Abstract
APOBEC1 is the catalytic subunit of the complex that edits ApolipoproteinB (ApoB) mRNA, which specifically deaminates cytidine 6666 to uracil in the human transcript. The editing leads to the generation of a stop codon, resulting in the synthesis of a truncated form of ApoB. We have developed a method to quantitatively assay ApoB RNA editing in live cells by using a double fluorescent mCherry-EGFP chimera containing a ∼300bp fragment encompassing the region of ApoB subject to RNA editing. Coexpression of APOBEC1 together with this chimera causes specific RNA editing of the ApoB fragment. The insertion of a stop codon between the mCherry and EGFP thus induces the loss of EGFP fluorescence. Using this method we analyze the dynamics of APOBEC1-dependent RNA editing under various conditions. Namely we show the interplay of APOBEC1 with known interactors (ACF, hnRNP-C1, GRY-RBP) in cells that are RNA editing-proficient (HuH-7) or -deficient (HEK-293T), and the effects of restricted cellular localization of APOBEC1 on the efficiency of the editing. Furthermore, our approach is effective in assaying the induction of RNA editing in Caco-2, a cellular model physiologically capable of ApoB RNA editing.
Keywords: AID/APOBECs, cytosine deaminase, lipid metabolism, mRNA, post-transcriptional modification, RNA editing
List of abbreviations
- APOBEC1
Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1
- ApoB
Apolipoprotein B
- ACF
APOBEC1 Complementation Factor
- ADAR
Adenosine Deaminase, RNA-specific
- ADAT
Adenosine Deaminase, tRNA-specific
- cds
coding sequence
- hnRNP-C1
heterogeneous nuclear ribonucleoprotein C1
- GRY-RBP
Glycine-Arginine-Tyrosine-rich RNA-binding protein
- EGFP
Enhanced Green Fluorescent Protein
- RBM47
RNA binding motif protein 47
- FBS
Fetal bovine serum
- FACS
Fluorescence activated cell sorting
Introduction
Post-transcriptional editing of various types of RNA in vertebrates is restricted to A>I editing by Adenosine Deaminases acting on mRNA and tRNA (ADARs and ADATs), and to C>U editing mediated by Apolipoprotein B mRNA editing enzyme, catalytic polypeptide 1 (APOBEC1), a poly(deoxy)nucleotide deaminase member of the AID/APOBEC gene family.1-5
APOBEC1 was identified as the catalytic subunit of the editing complex physiologically involved in the editing of C6666 of the human Apolipoprotein B (ApoB) mRNA in the small intestine.6,7 The editing causes the change of a CAA codon to a UAA codon, thereby inducing the translation of a truncated form of ApoB, which is important for the synthesis of chylomicrons. Subsequent studies have identified a number of other physiologically edited transcripts, mainly in the 3′ untranslated regions.8-10 While not directly affecting the coding sequence, these edited residues could affect the regulation of the transcript itself.9,10
Other roles not linked to APOBEC1 ability to edit specific mRNAs have been suggested: regulation of mRNA stability, restriction of retroviruses and mobile elements, and active demethylation of DNA.2,11,12 Moreover, its ability to target DNA has been linked to mutagenesis and human cancer.13-15
Higher expression levels of APOBEC1 induce hyperediting of physiological RNA targets at additional sites, as well as of other genes.e.g. 16-19 Such hyperediting has been linked to the onset of hepatocellular carcinomas in transgenic mice and rabbits overexpressing APOBEC1 in the liver.20 Moreover, it has been observed that deficiency of APOBEC1 decreases the number and size of polyps and tumors in cancer-prone mice.21
The main sequence determinant for the targeting of the editing complex is the so-called mooring sequence, an AU-rich motif lying immediately downstream to the edited site.22,23 In fact, the efficiency and specificity of APOBEC1-dependent RNA editing is determined by the editosome, the complex of proteins interacting with APOBEC1, some of which have been identified. Thus APOBEC1 Complementation Factor (ACF) is required for the specificity of the targeting through its interaction with the mooring sequence.24-30 Another molecule, RBM47, has been recently shown to be sufficient for the APOBEC1-mediated RNA editing.31 Other molecules, GRY-RBP and hnRNP-C1 among them, have been characterized as negative regulators of the editing process.32-38
The importance of APOBEC1 cofactors is evident when trying to reconstitute RNA editing: there are cells that become proficient after expression of APOBEC1, such as HepG2 and HuH-7 liver cell lines, others (e.g., Hela and HEK293T) in which overexpression of APOBEC1 is unable to support ApoB RNA editing,e.g. 39,40 and others - the Caco-2 - in which ApoB RNA editing can be induced. Caco-2 cells derive from colon adenocarcinoma, but present a number of features of the epithelium from small intestine.41 It has been shown that differentiation of Caco-2 leads to APOBEC1 expression and ApoB RNA editing.42,43 In addition ApoB RNA editing in Caco-2 cells can be induced by calcium or other stimuli.44,45
A number of assays have been developed to measure the efficiency of APOBEC1-dependent RNA editing in cells and organ extracts.39,46-50 The most commonly used assays are based either on sequence analysis or on poisoned primer extension of the edited site. Whereas Sanger sequencing provides a rough estimate of editing only in presence of a substantial percentage of edited molecules, the poisoned primer extension is far more sensitive, being able to discriminate the presence of a minimum of 0.3% edited fragment.50 The limits of these approaches lie in the assays starting material. Being based on PCR amplification of the transcript, amplification artifacts can affect the outcome. Moreover in experimental settings in which exogenous expression is required, these assays are significantly affected by the efficiency of transfection.
With the aim of overcoming these limits we developed a reporter system through which we can easily assess the levels of RNA editing in live cells through flow cytometry. Our system is based on a plasmid expressing a red-green fluorescent chimera in which APOBEC1-dependent RNA editing induces a stop codon that leads to the loss of the green fluorescence. Thus, when cells capable to support APOBEC1-dependent RNA editing are transfected with this construct, we observe a progressive decrease in the green fluorescence, proportional to the amount of RNA editing.
In order to validate our approach we tested it under conditions that affect the efficiency of APOBEC1-mediated editing. We thus assessed the effects of some of the known interactors of APOBEC1 in live cells by successfully complementing APOBEC1 with ACF in HEK293T cells, which do not normally support RNA editing. On the contrary other interactors, GRY-RBP and hnRNP-C1, do not specifically affect the editing process, while they do seem to influence the chimeric protein. We have also analyzed the efficiency of RNA editing using chimeras of APOBEC1 whose localization is mainly cytoplasmic or nuclear.
Finally, we show that our assay can be used to quantify the induction of RNA editing in Caco-2 cells treated with Forskolin, a Protein Kinase A activator.
