ 10.1074/jbc.M209299200. [DOI] [PubMed] [Google Scholar]
- 30.Reddy SV, Scarcez T, Windle JJ, Leach RJ, Hundley JE, Chirgwin JM, Chou JY, Roodman GD. Cloning and characterization of the 5′-flanking region of the mouse tartrate-resistant acid phosphatase gene. J Bone Miner Res 1993; 8:1263–70; PMID:8256664; http://dx.doi.org/ 10.1002/jbmr.5650081015. [DOI] [PubMed] [Google Scholar]
- 31.Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood 2011; 117:3648–57; PMID:21273303; http://dx.doi.org/ 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugatani T, Hruska KA. MicroRNA-223 is a key factor in osteoclast differentiation. J Cell Biochem 2007; 101:996–9; PMID:17471500; http://dx.doi.org/ 10.1002/jcb.21335. [DOI] [PubMed] [Google Scholar]
- 33.Mann M, Barad O, Agami R, Geiger B, Hornstein E. miRNA-based mechanism for the commitment of multipotent progenitors to a single cellular fate. Proc Natl Acad Sci U S A 2010; 107:15804–9; PMID:20720163; http://dx.doi.org/ 10.1073/pnas.0915022107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakasa T, Shibuya H, Nagata Y, Niimoto T, Ochi M. The inhibitory effect of microRNA-146a expression on bone destruction in collagen-induced arthritis. Arthritis Rheum 2011; 63:1582–90; PMID:21425254; http://dx.doi.org/ 10.1002/art.30321. [DOI] [PubMed] [Google Scholar]
- 35.Kagiya T, Nakamura S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor α and RANKL during osteoclast differentiation. J Periodontal Res 2013; 48:373–85; PMID:23078176; http://dx.doi.org/ 10.1111/jre.12017. [DOI] [PubMed] [Google Scholar]
- 36.Cheng P, Chen C, He HB, Hu R, Zhou HD, Xie H, Zhu W, Dai RC, Wu XP, Liao EY, et al.. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res 2013; 28:1180–90; PMID:23225151; http://dx.doi.org/ 10.1002/jbmr.1845. [DOI] [PubMed] [Google Scholar]
- 37.Cheng P, Chen C, He H-B, Hu R, Zhou H-D, Xie H, Zhu W, Dai R-C, Wu X-P, Liao E-Y, et al.. miR-148a regulates osteoclastogenesis by targeting V-maf musculoaponeurotic fibrosarcoma oncogene homolog B. J Bone Miner Res 2013; 28:1180–1190; PMID:23225151; http://dx.doi.org/ 10.1002/jbmr.1845. [DOI] [PubMed] [Google Scholar]
- 38.Desvignes T, Contreras A, Postlethwait JH. Evolution of the miR199-214 cluster and vertebrate skeletal development. RNA Biol 2014; 11:281–94; PMID:24643020; http://dx.doi.org/ 10.4161/rna.28141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al.. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res 2008; 68:425–33; PMID:18199536; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 40.Jindra PT, Bagley J, Godwin JG, Iacomini J. Costimulation-dependent expression of microRNA-214 increases the ability of T cells to proliferate by targeting Pten. J Immunol 2010; 185:990–7; PMID:20548023; http://dx.doi.org/ 10.4049/jimmunol.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao X. Targeting osteoclast-osteoblast communication. Nat Med 2011; 17:1344–6; PMID:22064408; http://dx.doi.org/ 10.1038/nm.2499. [DOI] [PubMed] [Google Scholar]
- 42.Matsuo K, Irie N. Osteoclast-osteoblast communication. Arch Biochem Biophys 2008; 473:201–9; PMID:18406338; http://dx.doi.org/ 10.1016/j.abb.2008.03.027. [DOI] [PubMed] [Google Scholar]
- 43.van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, van Eijndhoven MA, Pegtel DM, Stoorvogel W, Wurdinger T, et al.. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood 2013; 121:3997–4006, S1-15; http://dx.doi.org/ 10.1182/blood-2013-02-478925. [DOI] [PubMed] [Google Scholar]
- 44.Chen L, Charrier A, Zhou Y, Chen R, Yu B, Agarwal K, Tsukamoto H, Lee LJ, Paulaitis ME, Brigstock DR. Epigenetic regulation of connective tissue growth factor by MicroRNA-214 delivery in exosomes from mouse or human hepatic stellate cells. Hepatology 2014; 59:1118–29; PMID:24122827; http://dx.doi.org/ 10.1002/hep.26768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iwaniec UT, Wronski TJ, Liu J, Rivera MF, Arzaga RR, Hansen G, Brommage R. PTH stimulates bone formation in mice deficient in Lrp5. J Bone Miner Res 2007; 22:394–402; PMID:17147489; http://dx.doi.org/ 10.1359/jbmr.061118. [DOI] [PubMed] [Google Scholar]