Abstract
The endoplasmic reticulum (ER) is an extensive membrane system that serves as a platform for de novo phospholipid synthesis. The ER is partitioned into distinct functional and structural domains, the most notable of which is the nuclear envelope. Here we discuss the role of nuclear envelope localized CNEP-1Nem1 in spatial regulation of de novo phospholipid synthesis within the ER. CNEP-1Nem1 is an activator of lipinPah1, which is the key phosphatidic acid phosphatase that regulates the metabolic branch-point between the production of phosphatidylinositol (PtdIns) and major membrane phospholipids, phosphatidylcholine (PC) and phosphatidylethanolamine (PE). CNEP-1 activates lipin at the nuclear envelope to bias phospholipid flux toward PC and PE production and to limit PtdIns incorporation. Increased PtdIns causes the formation of ectopic ER sheets in the vicinity of the nucleus that wrap around the nuclear envelope and cause downstream defects in NE disassembly. We propose that spatial regulation of phospholipid flux promotes partitioning of the ER into distinct subdomains by generating a gradient of PtdIns incorporation.
Keywords: endoplasmic reticulum, lipin, nuclear envelope, nuclear envelope breakdown, phosphatidylinositol
Introduction
The ER is partitioned into 3 subdomains: the nuclear envelope (NE), the peripheral ER, and the cortical ER. The peripheral and cortical ER are required for synthesis of lipids and transmembrane proteins, whereas the NE provides a selective barrier between the nucleoplasm and cytoplasm.1,2 Despite its continuity with the cortical and peripheral ER, the NE maintains a unique protein composition within its polarized double membrane sheet.1 In part, this is accomplished by selectively enriching and tethering nuclear membrane proteins, which are synthesized in the peripheral ER, to the nuclear lamin matrix3- a filamentous network of lamins and lamin-associated proteins underlying the innermost nuclear membrane.3-5 The NE and ER are highly dynamic and undergo independent as well as coordinated reorganizations during the cell cycle. Perhaps the most dramatic changes that occur are in mitosis when the nuclear envelope must breakdown to allow for spindle assembly.6 During mitosis, the ER transitions from a mixture of membrane sheets and tubules to a mostly sheet-like structure, and nuclear membranes and associated proteins retract into the ER. Upon exit from mitosis, nuclear membranes must reemerge from the ER to form around decondensing chromatin.1,7-11 While the molecular mechanisms that promote the re-establishment of distinct NE and ER domains are poorly understood, the advent of sophisticated live imaging techniques has significantly advanced our ability to visualize NE and ER membrane structure and dynamics during these critical transition points.
While the protein composition of the NE is distinct from that of the ER, one outstanding question is whether the phospholipid composition differs between these two contiguous domains. A unique phospholipid signature at the NE may serve to specify NE identity and play structural and signaling roles during processes such as nuclear envelope breakdown (NEBD) and reformation, nuclear pore insertion, and unconventional nuclear transport of viruses or large ribonuclear particles.12,13 Because rapid diffusion of phospholipids occurs in the 2-dimensional plane of ER/NE membranes and the ER routinely exchanges phospholipids with other organelles, specialized mechanisms must be in place to prevent loss of phospholipids from the NE, maintaining its distinct phospholipid composition. Here we discuss how selective enrichment of CNEP-1Nem1 (CTD Nuclear Envelope Protein 1), an activator of the phosphatidic acid phosphatase, lipin,14-16 at the NE regulates phospholipid flux to control NE and ER dynamics.17 Spatial regulation of phospholipid synthesis enzymes within ER domains may serve as a general mechanism to maintain phospholipid homeostasis and membrane structure within different ER domains.
The reaction catalyzed by lipin is central to the de novo phospholipid synthesis pathway18,19 (Fig. 1). Phosphatidic acid is dephosphorylated by lipin for synthesis of major membrane phospholipids, phosphatidylcholine (PC) and phosphatidylethanolamine (PE), by a choline-ethanolamine phosphotransferase (CEPT).19-21 Phosphatidic acid is also converted to CDP-DAG by CDP-DAG synthase for the synthesis of phosphatidylinositol (PtdIns) by phosphatidylinositol synthase (PIS).21 Phosphatidylcholine and PE are transferred from the ER to other organelles where they play structural and signaling roles.22 Upon transfer of PtdIns to other organelles, PtdIns is modified by kinases and phosphatases at different positions of its head group to form phosphoinositides (PIPs)23 (Fig. 1A). Phosphatidylcholine and PE make up greater than 70% of phospholipids within ER membranes, whereas PtdIns exists in low abundance (<10%).22 Our work discussed here suggests that PtdIns synthesis is restricted to the peripheral and cortical ER by selective enrichment of the lipin activator CNEP-1 to the nuclear envelope.17
Figure 1.
