Abstract
Cryptosporidium parvum is a member of the Apicomplexa that lacks a plastid and associated nuclear-encoded genes, which has hampered its use in evolutionary comparisons with algae and eliminated a pool of potentially useful drug targets. Here we show that apicomplexan parasites possess an unusual family of class II histone deacetylase (HDAC) proteins with orthologues that are present in other chromalveolates and primitive algae. A striking feature of these HDAC proteins is the presence of ankyrin repeats in the amino-terminus that appear to be required for enzyme activity. In vitro and in vivo analyses of the C. parvum orthologue indicate that this subclass of chromatin-remodelling proteins is targeted by the anti-cancer drug suberoylanilide hydroxamic acid and that these proteins are most likely involved in the essential process of H4 histone deacetylation that coincides with DNA replication. We propose that members of this novel class of histone deacetylase can serve as promising new targets for treatments against debilitating diseases such as cryptosporidosis, toxoplasmosis and malaria.
Keywords: Apicomplexa, Diatom, Chlorophyte, Esterase, Histone deacetylase, Algae
1. Introduction
Cryptosporidium parvum is an opportunistic pathogen that causes enteric infections which, due to ineffective treatment options, may be life threatening to immunocompromised individuals (Abubakar et al., 2007). This parasite is a member of the phylum Apicomplexa which includes the causative agents of malaria and toxoplasmosis, as well as other parasites of medical and veterinary importance. Although treatments do exist for some apicomplexan diseases, the evolution of antibiotic-resistant parasites represents an impediment to existing therapies (McFadden et al., 2001, Mead, 2002, Daily, 2006 and Williams, 2006).
Many members of the Apicomplexa possess a non-photosynthetic plastid that has attracted significant attention, not only as a target for drug development, but also in attempts to decipher the evolutionary history of the apicomplexan lineage (Roos et al., 1999). Analyses of the plastid and of nuclear-encoded, plastid-targeted proteins suggest that the plastid was probably acquired through secondary symbiosis of a free-living photosynthetic organism and point to the unresolved possibility of either a chlorophyte (green alga) or a rhodophyte (red alga) origin for the ancestral apicomplexan plastid (Williamson et al., 1994, Kohler et al., 1997, Blanchard and Hicks, 1999, Fast et al., 2001, Funes et al., 2002, Cai et al., 2003 and Waller et al., 2003). However, a salient feature of C. parvum is that it has apparently lost the plastid and the nuclear genes required to support plastid maintenance (Zhu et al., 2000, Abrahamsen et al., 2004 and Keeling, 2004). Cryptosporidium spp. have, therefore, been excluded from analyses of apicomplexan relationships with algae when plastid-related sequences were used. Moreover, useful plastid-related targets are unavailable for chemotherapeutic interventions against C. parvum.
Given the paucity of available drugs to combat this pathogen, new therapeutic targets are being sought. Potential non-plastid targets include the chromatin-remodelling machinery (Darkin-Rattray et al., 1996, Andrews et al., 2000, Kwon et al., 2003, Mai et al., 2004 and Saksouk et al., 2005). This group of proteins includes the histone deacetylases (HDACs), which can catalyse the removal of acetyl groups from acetylated histone tails and can facilitate processes such as DNA replication, gene regulation and cell fate. Due to their role in determining cell fate, HDAC proteins have received considerable attention as targets for anti-cancer chemotherapy (Kim et al., 2006). Numerous HDAC inhibitors have been identified, some of which are specific to a particular family or class of HDAC. Recently the HDAC inhibitor suberoylanilide hydroxamic acid (SAHA) was approved for use in humans to treat cutaneous T-cell lymphoma (Mann et al., 2007).
HDAC proteins are diverse and can be divided into three unrelated groups based on primary structure. The HD2 family is reported to be a plant-specific group of nucleolar-localised HDAC (Dangl et al., 2001). NAD+-dependent enzymes related to the yeast Silent information regulator 2 protein (Sirtuins/class III) have also been described and are broadly distributed among prokaryotes, eukaryotes and archaea (Frye, 2000). A third group is structurally related to the yeast Reduced potassium dependency 3 protein (Rpd3 superfamily). The Rpd3 superfamily has a similar broad distribution across kingdoms and can be divided further into three main classes (designated as class I, II or IV) (Gregoretti et al., 2004). Strong statistical support of phylogenetic groupings and the presence of proteins from divergent taxa (e.g. invertebrates and vertebrates) served as a basis for the delineation of these three classes. Overall, the distribution of proteins in the class I, II or IV lineages in each of the three kingdoms indicates that the superfamily is an ancient group that probably pre-dates the evolution of histones and suggests that these proteins may have metabolic functions that are yet to be identified (Gregoretti et al., 2004). Recently, certain members of the Rpd3 superfamily were shown to possess significant esterase activity in vitro, but the biological significance of this remains to be elucidated (Moreth et al., 2007).
We describe herein the characterisation of CpHDAC3 – a member of a distinct group of Rpd3-like class II histone deacetylase proteins that has been identified in apicomplexan parasites and photosynthetic eukaryotes. Analysis of CpHDAC3 reveals that the proteins are replication-associated and nuclear-localised. Recombinant CpHDAC3 displays histone deacetylase activity with a significant preference toward N-terminal histone H4 peptides acetylated at specific lysine residues, but its ability to function as an esterase is limited. Additionally, CpHDAC3 is targeted by the anti-cancer HDAC inhibitor vorinostat. We suggest that this family of HDACs participates in the essential process of post-replication H4 histone deacetylation and offers a prospective opportunity for innovative chemotherapy.
2. Materials and methods
2.1. Bioinformatics
The Saccharomyces cerevisiae Rpd3 protein sequence (Vidal and Gaber, 1991) was used to identify genes encoding similar proteins in the C. parvum genome (Abrahamsen et al., 2004) by searching using the BLAST algorithm (Altschul et al., 1990). Three Rpd3-related protein-coding genes were identified (locus cgd6_80, locus cgd6_1380 and locus cgd8_480) and designated as CpHDAC1, CpHDAC2 and CpHDAC3, respectively, with the names based on the order of their chromosomal locations. The predicted C. parvum HDAC3 protein sequence was used as a basis for identifying related DNA or protein sequences present in various public databases in searches using the BLAST algorithm. Conserved domains or motifs were identified by comparisons with the conserved domains database (http://www.ncbi.nlm.nih.gov/Structure/), or by submitting unaligned sequences to the block maker tool (http://blocks.fhcrc.org/; Henikoff et al., 1995). Ankyrin repeats were also manually compared with an ankyrin-repeat consensus that was previously derived from alignments of over 4000 repeats (Mosavi et al., 2004). Preliminary phylogenetic analyses of HDAC domains were based on those used previously (Gregoretti et al., 2004) to classify Rpd3 HDACs into three major clades.