Results and Discussion
Live visualization of APOBEC1-dependent RNA editing
We prepared the double-fluorescent chimeric construct by fusing together a 5′ mCherry coding sequence (cds) with a 3′ EGFP cds through a 436bp region from ApoB mRNA encompassing the editable C6666 site (Fig. 1A). The three sequences were placed in frame and flow cytometry of cells transiently transfected with the construct reveals both red and green fluorescence (Fig. 1B, gates and ungated plots are shown in Fig. S1). APOBEC1-dependent RNA editing of the C6666 site in the chimeric transcript would induce a stop codon, thus preventing the translation of the EGFP portion of the chimera. The outcome of the RNA editing would therefore induce a stoichiometric imbalance between mCherry and EGFP that can be visualized through flow cytometry: the red fluorescence marks all transfected cells, and the amount of green fluorescence is indicative of the APOBEC1-dependent RNA editing.
Figure 1.
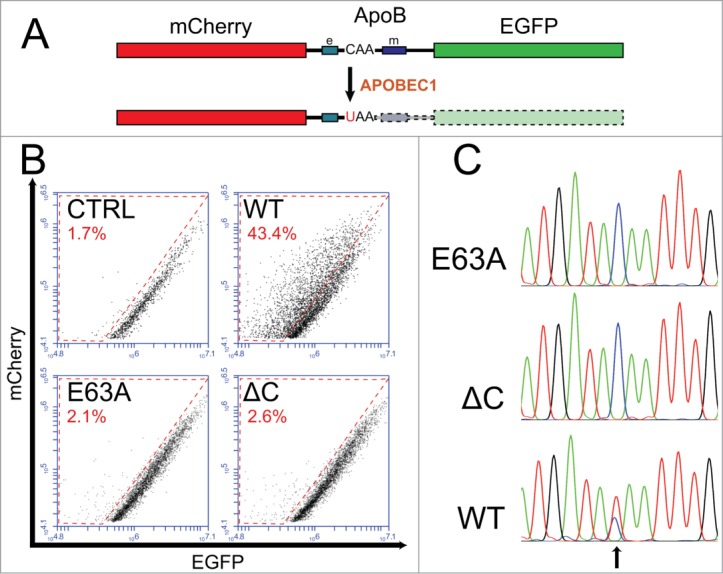
APOBEC1 edits the mRNA of a chimeric construct containing the ApoB region encompassing the editable site. (A) Schematic diagram of the chimeric transcript. The mCherry cds is placed in frame 5′ to the EGFP cds. The region of ApoB encompassing the C6666 editable site links the 2 coding sequences. APOBEC1-specific RNA editing induces the formation of a stop codon that blocks the translation of the EGFP coding sequence. The mCherry protein acts as a control for the transfection levels, while the loss of EGFP is indicative of the levels of RNA editing. The boxes on the side of the editable site indicate the efficiency and the mooring sequence (labeled e and m, respectively). (B) Representative FACS analysis of HuH-7 cells transiently transfected with the mCherry-ApoB-EGFP construct together with those expressing respectively rat APOBEC1, a catalytically inactive APOBEC1 mutant (E63A), a mutant lacking the C-terminal 40aa (ΔC), or control plasmids. Only rat APOBEC1 induces a shift from the mCherry/EGFP diagonal (events in the red-boxed area), indicating an overall decrease in the EGFP levels. The gates selected for the analysis, as well as ungated plots are shown in Supplementary Figure 1. (C) Representative chromatograms of the ApoB editing site of the mCherry-ApoB-EGFP transcript from cells transfected either with control plasmids or with the APOBEC1 expressing one. In the APOBEC1 sample the chromatogram shows a double peak corresponding to the C>U deamination (arrow).
We transiently cotransfected this construct in HuH-7 cells, an hepatocellular carcinoma cell line which is RNA editing-proficient upon transduction of APOBEC1,40 together with a plasmid expressing rat APOBEC1. FACS analysis of the transfected cells at 72 hours from transfection shows a relative decrease of green fluorescence compared to the red one (Fig. 1B), possibly indicative of RNA editing. We also tested a catalytically dead APOBEC1 mutant in which the glutamate acting as a proton donor in the deamination had been mutated (E63A), and a mutant lacking the last 40aa (ΔC). This mutant, similar to other ones previously described, is unable to dimerize and to support RNA editing.51 Indeed, neither of these mutants induces changes in the cellular fluorescence (Fig. 1B).
To confirm that the diminished green fluorescence in APOBEC1-transfected cells was due to editing of the ApoB C6666 residue, we recovered the chimeric transcripts from the transfected cells and we amplified them. Sequence analysis of the ApoB region confirmed the presence of a double peak at the C6666 residue only in the cells expressing APOBEC1 (Fig. 1C).
Next we analyzed the temporal dynamics of the RNA editing: starting from day 2 after transfection a steady increase in the RNA editing levels was evident (Fig. 2A). This is easily explained as both the chimeric transcript and APOBEC1 become available in the cell. Analogously, the amount of RNA editing was proportional to the amount of APOBEC1 plasmid used for the transfection (Fig. 2B).
Figure 2.
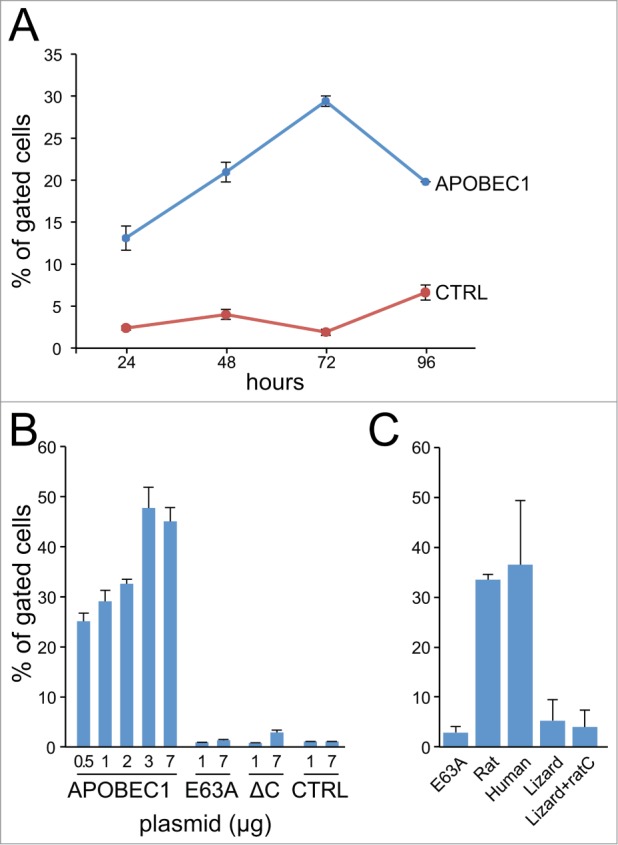
The amount of RNA editing depends on the exposure of the chimeric transcript to APOBEC1. HuH-7 cells were transiently transfected with the double-fluorescent chimera and the indicated plasmids. (A) RNA editing (percentage of gated cells) at different time points (hours) after transfection. (B) Titration of RNA editing in cells transiently transfected with different amounts of APOBEC1 expressing plasmid or with control plasmids. (C) The bar diagram shows the RNA editing in HuH-7 transiently cotransfected with plasmids encoding for either rat, human, lizard APOBEC1 or a chimera of lizard APOBEC1 fused with the C-terminal portion of the rat APOBEC1. The error bars represent the SEM from at least 3 experiments.
