Abstract
Long intergenic noncoding RNAs (lincRNAs) are one of the major unexplored components of genomes. Here we re-analyzed a published methylated DNA immunoprecipitation sequencing (MeDIP-seq) dataset to characterize the DNA methylation pattern of pig lincRNA genes in adipose and muscle tissues. Our study showed that the methylation level of lincRNA genes was higher than that of mRNA genes, with similar trends observed in comparisons of the promoter, exon or intron regions. Different methylation pattern were observed across the transcription start sites (TSS) of lincRNA and protein-coding genes. Furthermore, an overlap was observed between many lincRNA genes and differentially methylated regions (DMRs) identified among different breeds of pigs, which show different fat contents, sexes and anatomic locations of tissues. We identify a lincRNA gene, linc-sscg3623, that displayed differential methylation levels in backfat between Min and Large White pigs at 60 and 120 days of age. We found that a demethylation process occurred between days 150 and 180 in the Min and Large White pigs, which was followed by remethylation between days 180 and 210. These results contribute to our understanding of the domestication of domestic animals and identify lincRNA genes involved in adipogenesis and muscle development.
Long noncoding RNAs (lncRNAs) are a class of transcripts that are longer than 200 nt in length and do not encode proteins. Similar to mRNAs, lncRNAs are transcribed by RNA polymerase II and undergo splicing and polyadenylation. LncRNAs can be classified into antisense transcripts, long intronic noncoding RNAs and long intergenic noncoding RNAs (lincRNAs), according to their position relative to protein-coding genes. Some lincRNAs have been indicated to play important roles in a variety of biological processes, such as dosage compensation1,2,3,4, transcriptional regulation5,6,7, epigenetic regulation8,9 and pluripotency maintenance10. Previous studies have demonstrated that lincRNAs play a role in adipogenesis11,12 and muscle development13.
The pig is an emerging medical model for studying energy metabolism and obesity in humans, since both possess similar cardiovascular systems, metabolic features, and proportional organ size14. Thousands of years of selection on pigs have created abundant phenotypic variation, for example, different pig breeds show varying performance in adipose and lean meat production. Therefore, these breeds should be valuable models for studies of adipogenesis and muscle development.
In an earlier study, Li et al. performed an investigation on DNA methylation in eight different adipose and two distinct skeletal muscle tissues from three breeds of pig15, where they found differentially methylated regions in the promoters of the protein-coding genes are highly associated with the development of obesity15. In a different study, we identified 6,621 lincRNAs encoded by 4,515 gene loci in the pig genome16. Combining data from these two studies provides an opportunity to study the DNA methylation of lincRNAs loci in adipose and muscle tissues.
Here, we investigated the genome-wide levels of DNA methylation for lincRNA genes in adipose and muscle tissues from three breeds: Landrace, Rongchang and Tibetan. Comparison of the methylation patterns observed in protein-coding and lincRNA genes identified several distinctive methylation characteristics that differ between these classes of genes. We also analyzed differentially methylation regions (DMRs) that overlap lincRNA genes. This study contributes to our understanding of the DNA methylation of lincRNA genes and provides a valuable resource for the functional studies of lincRNAs that are associated with adipogenesis and muscle development.