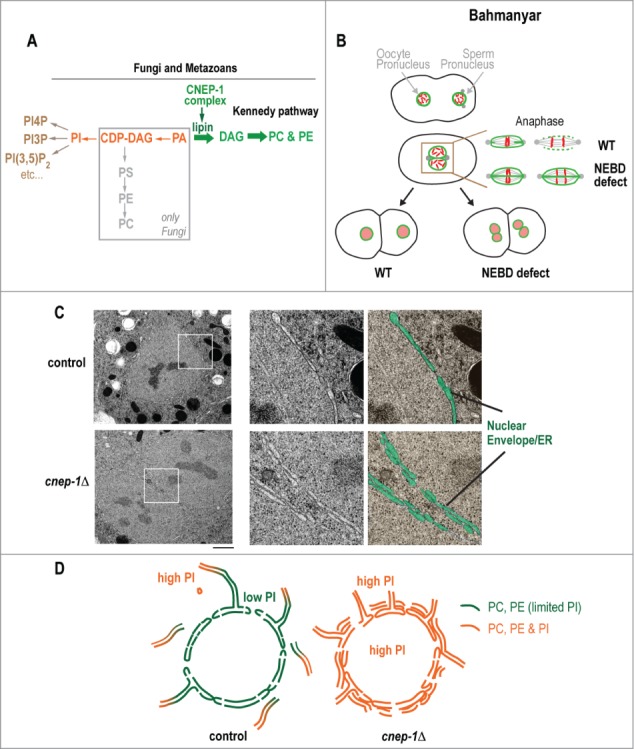
Endoplasmic reticulum and nuclear envelope structure are regulated by spatial activation of lipin at the nuclear envelope. (A) Schematic representing the de novo phospholipid pathway within the ER, which produces the major membrane phospholipids phosphatidylcholine and phosphatidylethanolamine (PC and PE, respectively, in green) and phosphatidylinositol (PI; orange). Activation of lipin results in increased phospholipid flux toward PC/PE limiting PtdIns at the nuclear envelope. Gray box highlights the fungi-specific CDP-DAG pathway to PE/PC production. (B) Schematic showing the first embryonic division in C. elegans, highlighting the membrane scission even that occurs at the anaphase onset. Defects in nuclear envelope breakdown (NEBD) defect cause failure of scission resulting in twinned nuclei at the 2-cell stage. (C) Electron micrographs of mitotic nuclei in wild type and cnep-1Δ worms. Magnified images of boxed regions are duplicated and pseudocolored to highlight nuclear envelope/ER lumens (purple) and cytoplasm/nucleoplasm regions (yellow). Bars, 1μm. (D) Schematic representing the gradient of PI that is created by CNEP-1 activation of lipin at the nuclear envelope (left). In the absence of CNEP-1 (right), PtdIns levels are high everywhere causing increased ER sheets in the vicinity of the nuclear envelope that wrap around the nuclear envelope at nuclear permeabilization and inhibit nuclear envelope breakdown.