We then assayed the efficiency of editing of the human APOBEC1, which - similarly to the rat homolog - induced a marked decrease in the EGFP fluorescence (Fig. 2C). On the contrary, an APOBEC1 homolog from green anole,52 an organism in which ApoB editing does not occur, is unable to induce RNA editing of the chimeric construct. Considering the importance of the C-terminal domain of mammalian APOBEC1 for RNA editing,51,53 we tested a fusion protein in which we stitched the rat C-terminal domain to the reptilian homolog. Also in this case there was no evidence of editing. Also Activation Induced Deaminase, a paralog of APOBEC1 physiologically targeting DNA54 is unable to induce changes (data not shown).
In principle our methodology could be modified to assay any target of RNA editing. Beyond the described construct, we also tested a construct in which the ApoB region connecting the mCherry and the EGFP was reduced to a 28 bp fragment encompassing the editable site flanked by the efficiency sequence and the mooring seaquence. This fragment has been considered the minimal stem-loop recognized by the editosome.29,55 However, this construct resulted completely uneditable in our system (data not shown), possibly due to secondary/tertiary structures in the transcript that masked the editing site.
Complementation of APOBEC1 in cells that do not support RNA editing
ACF, the first cofactor of APOBEC1 to be identified, is necessary for the efficiency and specificity of the RNA editing: ACF is responsible for the targeting of APOBEC1, as it shuttles into the nucleus with APOBEC1, and - once there - specifically recognizes the mooring sequence of the ApoB RNA.30,56,57 Reconstitution of APOBEC1-mediated ApoB RNA editing has been shown in vitro and in cells through overexpression of ACF.58-60
We thus tested whether we could use our system to visualize the complementation of APOBEC1 in cells that are not proficient for ApoB RNA editing. HEK293T cells don't support APOBEC1-mediated RNA editing as they do not express ACF.60 Indeed, transfection of our fluorescent chimera in HEK293T cells, in association or not with the construct encoding for rat APOBEC1, did not show evidence of RNA editing. On the other hand, cotransfection of a construct encoding for ACF induced a marked shift in the green fluorescence, thus implying the reconstitution of the editing complex and the targeting of APOBEC1 to the fluorescent chimera (Fig. 3).
Figure 3.
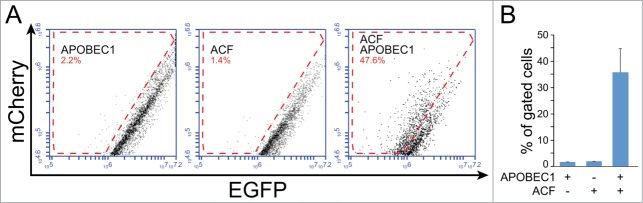
ACF is required for APOBEC1-mediated RNA editing. Cells were transiently transfected with plasmids encoding for mCherry-ApoB-EGFP, APOBEC1 and either ACF or a control plasmid as indicated. (A) Representative FACS analysis of HEK293T cells. (B) The bar diagram shows the percentage of gated cells from at least 3 experiments (the error bar indicates the SEM).
As in HuH-7 cells, the human APOBEC1 can support RNA editing also in HEK293T cells (Fig. S2A). The increased efficiency of the human homolog compared to the rat one is likely due to the human APOBEC1 being optimized for activity in human cells. While an increased ability to interact with ACF could be involved, the rat and human homologs of ACF are 94% identical, with most of the differences located in the C-terminal part, far from the region allegedly responsible for the interaction with APOBEC1.56 This could suggest that other unknown factors could positively effect the efficiency of the editing.
Finally, overexpression of ACF in HuH-7 leads to an increase in the RNA editing efficiency, suggesting that the rate of RNA editing in these cells is limited by the endogenous ACF (Fig. S2B).
Other interactors of A1
Next we tested the effects of other molecules that have been identified as interactors of APOBEC1: GRY-RBP and hnRNP-C1. These molecules have been shown through poisoned-primer extension assay to negatively affect the activity of the editosome as they compete for the mooring sequence and/or sequester APOBEC1.32,38,56 Surprisingly, coexpression of either of these proteins together with APOBEC1 and the fluorescent chimera did not result in a diminished efficiency of the editing either in HuH-7 cells or in HEK293T cells complemented with ACF (Fig. 4, Fig. S3A). We have tested whether increasing amounts of interactors (Fig. S3B) could exert any negative effect. We did not observe any decrease in RNA editing even with the highest amounts of the interactor.
Figure 4.
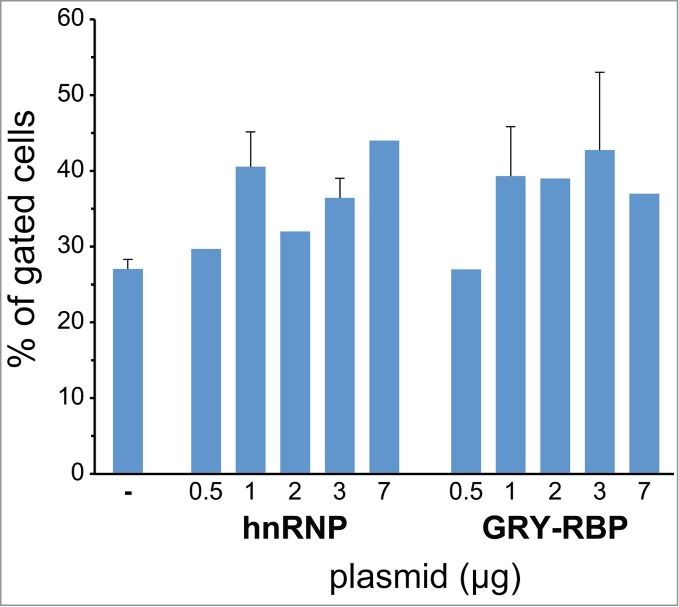
APOBEC1 interactors hnRNP and GRY-RBP do not affect the efficiency of RNA editing. The bar diagram shows the RNA editing (percentage of EGFP shift) in HuH-7 cells transiently cotransfected with plasmids encoding mCherry-ApoB-EGFP, APOBEC1 and increasing amounts of either hnRNP-C1 or GRY-RBP plasmids (as indicated in the diagram). The total amount of plasmid DNA was equal in all transfections. The error bars represent the SEM from at least 3 experiments.