Result
Global patterns of DNA methylation in lncRNA genes
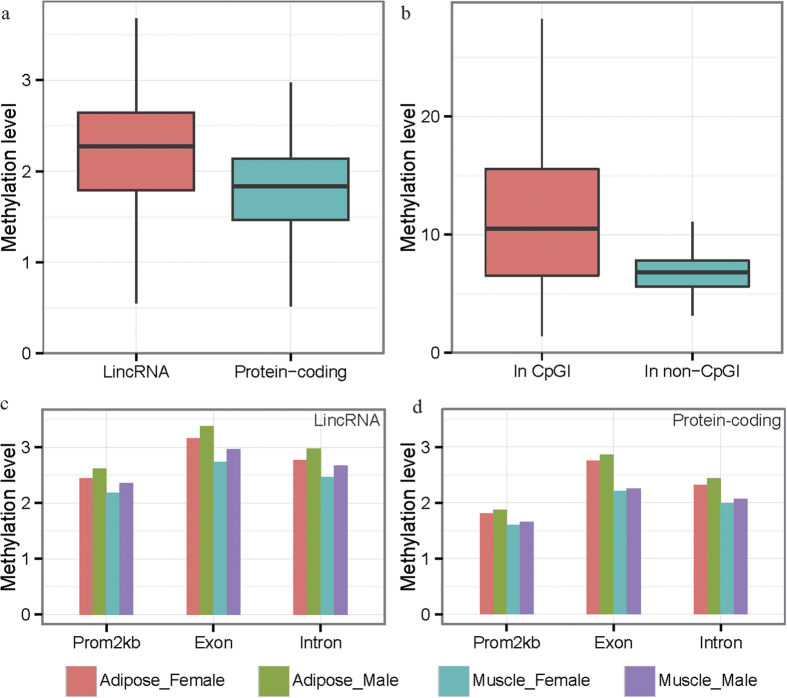
In our previous study, we found pig lincRNA genes have several characteristics which differ from those of mRNA genes such as their length, number of exons and level of expression16. Here, we found the GC content (0.37) and observed-over-expected number of CpG (CpGo/e) ratio (0.26) of lincRNA genes are similar to those of protein-coding genes (GC content: 0.38, CpGo/e: 0.29) in the genomic regions that span from 2 kb upstream of the transcription start sites (TSS) to the transcription end site (TES) for both of these types of genes. However, the methylation levels of the lincRNA genomic regions were significantly higher than that for the mRNA genes (Kolmogorov-Smirnov test, P = 2.621 × 10−10, Fig. 1a), which indicates that there is a differential methylation pattern between protein-coding and lincRNA genes. The results may be due to differential methylation regulation mechanisms between lincRNA and protein-coding genes.
Figure 1. DNA methylation levels in lincRNA and protein-coding genes.
(a) Box plots for the comparison of DNA methylation levels between lincRNA and protein-coding genes. (b) Methylation levels of CpG sites in CpGIs and non-CpGIs for lincRNA genes. (c) Average methylation levels within the promoters, exons, and introns of lincRNA genes. Different bins represent adipose in females (Adipose_Female), and males (Adipose_Male), and muscle in females (Muscle_Female) and males (Muscle_Male). (d) Average methylation levels within the promoters, exons, and introns of protein-coding genes. Bins are defined as in (c). Methylation levels were normalized for read depth using an overall average amount of reads form the 180 samples.
Among the 768,645 CpG sites in the lincRNA genomic region (−2K upstream TSS to TES) dataset, 85,012 CpG sites are located in CpGIs (CpG islands) and the remaining 683,633 CpG sites are not in CpGIs (non-CpGIs). Differing from the previous result of a microarray study17, the average DNA methylation level across all samples at CpGs in CpGIs was significantly higher those of non-CpGIs (Kolmogorov-Smirnov test, p < 2.2 × 10−16, Fig. 1b).
Methylation levels of exons, introns and promoters were compared between lincRNA and protein-coding genes for adipose and muscle tissues of both female and male pigs. Our analysis found that the methylation levels of exons, introns and promoters of the lincRNA genes were always higher than those of mRNA genes (Kolmogorov-Smirnov test, P < 2.2 × 10–16, Supplementary Fig. S1 and Fig. 1c,d). Consistent with Sati et al.18, we found that the methylation pattern of exons, introns and promoters of lncRNA genes were similar to those of protein-coding genes, with exons having higher methylation levels than introns or promoters (Fig. 1c,d).