Role for Lipin Activation in Nuclear Envelope Structure and Dynamics
Genetic evidence for regulation of phospholipid synthesis at the NE comes from a conserved role for lipin in NE structure and dynamics. In S. cerevisiae, deletion of the lipin homolog Pah1, or members of the activating protein phosphatase complex, Nem1/Spo7, results in large extrusions of the NE.24,25 In C. elegans embryos, which undergo an open mitosis, partial inhibition of the single lipin gene inhibits NEBD.17,26,27 In mammalian cells, co-inhibition of the 3 lipin genes (lipin 1-3) causes NEBD defects.28 While the reaction catalyzed by lipin is conserved in all cases, phospholipid synthesis pathways differ between metazoans and fungi18,21 (Fig. 1A). In fungi, the main pathway for PC and PE production is via PA conversion to CDP-DAG (Fig. 1A).19,20 Thus, nuclear membrane extrusions in the absence of lipin in budding yeast are likely a result of excess phospholipid synthesis via the CDP-DAG pathway.25,29 In metazoans, which do not display nuclear membrane extrusions upon lipin inhibition, the pathway that coverts CDP-DAG to PC/PE does not exist and lipin is essential for PC/PE synthesis.21
Lipin's essential role in PC/PE synthesis in metazoans makes it difficult to determine a specific function for lipin at the nuclear envelope. To understand the significance of lipin-mediated phospholipid synthesis at the nuclear envelope, we focused on the lipin-activating protein phosphatase CTD-NEP-1 (previously known as Dullard).14,15 CTD-NEP-1 is enriched at the NE in mammalian cells14 and in C. elegans.17 Unlike lipin, CNEP-1, the C. elegans ortholog of CTD-NEP-1, is not essential for embryo production.17 This suggests that CNEP-1 regulates a pool of lipin at the nuclear envelope.
To determine whether CNEP-1 was important for NE dynamics, we analyzed the NE dependent events of the first division in C. elegans embryos (Fig. 1B). After fertilization, maternal and paternal pro-nuclei expand and migrate to the center of the embryo as chromosomes condense in preparation for mitosis.30 Nuclear permeabilization is followed by a membrane scission event that removes membranes between maternal and paternal pronuclei. This event, which is coupled to lamin disassembly, facilitates fusion of maternal and paternal nuclei and chromosome mixing. Following chromosome segregation, a single nucleus forms in each daughter cell with the full complement of chromosomes. If membrane scission or lamin disassembly is inhibited, spindle microtubules penetrate the poles of unfused pronuclei after permeabilization and segregate duplicated chromosomes in a semi-open mitosis (Fig. 1B, bottom panel). At the 2-cell stage, maternal and paternal complements of chromosomes remain within separate nuclei31 (“twinned nuclei;” Fig. 1B, bottom panel). Previous work showed that partial inhibition of lipin results in twinned nuclei, suggesting a function for lipin in NEBD.26,27 Similarly, we found that deletion of CNEP-1 results in twinned and oblong shaped nuclei at the 2-cell stage.17 Timing of nuclear permeabilization and nuclear membrane protein dispersal in CNEP-1 mutants was normal and the kinetics of lamin disassembly kinetics was slowed but complete. The main defect in CNEP-1 mutants was delayed membrane scission, indicating that inhibition of membrane scission was likely the cause of abnormally shaped nuclei in 2-cell stage embryos.
To further probe the source of delayed membrane scission, we performed high pressure freezing and freeze substitution of CNEP-1 mutant embryos. Electron micrographs revealed an additional membrane sheet wrapped around each pronucleus upon entry into mitosis (Fig. 1C). In contrast to 4 membrane bilayers in control embryos, 8 membrane bilayers existed at the contact surface between pronuclei in CNEP-1 mutants. Our fluorescence microscopy data suggested that the source of the additional membrane sheet wrapped around each pronucleus was the peripheral ER, as CNEP-1 deleted embryos displayed a shift in the balance of ER tubules to sheets throughout interphase. Electron micrographs corroborated the observation of increased ER sheets in CNEP-1 mutants, particularly in the vicinity of nuclei. We propose that ectopic ER sheets in the vicinity of the nucleus wrap around the nuclear envelope in CNEP-1 mutants causing downstream defects in NEBD.
Because lipin lies at the branching point between PtdIns and PC/PE synthesis (Fig. 1A), we predicted that the absence of CNEP-1 would increase PA flux toward PI synthesis. To test this idea, we depleted enzymes required for PtdIns production, CDP-DAG synthase and phosphatidylinositol synthase, which rescued NE defects and ectopic ER sheets in CNEP-1 mutants. Similar results were found upon co-depletion of lipin. Radioactive labeling of total phospholipids in CNEP-1 deletion worms showed increased levels of PA and PI, which were reduced to wild type levels upon depletion of CDP-DAG synthase. Based on these results, we propose a model in which CNEP-1 at the NE increases the activity of lipin toward PA to limit PtdIns incorporation at the NE. Increased PtdIns at nuclear membranes diffuses to nearby ER membranes causing the formation of ectopic ER sheets that collapse onto the NE in mitosis and impede NEBD (Fig. 1D).