Indeed, we found that transient transfection of either GRY-RBP or hnRNP-C1 leads to decreased levels of fluorescence in transfected cells even in absence of APOBEC1 or of the ApoB sequence (Fig. S3C). This could point to an effect of these proteins to the overall efficiency of the transfection or to their involvement in the processing/degradation of the transcript. Since the FACS profiles show a shift of the fluorescent population rather than a decrease in the number of fluorescent cells the latter option seems more likely (Fig. S3D). In any case, whatever the molecular mechanisms involved, the effect is not specific to any determinant on the editable ApoB region.
RNA editing in nucleus and cytoplasm
ApoB RNA editing is physiologically a nuclear event in which APOBEC1 and ACF shuttle into the nucleus to target the ApoB transcript.17,30,61 Nonetheless, it has been shown that overexpression of APOBEC1 can induce editing also in the cytoplasm.62 Considering that both APOBEC1 and ACF appear localized in different compartments depending on the cells in which they are expressed,30,59,62-64 we wanted to assess the weight of the nuclear and of the cytoplasmic component of RNA editing in our model system.
Analogously to AID,65 we found that the position of the fluorescent tag markedly influences the intracellular localization of APOBEC1: an N-terminal EGFP predominantly locks APOBEC1 in the cytoplasm, whereas a C-terminal tag allows APOBEC1 to be present in both nucleus and cytoplasm, an N-terminal EGFP predominantly locks APOBEC1 in the cytoplasm66 (Fig. 5A).
Figure 5.
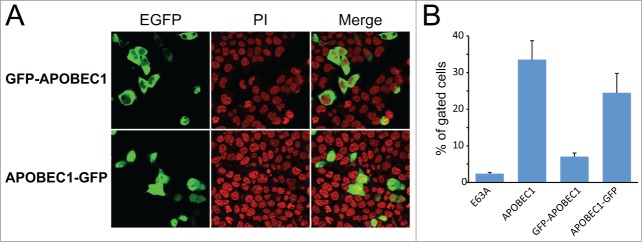
The intracellular localization of tagged APOBEC1 affects the efficiency of RNA editing. (A) The position of the fluorescent tag influences the localization of APOBEC1 chimeras. Representative confocal images of HEK293T cells transiently transfected with plasmids encoding EGFP-APOBEC1 or APOBEC1-EGFP chimeras. The nuclei were stained with Propidium Iodide (PI). (B) The bar diagram shows the efficiency of RNA editing in HEK293T cells transiently cotransfected with ACF and the APOBEC1 expressing plasmids. The EGFP-APOBEC1 and APOBEC1-EGFP constructs were mutated to inactivate the GFP to avoid interference with the assay. The error bars indicate the SEM from at least 3 experiments.
After inactivation of the EGFP chromophore through point mutation, we could use these constructs in our assay (the point mutation does not affect the intracellular localization of the chimeras, Fig. S4). Their cotransfection in HEK293T cells together with ACF shows that all constructs were able to support RNA editing (Fig. 5B), albeit with different efficiencies. The cytoplasmic APOBEC1 indeed displays a reduced ability to induce RNA editing, in accordance with the notion that physiological RNA editing is a mostly nuclear event.
Detection of RNA editing in induced Caco-2 cells
We have so far demonstrated that our assay is suitable in cells in which RNA editing is forced by the overexpression of the single components of the editosome. We next tested whether our assay could be used to detect RNA editing in cells with endogenous levels of APOBEC1 and ACF. Caco-2 cells, a colorectal adenocarcinoma cell line, have been widely used to study ApoB RNA editing. RNA editing in these cells can be modulated after cellular differentiation by a number of stimuli.45,67
We thus transduced Caco-2 cells with our fluorescent mCherry-ApoB-EGFP chimera through lentiviral infection and we induced RNA editing by treatment with Forskolin, a protein kinase activator which has been shown to induce RNA editing.45 Indeed treatment with 250 μM Forskolin induced a gradual increase in editing, proportional to the exposure (Fig. 6A). Similarly, the efficiency of the RNA editing was proportional to the concentration of the Forskolin used (Fig. 6B, C).
Figure 6.
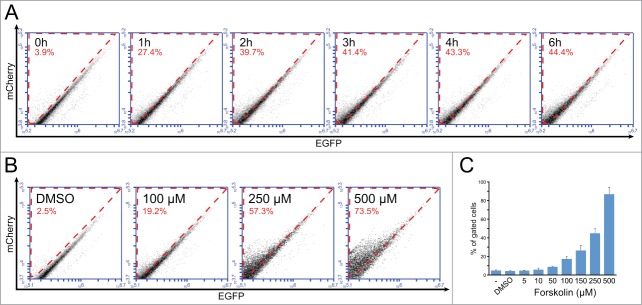
Visualization of RNA editing in Caco-2 cells after induction with Forskolin. (A) Increase of RNA editing in Caco-2 after induction with Forskolin. Cells transduced with an mCherry-ApoB-EGFP lentiviral construct were induced for the indicated times with 250 μM Forskolin. (B) Titration of RNA editing in cells transduced with an mCherry-ApoB-EGFP lentiviral construct after a 6 h treatment with Forskolin at the indicated concentrations. (C) The chart shows the percentage of gated cells in cells untreated, treated with Forskolin at the indicated concentrations or with DMSO alone (same amount as the highest concentration of Forskolin). The error bar indicates the SEM from 4 experiments.
In contrast to what observed in HuH-7 and HEK293T cells, in which APOBEC1 had been overexpressed, RNA editing in Caco-2 cells occurs rapidly, probably due to the fact that the modulation of RNA editing in treated cells is dependent on a post-translational mechanism.45 On the other hand, we observed that even at longer times, there is always a percentage of cells in which no RNA editing is measurable. This might suggest that either some cells are refractory to the treatment, or that the efficiency of RNA editing is not constant in time and might depend on other factors (e.g., state of the cell, phase of the cell cycle, etc.).
Conclusions
We set up a rapid flow cytometric assay to measure APOBEC1-specific RNA editing using a double fluorescent chimeric protein containing the canonical editing site for ApoB mRNA. The readout of the RNA editing in a cellular population is based on the loss of EGFP fluorescence - whose cds contains an editable site - compared to a stable mCherry fluorescence. From our experiments it seems to be an easy and robust method to study RNA editing in a number of different settings.
While the specificity of the assay is intrinsic to the system, its sensitivity depends on the conditions used. In contrast to a recently developed microscopy-assisted methodology to visualize ADAR-mediated A>I editing in individual cells,68 our assay is based on loss of signal and it is thus better suited to analyze RNA editing in the whole cellular population rather than in individual cells.
The methodology measures a dynamic process and a number of concatenated events are involved in the outcome: plasmids transcription, RNA editing, translation, protein degradation. This makes it difficult to set up experiments in which abrupt changes in RNA editing are expected.