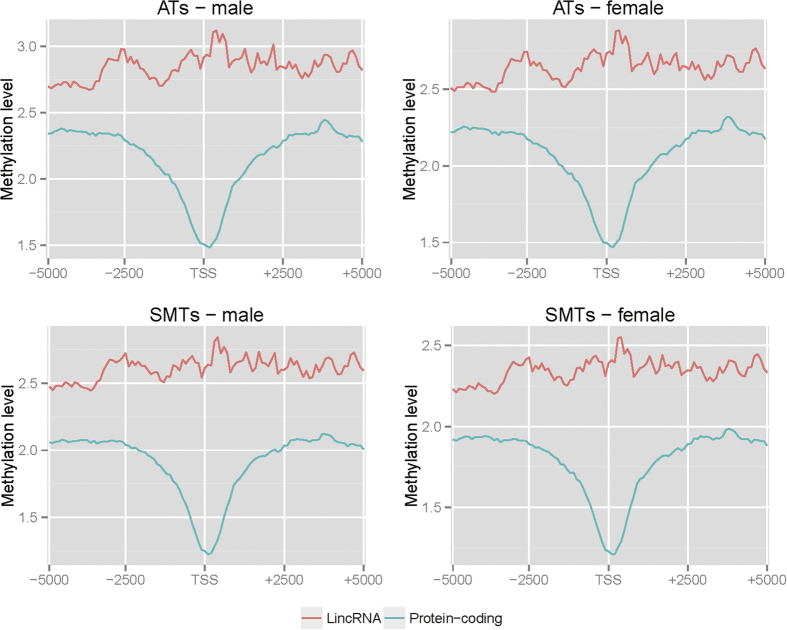
The patterns of DNA methylation across TSS of lincRNA genes
In humans, the methylation pattern across the TSS of lincRNAs is different from that of protein-coding genes18. In pigs, we found similar methylation patterns in male and female, for both adipose and muscle tissues, with the methylation level across the TSS of lincRNA genes being higher than for protein-coding genes (Fig. 2). The TSS of protein-coding genes showed a V-shaped curve for methylation level indicating a relative lowing of the methylation density (Fig. 2), in concordance with a previous report18. In contrast to mRNA genes, we found a slightly increased methylation level around the TSS of lncRNA genes (Fig. 2). In an earlier study18, Sati et al. found a sharp peak immediately downstream of the TSS of lincRNA genes in humans, whereas no similar results were found for the pig genes in this study. These observations indicate a different pattern of methylation occurs around the TSS of lincRNA and protein-coding genes. In this study, only lincRNA genes that are located at least 500bp away from a protein-coding or a house-keeping gene were included, to reduce the influence due to potential overlap between the exons of a lincRNA and a protein-coding gene.
Figure 2. DNA methylation patterns around the TSS of lincRNA and protein-coding genes.
Distribution of the methylation level was calculated in 100-bp sliding windows, 5-kb upstream and downstream from the TSS. ATs–male: male adipose tissues, ATs–female: female adipose tissues, SMTs–male: male skeletal muscle tissues, SMTs–female: female skeletal muscle tissues.
GC content and CpGo/e across TSSs were compared between pig lincRNA and protein-coding genes. Both lincRNA and protein-coding genes showed higher GC content and CpGo/e near their TSSs (Fig. 3). When this result is combined with our observations on the methylation levels around TSS, we concluded that the differential methylation pattern seen between lincRNA and mRNA genes is most likely due to differential regulation of DNA methylation, rather than nucleotide composition. The above observations could also partly explain why the majority of lincRNA genes show tissue-specific and developmental stage-specific expression.
Figure 3. Distribution of the GC content and CpGo/e ratio around the TSS of lincRNA and protein-coding genes.