Partitioning ER Domains with a Phospholipid Gradient
We propose a model whereby spatial regulation of lipin biases the distribution of phospholipids within the nuclear envelope and peripheral ER. This suggests that a gradient of PtdIns exists within ER membranes (Fig. 1D), where low levels of PI are incorporated at and near the NE, and high PtdIns levels are incorporated near the cortex. In support of this idea, ER-plasma membrane contacts are sites of non-vesicular transport of lipids, particularly of those that are derived from phosphatidylinositol.32 We argue that deletion of CNEP-1 eliminates the PtdIns gradient by shifting phospholipid flux at the nuclear envelope toward PtdIns synthesis and allowing PtdIns incorporation within the nuclear envelope domain. Increased PtdIns incorporation at this site inhibits proper partitioning of ER domains, resulting in ectopic ER sheet formation and aberrant wrapping of the ER around the NE. To test whether spatial regulation of PtdIns synthesis at the nuclear envelope, and thereby a gradient of PtdIns, is important for partitioning the ER into separate domains, the outcome of targeting CNEP-1 exclusively to the peripheral ER should be determined.
How PtdIns induces ER sheet formation and whether the key lipid is PtdIns itself or a PtdIns-derived phosphoinositide remain open questions. Asymmetric distribution of curvature inducing lipids within the monolayers in a bilayer can generate local curvature in membranes.33 However, this is unlikely because theoretical work shows that compositional asymmetry that would result in high-curvature membranes like that in ER tubules or at the edges of ER sheets is energetically highly unfavorable.34,35 An unexplored possibility is that lipid composition within ER domains controls the recruitment or activity of ER shaping proteins or the ability of ER membranes to associate with the cytoskeleton. Membrane-associated proteins that structure membranes into tubules and sheets are known to dictate ER morphology34,36 and may be regulated by the lipids they bind. Mechanisms that induce membrane sheet formation in the ER differ from those at the nuclear envelope. For example, CLIMP-63 forms ER-sheets and is specifically excluded from the nuclear envelope.37 This suggests a model in which CLIMP-63 binds or is activated within PtdIns-rich regions in the peripheral ER to induce sheet-formation. Thus, spatial regulation of phospholipid synthesis may partition the ER into domains by directly or indirectly recruiting or excluding membrane-shaping proteins.
Conclusions and Future Directions
Our study shows that regulation of phospholipid flux within the NE creates a compositional gradient of phospholipids within ER subdomains, which are important for ER structure and NE dynamics.17 This is achieved through regulation of the activity of a central enzyme in the phospholipid synthesis pathway, lipin, by its activator, CNEP-1, at the nuclear envelope. This work reveals a new mechanism of spatial regulation of phospholipid flux for partitioning of the ER into distinct subdomains. Given experimental limitations in biochemical isolation of nuclear envelope membranes from peripheral ER membranes, this work greatly advances our understanding of how the phospholipid composition of ER subdomains might be controlled.
How the NE maintains a distinct phospholipid composition within the context of the greater ER remains a compelling open question. Future directions might include the exploration of possible phospholipid diffusion barriers or flippases that could control the diffusion of undesirable lipids into the NE domain. Furthermore, a clear next step is to understand whether the inner and outer nuclear envelope are compositionally different and whether this is important for nuclear lamina and nuclear membrane protein function. The level of compositional diversity may not be limited to phospholipid head groups, but may include fatty acid chains that have recently been shown to be important for lipid function. While many challenges exist in studying the cell biology of lipids, recent advances in imaging techniques, genetic manipulation of lipid species, and lipid identification38 will be important for extending our understanding of the role of phospholipids in compartmentalizing the NE and other subdomains within the greater ER.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
I am grateful to Becky Green and Sophie Miller for their critical reading of this review.