Since only the fraction of transfected cells is used for the analysis, the assay allows the quantification of RNA editing in a manner independent from the efficiency of the transfection of exogenous factors. This represents an advantage over other commonly used methods, in which the totality of cells is analyzed.
Our assay can be used to visualize APOBEC1-dependent RNA editing in cellular models in which APOBEC1 and ACF are expressed either artificially or physiologically. The assay eliminates the need for PCR amplification steps as it is based on live cells analysis. Moreover the possibility to identify RNA-editing(+) cells could be used for downstream applications.
Finally, while tailored for APOBEC1-mediated ApoB RNA editing, our approach could be easily adapted to test other APOBEC1 targets or to assay other types of RNA editing.
Materials and Methods
Plasmids
The mCherry cds was amplified from pmCherry using primers 1 and 2 and cloned into the NheI/HindIII restriction sites of pEGFP-N1 to obtain the mCherry-EGFP plasmid. The mCherry-ApoB-EGFP plasmid was obtained by inserting a PCR fragment from human ApoB (426 bp from HepG2 cDNA, primers 3 and 4) into the HindIII/EcoRI sites of the mCherry-EGFP plasmid. A minimal editable site including the mooring sequence (28 bp) was inserted in the HindIII/EcoRI sites of the mCherry-EGFP plasmid using the oligonucleotides 5 and 6. The mCherry-ApoB-EGFP cds was subcloned into the lentiviral CSGW vector69 (blunted BamHI/NotI) after excision with NheI/NotI and blunting of the 5′ end.
The human and rat APOBEC1 expression vectors described in Saraconi et al.15 were digested with BglII/BsrGI to remove the IRES-EGFP sequence and ligated back after blunting the DNA ends. The APOBEC-1ΔC construct was obtained by PCR with primers 7 and 8 and cloned back into the pAIDexpressPuro2 backbone.70 The E63A mutation was introduced in the rat APOBEC1 cds by site directed mutagenesis (Stratagene - Catalog #200523) using the primers 9 and 10 and cloned back with primers 7 and 11. The lizard APOBEC1 cds was amplified using primers 12 and 13 from the pTrc99 vector detailed in Severi et al.,52 digested with NheI/BamHI restriction site and cloned into the NheI/BglII site of pAIDexpressPuro2. In all constructs the IRES-EGFP sequence was removed as described. The chimera of the lizard APOBEC1 and the C-terminus of rat APOBEC1 was obtained by PCR amplification using the primers 12and14 and 15 and 16, and subsequent extension using the primers 12 and 16. The PCR product was then cloned NheI/blunt in pAIDexpressPuro2 (opened NheI/BsrGI).
The GRY-RBP, ACF and hnRNP-C1 coding sequences were amplified from HuH-7 cDNA using primer pairs 17 and 18 (SacI/ ApaI), 19 and 20 (NheI/BglII), and 21 and 22 (HindIII/ApaI), respectively. The fragments were then cloned into a pEGFP-C3 plasmid in which the EGFP sequence had been replaced by a FLAG sequence.71 As control plasmids were used either vectors encoding for AID or CTNNBL1, or an empty vector.71
The EGFP-APOBEC1 construct was detailed in Conticello et al.71 The APOBEC1-EGFP construct was built by cloning the APOBEC1 cds (primers 23 and 24) into BglII/EcoRI restriction sites of the pEGFP-N1 expression vector. In order to produce non fluorescent chimeras, the EGFP chromophore was inactivated inserting a Y66H mutation in the EGFP cds by site-specific mutagenesis (primers 25 and 26 and 27 and 28). The mutated PCR products were then cloned back to replace the functional EGFP in the EGFP-APOBEC1 and APOBEC1-EGFP constructs (NheI/EcoRI and NheI/NotI sites, respectively).
All oligonucleotides, including PCR primers and sequencing primers are listed in the Additional file 1: Supplementary Table 1.
Cells, flow cytometry, and microscopy
HEK293T, HuH-7 and Caco-2 cells were maintained in DMEM supplemented with 10% FBS at 37°C in 5% CO2. Transient transfections were performed using Lipofectamine LTX (Invitrogen - Catalog #15338100) and GeneJuice (Novagen -Catalog #70967-3) according to manufacturer's instructions. Lentiviral infections of Caco-2 cells were performed as detailed in Langlois et al.72 All transfections with multiple plasmids were performed using an equal amount of overall plasmid DNA. Differences in the amount of DNA from experimental constructs were filled with an empty construct.
FACS analysis was performed on a Accuri C6 cytometer (BD) after transfection/infection of the cells, at the times indicated in the figure legends.
Cell visualization was performed on an Eclipse TE2000-E confocal (Nikon) confocal microscope (Fig. 5), or on a Axio Observer Z1 (Zeiss) inverted microscope (Fig. S4). Transiently transfected cells were replated on coverslips after 24 h from transfection, and then fixed by 4% Formaldehyde and stained at 48 hr. Immunocgemistry for EGFP was carried out using a rabbit anti-GFP antibody (1:500; Molecular Probes – Catalog #A11122) and a AlexaFluor488 goat anti-rabbit IgG secondary antibody (1:500; Molecular Probes – Catalog #A11008).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Riccardo Pecori, Rosario Notaro, Lucio Luzzatto, Javier Di Noia and Svend Petersen-Mahrt for the discussion and the valuable comments on the manuscript.