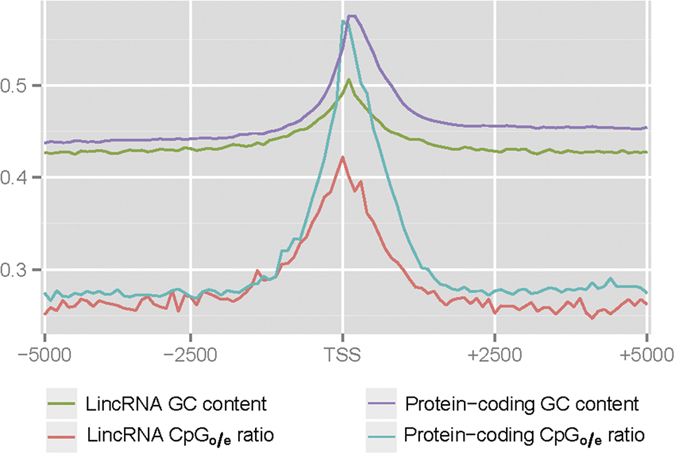
Distributions were calculated in 100-bp sliding windows, 5-kb upstream and downstream from the TSS.
DMRs located in lincRNA genes
The study by Li et al. on genome-wide DNA methylation levels was performed on three breeds of pigs (Landrace, Rongchang and Tibetan) that show different obesity and muscle-related phenotypes15. To study the regulation of adipocytes and muscle development, they adipose tissue (AT) was sampled from 8 diverse anatomic locations as well as two skeletal muscle tissues (SMT), white longissimus dorsi muscle (LDM) and red psoas major muscle (PMM)15. Phenotypic differences were seen in the sampled adipose and skeletal muscle tissues between breeds, sexes and anatomic locations15.
To identify lincRNA genes associated with adipocyte regulation and muscle development, we re-analyzed the methylation datasets from Li et al.15 using MEDIPS package19 and found a considerable number of differentially methylated regions (DMRs) that overlap with lincRNA gene regions (Table 1).
Table 1. Summary of differentially methylated regions (DMRs) identified by the MEDIPS software.
| DMRs type | Number of DMRs | DMRs in lincRNA regions |
|---|---|---|
| Adipose S-DMRs (n = 72 per sex) | 44,664 | 983 |
| Muscle S-DMRs (n = 18 per sex) | 4,861 | 109 |
| Adipose T-DMRs (n = 18 per tissue) | 163,995 | 1,203 |
| Muscle T-DMRs (n = 18 per tissue) | 491 | 65 |
| Adipose B-DMRs (n = 48 per breed) | 477,131 | 33,602 |
| Muscle B-DMRs (n = 12 per breed) | 415,929 | 28,304 |
| Adipose (n = 144) versus muscle (n = 36) T-DMRs | 108,361 | 9,316 |
Interestingly, we found that Xist, a lncRNA associated with X chromosome inactivation2, is located in a DMR between sexes for adipose. DMRs in lincRNA gene regions have a higher GC content (0.47) and CpGo/e ratios (0.39) than the average for lincRNA genes. A total of 4,139 DMRs were found to be located in the promoter regions of lincRNA genes. We then classified lincRNA promoters into three classes: HCPs (high-CpG promoters), ICPs (intermediate CpG promoters) and LCPs (low-CpGs promoters) according to their CpG profiles as previously described20. Similar to protein-coding genes15, DMRs of lincRNA genes are most frequently located in ICP than in HCP or LCP (hypergeometric test, P < 2.2 × 10−16). Consistent with previous studies15,21,22,23, we found that most DMRs associated with lncRNA genes are located in CpGIs rather than non-CpGIs (hypergeometric test, P < 2.2 × 10−16).