2.2. Molecular manipulations
Genomic locus cgd8_480 (GenBank Accession No. XP_625509) was chosen for examination. There is an in-frame stop five amino acids upstream of the predicted CpHDAC3 start site, and the remaining intergenic region does not contain obvious candidates for other start sites. The next nearest gene upstream faces in the opposite direction. Additionally, the CpHDAC3 protein is well represented in mass spectrum data derived from proteomics analyses of parasites (Snelling et al., 2007), with nine peptides being identified that cover most of the length of the predicted protein, including one peptide just five amino acids from the predicted N-terminus (at the first predicted trypsin cleavage site). No peptides have been discovered that extend beyond the predicted CpHDAC3 open reading frame (e.g. into the predicted intergenic regions), suggesting that the prediction is correct. Reverse-transcriptase PCR was performed using the ProStar Ultra HF RT-PCR system (Stratagene) and the primers listed below to confirm the size of the predicted open reading frame. For cloning, however, the open reading frame corresponding to CpHDAC3 was amplified from genomic DNA using a high fidelity polymerase (PfuTurbo, Stratagene) and the primers 5′-CGggatccATGGACCCGGACAAAAGTC-3′ and 5′-CGtctagaTCAAATATTTTCGCTCAAG-3′ which added BamHI and XbaI restriction enzyme sites (lower case) to the 5′ and 3′ termini, respectively. PCR products were cloned using the ZeroBlunt TOPO kit (Invitrogen), sequenced and transferred to the pMAL-c2E vector (New England Biolabs) at the BamHI and XbaI sites using standard procedures. The final expression construct (designated pDR59) was verified by sequencing. CpHDAC3 protein was expressed in Escherichia coli harbouring the pDR59 vector. Medium consisted of lysogeny broth containing 15 mM glucose. Five individual 5 ml cultures were grown overnight and used to inoculate 1 L of medium. Growth was allowed to continue for 5 h at 37 °C followed by isopropyl β-d-1-thiogalactopyranoside induction (1 mM) for 2.5 h prior to harvesting. Cell paste was suspended in 200 mM NaCl, 10 mM Tris pH 7.4 containing bacterial protease inhibitors (Sigma) and stored overnight at −80 °C. Thawed cells were then lysed by sonication and recombinant protein was affinity purified as described previously (Rider et al., 2005) using amylose resin (New England Biolabs). Purified protein concentration was estimated using the Bio-Rad protein assay reagent (Bio-Rad) and BSA (fraction IV) as a standard. Except for the use of a different forward or reverse primer, construct building, expression and purification for truncated CpHDAC3 proteins was done as described above. The reverse primer used for the C-terminal deletion (Δ HDAC) was 5′-GTCTAGATTACACTAATTTTGAACGTCTAA-3′ and the expression plasmid was designated pDR84. The forward primer used for the N-terminal deletion (Δ Ankyrin) was 5′-CGGATCCATGATTTCTGAGAGTGAATATGA-3′ and the expression plasmid was designated pDR89. For studies with CpHDAC3 lacking the maltose binding protein (MBP) fusion, calcium chloride was added to a final concentration of 2 mM and Enterokinase (New England Biolabs) was added to cleave the MBP from the fusion at 4 °C for 48 h. Enterokinase was used at 4 ng for each 600 µg fusion protein in a total volume of 1 ml. A separate sample of purified fusion protein was treated with calcium chloride (4 °C for 48 h) to serve as an uncut control. Complete cleavage was assessed by coomassie staining of cut or uncut samples (10 µg) separated by SDS–PAGE.
2.3. Expression analysis
Parasite growth in vitro was done as described previously (Upton et al., 1995) using Iowa strain parasites obtained from Bunch Grass Farms (Deary, ID). Transcript analysis was done as described previously (Rider et al., 2005). The primers used for monitoring CpHDAC3 transcript were 5′-AATGCCGTTGATTGTGTCTGC-3′ and 5′-CCATGTTCCAAGATGATGTCCAG-3′. Ribosomal RNA served as a normalisation control and the primers used were 5′-CTCCACCAACTAAGAACGGCC-3′ and 5′-TAGAGATTGGAGGTTGTTCCT-3′. The expression level in oocysts was set to 1 for comparison with samples from intracellular parasites harvested at 24 h post-inoculation. Anti-CpHDAC3 polyclonal antibodies for immunofluorescence and Western blotting were generated in a rabbit using recombinant MBP-CpHDAC3 as the immunogen (Lampire Biological Laboratories, Pipersville, PA). All animal protocols were approved by an institutional animal care and use committee and were performed in a facility accredited by the Association for Assessment and Accreditation of Laboratory Animal Care. The rabbit was chosen from among several whose pre-immune sera displayed no reactivity to sporozoite proteins when used at a dilution of 1:100 (data not presented). Pre-immune sera also did not react with in vitro cultivated parasites when used at 1:2000 with immunofluorescence microscopy. Western blots were performed with each lane representing the total protein extracted from ~107 parasites and polyclonal antisera at a 1:10,000 dilution. Immunofluorescence was performed using polyclonal anti-CpHDAC3 antisera at a 1:2000 dilution. Bromodeoxyuridine (BrdU) incorporation was performed as described previously (Striepen et al., 2004) except that a more mild treatment with 2 N HCl (20 min at 20 °C) facilitated improved visualisation of DAPI-stained nuclei. BrdU and anti-BrdU antibody were purchased from Sigma Chemicals.
2.4. Enzyme reactions
The 4-methylumbelliferyl-acetate, 4-methylumbelliferyl-butyrate, 4-methylumbelliferone, depudecin, valproic acid, sodium butyrate, APHA 8 and Pseudomonas fluorescens lipase were purchased from Sigma Chemicals (St. Louis, MO, USA). The 4-methylumbelliferyl-propionate was purchased from Research Organics (Cleveland, OH, USA). Apicidin, purified from Fusarium spp., was obtained from EMD Biosciences (Gibbstown, NJ, USA). Vorinostat (SAHA) was purchased from Cayman Chemicals (Ann Arbor, MI, USA). Fluor de Lys substrate, deacetylated Fluor de Lys standard, Trichostatin A, developer and HeLa cell nuclear extract were purchased from BioMol (Plymouth Meeting, PA, USA). MBP2 was purchased from New England Biolabs (Ipswich, MA). H4 peptides were purchased from Upstate/Millipore (Billerica, MA, USA).
Esterase activities were determined using various dilutions of 4-methylumbelliferyl-acetate or 4-methylumbelliferyl-butyrate prepared in ice-cold assay buffer immediately before use. P. fluorescens lipase (Pf lipase), recombinant MBP-CpHDAC3 or MBP2 were prepared in assay buffer containing 50% glycerol and stored at −20 °C. The CpHDAC3 and MBP2 were at the same molar concentration so that direct comparisons could be made. The fluorescence of the 4-methylumbelliferone produced was monitored over time at 1 min intervals for up to 1 h using a Fluoroskan Ascent fluorescence plate reader (Thermoelectron, Hudson, NH, USA) fitted with bandpass interference filters (Ex = 355/20, Em = 444/6). Concentrations of 4-methylumbelliferone (the product of the esterase reactions) were determined from a standard curve generated using dilutions made in assay buffer immediately before reactions were initiated. Each reaction was performed at 25 °C and consisted of 5 µl protein and 25 µl of diluted substrate in a final volume of 50 µl. The final concentration of protein in each reaction was 112 nM for CpHDAC3 and MBP or 4.6 nM for Pf lipase. Control reactions without protein were also performed to monitor autohydrolysis of the esterase substrates and for background subtractions prior to kinetic analyses. The 10 min time points were used for kinetic analyses. Esterase assay buffer consisted of 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2 and 50 mM Tris/Cl, pH 6.8.
