Abstract
Upon pathogen invasion, bacteria and archaea activate an RNA-interference-like mechanism termed CRISPR (clustered regularly interspaced short palindromic repeats). A large family of Cas (CRISPR-associated) proteins mediates the different stages of this sophisticated immune response. Bioinformatic studies have classified the Cas proteins into families, according to their sequences and respective functions. These range from the insertion of the foreign genetic elements into the host genome to the activation of the interference machinery as well as target degradation upon attack. Cas7 family proteins are central to the type I and type III interference machineries as they constitute the backbone of the large interference complexes. Here we report the crystal structure of Thermofilum pendens Csc2, a Cas7 family protein of type I-D. We found that Csc2 forms a core RRM-like domain, flanked by three peripheral insertion domains: a lid domain, a Zinc-binding domain and a helical domain. Comparison with other Cas7 family proteins reveals a set of similar structural features both in the core and in the peripheral domains, despite the absence of significant sequence similarity. T. pendens Csc2 binds single-stranded RNA in vitro in a sequence-independent manner. Using a crosslinking - mass-spectrometry approach, we mapped the RNA-binding surface to a positively charged surface patch on T. pendens Csc2. Thus our analysis of the key structural and functional features of T. pendens Csc2 highlights recurring themes and evolutionary relationships in type I and type III Cas proteins.
Keywords: RRM domain, CRISPR, prokaryotic immune system, Cas7, RNA binding
Abbreviations
- CRISPR
Clustered regulatory short interspaced palindromic repeats
- Cas
CRISPR-associated
- dCASCADE
interference complex subtype I-D
- eCASCADE
interference complex subtype I-E
- crRNA
CRISPR RNA
- RAMP
Repeat associated mysterious protein
- RRM
RNA recognition motif
- RNAi
RNA interference
- H1 and H2 and H1-2
β-hairpins of insertion domain 1 (or lid domain)
- Tp
Thermofilum pendens
- Ss
Sulfolobus solfataricus
- Mk
Methanopyrus kandleri
- Rmsd
Root mean square deviation
- SAD
Single-wavelength anomalous dispersion
Introduction
CRISPR (clustered regularly interspaced short palindromic repeats) confer an adaptive prokaryotic defense mechanism that recognizes and inactivates foreign genetic elements,1 a mechanism that is functionally reminiscent of the eukaryotic RNA interference (RNAi) pathway.2,3 In contrast to RNAi, CRISPR establishes a genetic memory of previously encountered pathogens that is accessed upon re-infection. Foreign nucleic acid sequences (spacers) derived from viruses or conjugative plasmids are integrated into the host genome.4,5 The unique spacers are located within a CRISPR locus and interspersed by a series of identical host repeat sequences.6 The CRISPR locus is transcribed into a precursor RNA that is subsequently processed to yield the mature functional crRNAs.7 Adjacent to the CRISPR locus are genes encoding the protein machinery behind this response.8 Upon infection, Cas (CRISPR-associated) proteins mediate spacer acquisition,4,5 crRNA biogenesis,9 target interference and degradation.10
The CRISPR-Cas systems have been classified into three major types (I, II and III), that can be further divided into at least 10 subtypes.11 The classification of Cas proteins is hampered by the fact that even proteins with the same function have very little sequence similarity.12,13 Therefore structural data are indispensible for accurate classification. The diversity within the CRISPR protein machinery is believed to have evolved out of the demand to respond to the specific nature of the pathogen as well as the environment of the host cell (such as thermophilic, mesophilic, halophilic, etc.). The majority of the Cas proteins contain RAMP (repeat-associated mysterious protein) domains.14 These domains are based on a ferredoxin-like fold,15 similar to that of an RRM (RNA-recognition motif), a ubiquitous RNA-binding domain.16 However, the RRM-like domains in Cas proteins differ from those of canonical RRM domains.16,17 First, they generally do not share the conserved consensus sequences that are involved in RNA binding in canonical RRM domains.16-19 Second, they are structurally much more variable,20 as they feature longer insertions18,19,21 and extensions at the N- and C-termini.22,23 This variation is reflected in their different functions, which range from having a structural role18 to harboring catalytic activity.21,24-26
Insight into how Cas proteins assemble the functional interference complexes has been provided by electron-microscopy studies of type I-A18/I-E27/I-F28 and III-A29/B30,31 interference complexes and high resolution X-ray crystallography structures of single proteins.20 Characteristic of type I and type III systems are members of the Cas7 family of proteins, which constitute the core subunit of the interference complexes. Multiple copies of Cas7 assemble in a helical fashion around the processed crRNA18,27,28,32 and mediate interactions with further factors, which ultimately define complex length, activity and target recognition. To date, little information is available on the subtype I-D proteins and the associated dCASCADE interference complex. Here, we have studied Thermofilum pendens Csc2, a subtype I-D protein of the Cas7 family. Subtype I-D is commonly present in Archaea33 and Cyanobacteria.34 It harbors characteristic features of both subtypes I and III: a type I HD nuclease domain is fused to Cas10, the signature protein of type III. The general domain organization of CASCADE proteins is predicted to resemble type III proteins,33 emphasizing the prominent role of this subtype as an evolutionary link between types I and III. We report the insights we obtained from the crystal structure and biochemical analysis of Thermofilum pendens (Tp) Csc2. The comparison of type I-D TpCsc2 with type I-A Sulfolobus solfataricus (Ss) Csa218 and type III-A Methanopyrus kandleri (Mk) Csm319 allows building a comprehensive picture of the Cas7 protein family and its conserved RNA-binding properties.