Candidate lincRNA genes associated with adipogenesis in the pig
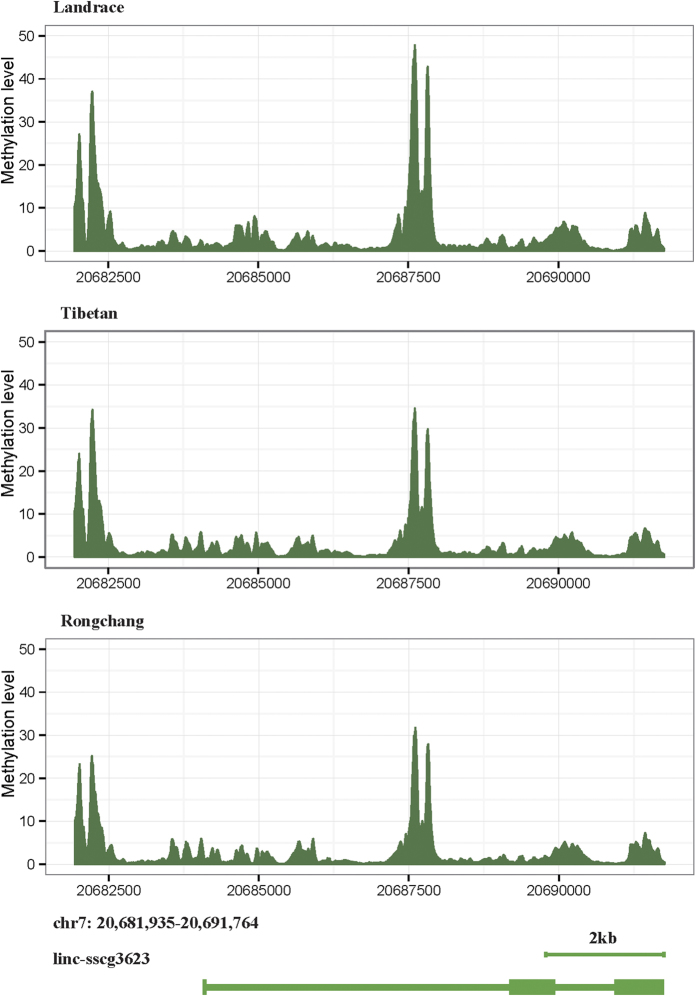
In eight adipose tissues from different body sites, linc-sscg3623 shows differences in level of methylation (located in DMRs) between the Landrace and Rongchang breed of 210-day-old pigs, with the methylation level of linc-sscg3623 being higher in Landrace than in Rongchang breed (Fig. 4), which is consistent with the difference in fat deposition between these two breeds15. As the Fig. 1a in Li et al.15, the ability to deposit fat is Rongchang>Tibetan>Landrace, which is contrary to the methylation level of linc-sscg3623 among the three breeds: Rongchang<Tibetan<Landrace (Fig.4).
Figure 4. Comparison of the methylation levels for eight adipose tissues from differential body sites of Rongchang, Tibetan and Landrace breeds for linc-sscg3623 gene.
Dynamics of DNA methylation of lincRNA gene in backfat tissue
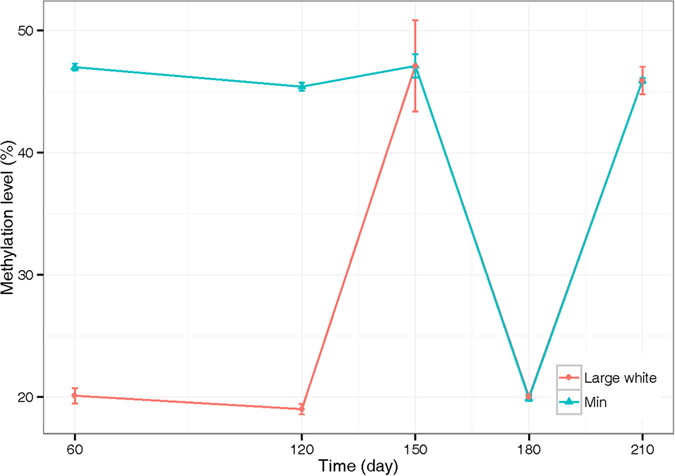
To further investigate if the linc-sscg3623 gene is involved in adipogenesis, we used bisulfite pyrosequencing to investigate the methylation level of two GC sites (located in DMRs) in this gene in backfat from five developmental stages (60, 120, 150, 180 and 210 days) in Min and Large White pigs. Min pigs have a higher methylation level than the Large White pigs at 60 and 120 days, which is consistent with the differential fat deposition between the two breeds (Fig. 5, Supplementary Fig. S2 and Table S2). At 150 days and later, similar methylation patterns are observed between the Min and Large White pigs (Fig. 5, Supplementary Fig. S2 and Table S2). The data suggest a demethylation process occurred between days 150 and 180 in both pigs, which was followed by a remethylation process between days 180 and 210 (Fig. 5, Supplementary Fig. S2 and Table S2). The demethylation and remethylation processes may be associated with adipocyte differentiation. At day 210, we could not find any difference in the methylation level between the Min and Large White breeds for backfat tissue. This may be due to differential fat deposition mechanisms in differential adipose sites. These observations suggest that linc-sscg3623 may show differences in expression level between differential breeds or development stages and affect adipogenesis.