Deacetylase assays were similarly performed using various concentrations of Fluor de Lys substrate. Concentrations of deacetylated product were determined using a standard curve generated with dilutions of deacetylated Fluor de Lys standard made in assay buffer and mixed with Fluor de Lys developer. Production of the fluorophore requires the use of a trypsin-containing developer that can digest CpHDAC3. Thus, reactions cannot be reliably monitored in a continuous fashion. Preliminary tests indicated that the linear phase of the reaction occurs up to approximately 20–30 min at 20 °C. Each HDAC reaction consisted of 5 µl of protein, 25 µl of diluted substrate and 10 µl of diluted competitor/inhibitor (if used) in a final volume of 50 µl. The final concentration of protein in each reaction was 112 nM for CpHDAC3, MBP2 or CpHDAC3 truncations. After a 10 min incubation at 20 °C, reactions were stopped by the addition of 50 µl of developer solution containing 400 nM trichostatin A. Deacetylase assay buffer was similar to the esterase assay buffer except that the buffer had a pH of 8.1. For competition and inhibitor studies, Fluor de Lys substrate was used at a final concentration of 5 µM during the reaction step, H4 peptides (if used) were present at 15 µM and no-enzyme controls were used for subtracting background fluorescence. Student’s t-test was used to determine the P-value in comparisons between acetylated peptides and the deacetylated H4 peptide. Chemical Inhibitor tests included a 20 min pre-incubation of inhibitor with enzyme prior to the addition of substrate. Concentrations of chemical inhibitors ranged from 0 to 2 mM. The broad-spectrum histone deacetylase inhibitors that have been shown to be effective against apicomplexans or their HDACs included apicidin (Darkin-Rattray et al., 1996), APHA compound 8 (Mai et al., 2004), trichostatin A (Andrews et al., 2000), Depudecin (Isaka et al., 2000 and Kwon et al., 2003), Valproic acid (Jones-Brando et al., 2003 and Gurvich et al., 2004) and sodium butyrate (Saksouk et al., 2005).
Reactions were performed in duplicate or triplicate. Kinetic parameters (Km, kcat) for HDAC or esterase activity, and IC50 values (half maximal inhibition) for inhibitors were determined by non-linear regression and fitting the dose–response data to hyperbolic or sigmoidal models using GraphPad Prism 4.0c Software.
3. Results
3.1. Related proteins
Cryptosporidium parvum possesses a unique protein with similarity to HDAC-like proteins from proteobacteria but with N-terminal ankyrin repeats (Fig. 1A). At least two of the five identifiable ankyrin repeats present in the C. parvum protein are highly divergent and contain insertions or partial deletions (Fig. 1A). Orthologous genes were identified in all other apicomplexan genera examined, including Plasmodium, Toxoplasma, Babesia, Eimeria, Neospora and Theileria, implying that this protein was present in the ancestral apicomplexan, rather than the result of a more recent horizontal genetic transfer. Protein sequence analyses place this group of proteins clearly within the Rpd3-like HDACs and these proteins include a class II signature (N-RPPGHH-C) upstream of the predicted active site within the HDAC domain. Surprisingly, related ankyrin-repeat-containing HDAC proteins were also identified in the recently completed genomes of several photosynthetic eukaryotes: the diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum (Bacillariophyta), the coccolithophore Emiliania huxleyi (Haptophyta); several extremely primitive unicellular chlorophyte algae: Ostreococcus tauri, Ostreococcus lucimarinus and Micromonas pusilla (Chlorophyta: Prasinophyceae) and the endosymbiotic Chlorella strain NC64A (Chlorophyta: Trebouxiophyceae). Our observations of ankyrin-repeat proteins being encoded within the Ostreococcus and Thalassiosira genomes were recently confirmed (Iyer et al., 2008). However, searches for an orthologous gene in rhodophytes were unsuccessful. Specifically, examination of the genomes of Cyanidioschyzon merolae and Galdieria sulphuraria (rhodophytes whose genomes have been fully sequenced) revealed only HDAC domains or ankyrin repeats, but no genomic regions could be found that may encode single proteins containing both domains (Matsuzaki et al., 2004 and Barbier et al., 2005). Given the taxonomic distribution and phylogenetic position of the ankyrin-repeat HDAC proteins, we propose that the ankyrin-repeat-containing HDAC-like proteins represent a newly defined subclass of class II HDACs.
Fig. 1.
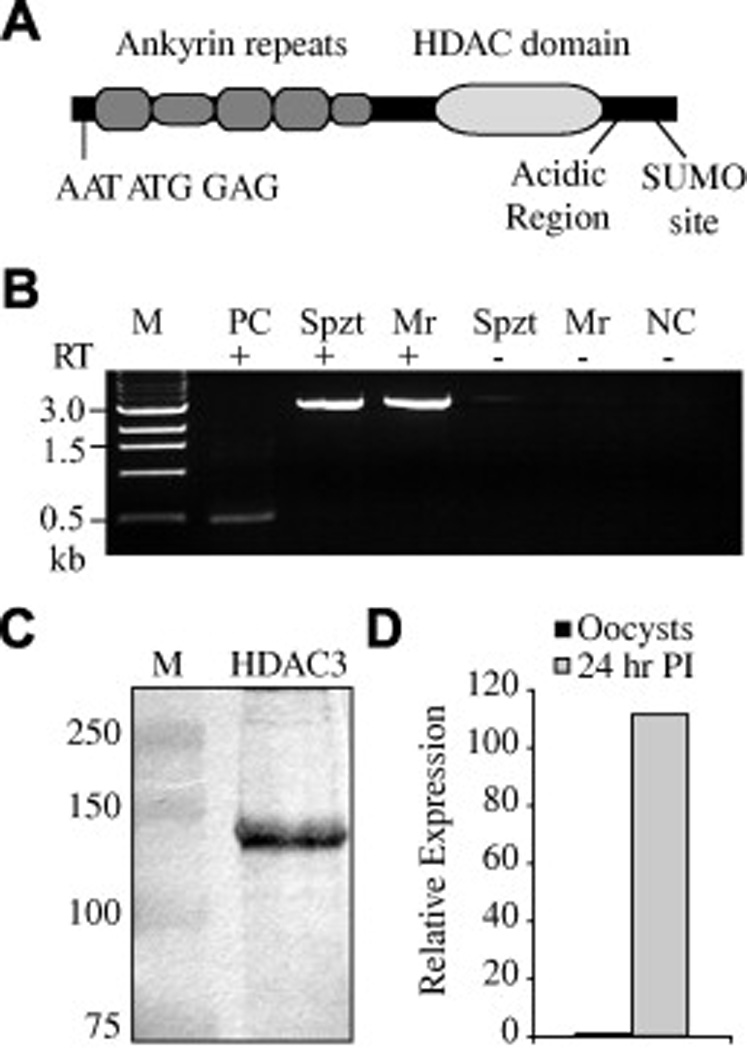
The Cryptosporidium parvum ankyrin histone deacetylase. (A) Schematic of the ankyrin-containing histone deacetylase (HDAC) from the parasitic protozoa C. parvum. Well conserved ankyrin repeats are presented as taller than poorly conserved or partial repeats. The C. parvum open reading frame contains a second, in-frame, initiation sequence AATATGGAG, an acidic region and a putative small ubiquitin-like modifier site that could cause slower than expected migrations on SDS–PAGE gels. (B) Reverse-transcriptase (RT) PCR of Sporozoite (Spzt) and developing meront (Mr) RNA (24 h p.i.). The predicted size of the open reading frame is 2.9 kb. Controls without RT, an unrelated 0.5 kb RT-PCR positive control (PC) and a negative no-template PCR control (NC) were also included. Size estimates for the marker (M) are to the left of the image. (C) Western blot of sporozoite proteins showing a band at approximately the predicted size (~108 kDa) for CpHDAC3; M is the marker lane with size estimates given at the left. (D) Quantitative PCR of parasite cDNA from oocysts and intracellular parasites indicated a strong up-regulation of CpHDAC3 transcript during intracellular development.