Funding
This work was supported by an institutional grant of the Istituto Toscano Tumori, and by the Italian Ministry of Health [GR-2008-1141464]. Funding for open access charge: Istituto Toscano Tumori.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci 2001; 26:376-84; PMID:11406411 [DOI] [PubMed] [Google Scholar]
- 2. Blanc V, Davidson NO. APOBEC-1-mediated RNA editing. Wiley Interdiscip. Rev Syst Biol Med 2010; 2:594-602; PMID:20836050; http://dx.doi.org/ 10.1002/wsbm.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Savva YA, Rieder LE, Reenan RA. The ADAR protein family. Genome Biol 2012; 13:252; PMID:23273215; http://dx.doi.org/ 10.1186/gb-2012-13-12-252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Conticello SG. Creative deaminases, self-inflicted damage, and genome evolution. Ann N Y Acad Sci 2012; 1267:79-85; PMID:22954220; http://dx.doi.org/ 10.1111/j.1749-6632.2012.06614.x [DOI] [PubMed] [Google Scholar]
- 5. Prohaska KM, Bennett RP, Salter JD, Smith HC. The multifaceted roles of RNA binding in APOBEC cytidine deaminase functions. WIREs RNA 2014; 5:493-508; PMID:24664896; http://dx.doi.org/ 10.1002/wrna.1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Navaratnam N, Morrison JR, Bhattacharya S, Patel D, Funahashi T, Giannoni F, Teng BB, Davidson NO, Scott J. The p27 catalytic subunit of the apolipoprotein B mRNA editing enzyme is a cytidine deaminase. J Biol Chem 1993; 268:20709-12; PMID:8407891 [PubMed] [Google Scholar]
- 7. Teng B, Burant CF, Davidson NO. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science 1993; 260:1816-9; PMID:8511591 [DOI] [PubMed] [Google Scholar]
- 8. Skuse GR, Cappione AJ, Sowden M, Metheny LJ, Smith HC. The neurofibromatosis type I messenger RNA undergoes base-modification RNA editing. Nucleic Acids Res 1996; 24:478-85; PMID:8602361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rosenberg BR, Hamilton CE, Mwangi MM, Dewell S, Papavasiliou FN. Transcriptome-wide sequencing reveals numerous APOBEC1 mRNA-editing targets in transcript 3' UTRs. Nat Struct Mol Biol 2011; 18:230-6; PMID:21258325; http://dx.doi.org/ 10.1038/nsmb.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Blanc V, Park E, Schaefer S, Miller M, Lin Y, Kennedy S, Billing AM, Ben Hamidane H, Graumann J, Mortazavi A, et al. Genome-wide identification and functional analysis of Apobec-1 mediated C-to-U RNA editing in mouse small intestine and liver. Genome Biol 2014; 15:R79; PMID:24946870; http://dx.doi.org/ 10.1186/gb-2014-15-6-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol 2013; 4:28; PMID:23431045; http://dx.doi.org/ 10.3389/fmicb.2013.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Franchini DM, Chan CF, Morgan H, Incorvaia E, Rangam G, Dean W, Santos F, Reik W, Petersen-Mahrt SK. Processive DNA demethylation via DNA deaminase-induced lesion resolution. PLoS ONE 2014; 9:e97754; PMID:25025377; http://dx.doi.org/ 10.1371/journal.pone.0097754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Harris RS, Petersen-Mahrt SK, Neuberger MS. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol Cell 2002; 10:1247-53; PMID:12453430 [DOI] [PubMed] [Google Scholar]
- 14. Petersen-Mahrt SK, Neuberger MS. In vitro deamination of cytosine to uracil in single-stranded DNA by apolipoprotein B editing complex catalytic subunit 1 (APOBEC1). J Biol Chem 2003; 278:19583-6; PMID:12697753; http://dx.doi.org/ 10.1074/jbc.C300114200 [DOI] [PubMed] [Google Scholar]
- 15. Saraconi G, Severi F, Sala C, Mattiuz G, Conticello SG. The RNA editing enzyme APOBEC1 induces somatic mutations and a compatible mutational signature is present in esophageal adenocarcinomas. Genome Biol 2014; 15:417; PMID:25085003; http://dx.doi.org/ 10.1186/PREACCEPT-3516273912883547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yamanaka S, Poksay KS, Driscoll DM, Innerarity TL. Hyperediting of multiple cytidines of apolipoprotein B mRNA by APOBEC-1 requires auxiliary protein(s) but not a mooring sequence motif. J Biol Chem 1996; 271:11506-10; PMID:8626710 [DOI] [PubMed] [Google Scholar]
- 17. Sowden M, Hamm JK, Smith HC. Overexpression of APOBEC-1 results in mooring sequence-dependent promiscuous RNA editing. J Biol Chem 1996; 271:3011-7; PMID:8621694 [DOI] [PubMed] [Google Scholar]
- 18. Yamanaka S, Poksay KS, Arnold KS, Innerarity TL. A novel translational repressor mRNA is edited extensively in livers containing tumors caused by the transgene expression of the apoB mRNA-editing enzyme. Genes Dev 1997; 11:321-33; PMID:9030685 [DOI] [PubMed] [Google Scholar]
- 19. Chen Z, Eggerman TL, Bocharov AV, Baranova IN, Vishnyakova TG, Kurlander RJ, Csako G, Patterson AP. Hypermutation of ApoB mRNA by rat APOBEC-1 overexpression mimics APOBEC-3 hypermutation. J Mol Biol 2012; 418:65-81; PMID:22326345; http://dx.doi.org/ 10.1016/j.jmb.2012.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamanaka S, Balestra ME, Ferrell LD, Fan J, Arnold KS, Taylor S, Taylor JM, Innerarity TL. Apolipoprotein B mRNA-editing protein induces hepatocellular carcinoma and dysplasia in transgenic animals. Proc Natl Acad Sci U S A 1995; 92:8483-7; PMID:7667315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blanc V, Henderson JO, Newberry RD, Xie Y, Cho SJ, Newberry EP, Kennedy S, Rubin DC, Wang HL, Luo J, et al. Deletion of the AU-rich RNA binding protein Apobec-1 reduces intestinal tumor burden in Apc(min) mice. Cancer Res 2007; 67:8565-73; PMID:17875695; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-1593 [DOI] [PubMed] [Google Scholar]
- 22. Shah RR, Knott TJ, Legros JE, Navaratnam N, Greeve JC, Scott J. Sequence requirements for the editing of apolipoprotein B mRNA. J Biol Chem 1991; 266:16301-4; PMID:1885564 [PubMed] [Google Scholar]
- 23. Backus JW, Smith HC. Three distinct RNA sequence elements are required for efficient apolipoprotein B (apoB) RNA editing in vitro. Nucleic Acids Res 1992; 20:6007-14; PMID:1461733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mehta A, Banerjee S, Driscoll DM. Apobec-1 interacts with a 65-kDa complementing protein to edit apolipoprotein-B mRNA in vitro. J Biol Chem 1996; 271:28294-9; PMID:8910449 [DOI] [PubMed] [Google Scholar]