Figure 5. Comparison of the methylation levels for the GC site (chr7:20684724) in linc-sscg3623 gene from genomic DNA from backfat tissue at different developmental stages in Min and Large White breeds of pig.
Discussion
Several studies have indicated that DNA methylation plays important roles in stem cell differentiation24 and embryonic development25, and that alternations in DNA methylation are associated with disease26. These studies, though, mainly focused on protein-coding genes. In this study, we characterized the DNA methylation patterns of pig lincRNA genes in adipose and muscle tissues.
To compare DNA methylation pattern between lincRNA and protein-coding genes, we focused on the genomic regions consisting of their promoters and their gene bodies, and found that lincRNA genes have higher DNA methylation levels than protein-coding genes despite lincRNA genes having similar GC content and CpGo/e as protein-coding genes. When we considered promoters, exons or introns separately, the level of methylation for lincRNA genes is higher for each region compared to the corresponding parts of the protein-coding genes. We also found that the methylation level of CpG sites in CpGIs was higher than for non-CpGIs for lincRNA genes, in contrast to previous studies of genomic CpG sites17, which indicated that the methylation levels of CpG sites in CpGIs was lower than that of non-CpGIs. GC content and CpGo/e across TSS showed similar patterns between protein-coding and lincRNA genes, whereas the methylation levels across TSS of these genes display differences. These features imply that DNA methylation is differentially regulated between lincRNA and protein-coding genes and may partly explain why the expression level of lincRNA genes is lower than that of protein-coding genes.
In this study, we only focused on the pattern of methylation of lincRNA genes in adipose and muscle tissues of pigs, and these results should contribute to our understanding of the roles of lincRNAs in tissue-specific regulatory mechanisms, including those used in humans and mouse. In our previous studies of pig lincRNA genes, we indicated that these genes might have contributed to the domestication of the pig16. Fatness and lean muscle growth are two phenotypes that have experienced strong artificial selection in pigs. Here, we found many lincRNA genes are located in DMRs, with many of the DMRs overlapping with the promoters of these genes. Recently, a limited number of lincRNAs were identified to be involved in adipogenesis11,12. Interestingly, many lincRNA genes were found to overlap with DMRs. For example, linc-sscg3623 showed different methylation levels in adipose tissues between the Landrace and Rongchang breeds of pigs that differ in fatness. The Min and Large White breeds also showed different methylation levels in backfat tissues at two developmental stages, 60 and 120 days, and then displayed a demethylation process and a remethylation process between days 150 and 210. These results imply that lincRNA genes contribute to fatness and lean growth in pigs, and specific alleles may have been selected in different breeds. It is believed that DNA methylation in promoters is one of the regulatory mechanisms that influence gene expression. Li et al. had used a gene expression microarray to measure the expression levels of genes15, however their microarray contained only a small number of lincRNA genes (data not shown), thus, it is impossible for us to calculate any correlations between methylation levels in lincRNA promoters and the expression levels of the associated lincRNA genes using the MeDIP-seq data they generated15. Additional studies are needed to investigate the mechanisms controlling DNA methylation in lincRNA gene promoters for regulating their gene expression.