Structure Determination of Csc2
We expressed full-length T. pendens (Tp) Csc2 (374 residues) in E. coli and purified it to homogeneity. Tp Csc2 yielded crystals in an orthorhombic space group (P222), containing one molecule per asymmetric unit and diffracting beyond 1.8 Å resolution. X-ray fluorescence scans on the crystals showed a peak at the Zinc excitation, suggesting the presence of a bound zinc ion in the crystallized protein. We obtained phases by crystallizing the selenomethionine derivatized protein and solved the structure by single-wavelength anomalous dispersion method (SAD). The structure was refined at 1.8 Å resolution to an Rfree/Rwork of 21/18%. The final atomic model has good stereochemistry (Table 1) and includes most of the polypeptide. Disordered regions include a loop between residues Gln134 and Gly147, the four N-terminal residues and 21 C-terminal residues.
Table 1.
Data Collection and Structure Refinement Statstics of Tp Csc2
| Data collection | ||
| Native Tp Csc2 | SeMet Tp Csc2 | |
| Space group | P 2 21 21 | P 2 21 21 |
| Unit cell (Å)a | a = 60.47 b = 80.95 c = 112.60 |
a = 60.81 b = 81.24 c = 114.02 |
| Resolution range (Å)a | 46.22–1.82 (1.88–1.82) | 48.68–2.37 (2.46–2.37) |
| Unique reflectionsa | 50416 (7188) | 23518 (2402) |
| I/σ (I)a | 17.8 (1.6) | 31.9 (6.4) |
| Multiplicitya | 6.5 (6.0) | 13.1 (12.6) |
| Rmerge (%)a | 6.7 (97.7) | 7.3 (43.4) |
| CC(1/2) (%)a | 99.9 (50.5) | 99.9 (95.4) |
| Refinement | ||
| Average B-factor | 32.70 | 34.28 |
| Rwork (%) | 18.15 (31.75) | 20.85 (24.14) |
| Rfree (%) | 21.21 (34.64) | 23.72 (25.13) |
| Rmsd bonds (Å) | 0.017 | 0.004 |
| Rmsd angles (°) | 1.36 | 0.789 |
| Ramachandran favored (%) | 97.0 | 96.7 |
| Ramachandran outliers (%) | 0.0 | 0.0 |
Values in parentheses correspond to the highest resolution shell; SeMet: Selenomethionine derivatized protein.
Csc2 has a Central RRM-Like Core Domain with Three Elaborate Insertion Domains
The overall architecture of Tp Csc2 can be described as composed of four domains (Fig. 1A). At the core of the molecule is a domain with β–α–β–β–α–β topology reminiscent of a RRM fold (Fig. 1B). The four β-strands form a twisted β-sheet, with two α–helices (α1, α2) resting against a concave groove. Strands β1 and β3 of the core domain lack residues of the so-called RNP2 and RNP1 motifs, which are required for RNA binding in canonical RRM domains. In addition, the canonical RNA-interacting interface of the RRM fold is obstructed from the solvent by an α helix (αE). Overall, a large part of the core domain is inaccessible to solvent. The most exposed structural element is helix α1. Helix α1 contains conserved residues and contacts a conserved glycine-rich loop between helix α2 and strand β4. The presence of a rather flexible glycine-rich loop at this structural position is a characteristic feature in the non-canonical RRM folds of the Cas superfamily, although its exact function remains elusive.
Figure 1.

Crystal Structure of Thermofilum pendens Csc2. (A) Structure of Tp Csc2 can be divided into four distinct domains: a core domain (green), a lid domain (insertion 1, blue), a metal-binding domain (insertion 2, red) and a helical domain (insertion 3, yellow). Secondary structure elements of the core adopt a ferredoxin-like fold with β-α-β-β-α-β arrangement. Multiple insertions within the core define the accessory domains. Dashed lines indicate the disordered loops. The inset shows a detailed view of the zinc ion (gray sphere) with coordinating residues. (B) Topology diagram of TpCsc2. α-Helices are represented as circles and β-strands arrows. The secondary structure elements have been labeled numerically maintaining the nomenclature of RRM domains. The hairpins of insertion domain 1 are labeled as described in the text (H1, H2 and H1–2). The α-helices of in the insertion domains are labeled with letters (αA to αH).
The core domain is flanked by three peripheral domains that are composed of elaborate insertions originating from the secondary structure elements of the core. The first insertion domain (insertion domain 1 or lid domain) is formed by three β-hairpins (H1, H2 and H1–2) that create a lid on top of the core (Fig. 1A and 1B). Hairpin H1 is formed between the β1–α1 elements of the RRM-like domain and contacts both strand β2 of the core and helix αE. Hairpin H2 is formed between the β2–β3 elements of the RRM-like domain. H2 features a sharp bend at the tip (where Gly205 is located) and a hinge at the bottom (where the residues Pro196 and Gly211 are located). The bottom of H2 packs against hairpin H1–2. Hairpin H1–2 constitutes the base of the lid domain and is formed by both the β1–α1 and β2–β3 segments of the RRM-like domain. This hairpin effectively extends the β-strands β1 and β3 of the core, after a sharp bend created by the conserved residues Pro222 and Gly223 (Fig. 2).