References
- 1. Hetzer MW. The nuclear envelope. Cold Spring Harb Perspect Biol 2010; 2:a000539; PMID:20300205; http://dx.doi.org/ 10.1101/cshperspect.a000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. English AR, Voeltz GK. Endoplasmic reticulum structure and interconnections with other organelles. Cold Spring Harb Perspect Biol 2013; 5:a013227-7; PMID:23545422; http://dx.doi.org/ 10.1101/cshperspect.a013227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic “network of networks.” Nature Publishing Group 2011; 12:695-708; PMID:21971041 [DOI] [PubMed] [Google Scholar]
- 4. Hetzer MW. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum. Nature Cell Bio 2007; 9:1160-66. [DOI] [PubMed] [Google Scholar]
- 5. Gerace L, Huber MD. Nuclear lamina at the crossroads of the cytoplasm and nucleus. J Struct Biol 2012; 177:24-31; PMID:22126840; http://dx.doi.org/ 10.1016/j.jsb.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Güttinger S, Laurell E, Kutay U. Orchestrating nuclear envelope disassembly and reassembly during mitosis. Nature Publishing Group 2009; 10:178-91; PMID:19234477 [DOI] [PubMed] [Google Scholar]
- 7. Lu L, Ladinsky MS, Kirchhausen T. Cisternal organization of the endoplasmic reticulum during mitosis. Mol Biol Cell 2009; 20:3471-80; PMID:19494040; http://dx.doi.org/ 10.1091/mbc.E09-04-0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang S, Romano FB, Field CM, Mitchison TJ, Rapoport TA. Multiple mechanisms determine ER network morphology during the cell cycle in Xenopus egg extracts. J Cell Biol 2013; 203:801-14; PMID:24297752; http://dx.doi.org/ 10.1083/jcb.201308001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poteryaev D, Squirrell JM, Campbell JM, White JG, Spang A. Involvement of the actin cytoskeleton and homotypic membrane fusion in ER dynamics in Caenorhabditis elegans. Mol Biol Cell 2005; 16:2139-53; PMID:15716356; http://dx.doi.org/ 10.1091/mbc.E04-08-0726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lu L, Ladinsky MS, Kirchhausen T. Formation of the postmitotic nuclear envelope from extended ER cisternae precedes nuclear pore assembly. J Cell Biol 2011; 194:425-40; PMID:21825076; http://dx.doi.org/ 10.1083/jcb.201012063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellenberg J, Siggia ED, Moreira JE, Smith CL, Presley JF, Worman HJ, Lippincott-Schwartz J. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J Cell Biol 1997; 138:1193-206; PMID:9298976; http://dx.doi.org/ 10.1083/jcb.138.6.1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson DC, Baines JD. Herpesviruses remodel host membranes for virus egress. Nat Rev Microbiol 2011; 9:382-94; PMID:21494278; http://dx.doi.org/ 10.1038/nrmicro2559 [DOI] [PubMed] [Google Scholar]
- 13. Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang Y-T, Li Q, et al. Nuclear Envelope Budding Enables Large Ribonucleoprotein Particle Exportduring Synaptic Wnt Signaling. Cell 2012; 149:832-46; PMID:22579286; http://dx.doi.org/ 10.1016/j.cell.2012.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kim Y, Gentry MS, Harris TE, Wiley SE, Lawrence JC, Dixon JE. A conserved phosphatase cascade that regulates nuclear membrane biogenesis. Proc Natl Acad Sci USA 2007; 104:6596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han S, Bahmanyar S, Zhang P, Grishin N, Oegema K, Crooke R, Graham M, Reue K, Dixon JE, Goodman JM. Nuclear envelope phosphatase 1-regulatory subunit 1 (Formerly TMEM188) is the metazoan Spo7p ortholog and functions in the lipin activation pathway. J Biol Chem 2012; 287:3123-37; PMID:22134922; http://dx.doi.org/ 10.1074/jbc.M111.324350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Hara L, Han G-S, Peak-Chew S, Grimsey N, Carman GM, Siniossoglou S. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J Biol Chem 2006; 281:34537-48; PMID:16968695; http://dx.doi.org/ 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bahmanyar S, Biggs R, Schuh AL, Desai A, Müller-Reichert T, Audhya A, Dixon JE, Oegema K. Spatial control of phospholipid flux restricts endoplasmic reticulum sheet formation to allow nuclear envelope breakdown. Genes Dev 2014; 28:121-6; PMID:24449268; http://dx.doi.org/ 10.1101/gad.230599.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carman GM, Han G-S. Roles of phosphatidate phosphatase enzymes in lipid metabolism. Trends Biochem Sci 2006; 31:694-9; PMID:17079146; http://dx.doi.org/ 10.1016/j.tibs.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Siniossoglou S. Phospholipid metabolism and nuclear function: roles of the lipin family of phosphatidic acid phosphatases. Biochim Biophys Acta 2013; 1831:575-81; PMID:23026159; http://dx.doi.org/ 10.1016/j.bbalip.2012.09.014 [DOI] [PubMed] [Google Scholar]