In summary, we found several differences in the methylation patterns between lincRNA and protein-coding genes, including differences in methylation levels and the pattern of methylation around their TSS. These results provide avenues for more in-depth research into the methylation patterns of lincRNA genes. Furthermore, we identified many lincRNA genes that are overlapped with DMRs which may help uncover the molecular basis of adipogenesis and muscle development and the further our understanding of the domestication of domestic animals.
Material and Methods
All experimental protocols were approved by the Kunming Institute of Zoology, Chinese Academy of Sciences and Institute of Animal Science of the Chinese Academy of Agricultural Sciences.
Data used
The methylated DNA immunoprecipitation sequencing (MeDIP-seq) dataset, which sampled eight adipose and two muscle tissues from three pig breeds including 180 samples15, was downloaded from the NCBI GEO database (GSE30344). Raw sequence reads were filtered as Li et al.15 and were aligned to the Sus scrofa 10.2 genome sequence using bwa aln (version 0.7.8-r455) with default parameters27. Protein-coding gene annotation was downloaded from the Ensembl database (version 73). The lincRNA annotation that we used in this study was generated from our previous study16. Pig CpG island (CpGI) positions were retrieved from the UCSC Genome Browser for the pig 10.2 genome. CpGI shores are located within 2 kb of CpGIs.
Definition of promoters
In this study, we defined genomic region from −2000 to the TSS as the promoter for 6,572 lincRNA transcripts. These promoters were classified into three types according to CpG ratio as in a previous study20. There are 1,100 HCPs, 2,711 ICPs and 2,761 LCPs for the lincRNA transcripts.
Identification of DMRs
To identify DMRs among the different breeds, sexes and anatomic locations, we used edgeR integrated in the Bioconductor package MEDIPS at genome-wide 250 bp bins19. MEDIPS inferred differential methylation for the sample groups by calculating Wilcoxon rank tests for the reads per million (rpm) values of each window19. DMRs were filtered for windows with adjusted P < 0.1 (exact test for negative binomial distribution, using edgeR integrated in the Bioconductor package MEDIPS).
Bisulfite pyrosequencing
Animals: Three biological replicates of 60, 120, 150, 180 and 210 day old Min and Large White pigs were used in this experiment (Supplementary Table S2). All animals were females and were fed under the identical normal conditions.
Tissue preparation: Animals were humanely killed in accordance with the guidelines of the Good Experimental Practices adopted by the Institute of Animal Science of the Chinese Academy of Agricultural Sciences. All backfat samples were collected between the third and fourth ribs, and were maintained in liquid nitrogen.
DNA methylation sequencing: Genomic DNA was isolated with the QIAamp DNA mini kit (Qiagen) and treated with bisulphite using EZ DNA methylation Gold kit (Zymo Research) according the manufacturer’s instructions. Detailed information regarding primer sequences is given in Supplementary Table S1. PCR amplification of interest regions was performed with a total reaction volume of 50 μl, using 10 μl 5Xbuffer (KAPA), 1 ul dNTP (10 mM/each), 1 ul forward primer (50pM/μl), 1 μl reverse primer (50pM/μl), 2 ul bisulfite-treated genomic DNA and water. PCR products were purified and sequenced by BGI Tech Solutions (Liuhe Beijing) Co., Limited using the PyroMark Q96 ID Pyrosequencing System (Qiagen). The methylation level was expressed as a percentage of the methylated cytosines over the sum of the methylated and unmethylated cytosines.
URLs. Differentially methylated regions (DMRs) identified in this study are online, http://res.xaut.edu.cn/aldb/download.html.
Additional Information
How to cite this article: Zhou, Z.-Y. et al. DNA methylation signatures of long intergenic noncoding RNAs in porcine adipose and muscle tissues. Sci. Rep. 5, 15435; doi: 10.1038/srep15435 (2015).