Figure 2.
Structure-based sequence alignment of Tp Csc2. The alignment includes four sequences from representative species of the Csc2 family, based on a comprehensive alignment. Secondary structure elements are indicted by the cartoon above the sequences, color-coded and labeled according to Figure 1A. Colors represent the percentage of sequence identity (dark > 60%, light 60–30%). U15 cross-linked residues are highlighted with yellow dots. Blue dots above the K179 and R183 mark the mutated amino acids, brackets indicate the boundaries of the sequence spanning (P197-L214), which was replaced by (GS)3 (Δloop mutant).
The insertion domain 2 (or metal-binding domain) is defined by an 80 amino-acid long segment between α1–β2 and the very C-terminal helix αH (Fig. 1B). αH is an elongated helix, embedded within a predominantly hydrophobic cavity lined by the helices αC and αD. This domain coordinates a Zinc ion via the residues Cys131, Cys153, His155, Cys156 and in addition Asp150 (Fig. 1A). Metal binding appears to have a structural role, maintaining the close packing within the domain. The insertion domain 3 is a helical domain formed by three helices (αE, αF and αG) that are between the last β-strand of the core domain and the C-terminal helix αH. Insertion domain 3 contacts secondary structure elements of the core domain and, together with the small N-terminal helices αA and αB, it wraps around the convex surface of the β-sheets and helix α2 (Fig. 1A).
Csc2: a Cas7 Family Protein
Bioinformatic predictions categorized Tp Csc2 within the Cas7 protein family and suggested it as the Cas7 homolog in subtype I-D interference complexes.11 We compared the structure of Tp Csc2 with those of Sulfolobus solfataricus (Ss) Csa2 (3PS0)18 and Methanopyrus kandleri (Mk) Csm3 (4NOL)19 (Fig. 3A and 3B). Ss Csa2 and Mk Csm3 are Cas7 homologs in the subtypes I-A and III-A and share a sequence identity of 9% and 20%, respectively, with Tp Csc2. The RRM-like fold of Tp Csc2 (76 amino acids) superposes with the respective domains with a root mean square deviation (rmsd) of 1.5 Å for Mk Csm3 and 3.0 Å for Ss Csa2. The main difference in the core domains is that Tp Csc2 lacks the fifth β-strand that is characteristic of the β-sheet of Mk Csm3 and Ss Csa2 (Fig. 3A).
Figure 3.
Structural comparison of Cas7 proteins. (A) Topology diagrams of Tp Csc2, Ss Cas7 and Mk Csm3 highlight the high structural conservation within the core RRM-like fold (boxed in gray) and show the connectivity of the insertion domains. The topological arrangement of the insertions 1–3 is similar in all proteins. Variations within secondary structure elements of the three proteins reflect subtype specificities. (B) Crystal structures of Cas7 orthologs, Tp Csc2, Ss Csa2, Mk Csm3, depicted according to the orientation in Figure 1A after optimal superposition of their RRM-like domains. The molecules are overall colored in gray. Significant structural similarities are colored according to the color-code of the respective proteins, Tp Csc2 (salmon), Ss Csa2 (orange), Mk Csm3 (blue). Numbers (1–5) refer to the significant structural elements discussed in the text. Dashed lines indicate the structurally unresolved loops. (C) Boxes highlight the structurally and sequence-conserved basic residues along β2 and the preceding insertion.
Tp Csc2, Ss Csa2 and Mk Csm3 have insertions at equivalent positions within the core domain. Structure-based comparisons suggest that the peripheral insertion domains also share similarities. The lid domain (insertion 1) is in all cases the most flexible part of the molecule (Fig. 3B). A common structural feature of the lid domain is the β-hairpin corresponding to Tp Csc2 H1 (structural element 1 in Figure 3B), which protrudes toward the front of the RRM-like core. Insertion domain 2 is in all cases a predominantly α-helical domain. In both Tp Csc2 and Mk Csm3, this domain contains a structural Zinc ion (Fig. 3A and 3B). In the case of Ss Csa2, insertion domain 2 does not require a metal ion to be folded. A common structural feature of insertion domain 2 in the three structures is helix αD, which is buried in the heart of the proteins (structural element 2 in Figure 3B). Another conserved feature is a helix that connects this insertion domain to the core domain (structural element 3 in Figure 3B). In the case of Tp Csc2, this structural element corresponds to the C-terminal helix αH. In the case of Mk Csm3 and Ss Csa2, this structural element is derived from a topologically different region. Similarly, helices at the base of the core domain are present in all three proteins but are derived from different elements (structural element 4 in Figure 3B). Generally, the structural conservation among the Cas7 proteins is not reflected in high sequence similarities. The exception is a solvent-exposed platform formed by the segment preceding β2 at the interface between the core domain and insertion domain 2 (structural element 5 in Figure 3C), which is not only conserved at the structural but also at the sequence level.