- 20. Carman GM, Han G-S. Phosphatidic acid phosphatase, a key enzyme in the regulation of lipid synthesis. J Biol Chem 2009; 284:2593-7; PMID:18812320; http://dx.doi.org/ 10.1074/jbc.R800059200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fagone P, Jackowski S. Membrane phospholipid synthesis and endoplasmic reticulum function. J Lipid Res 2009; 50 Suppl:S311-6; PMID:18952570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nature Publishing Group 2008; 9:112-24; PMID:18216768;; http://dx.doi.org/ 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Balla T. Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 2013; 93:1019-137; PMID:23899561; http://dx.doi.org/ 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J 1998; 17:6449-64; PMID:9822591; http://dx.doi.org/ 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J 2005; 24:1931-41; PMID:15889145; http://dx.doi.org/ 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Golden A, Liu J, Cohen-Fix O. Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Biol 2009; 122:1970-8; PMID:19494126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gorjánácz M, Mattaj IW. Lipin is required for efficient breakdown of the nuclear envelope in Caenorhabditis elegans. J Cell Biol 2009; 122:1963-9; PMID:19494125 [DOI] [PubMed] [Google Scholar]
- 28. Mall M, Walter T, Gorjánácz M, Davidson IF, Nga Ly-Hartig TB, Ellenberg J, Mattaj IW. Mitotic lamin disassembly is triggered by lipid-mediated signaling. J Cell Biol 2012; 198:981-90; PMID:22986494; http://dx.doi.org/ 10.1083/jcb.201205103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han G-S, Wu W-I, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem 2006; 281:9210-8; PMID:16467296; http://dx.doi.org/ 10.1074/jbc.M600425200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oegema K, Hyman A. A. Cell division. WormBook, ed. The C. elegans Research Community, WormBook 2006; doi/ 10.1895/wormbook.1.72.1, http://www.wormbook.org. [DOI] [Google Scholar]
- 31. Audhya A, Desai A, Oegema K. A role for Rab5 in structuring the endoplasmic reticulum. J Cell Biol 2007; 178:43-56; PMID:17591921; http://dx.doi.org/ 10.1083/jcb.200701139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, Mao Y, Emr SD. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 2011; 144:389-401; PMID:21295699; http://dx.doi.org/ 10.1016/j.cell.2010.12.034 [DOI] [PubMed] [Google Scholar]
- 33. Bigay J, Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev Cell 2012; 23:886-95; PMID:23153485; http://dx.doi.org/ 10.1016/j.devcel.2012.10.009 [DOI] [PubMed] [Google Scholar]
- 34. Shibata Y, Hu J, Kozlov MM, Rapoport TA. Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol 2009; 25:329-54; PMID:19575675; http://dx.doi.org/ 10.1146/annurev.cellbio.042308.113324 [DOI] [PubMed] [Google Scholar]
- 35. Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 2006; 7:9-19; PMID:16365634; http://dx.doi.org/ 10.1038/nrm1784 [DOI] [PubMed] [Google Scholar]
- 36. Goyal U, Blackstone C. Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta 2013; 1833:2492-8; PMID:23602970; http://dx.doi.org/ 10.1016/j.bbamcr.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA. Mechanisms determining the morphology of the peripheral ER. Cell 2010; 143:774-88; PMID:21111237; http://dx.doi.org/ 10.1016/j.cell.2010.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Muro E, Atilla-Gokcumen GE, Eggert US. Lipids in cell biology: how can we understand them better? Mol Biol Cell 2014; 25:1819-23; PMID:24925915; http://dx.doi.org/ 10.1091/mbc.E13-09-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]