Supplementary Material
Acknowledgments
This work was supported by grants from the National 863 Program of China (2011AA100304-5), the Ministry of Agriculture of China (2014ZX08009003-006), the National 973 Program of China (2013CB835203), the National Natural Science Foundation of China (31061160189), the Yunnan Provincial Science and Technology Department (2011AB008), the Agricultural Science and Technology Innovation Program (ASTIP-IAS02), and the Animal Branch of the Germplasm Bank of Wild Species, Chinese Academy of Sciences (the Large Research Infrastructure Funding).
Footnotes
Author Contributions Y.P.Z. and H.B.X. supervised this work. Z.Y.Z. designed the research. Z.Y.Z., A.M.L. and Y.H.L. performed data collection and data analysis. L.G.W. and L.X.W. provided samples. D.X., S.F.W. and X.M.H. helped with the experiments. Z.Y.Z., A.M.L., D.M.I., L.W., H.B.X. and Z.P.Z. wrote and revised the manuscript.
References
- Borsani G. et al. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351, 325–329 (1991). [DOI] [PubMed] [Google Scholar]
- Brockdorff N. et al. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71, 515–526 (1992). [DOI] [PubMed] [Google Scholar]
- Brown C. J. et al. The human XIST gene: Analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71, 527–542 (1992). [DOI] [PubMed] [Google Scholar]
- Payer B. & Lee J. T. X chromosome dosage compensation: how mammals keep the balance. Annual review of genetics 42, 733–772 (2008). [DOI] [PubMed] [Google Scholar]
- Orom U. A. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 143, 46–58 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huarte M. et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T. et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nature genetics 43, 621–629 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Ramadass A., Barros A. S., Chow N. & Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666–670 (2007). [DOI] [PubMed] [Google Scholar]
- Rinn J. L. et al. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. Cell 129, 1311–1323 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477, 295–300 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L. et al. Long noncoding RNAs regulate adipogenesis. Proceedings of the National Academy of Sciences 110, 3387–3392 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.-Y., Li S., Wang G.-X., Yu Q. & Lin J. D. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Molecular cell 55, 372–382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesana M. et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurlock M. E. & Gabler N. K. The development of porcine models of obesity and the metabolic syndrome. The Journal of nutrition 138, 397–402 (2008). [DOI] [PubMed] [Google Scholar]
- Li M. et al. An atlas of DNA methylomes in porcine adipose and muscle tissues. Nature communications 3, 850 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z.-Y. et al. Genome-wide identification of long intergenic noncoding RNA genes and their potential association with domestication in pigs. Genome Biology and Evolution 6, 1387–1392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata S. et al. DNA methylation signatures in development and aging of the human prefrontal cortex. The American Journal of Human Genetics 90, 260–272 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sati S., Ghosh S., Jain V., Scaria V. & Sengupta S. Genome-wide analysis reveals distinct patterns of epigenetic features in long non-coding RNA loci. Nucleic acids research 40, 10018–10031 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienhard M., Grimm C., Morkel M., Herwig R. & Chavez L. MEDIPS: genome-wide differential coverage analysis of sequencing data derived from DNA enrichment experiments. Bioinformatics 30, 284–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M. et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nature genetics 39, 457–466 (2007). [DOI] [PubMed] [Google Scholar]
- Doi A. et al. Differential methylation of tissue-and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nature genetics 41, 1350–1353 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R. A. et al. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nature genetics 41, 178–186 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H. et al. Comprehensive methylome map of lineage commitment from haematopoietic progenitors. Nature 467, 338–342 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nature biotechnology 28, 1079–1088 (2010). [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 447, 425–432 (2007). [DOI] [PubMed] [Google Scholar]
- Feinberg A. P. Phenotypic plasticity and the epigenetics of human disease. Nature 447, 433–440 (2007). [DOI] [PubMed] [Google Scholar]
- Li H. & Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.