- 25. Mehta A, Driscoll DM. A sequence-specific RNA-binding protein complements apobec-1 To edit apolipoprotein B mRNA. Mol Cell Biol 1998; 18:4426-32; PMID:9671452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mehta A, Kinter MT, Sherman NE, Driscoll DM. Molecular cloning of apobec-1 complementation factor, a novel RNA-binding protein involved in the editing of apolipoprotein B mRNA. Mol Cell Biol 2000; 20:1846-54; PMID:10669759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lellek H, Kirsten R, Diehl I, Apostel F, Buck F, Greeve J. Purification and molecular cloning of a novel essential component of the apolipoprotein B mRNA editing enzyme-complex. J Biol Chem 2000; 275:19848-56; PMID:10781591; http://dx.doi.org/ 10.1074/jbc.M001786200 [DOI] [PubMed] [Google Scholar]
- 28. Lellek H, Welker S, Diehl I, Kirsten R, Greeve J. Reconstitution of mRNA editing in yeast using a Gal4-apoB-Gal80 fusion transcript as the selectable marker. J Biol Chem 2002; 277:23638-44; PMID:11976346; http://dx.doi.org/ 10.1074/jbc.M203517200 [DOI] [PubMed] [Google Scholar]
- 29. Maris C, Masse J, Chester A, Navaratnam N, Allain FH. NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor. RNA 2005; 11:173-86; PMID:15659357; http://dx.doi.org/ 10.1261/rna.7190705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chester A, Somasekaram A, Tzimina M, Jarmuz A, Gisbourne J, O'Keefe R, Scott J, Navaratnam N. The apolipoprotein B mRNA editing complex performs a multifunctional cycle and suppresses nonsense-mediated decay. EMBO J 2003; 22:3971-82; PMID:12881431; http://dx.doi.org/ 10.1093/emboj/cdg369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fossat N, Tourle K, Radziewic T, Barratt K, Liebhold D, Studdert JB, Power M, Jones V, Loebel DA, Tam PP. C to U RNA editing mediated by APOBEC1 requires RNA-binding protein RBM47. EMBO Rep 2014; 15:903-10; PMID:24916387; http://dx.doi.org/ 10.15252/embr.201438450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Greeve J, Lellek H, Rautenberg P, Greten H. Inhibition of the apolipoprotein B mRNA editing enzyme-complex by hnRNP C1 protein and 40S hnRNP complexes. Biol Chem 1998; 379:1063-73; PMID:9792439 [DOI] [PubMed] [Google Scholar]
- 33. Blanc V, Navaratnam N, Henderson JO, Anant S, Kennedy S, Jarmuz A, Scott J, Davidson NO. Identification of GRY-RBP as an apolipoprotein B RNA-binding protein that interacts with both apobec-1 and apobec-1 complementation factor to modulate C to U editing. J Biol Chem 2001; 276:10272-83; PMID:11134005; http://dx.doi.org/ 10.1074/jbc.M006435200 [DOI] [PubMed] [Google Scholar]
- 34. Lau PP, Villanueva H, Kobayashi K, Nakamuta M, Chang BH, Chan L. A DnaJ protein, apobec-1-binding protein-2, modulates apolipoprotein B mRNA editing. J Biol Chem 2001; 276:46445-52; PMID:11584023; http://dx.doi.org/ 10.1074/jbc.M109215200 [DOI] [PubMed] [Google Scholar]
- 35. Lau PP, Chang BH, Chan L. Two-hybrid cloning identifies an RNA-binding protein, GRY-RBP, as a component of apobec-1 editosome. Biochem Biophys Res Commun 2001; 282:977-83; PMID:11352648; http://dx.doi.org/ 10.1006/bbrc.2001.4679 [DOI] [PubMed] [Google Scholar]
- 36. Anant S, Henderson JO, Mukhopadhyay D, Navaratnam N, Kennedy S, Min J, Davidson NO. Novel role for RNA-binding protein CUGBP2 in mammalian RNA editing. CUGBP2 modulates C to U editing of apolipoprotein B mRNA by interacting with apobec-1 and ACF, the apobec-1 complementation factor. J Biol Chem 2001; 276:47338-51; PMID:11577082; http://dx.doi.org/ 10.1074/jbc.M104911200 [DOI] [PubMed] [Google Scholar]
- 37. Lau PP, Chan L. Involvement of a chaperone regulator, Bcl2-associated athanogene-4, in apolipoprotein B mRNA editing. J Biol Chem 2003; 278:52988-96; PMID:14559896; http://dx.doi.org/ 10.1074/jbc.M310153200 [DOI] [PubMed] [Google Scholar]
- 38. Chen Z, Eggerman TL, Patterson AP. ApoB mRNA editing is mediated by a coordinated modulation of multiple apoB mRNA editing enzyme components. Am J Physiol Gastrointest Liver Physiol 2007; 292:G53-G65; PMID:16920700; http://dx.doi.org/ 10.1152/ajpgi.00118.2006 [DOI] [PubMed] [Google Scholar]
- 39. Davies MS, Wallis SC, Driscoll DM, Wynne JK, Williams GW, Powell LM, Scott J. Sequence requirements for apolipoprotein B RNA editing in transfected rat hepatoma cells. J Biol Chem 1989; 264:13395-8; PMID:2760026 [PubMed] [Google Scholar]
- 40. Greeve J, Jona VK, Chowdhury NR, Horwitz MS, Chowdhury JR. Hepatic gene transfer of the catalytic subunit of the apolipoprotein B mRNA editing enzyme results in a reduction of plasma LDL levels in normal and watanabe heritable hyperlipidemic rabbits. J Lipid Res 1996; 37:2001-17; PMID:8895066; http://dx.doi.org/Greeve 1996 [PubMed] [Google Scholar]
- 41. Fogh J, Fogh JM, Orfeo T. One hundred and twenty-seven cultured human tumor cell lines producing tumors in nude mice. J Natl Cancer Inst 1977; 59:221-226; PMID:327080 [DOI] [PubMed] [Google Scholar]
- 42. Jiao S, Moberly JB, Schonfeld G. Editing of apolipoprotein B messenger RNA in differentiated Caco-2 cells. J Lipid Res 1990; 31:695-700; PMID:2351874 [PubMed] [Google Scholar]
- 43. Wagner RD, Krul ES, Moberly JB, Alpers DH, Schonfeld G. Apolipoprotein expression and cellular differentiation in Caco-2 intestinal cells. Am J Physiol 1992; 263:E374-82; PMID:1514621 [DOI] [PubMed] [Google Scholar]
- 44. Chen Z, Eggerman TL, Potosky D, Arborati M, Patterson AP. Calcium increases apolipoprotein B mRNA editing. Biochem Biophys Res Commun 2000; 277:221-7; PMID:11027667; http://dx.doi.org/ 10.1006/bbrc.2000.3668 [DOI] [PubMed] [Google Scholar]
- 45. Chen Z, Eggerman TL, Patterson AP. Phosphorylation is a regulatory mechanism in apolipoprotein B mRNA editing. Biochem J 2001; 357:661-72; PMID:11463337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davidson NO, Powell LM, Wallis SC, Scott J. Thyroid hormone modulates the introduction of a stop codon in rat liver apolipoprotein B messenger RNA. J Biol Chem 1988; 263:13482-5; PMID:3417667 [PubMed] [Google Scholar]
- 47. Driscoll DM, Wynne JK, Wallis SC, Scott J. An in vitro system for the editing of apolipoprotein B mRNA. Cell 1989; 58:519-25; PMID:2758465 [DOI] [PubMed] [Google Scholar]