In order to confirm our classification and pervious bioinformatic analysis, we compared Tp Csc2 to well-studied Cas protein families (Cas6 and Cas5), based on the structural analysis by Reeks et al. Although Cas5 and Cas6 proteins also contain an RRM-like domain as a core structural element and a glycine-rich flexible region between α2 and β4, the peripheral domains diverge. First, the β hairpin between β2-β3 is part of the lid domain in Tp Csc2 and other Cas7 family members, while in Cas5d it can be seen as an extension of the RRM β strands, in Cas6 it is present in both RRM domains. Second, the insertion domain 2 that is present between α1 and β2 in Tp Csc2 and other Cas7 proteins is absent in the Cas6 and Cas5 family. Third are the structurally variable C-terminal domains, which consist of a second RRM domain in the cases of most Cas6 proteins and an extended β-hairpin in Cas5d representatives. Despite the unifying RRM-like core, the structural variation of the peripheral domains might reflect the different RNA binding requirements.
Mapping the RNA-Binding Interface
Proteins of the Cas7-family assemble around processed crRNA and constitute the backbone of the interference complex.27,29,31 Several Cas7 monomers are involved in binding to the variable pathogen derived spacer region, exposing it to the complementary target DNA.27 We sought to determine the RNA-binding interface in Tp Csc2. In electrophoretic mobility shift assays (EMSAs), Tp Csc2 bound a polyU RNA of 15 nt length (poly(U)15 and poly(A)15, (surrogates of the variable spacer sequence) in a comparable fashion as previously shown for Mk Csm319 (Fig. 4A, left panel). At increasing concentrations, Tp Csc2 binds likely in multiple copies on the 15-mer RNA oligonucleotides, as shown by the supershift in the gel (Fig. 4A, left panel, concentration 25 μM and 50 μM). A similar behavior was observed upon binding to the Tp crRNA (Fig. 4A, right panel).
Figure 4.
For figure legend, see page 1078.Figure 4 (See previous page). Mapping the RNA-binding surface of Thermofilum pendens Csc2. (A) Electrophoretic mobility shift assays (EMSA) with wild-type Tp Csc2. Left panel: EMSA were performed with 32P -5′-end labeled poly(U)15 or poly(U)15 RNAs and increasing concentrations of Tp Csc2 (0, 5, 25, 50μM). The positions of the free RNA probe (arrow head) and of the RNA-bound complexes (asterisks) are shown on the right. Right panel: EMSA assay with Tp Csc2 and 32P -5′-end labeled crRNA. (B) MS/MS mass spectra of Tp Csc2 peptides, carrying an additional mass corresponding to one (panel two and three) or two (panel one) uracil nucleotides associated with the respective amino-acid. Peptide sequence and the fragment ions are indicated on top. The direcly crosslinked residues are colored yellow. The peptide fragmentation occurs with the cleavage of amide bonds resulting in b-ions and y-ions when the charge is retained by the N-terminal and C-terminal fragments, respectively. #, #,1 #2 and #3 indicate the b- and y-ions that were observed with a mass shift corresponding to U’, U-H3PO4, U-H2O and U, respectively. IM: Immonium ions. U’: U marker ion adduct of 112.0273 Da. (C) Mapping RNA-binding properties on the Tp Csc2 crystal structure. Upper panel: a cartoon representation of Tp Csc2 is shown in gray (in the same orientation as in Figure 1A) with the crosslinked residues colored in yellow (stick representation) and regions targeted for mutagenesis colored in blue (K178E/R183E and Δloop, indicated with scissors, stick representation). Lower panel: surface representation of Tp Csc2 (in the same orientation as in panel C) depicting the electrostatic potential (red for electronegative and blue for electropositive). (D) Quantitative measurements of RNA-binding affinities. Upper panel: 13% SDS-PAGE with the wild-type (WT) an mutant proteins used in the fluorescence anisotropy (FA) assay and a table with the Kd values obtained. The Δloop mutant was engineered by replacing the segment between Pro197 and Leu214 with a (GS)3 sequence. Lower panel: FA measurements of WT and mutant Tp Csc2 with a 5′-6-carboxy-fluorescein-labeled poly(U)15 -RNA.
Next, we sought to identify RNA binding interface of the protein using a crosslinking - mass spectrometry approach. We incubated Tp Csc2 with poly(U)15 RNA, and cross-linked the complex by subjecting it to UV irradiation at 254 nm. We used LC-MS/MS mass spectrometry to detect and sequence peptides cross-linked to an RNA nucleotide as previously described.35 UV crosslinks favorably occur with sulfur-containing or aromatic side chains that are in close proximity to the nucleic acid, although not necessarily in direct contact. The mass spectrometric analysis identified three modified peptides (Fig. 4B). The reactive residues that were directly conjugated to a uridine (Cys131, Met83 and Trp346, Figure 4C, upper panel) encircle a central positively-charged patch on the surface of the protein, suggesting that this region mediates RNA binding (Fig. 4C, lower panel). Moreover, this patch is adjacent to the conserved segment preceding β2 that is enriched in lysine and arginine residues (structural element 5 in Figure 3C).