3.2. Developmentally regulated expression
In an effort to determine the possible roles these proteins may play in apicomplexan parasites, the ankyrin HDAC from C. parvum (CpHDAC3) was examined further. Consistent with the majority of C. parvum loci, reverse transcriptase PCR indicated that the 2.9 kb coding region does not contain introns (Fig. 1B). Multiple Western blots of sporozoite proteins showed the presence of a clear band approximate to the predicted size of CpHDAC3 (Fig. 1C). Examination of the CpHDAC3 predicted open reading frame indicates that it contains two nearly identical in-frame sequences: AAATATGGAC and AAATATGGAG (each putative initiation codon is underlined) that conform to the Kozak consensus (Kozak, 1987). These start sites follow the “N-end rule” (M followed by A, D or E) observed in the related apicomplexan Toxoplasma gondii and may be required for efficient translation initiation (Matrajt et al., 2002). Although not clear at the present time, it is possible that two alternate translation start sites may be used by the parasite. While CpHDAC3 expression could be detected by Western blots of bulked sporozoite proteins, the level of expression for these proteins within individual sporozoites was below the limits of detection when examined by immunofluorescence microscopy (Fig. 2A).
Fig. 2.
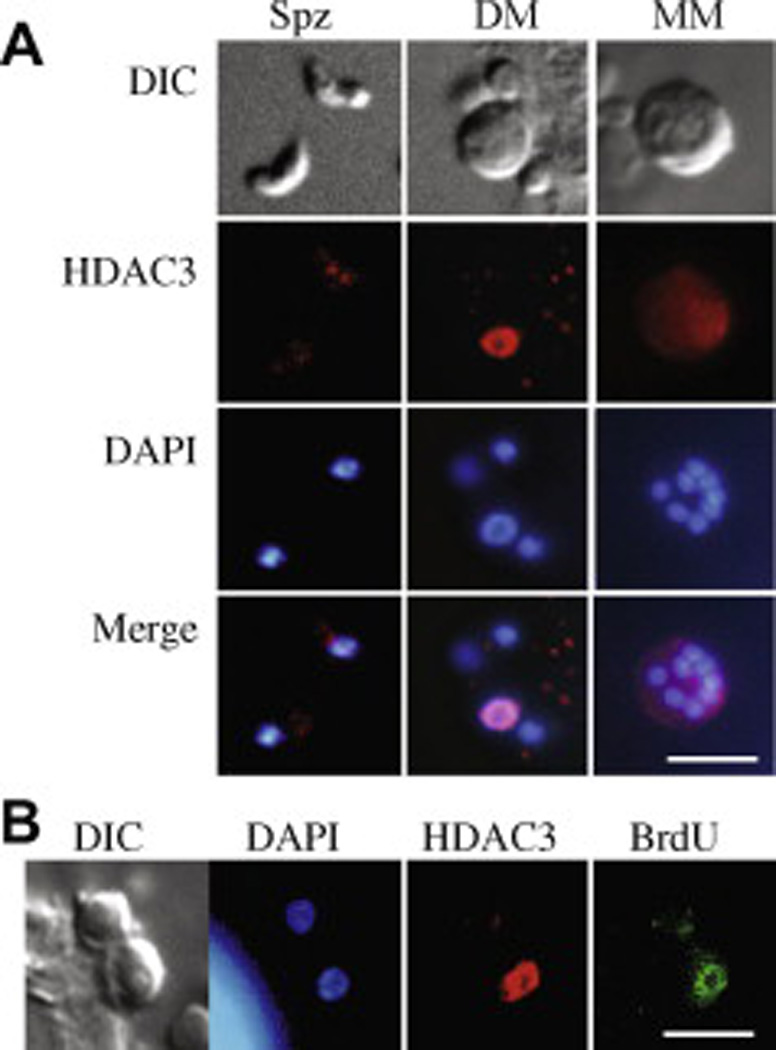
Immunolocalisation of Cryptosporidium parvum histone deacetylase 3 (CpHDAC3). (A) Different life stages include sporozoites (Spz), developing meronts (DM) and a mature type I meront (MM). Developing meronts whose nucleus adopted a toroidal appearance expressed CpHDAC3 at high levels and this signal overlapped that of the DAPI stain (evident in the merged images). (B) Immunolocalisation of recently incorporated bromodeoxyuridine (BrdU, green) indicated that DNA replication occurred in the meront that displayed high levels of CpHDAC3 but not the adjacent meront. The bright signal in the lower left corner of the DAPI panel was from a human host cell. DIC = differential interference contrast. The white bars represent 5 µm.
Cryptosporidium undergoes a complex replication process. In the initial phase of replication after invasion (called type I merogony) a single sporozoite produces an unmistakable mature meront containing eight merozoites. This regulated process includes obvious increases in parasite size, as well as morphological alterations and gross changes in nuclear architecture. During intracellular development with HCT8 host cells, CpHDAC3 was up-regulated at the transcript level (Fig. 1D). Regulation was also evident at the protein level as indirect immunofluorescence microscopy revealed that CpHDAC3 is highly expressed and associated with the nucleus in a developmentally controlled manner (Fig. 2A). Specifically, while CpHDAC3 is near background levels in most developmental stages, high-level CpHDAC3 expression is restricted to parasites whose DAPI-stained nucleus appears toroidal-shaped due to an enlarged nucleolus (Fig. 2A). Previous studies indicated that DNA replication occurs near this time in type I meronts (Rider and Zhu, 2008). Examination of BrdU that was incorporated into DNA during genome replication further revealed that CpHDAC3 was associated with recently replicated parasite DNA (Fig. 2B).