- 48. Tennyson GE, Sabatos CA, Higuchi K, Meglin N, Brewer HB. Expression of apolipoprotein B mRNAs encoding higher- and lower-molecular weight isoproteins in rat liver and intestine. Proc Natl Acad Sci U S A 1989; 86:500-4; PMID:2911593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu JH, Semenkovich CF, Chen SH, Li WH, Chan L. Apolipoprotein B mRNA editing. Validation of a sensitive assay and developmental biology of RNA editing in the rat. J Biol Chem 1990; 265:12312-6; PMID:2373694 [PubMed] [Google Scholar]
- 50. Smith HC. Measuring editing activity and identifying cytidine-to-uridine mRNA editing factors in cells and biochemical isolates. Methods Enzymol 2007; 424:389-416; PMID:17662851; http://dx.doi.org/ 10.1016/S0076-6879(07)24018-2 [DOI] [PubMed] [Google Scholar]
- 51. Teng BB, Ochsner S, Zhang Q, Soman KV, Lau PP, Chan L. Mutational analysis of apolipoprotein B mRNA editing enzyme (APOBEC1). structure-function relationships of RNA editing and dimerization. J Lipid Res 1999; 40:623-35; PMID:10191286 [PubMed] [Google Scholar]
- 52. Severi F, Chicca A, Conticello SG. Analysis of Reptilian APOBEC1 Suggests that RNA Editing May Not Be Its Ancestral Function. Mol Biol Evol 2011; 28:1125-9; PMID:21172829; http://dx.doi.org/ 10.1093/molbev/msq338 [DOI] [PubMed] [Google Scholar]
- 53. Oka K, Kobayashi K, Sullivan M, Martinez J, Teng BB, Ishimura-Oka K, Chan L. Tissue-specific inhibition of apolipoprotein B mRNA editing in the liver by adenovirus-mediated transfer of a dominant negative mutant APOBEC-1 leads to increased low density lipoprotein in mice. J Biol Chem 1997; 272:1456-60; PMID:8999814 [DOI] [PubMed] [Google Scholar]
- 54. Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 2007; 76:1-22; PMID:17328676; http://dx.doi.org/ 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- 55. Richardson N, Navaratnam N, Scott J. Secondary structure for the apolipoprotein B mRNA editing site. Au-binding proteins interact with a stem loop. J Biol Chem 1998; 273:31707-17; PMID:9822632 [DOI] [PubMed] [Google Scholar]
- 56. Blanc V, Henderson JO, Kennedy S, Davidson NO. Mutagenesis of apobec-1 complementation factor reveals distinct domains that modulate RNA binding, protein-protein interaction with apobec-1, and complementation of C to U RNA-editing activity. J Biol Chem 2001; 276:46386-93; PMID:11571303; http://dx.doi.org/ 10.1074/jbc.M107654200 [DOI] [PubMed] [Google Scholar]
- 57. Mehta A, Driscoll DM. Identification of domains in apobec-1 complementation factor required for RNA binding and apolipoprotein-B mRNA editing. RNA 2002; 8:69-82; PMID:11871661; http://dx.doi.org/10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chester A, Weinreb V, Carter CW, Navaratnam N. Optimization of apolipoprotein B mRNA editing by APOBEC1 apoenzyme and the role of its auxiliary factor, ACF. RNA 2004; 10:1399-411; PMID:15273326; http://dx.doi.org/ 10.1261/rna.7490704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lehmann DM, Galloway CA, Sowden MP, Smith HC. Metabolic regulation of apoB mRNA editing is associated with phosphorylation of APOBEC-1 complementation factor. Nucleic Acids Res 2006; 34:3299-308; PMID:16820530; http://dx.doi.org/ 10.1093/nar/gkl417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Galloway CA, Kumar A, Krucinska J, Smith HC. APOBEC-1 Complementation Factor (ACF) forms RNA-Dependent Multimers. Biochem Biophys Res Commun 2010; 398:38-43; PMID:20541536; http://dx.doi.org/ 10.1016/j.bbrc.2010.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lau PP, Xiong WJ, Zhu HJ, Chen SH, Chan L. Apolipoprotein B mRNA editing is an intranuclear event that occurs posttranscriptionally coincident with splicing and polyadenylation. J Biol Chem 1991; 266:20550-4; PMID:1939106 [PubMed] [Google Scholar]
- 62. Yang Y, Sowden MP, Smith HC. Induction of cytidine to uridine editing on cytoplasmic apolipoprotein B mRNA by overexpressing APOBEC-1. J Biol Chem 2000; 275:22663-9; PMID:10833526; http://dx.doi.org/ 10.1074/jbc.M910406199 [DOI] [PubMed] [Google Scholar]
- 63. Yang Y, Smith HC. Multiple protein domains determine the cell type-specific nuclear distribution of the catalytic subunit required for apolipoprotein B mRNA editing. Proc Natl Acad Sci U S A 1997; 94:13075-80; PMID:9371802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Blanc V, Kennedy S, Davidson NO. A novel nuclear localization signal in the auxiliary domain of apobec-1 complementation factor regulates nucleocytoplasmic import and shuttling. J Biol Chem 2003; 278:41198-204; PMID:12896982; http://dx.doi.org/ 10.1074/jbc.M302951200 [DOI] [PubMed] [Google Scholar]
- 65. Patenaude AM, Orthwein A, Hu Y, Campo VA, Kavli B, Buschiazzo A, Di Noia JM. Active nuclear import and cytoplasmic retention of activation-induced deaminase. Nat Struct Mol Biol 2009; 16:517-27; PMID:19412186; http://dx.doi.org/ 10.1038/nsmb.1598 [DOI] [PubMed] [Google Scholar]
- 66. Siddiqui JF, Van Mater D, Sowden MP, Smith HC. Disproportionate relationship between APOBEC-1 expression and apolipoprotein B mRNA editing activity. Exp Cell Res 1999; 252:154-64; PMID:10502408; http://dx.doi.org/ 10.1006/excr.1999.4598 [DOI] [PubMed] [Google Scholar]
- 67. Chen Z, Eggerman TL, Potosky D, Arborati M, Patterson AP. Calcium increases apolipoprotein B mRNA editing. Biochem Biophys Res Commun 2000; 277:221-7; PMID:11027667; http://dx.doi.org/ 10.1006/bbrc.2000.3668 [DOI] [PubMed] [Google Scholar]
- 68. Garrncarz W, Tariq A, Handl C, Pusch O, Jantsch MF. A high throughput screen to identify enhancers of ADAR-mediated RNA-editing. RNA Biol 2013; 10; 192-204; PMID:23353575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, Grez M, Thrasher AJ. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther 2002; 13:803-13; PMID:11975847; http://dx.doi.org/ 10.1089/10430340252898984 [DOI] [PubMed] [Google Scholar]
- 70. Arakawa H, Hauschild J, Buerstedde JM. Requirement of the activation-induced deaminase (AID) gene for immunoglobulin gene conversion. Science 2002; 295:1301-6; PMID:11847344; http://dx.doi.org/ 10.1126/science.1067308 [DOI] [PubMed] [Google Scholar]
- 71. Conticello SG, Ganesh K, Xue K, Lu M, Rada C, Neuberger MS. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol Cell 2008; 31:474-84; PMID:18722174; http://dx.doi.org/ 10.1016/j.molcel.2008.07.009 [DOI] [PubMed] [Google Scholar]
- 72. Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res 2005; 33:1913-23; PMID:15809227; http://dx.doi.org/ 10.1093/nar/gki343 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