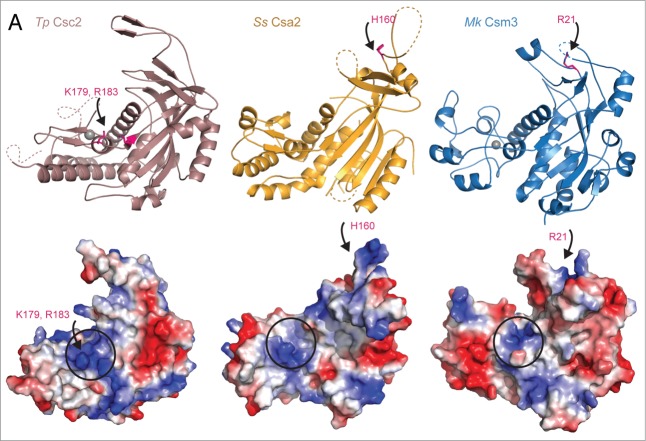
We targeted surface exposed regions of Tp Csc2 for mutagenesis and used quantitative RNA-binding experiments to compare the mutants to the wild-type protein (Fig. 4D). In fluorescence anisotropy experiments, Tp Csc2 bound a poly(U)15 RNA with an affinity in the low micro-molar range (Fig. 4D, black curve in lower panel). Reverse-charge mutations of two positively charged residues in structural element 5 (K179E/R183E) resulted in a 6-fold reduction of RNA-binding affinity as compared with the wild-type protein (Fig. 4D, light blue curve in the lower panel), consistently with the information form the structural and mass-spectrometry analyses. Importantly, a positively charged surface groove is present at the equivalent structural position (between the core RRM-like domain and insertion domain 2) in the structures of the Cas7 family proteins Ss Csa2 and Mk Csm3 (Fig. 5). This groove corresponds to the predicted site for crRNA binding on the Cas7 family protein CasC deduced from the interpretation of the cryo-EM structure of the eCASCADE27 complex (Fig. S1).
Figure 5.
A structurally and functionally conserved surface groove in the Cas7 protein family. Upper panels: cartoon representation of the structures of the Cas7-like proteins Tp Csc2 (salmon), Ss Csa2 (orange), Mk Csm3 (blue) (in the same orientations as in Figure 3B). Lower panels: corresponding surface representations showing the electrostatic potential (red for electronegative and blue for electropositive). For all proteins positively charged patches are present at the interface between the core and insertion domain 2 (identified with a circle). Conserved lysines and arginines contribute to these patches (Fig. 2) and in Tp Csc2 (arrows point to pink residues) are involved in RNA binding (Fig. 4). Residues reported to have an effect on RNA binding in Ss Csa2 and Mk Csm3 (arrows point to pink residues) are located within positively charged surfaces of the respective lid domains.
In the case of Ss Csa218 and Mk Csm3,19 the lid domain is involved in nucleic-acid binding (Fig. 5). For Tp Csc2 we did not identify direct RNA-binding residues from the lid by mass-spectrometry. Replacing the 18 residues that shape the tip of the lid domain (between Pro197 and Leu214) with a generic (Gly-Ser)3 linker (Fig. 4C, upper panel) resulted in a modest reduction of poly(U)15 RNA-binding affinity as compared with the wild-type protein (Fig. 4D, dark blue curve in the lower panel). We conclude that the influence of the lid domain in crRNA recognition or crRNA directed target DNA recognition depends on the specific Cas7 protein family, while the positively-charged surface groove between the core and the insertion 2 domain appears to be a conserved functional site.
Conclusions
A common structural feature of many Cas proteins is the central RRM-like fold and the presence of peripheral insertion domains. The structural diversity within these peripheral domains is thought to be responsible for the multitude of Cas protein functions.20 Computational12,17 and biochemical18,19 studies have contributed to classifying the Cas7 proteins. Nevertheless weak sequence homology makes structural data indispensable to enable a complete understanding of their structure-to-function relationship. In this study, we solved the structure of Tp Csc2 and confirmed the classification of the Csc2 protein as a Cas7 protein of the subtype I-D. The structural similarity among Tp Csc2 and known Cas7 proteins, such as Mk Csm3 and Ss Csa2, encompasses the central RRM-like core domain as well as the arrangement of the insertion domains. Tp Csc2 and Mk Csm3 share higher sequence and structural similarities. This is in line with previous reports suggesting that Csc2 proteins resemble their type III Cas7 counterparts.15 Despite lacking significant sequence similarity, the structural features as well as the charge distribution are strongly conserved within type I orthologs. Subtype specificities are reflected by the variations within the topology, structural composition and flexibility of the peripheral domains, such as the absence or presence of metal coordination and different secondary structure elements within the lid domain.
The basic physiological role of Cas7-like proteins of type I interference complexes18 as well as its homologs, Csm3 in type III-A and Cmr4 in type III-B systems,29,36 is to bind the variable crRNA spacer sequence and with it constitute a platform for stable binding of target DNA. In this study, we have defined the RNA binding interface of Tp Csc2. We show that Tp Csc2 binds variable sequences of ssRNA. The affinity toward the RNA is within the low micro-molar range, weak yet significant, and in line with the protein's physiological function. We further investigated the RNA-binding properties of Tp Csc2 using crosslinking mass spectrometry and structure-based mutation analyses. Here, we identified the RNA-protein interface and pinpointed functionally relevant residues. We show that Tp Csc2 possesses a critical, positively charged groove formed by conserved residues in the interface between the core and the second insertion domain. This feature is largely conserved among characterized protein family members. Our findings are in accordance to the predicted crRNA-binding interface of CasC, the E. coli Cas7 ortholog.27 Therefore we speculate that upon dCASCADE assembly, multiple copies of Tp Csc2 may adopt a similar arrangement within the complex as CasC in the eCASCADE, defining a channel and exposing the central positively charged groove toward the solvent (Fig. S1).