3.3. Biochemical functions
To determine whether CpHDAC3 could function as an HDAC, we examined the biochemical functions of a recombinant MBP tagged version of the CpHDAC3 protein (Fig. 3). Concordant with its predicted activity, recombinant CpHDAC3 functioned in vitro as a HDAC. It acted as an amidohydrolase that removed acetyl groups from acetylated lysine residues present in synthetic fluorogenic peptides (Fig. 3B). Dose–response curves of recombinant proteins with the tag removed (CpHDAC3) were not significantly different from those generated by the intact (uncut) MBP-CpHDAC3 fusion protein (Fig. 3C), indicating that CpHDAC3 was not affected by the presence of the MBP tag. The Km for MBP-CpHDAC3 and CpHDAC3 were 61 ± 3.4 µM and 65 ± 5.0 µM, respectively, while the kcat for the respective proteins were 0.25 ± 0.010 s−1 and 0.23 ± 0.010 s−1. These values are within the range reported for the human class II proteins HDAC6 and HDAC10 using a similar substrate (Schultz et al., 2004).
Fig. 3.
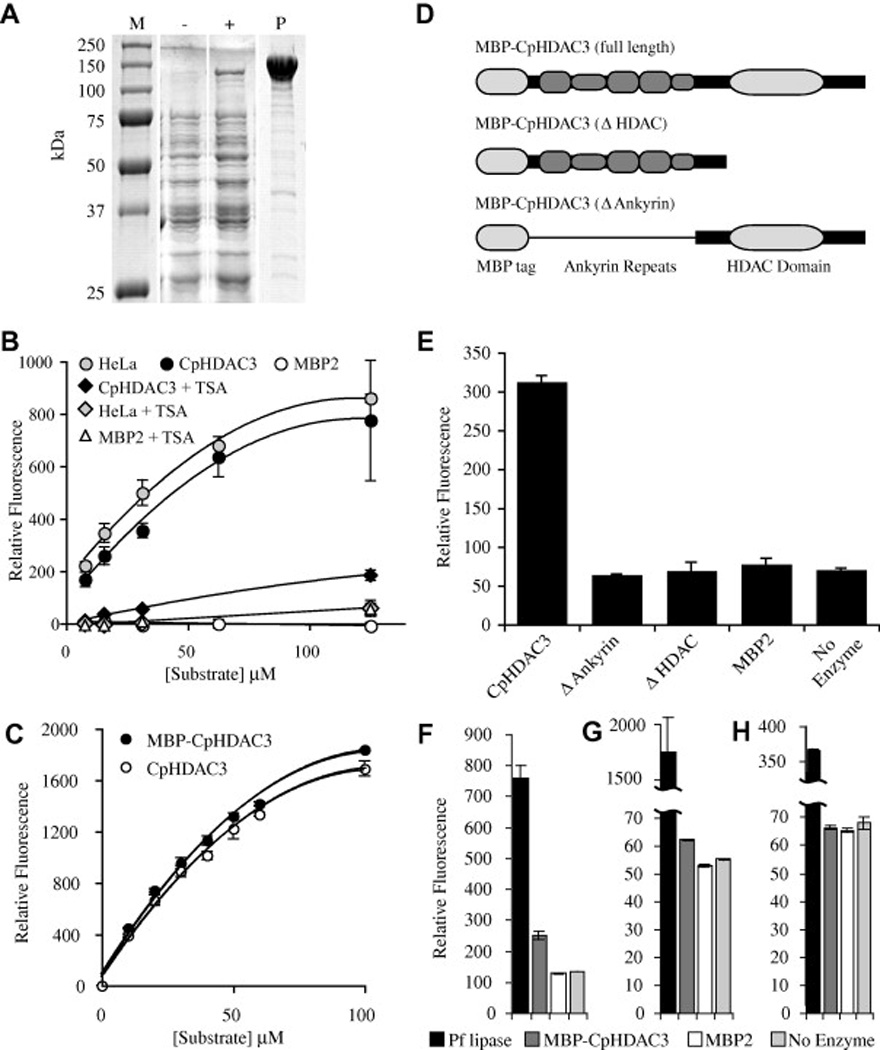
Activity of recombinant Cryptosporidium parvum histone deacetylase 3 (CpHDAC3). (A) Protein gel of recombinant CpHDAC3. Lane M contains protein marker with sizes indicated to the left of the figure. Whole cell lysates are presented for bacteria harbouring an expression plasmid encoding a maltose binding protein fusion to CpHDAC3 before (−) and after (+) induction with isopropyl β-d-1-thiogalactopyranoside. Lane P contains affinity-purified recombinant maltose binding protein (MBP)-CpHDAC3. (B) In vitro assays with a fluorogenic HDAC substrate were compared with human HeLa cell nuclear extract as a positive control and MBP2 as a negative control. With the broad-spectrum HDAC inhibitor trichostatin A, CpHDAC3 was suppressible to levels near that of a no-enzyme control. (C) Comparison of equimolar amounts of MBP-CpHDAC3 fusion (uncut) and CpHDAC3 (cut) indicated equivalent activities for both proteins. Kinetic data are presented in the main text. (D) Schematic representations of full-length and truncated MBP fusions of CpHDAC3. A thin line running through the N-terminal truncation is to indicate that the MBP is linked to the C-terminal region containing only the HDAC domain. (E) HDAC Activity of MBP-CpHDAC3 and truncated versions together with MBP2 and no-enzyme negative controls. For simplicity, only a single concentration of substrate is presented (10 µM). (F–H) In vitro assays with (F) 250 µM 4-methylumbelliferyl acetate, (G) 4-methylumbelliferyl propionate or (H) 4-methylumbelliferyl butyrate as esterase substrates compared with Pseudomonas fluorescens lipase (Pf lipase) as a positive control. MBP-CpHDAC3 displayed activity against acetyl and propionyl esters (but not butyryl esters) that was significantly higher than MBP2 or no-enzyme controls. Kinetic data are presented in Table 1.
In an effort to shed light on the roles of the ankyrin repeats in the function of the CpHDAC3 protein, deletion mutants were generated that lacked either the HDAC domain, or that lacked the ankyrin repeats (Fig. 3D). Although MBP-CpHDAC3 possessed intrinsic HDAC activity, neither deletion protein was able to deacetylate the fluorogenic HDAC substrate (Fig. 3E). This suggests that both the ankyrin repeats and the deacetylase domain are necessary for proper enzymatic function of the CpHDAC3 protein. Further, it revealed that neither domain alone is sufficient to produce the observed HDAC activity of the intact CpHDAC3.
Several HDAC-like proteins have recently been shown to possess significant esterase activity with specificity for esters of different chain lengths (Moreth et al., 2007). In vitro, recombinant CpHDAC3 was also capable of functioning as an esterase when acting on synthetic fluorogenic substrates (Fig. 3F–H). As an ester hydrolase, this protein acted on acetyl and propionyl, but not on butyryl esters. However, the kcat for CpHDAC3 esterase activity was over 100 times lower than that of a control esterase from P. fluorescens (Pf lipase; Table 1), and potential endogenous and physiologically relevant ester substrates remain undiscovered. As recombinant CpHDAC3 esterase activity was weak and native CpHDAC3 was nuclear-localised, we sought to identify a potential chromatin-related function for this protein.
Table 1.
Kinetics of Cryptosporidium parvum histone deacetylase 3 (CpHDAC3) esterase activity compared with that of Pseudomonas fluorescens lipase (Pf lipase).
| Enzyme | Acetyl esterase | Propionyl esterase | Butyryl esterase | |||
|---|---|---|---|---|---|---|
| Km (µM) | kcat (s−1) | Km (µM) | kcat (s−1) | Km (µM) | kcat (s−1) | |
| Pf lipase | 230 ± 48 | 1.4 ± 0.079 | 710 ± 390 | 9.6 ± 4.1 | 260 ± 30 | 0.85 ± 0.026 |
| CpHDAC3 | 89 ± 18 | 0.010 ± 0.00089 | 77 ± 30 | 0.00058 ± 0.000092 | Undetectable | Undetectable |
In many organisms, newly synthesised histones are acetylated at multiple lysines before they are incorporated into chromatin during genome replication (Sobel et al., 1995, Ma et al., 1998 and Dion et al., 2005). This essential replication-associated acetylation pattern is removed by HDACs once DNA replication is completed (Allis et al., 1985). To determine whether CpHDAC3 could participate in this replication-associated activity, we examined recombinant CpHDAC3 activity against H4 peptides. H4 was chosen due to the perfect conservation between C. parvum H4 tails and the H4 tails from other organisms (the other C. parvum histones are less well conserved). Competition studies were performed using synthetic peptides corresponding to the N-terminal tail of histone H4. The synthetic peptides successfully competed against a fluorogenic HDAC substrate if they contained acetylated lysines, while control peptides without acetylated lysine showed no effect. Moreover, lysine acetylation at either position 8 or 12 was sufficient for competition, but lysine acetylation at position 5 or 16 had no effect (Fig. 4A). Thus, recombinant CpHDAC3 displays in vitro specificity toward acetylated lysines 8 and 12 of histone H4 tails.
Fig. 4.
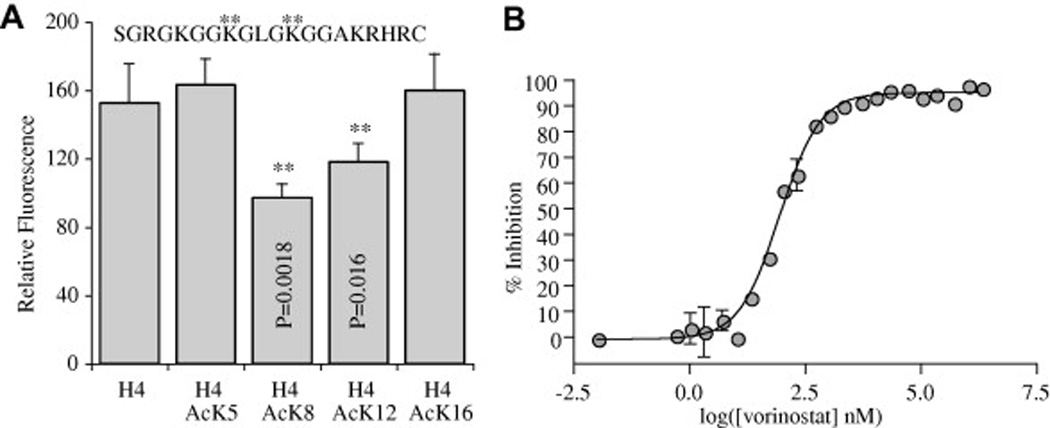
Substrate and inhibitor assays. (A) Competition assays with H4 peptides (15 µM) acetylated at positions 8 and 12 (asterisks) competed with the fluorogenic substrate (5 µM) and significantly reduced the fluorescence compared with unacetylated H4 peptide. The peptide sequence used was identical to the Cryptosporidium parvum H4 tail and is presented at the top of the figure. Asterisks indicate sites preferred by the maltose binding protein fusion of Cryptosporidium parvum histone deacetylase 3 (MBP-CpHDAC3). (B) Dose–response curve for vorinostat inhibition of MBP-CpHDAC3. Error bars are SD, n = 2.
3.4. Pharmacological inhibitors of CpHDAC3
Several HDAC inhibitors have been shown to have anti-apicomplexan activity (Darkin-Rattray et al., 1996, Andrews et al., 2000, Isaka et al., 2000, Jones-Brando et al., 2003, Kwon et al., 2003, Gurvich et al., 2004, Mai et al., 2004 and Saksouk et al., 2005). To determine whether any of the anti-apicomplexan HDAC inhibitors may be targeting the ankyrin-repeat-containing class of HDAC, we tested those inhibitors against recombinant CpHDAC3 in vitro. Many of the compounds tested displayed strong inhibition (nanomolar IC50) of CpHDAC3 deacetylase activity (Table 2). Other compounds known to be anti-parasitic and to disrupt HDAC activity showed no inhibitory effects on recombinant CpHDAC3 (eg. valproic acid). Thus, some broad-spectrum HDAC inhibitors that were previously established as having anti-apicomplexan activity are capable of disrupting CpHDAC3 function. Additionally, CpHDAC3 was validated as a target of vorinostat (Fig. 4B and Table 2), an HDAC inhibitor that has been successfully used as a drug in humans (Mann et al., 2007).
Table 2.
Quantitative data for inhibitors used against the Cryptosporidium parvum histone deacetylase 3 protein.
| Compound | Class | IC50 in nM | Maximum inhibition |
|---|---|---|---|
| APHA 8 | Hydroxamic acid | 310 ± 1.2 | 99% |
| Apicidin | Cyclic peptide | 2100 ± 2.9 | 63% |
| Butyrate | Fatty acid | Undetectable | Undetectable |
| Depudecin | Epoxide | 86 ± 2.4 | 47% |
| Trichostatin A | Hydroxamic acid | 18 ± 1.3 | 97% |
| Valproic acid | Fatty acid | Undetectable | Undetectable |
| Vorinostat | Hydroxamic acid | 89 ± 7.9 | 96% |
4. Discussion
Although the existence of few fully sequenced genomes limits current analyses, the presence of the ankyrin-repeat-containing HDACs in chromalveolates (apicomplexans, diatoms and coccolithophores) and primitive chlorophyte algae (prasinophytes and trebouxiophytes), together with the apparent lack of similar proteins in rhodophyte algae and higher plants are compelling. This data suggests that continued examination of genes such as the ankyrin HDAC could provide an opportunity to add significant additional clues regarding the early evolution of the apicomplexan lineage and the complex relationships between apicomplexans and algae.
The CpHDAC3 protein from parasites consistently runs between what we believe to be approximately 100 and 150 kDa, and although slightly larger than expected, is in reasonable agreement with the predicted protein size. SDS–PAGE is capable of only approximating protein mass. This does not preclude the possibility of an error in judging the identity of the 5′ and 3′ ends of the gene or that anomalously slow migration is due to post-translational modifications, materials present in the parasite-derived sample that are absent from the marker or for other reasons.
A considerable body of the literature exists indicating that some proteins do migrate anomalously slowly without post-translational modifications. We examined the predicted sequence for anything that might cause a shift similar to what is observed on protein blots. It is unlikely to be due to glycosylation since the predicted protein does not contain a predicted signal peptide and the protein is at least nuclearly localised in the parasite. However, the predicted protein contains two regions that may explain slightly slower migrations. One is an extended acidic region that is also hydrophilic (these have been shown to cause larger than expected apparent masses) and one is a putative small ubiquitin-like modifier (SUMO) modification site very near the C-terminus (Graceffa et al., 1992 and Xue et al., 2006). If this modification occurs in C. parvum, it would add about 13 kDa to the mass of the protein.