Taken together our study highlights the evolutionary relationship within the Cas7 protein family and helps to better understand the RNA-interacting features that are conserved among the Cas7 proteins. Further structural studies will identify the contribution of the insertion domains on protein interactions during dCASCADE assembly.
Accession Numbers
The coordinates and the structure factors of Thermofilum pendens Csc2 have been deposited in the protein Data Bank with the accession code 4TXD.
Experimental Procedures
Protein expression and purification
The gene for the Csc2 from T. pendens was ordered as a synthetic construct (GeneArt, Life technologies). His- and His-SUMO tagged Tp Csc2 proteins were expressed using E. coli BL21-Gold (DE3) Star pRARE cells (Stratagene) grown in TB medium and induced overnight at 18°C. The cells were lysed in buffer A (50 mM TRIS-HCl pH 7.5, 1 M NaCl, 10 mM imidazole, 10% glycerol) supplemented with protease inhibitors (Roche). The lysate was heated to a temperature of 55°C for 10 min and subsequently centrifuged at 25000 g. The protein was purified from the resulting supernatant at room-temperature by Ni2+- affinity chromatography as an initial step and further purified over a HiTrap Heparin column (GE Healthcare) to remove minor contaminants. The His-tag was removed by treatment with SUMO protease. Size-exclusion chromatography (SEC) on a Superdex 75 column (GE Healthcare) was performed as a final step of purification using buffer B (20 mM HEPES pH 7.5, 150 mM NaCl and 5 mM DTT and 10% glycerol). Selenomethionine derivatized protein was purified as described above from E. coli grown in M9 media complemented with the essential amino acids and selenomethionine.37
Crystallization and structure determination
Native crystals of Tp Csc2 were grown at 20 °C by sitting-drop vapor diffusion from drops formed by equal volumes of protein (at 9.5 mg/ml) and of crystallization solution containing 0.05 M Mes pH 5.6, 0.2 M KCl, 0.01 M MgCl2, 5% Peg 8000 and 17% glycerol. Crystals were cryoprotected with a final concentration of 20% glycerol prior to data collection. Selenomethionine derivatized crystals were obtained in similar conditions and cryo-protected as described above.
Native and SAD data were collected at the PXII and PXIII beamlines of the Swiss Light Source (SLS) (Villigen, Switzerland), respectively. Data were processed with XDS38 and scaled using Aimless.39 Selenium sites were located and experimental phases were calculated using the AutoSol pipeline in Phenix.40 Model building and refinement were performed with COOT41 and Phenix and the final model was validated using Molprobity.42
Biochemical assays
The RNA molecules poly(U)15, poly(A)15 and the crRNA (spacer 1 of locus 2, sequence: ACUAAGAGCC UCCUUUGCCC ACGGCAUCGG UAGGUCAGGU CCACGUUCAA AAUCAGCAAG)43 were synthesized by Purimex. The poly(U)15, and poly(A)15 RNA were 5′ labeled with T4 polynucleotide kinase (New England Biolabs) and γ-32P ATP (Perkin-Elmer), the cRNA was 3′ labeled with a-32P-pCp (Hartmann Analytic) and T4 RNA ligase (Fermentas, Thermofisher Scientific). For the gel-shift assays using poly(U)15 and poly(A)15, 0.5 pmol labeled RNA was mixed with 5 μM, 25 μM, and 50 μM Tp Csc2 protein in 10 μL reaction volume containing 20 mM Hepes at pH 7.5, 100 mM KOAc, 4 mM Mg(OAc)2, 0.1% (vol/vol) NP-40 and 2 mM DTT. For crRNA gel-shift assays, 0.14 pmol labeled RNA was mixed with 1 μM, 20 μM and 200μM Tp Csc2 protein.
The mixtures were incubated for 20 min at 55°C before adding 2 μL 50% (vol/vol) glycerol containing 0.25% (w/vol) xylene cyanole. Samples were separated on a 8% (w/vol) polyacrylamide gel at 4°C and visualized by phospho-imaging (GE Healthcare).
Fluorescence anisotropy
Fluorescence anisotropy measurements were performed with a 5′-6-carboxy-fluorescein-labeled poly(U)15 RNA at 20°C in 50 μl reactions on a Genios Pro (Tecan). The RNA was dissolved to a concentration of 10 nM and incubated with Tp Csc2 for 20 min at 55°C before adding upon measurement. The excitation and emission wavelengths were 485 nm and 535 nm, respectively. Each titration was measured three times using ten reads with an integration time of 40 μs. The data were analyzed by nonlinear regression fitting using the BIOEQS software.44
Crosslinking–mass spectrometry analysis
Tp Csc2 – poly(U)15 contacts sites were investigated with mass spectrometry after UV-induced protein–RNA crosslinking as described.35,45 The purified crosslinks were analyzed using Top10HCD method on an Orbitrap Velos instrument and the data were analyzed using OpenMS and OMSSA as a search engine (see Supplementary Experimental Procedures).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank O. Alkhnbasi and R. Backofen for bioinformatics advice; J. Basquin, K. Valer-Saldana, and S. Pleyer at the MPI-Martinsried Crystallization Facility for crystallization experiments; the staff of the PXII and PXIII beamline at the Swiss Light Source (Villigen, Switzerland) for assistance during data collection; M. Hein and members of our labs for critical reading of the manuscript.
Funding
This study was supported by the Max Planck Gesellschaft, the DFG Research Group 1680 (FOR1680) to E.C., H.U and A.M, and CIPSM to E.C.
Author Contributions
Author contributions: A.H. performed the structural and biochemical analyses; L.M. cloned WT Tp Csc2 and performed the crRNA experiment in Figure 4A; J.E. obtained native crystals; C.B. performed fluorescence anisotropy experiments; K.S. performed the mass-spec experiment in Figure 4B; E.C, A.M and H.U. supervised research; A.H. and E.C wrote the manuscript.
Supplemental Materials
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 2010; 463:568-71; PMID:20072129; http://dx.doi.org/ 10.1038/nature08703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Makarova KS, Wolf YI, van der Oost J, Koonin EV. Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol Direct 2009; 4:29; PMID:19706170; http://dx.doi.org/ 10.1186/1745-6150-4-29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/ 10.1016/j.cell.2009.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007; 315:1709-12; PMID:17379808; http://dx.doi.org/ 10.1126/science.1138140 [DOI] [PubMed] [Google Scholar]
- 5. Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010; 468:67-71; PMID:21048762; http://dx.doi.org/ 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- 6. Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): the hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem 2011; 392:277-89; PMID:21294681; http://dx.doi.org/ 10.1515/bc.2011.042 [DOI] [PubMed] [Google Scholar]
- 7. Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008; 321:960-4; PMID:18703739; http://dx.doi.org/ 10.1126/science.1159689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 2002; 43:1565-75; PMID:11952905; http://dx.doi.org/ 10.1046/j.1365-2958.2002.02839.x [DOI] [PubMed] [Google Scholar]
- 9. Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 2010; 329:1355-8; PMID:20829488; http://dx.doi.org/ 10.1126/science.1192272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 2011; 30:1335-42; PMID:21343909; http://dx.doi.org/ 10.1038/emboj.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 2011; 9:467-77; PMID:21552286; http://dx.doi.org/ 10.1038/nrmicro2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct 2006; 1:7; PMID:16545108; http://dx.doi.org/ 10.1186/1745-6150-1-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brendel J, Stoll B, Lange SJ, Sharma K, Lenz C, Stachler AE, Maier LK, Richter H, Nickel L, Schmitz RA, et al. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of crRNAs in Haloferax volcanii. J Biol Chem 2014;In press; PMID:24459147; http://dx.doi.org/ 10.1074/jbc.M113.508184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang R, Li H. The mysterious RAMP proteins and their roles in small RNA-based immunity. Protein Sci 2012; 21:463-70; PMID:22323284; http://dx.doi.org/ 10.1002/pro.2044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 2011; 6:38; PMID:21756346; http://dx.doi.org/ 10.1186/1745-6150-6-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 2005; 272:2118-31; PMID:15853797; http://dx.doi.org/ 10.1111/j.1742-4658.2005.04653.x [DOI] [PubMed] [Google Scholar]
- 17. Koonin EV, Makarova KS. CRISPR-Cas: evolution of an RNA-based adaptive immunity system in prokaryotes. RNA Biol 2013; 10:679-86; PMID:23439366; http://dx.doi.org/ 10.4161/rna.24022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lintner NG, Kerou M, Brumfield SK, Graham S, Liu H, Naismith JH, Sdano M, Peng N, She Q, Copié V, et al. Structural and functional characterization of an archaeal clustered regularly interspaced short palindromic repeat (CRISPR)-associated complex for antiviral defense (CASCADE). J Biol Chem 2011; 286:21643-56; PMID:21507944; http://dx.doi.org/ 10.1074/jbc.M111.238485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hrle A, Su AA, Ebert J, Benda C, Randau L, Conti E. Structure and RNA-binding properties of the type III-A CRISPR-associated protein Csm3. RNA Biol 2013; 10:1670-8; PMID:24157656; http://dx.doi.org/ 10.4161/rna.26500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reeks J, Naismith JH, White MF. CRISPR interference: a structural perspective. Biochem J 2013; 453:155-66; PMID:23805973; http://dx.doi.org/ 10.1042/BJ20130316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. Cas5d protein processes pre-crRNA and assembles into a cascade-like interference complex in subtype I-C/Dvulg CRISPR-Cas system. Structure 2012; 20:1574-84; PMID:22841292; http://dx.doi.org/ 10.1016/j.str.2012.