The biochemical evidence presented herein is consistent with the hypothesis that CpHDAC3 is targeted to chromatin to facilitate the removal of acetyl groups from newly deposited H4 as DNA replication is completed. However, this speculation does not preclude the possibility that the protein may function in various contexts as a transcriptional regulator or as an ester hydrolase. It is anticipated that additional experimentation will identify new roles for this protein throughout the C. parvum life cycle. The deletion mutants of the recombinant CpHDAC3 indicate a need for the ankyrin repeats in the function of this protein. This could be something as simple as protein misfolding due to the truncation. However, further work beyond the scope of the present study, together with additional mutant proteins, will be needed to elucidate the function of the ankyrin repeats.
Because continuous cultivation, routine axenic growth and genetic manipulation of C. parvum are beyond current technologies, biochemical comparisons with ankyrin-containing HDACs from other species may be needed to reveal conserved functions among these proteins. This could have important implications in the drug development process, as CpHDAC3 inhibitors that are effective against cryptosporidiosis might be useful against other apicomplexan parasites.
The compounds tested against CpHDAC3, including SAHA, are broad-spectrum inhibitors that can affect human HDAC activity. Thus, there is a need for inhibitors specific to the ankyrin-repeat-containing HDAC family. Such inhibitors will be useful in teasing out the contribution of the ankyrin-repeat-containing HDAC proteins to apicomplexan biology.
CpHDAC3 is unlike the other putative HDACs encoded by either the C. parvum or human genomes and its highly divergent HDAC domain makes it an attractive candidate for future drug discovery efforts. The differential inhibition of CpHDAC3 by diverse compounds suggests that identifying CpHDAC3-specific inhibitors with anti-cryptosporidial activity is a plausible route toward developing novel anti-cryptosporidial chemotherapies.
Acknowledgements
This research was kindly funded by the National Institutes of Health through the National Institute of Allergy and Infectious Diseases with Grant AI068461 to S.D.R. and G.Z. The authors also thank Xiaomin Cai for technical assistance and Dr. Heather A. Hostetler for early comments on the article.
References
- Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science. 2004;304:441–445. doi: 10.1126/science.1094786. Abrahamsen et al. 2004. [DOI] [PubMed] [Google Scholar]
- Abubakar I, Aliyu SH, Arumugam C, Hunter PR, Usman NK. Prevention and treatment of cryptosporidiosis in immunocompromised patients. Cochrane Database Syst. Rev. 2007;1:CD004932. doi: 10.1002/14651858.CD004932.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis CD, Chicoine LG, Richman R, Schulman IG. Deposition-related histone acetylation in micronuclei of conjugating tetrahymena. Proc. Natl. Acad. Sci. USA. 1985;82:8048–8052. doi: 10.1073/pnas.82.23.8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Andrews KT, Walduck A, Kelso MJ, Fairlie DP, Saul A, Parsons PG. Anti-malarial effect of histone deacetylation inhibitors and mammalian tumour cytodifferentiating agents. Int. J. Parasitol. 2000;30:761–768. doi: 10.1016/s0020-7519(00)00043-6. [DOI] [PubMed] [Google Scholar]
- Barbier G, Oesterhelt C, Larson MD, Halgren RG, Wilkerson C, Garavito RM, Benning C, Weber AP. Comparative genomics of two closely related unicellular thermo-acidophilic red algae, Galdieria sulphuraria and Cyanidioschyzon merolae, reveals the molecular basis of the metabolic flexibility of Galdieria sulphuraria and significant differences in carbohydrate metabolism of both algae. Plant Physiol. 2005;137:460–474. doi: 10.1104/pp.104.051169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard JL, Hicks JS. The non-photosynthetic plastid in malarial parasites and other apicomplexans is derived from outside the green plastid lineage. J. Eukaryot. Microbiol. 1999;46:367–375. doi: 10.1111/j.1550-7408.1999.tb04615.x. [DOI] [PubMed] [Google Scholar]
- Cai X, Fuller AL, McDougald LR, Zhu G. Apicoplast genome of the coccidian Eimeria tenella. Gene. 2003;321:39–46. doi: 10.1016/j.gene.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Daily JP. Antimalarial drug therapy: the role of parasite biology and drug resistance. J. Clin. Pharmacol. 2006;46:1487–1497. doi: 10.1177/0091270006294276. [DOI] [PubMed] [Google Scholar]
- Dangl M, Brosch G, Haas H, Loidl P, Lusser A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta. 2001;213:280–285. doi: 10.1007/s004250000506. [DOI] [PubMed] [Google Scholar]
- Darkin-Rattray SJ, Gurnett AM, Myers RW, Dulski PM, Crumley TM, Allocco JJ, Cannova C, Meinke PT, Colletti SL, Bednarek MA, Singh SB, Goetz MA, Dombrowski AW, Polishook JD, Schmatz DM. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc. Natl. Acad. Sci. USA. 1996;93:13143–13147. doi: 10.1073/pnas.93.23.13143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. USA. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fast NM, Kissinger JC, Roos DS, Keeling PJ. Nuclear-encoded, plastid-targeted genes suggest a single common origin for apicomplexan and dinoflagellate plastids. Mol. Biol. Evol. 2001;18:418–426. doi: 10.1093/oxfordjournals.molbev.a003818. [DOI] [PubMed] [Google Scholar]
- Frye RA. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000;273:793–798. doi: 10.1006/bbrc.2000.3000. [DOI] [PubMed] [Google Scholar]
- Funes S, Davidson E, Reyes-Prieto A, Magallon S, Herion P, King MP, Gonzalez-Halphen D. A green algal apicoplast ancestor. Science. 2002;298:2155. doi: 10.1126/science.1076003. [DOI] [PubMed] [Google Scholar]
- Graceffa P, Jancso A, Mabuchi K. Modification of acidic residues normalizes sodium dodecyl sulfate–polyacrylamide gel electrophoresis of caldesmon and other proteins that migrate anomalously. Arch. Biochem. Biophys. 1992;297:46–51. doi: 10.1016/0003-9861(92)90639-e. [DOI] [PubMed] [Google Scholar]
- Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J. Mol. Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Alford WJ, Pietrokovski S. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 1995;163:GC17–GC26. doi: 10.1016/0378-1119(95)00486-p. [DOI] [PubMed] [Google Scholar]
- Isaka M, Jaturapat A, Kladwang W, Punya J, Lertwerawat Y, Tanticharoen M, Thebtaranonth Y. Antiplasmodial compounds from the wood-decayed fungus Xylaria sp. BCC 1067. Planta Med. 2000;66:473–475. doi: 10.1055/s-2000-8588. [DOI] [PubMed] [Google Scholar]