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shao Y, Cocozaki AI, Ramia NF, Terns RM, Terns MP, Li H. Structure of the Cmr2-Cmr3 subcomplex of the Cmr RNA silencing complex. Structure 2013; 21:376-84; PMID:23395183; http://dx.doi.org/ 10.1016/j.str.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carte J, Pfister NT, Compton MM, Terns RM, Terns MP. Binding and cleavage of CRISPR RNA by Cas6. RNA 2010; 16:2181-8; PMID:20884784; http://dx.doi.org/ 10.1261/rna.2230110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Garside EL, Schellenberg MJ, Gesner EM, Bonanno JB, Sauder JM, Burley SK, Almo SC, Mehta G, MacMillan AM. Cas5d processes pre-crRNA and is a member of a larger family of CRISPR RNA endonucleases. RNA 2012; 18:2020-8; PMID:23006625; http://dx.doi.org/ 10.1261/rna.033100.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haurwitz RE, Sternberg SH, Doudna JA. Csy4 relies on an unusual catalytic dyad to position and cleave CRISPR RNA. EMBO J 2012; 31:2824-32; PMID:22522703; http://dx.doi.org/ 10.1038/emboj.2012.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 2011; 19:257-64; PMID:21300293; http://dx.doi.org/ 10.1016/j.str.2010.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJ, van der Oost J, Doudna JA, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature 2011; 477:486-9; PMID:21938068; http://dx.doi.org/ 10.1038/nature10402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wiedenheft B, van Duijn E, Bultema JB, Waghmare SP, Zhou K, Barendregt A, Westphal W, Heck AJ, Boekema EJ, Dickman MJ, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A 2011; 108:10092-7; PMID:21536913; http://dx.doi.org/ 10.1073/pnas.1102716108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol Cell 2013; 52:124-34; PMID:24119402; http://dx.doi.org/ 10.1016/j.molcel.2013.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell 2013; 52:146-52; PMID:24119404; http://dx.doi.org/ 10.1016/j.molcel.2013.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Staals RH, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell 2013; 52:135-45; PMID:24119403; http://dx.doi.org/ 10.1016/j.molcel.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Duijn E, Barbu IM, Barendregt A, Jore MM, Wiedenheft B, Lundgren M, Westra ER, Brouns SJ, Doudna JA, van der Oost J, et al. Native tandem and ion mobility mass spectrometry highlight structural and modular similarities in clustered-regularly-interspaced shot-palindromic-repeats (CRISPR)-associated protein complexes from Escherichia coli and Pseudomonas aeruginosa. Mol Cell Proteomics 2012; 11:1430-41; PMID:22918228; http://dx.doi.org/ 10.1074/mcp.M112.020263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Staals RH, Brouns SJ. Distribution and Mechanism of the type I CRISPR-Cas systems, (Barrangou R. and van de Oost J. eds.), pp115-144, Berlin-Heidelberg: Springer, 2013. [Google Scholar]
- 34. Cai F, Axen SD, Kerfeld CA. Evidence for the widespread distribution of CRISPR-Cas system in the Phylum Cyanobacteria. RNA Biol 2013; 10:687-93; PMID:23628889; http://dx.doi.org/ 10.4161/rna.24571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sharif H, Ozgur S, Sharma K, Basquin C, Urlaub H, Conti E. Structural analysis of the yeast Dhh1-Pat1 complex reveals how Dhh1 engages Pat1, Edc3 and RNA in mutually exclusive interactions. Nucleic Acids Res 2013; 41:8377-90; PMID:23851565; http://dx.doi.org/ 10.1093/nar/gkt600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhang J, Rouillon C, Kerou M, Reeks J, Brugger K, Graham S, Reimann J, Cannone G, Liu H, Albers SV, et al. Structure and mechanism of the CMR complex for CRISPR-mediated antiviral immunity. Mol Cell 2012; 45:303-13; PMID:22227115; http://dx.doi.org/ 10.1016/j.molcel.2011.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bergfors T. Protein Crystallization: Second Edition. International University Line, 2009. [Google Scholar]
- 38. Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr 2010; 66:125-32; PMID:20124692; http://dx.doi.org/ 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr 2013; 69:1204-14; PMID:23793146; http://dx.doi.org/ 10.1107/S0907444913000061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 2010; 66:213-21; PMID:20124702; http://dx.doi.org/ 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 2004; 60:2126-32; PMID:15572765; http://dx.doi.org/ 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 42. Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 2007; 35:W375-83; PMID:17452350; http://dx.doi.org/ 10.1093/nar/gkm216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lange SJ, Alkhnbashi OS, Rose D, Will S, Backofen R. CRISPRmap: an automated classification of repeat conservation in prokaryotic adaptive immune systems. Nucleic Acids Res 2013; 41:8034-44; PMID:23863837; http://dx.doi.org/ 10.1093/nar/gkt606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Royer CA. Improvements in the numerical analysis of thermodynamic data from biomolecular complexes. Anal Biochem 1993; 210:91-7; PMID:8489028; http://dx.doi.org/ 10.1006/abio.1993.1155 [DOI] [PubMed] [Google Scholar]
- 45. Schmidt C, Kramer K, Urlaub H. Investigation of protein-RNA interactions by mass spectrometry–Techniques and applications. J Proteomics 2012; 75:3478-94; PMID:22575267; http://dx.doi.org/ 10.1016/j.jprot.2012.04.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.