- Iyer L, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Jones-Brando L, Torrey EF, Yolken R. Drugs used in the treatment of schizophrenia and bipolar disorder inhibit the replication of Toxoplasma gondii. Schizophr. Res. 2003;62:237–244. doi: 10.1016/s0920-9964(02)00357-2. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. Reduction and compaction in the genome of the apicomplexan parasite Cryptosporidium parvum. Dev. Cell. 2004;6:614–616. doi: 10.1016/s1534-5807(04)00135-2. [DOI] [PubMed] [Google Scholar]
- Kim TY, Bang YJ, Robertson KD. Histone deacetylase inhibitors for cancer therapy. Epigenetics. 2006;1:14–23. doi: 10.4161/epi.1.1.2644. [DOI] [PubMed] [Google Scholar]
- Kohler S, Delwiche CF, Denny PW, Tilney LG, Webster P, Wilson RJ, Palmer JD, Roos DS. A plastid of probable green algal origin in apicomplexan parasites. Science. 1997;275:1485–1489. doi: 10.1126/science.275.5305.1485. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HJ, Kim JH, Kim M, Lee JK, Hwang WS, Kim DY. Anti-parasitic activity of depudecin on Neospora caninum via the inhibition of histone deacetylase. Vet. Parasitol. 2003;112:269–276. doi: 10.1016/s0304-4017(03)00035-9. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Wu J, Altheim BA, Schultz MC, Grunstein M. Deposition-related sites K5/K12 in histone H4 are not required for nucleosome deposition in yeast. Proc. Natl. Acad. Sci. USA. 1998;95:6693–6698. doi: 10.1073/pnas.95.12.6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai A, Cerbara I, Valente S, Massa S, Walker LA, Tekwani BL. Antimalarial and antileishmanial activities of aroyl-pyrrolyl-hydroxyamides, a new class of histone deacetylase inhibitors. Antimicrob. Agents Chemother. 2004;48:1435–1436. doi: 10.1128/AAC.48.4.1435-1436.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- Matrajt M, Nishi M, Fraunholz MJ, Peter O, Roos DS. Amino-terminal control of transgenic protein expression levels in Toxoplasma gondii. Mol. Biochem. Parasitol. 2002;120:285–289. doi: 10.1016/s0166-6851(02)00014-2. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, Misumi O, Shin IT, Maruyama S, Takahara M, Miyagishima SY, Mori T, Nishida K, Yagisawa F, Nishida K, Yoshida Y, Nishimura Y, Nakao S, Kobayashi T, Momoyama Y, Higashiyama T, Minoda A, Sano M, Nomoto H, Oishi K, Hayashi H, Ohta F, Nishizaka S, Haga S, Miura S, Morishita T, Kabeya Y, Terasawa K, Suzuki Y, Ishii Y, Asakawa S, Takano H, Ohta N, Kuroiwa H, Tanaka K, Shimizu N, Sugano S, Sato N, Nozaki H, Ogasawara N, Kohara Y, Kuroiwa T. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- McFadden DC, Camps M, Boothroyd JC. Resistance as a tool in the study of old and new drug targets in Toxoplasma. Drug Resist. Updat. 2001;4:79–84. doi: 10.1054/drup.2001.0184. [DOI] [PubMed] [Google Scholar]
- Mead JR. Cryptosporidiosis and the challenges of chemotherapy. Drug Resist. Updat. 2002;5:47–57. doi: 10.1016/s1368-7646(02)00011-0. [DOI] [PubMed] [Google Scholar]
- Moreth K, Riester D, Hildmann C, Hempel R, Wegener D, Schober A, Schwienhorst A. An active site tyrosine residue is essential for amidohydrolase but not for esterase activity of a class 2 histone deacetylase-like bacterial enzyme. Biochem. J. 2007;401:659–665. doi: 10.1042/BJ20061239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider SD, Jr., Cai X, Sullivan WJ, Jr., Smith AT, Radke J, White M, Zhu G. The protozoan parasite Cryptosporidium parvum possesses two functionally and evolutionarily divergent replication protein A large subunits. J. Biol. Chem. 2005;280:31460–31469. doi: 10.1074/jbc.M504466200. [DOI] [PubMed] [Google Scholar]
- Rider SD, Jr., Zhu G. Differential expression of the two distinct replication protein A subunits from Cryptosporidium parvum. J. Cell. Biochem. 2008;104:2207–2216. doi: 10.1002/jcb.21784. [DOI] [PubMed] [Google Scholar]
- Roos DS, Crawford MJ, Donald RG, Kissinger JC, Klimczak LJ, Striepen B. Origin, targeting, and function of the apicomplexan plastid. Curr. Opin. Microbiol. 1999;2:426–432. doi: 10.1016/S1369-5274(99)80075-7. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ, Jr., Cesbron-Delauw MF, Hakimi MA. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol. Cell. Biol. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz BE, Misialek S, Wu J, Tang J, Conn MT, Tahilramani R, Wong L. Kinetics and comparative reactivity of human class I and class IIb histone deacetylases. Biochemistry. 2004;43:11083–11091. doi: 10.1021/bi0494471. [DOI] [PubMed] [Google Scholar]
- Snelling WJ, Lin Q, Moore JE, Millar BC, Tosini F, Pozio E, Dooley JS, Lowery CJ. Proteomics analysis and protein expression during sporozoite excystation of Cryptosporidium parvum (coccidia, apicomplexa) Mol. Cell. Proteomics. 2007;6:346–355. doi: 10.1074/mcp.M600372-MCP200. [DOI] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Striepen B, Pruijssers AJ, Huang J, Li C, Gubbels MJ, Umejiego NN, Hedstrom L, Kissinger JC. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc. Natl. Acad. Sci. USA. 2004;101:3154–3159. doi: 10.1073/pnas.0304686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton SJ, Tilley M, Brillhart DB. Effects of select medium supplements on in vitro development of Cryptosporidium parvum in HCT-8 cells. J. Clin. Microbiol. 1995;33:371–375. doi: 10.1128/jcm.33.2.371-375.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Gaber RF. RPD3 encodes a second factor required to achieve maximum positive and negative transcriptional states in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:6317–6327. doi: 10.1128/mcb.11.12.6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller RF, Keeling PJ, van Dooren GG, McFadden GI. Comment on “A green algal apicoplast ancestor”. Science. 2003;301:49. doi: 10.1126/science.1084684. [DOI] [PubMed] [Google Scholar]
- Williams RB. Tracing the emergence of drug-resistance in coccidia (Eimeria spp.) of commercial broiler flocks medicated with decoquinate for the first time in the United Kingdom. Vet. Parasitol. 2006;135:1–14. doi: 10.1016/j.vetpar.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Gardner MJ, Preiser P, Moore DJ, Rangachari K, Wilson RJ. The evolutionary origin of the 35 kb circular DNA of Plasmodium falciparum: new evidence supports a possible rhodophyte ancestry. Mol. Gen. Genet. 1994;243:249–252. doi: 10.1007/BF00280323. [DOI] [PubMed] [Google Scholar]
- Xue Y, Zhou F, Fu C, Xu Y, Yao X. SUMOsp: a web server for sumoylation site prediction. Nucleic Acids Res. 2006;34:W254–W257. doi: 10.1093/nar/gkl207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Marchewka MJ, Keithly JS. Cryptosporidium parvum appears to lack a plastid genome. Microbiology. 2000;146:315–321. doi: 10.1099/00221287-146-2-315. [DOI] [PubMed] [Google Scholar]


