Supplemental Digital Content is available in the text
Abstract
Recently, common variants within or near ATG5, which is a key autophagy gene required for the formation of autophagosomes, have been identified as a candidate gene of systemic lupus erythematosus (SLE) by several genome-wide association studies. Moreover, elevated ATG5 expression was observed in SLE as well as other autoimmune diseases. However, no significant associations between variants within ATG5 and SLE were identified in several Chinese populations. The present study was conducted to further check the genetic role of ATG5 by associating both common and rare variants of ATG5 in Chinese patients with lupus nephritis (LN), a major phenotype with poor prognosis in SLE.
To detect the association of common variants of ATG5 with LN, 7 tagging single nucleotide polymorphisms (SNPs) designed in immunochip and 4 SNPs reported to be associated with SLE were genotyped in 500 LN patients and 500 healthy controls. Furthermore, direct sequencing of exons and their flanking regions in 90 LN patients, 30 SLE patients, and 60 healthy controls were performed. Functional genomic annotation was performed by using public databases.
None of the 11 tagging SNPs was observed to be associated with LN. By sequencing, 13 variants were identified, including 5 common SNPs, 7 not previously described, and 1 reported as rare variants (<1%) in the Single Nucleotide Polymorphism Database or the 1000 Genome project. None of the 5 common SNPs showed significant association between patients and controls, whereas increased frequencies of rare or novel variants were observed in patients compared with healthy controls, with 6/90 in LN patients, 2/30 in SLE patients, and 1/163 in healthy controls. Although these rare variants were observed to be located in the flanking regions of exons instead of missense mutations, patients carrying them tended to have severe clinical phenotype, and in silicon analysis suggested their regulatory effects.
Increased frequencies of rare variants of ATG5 were identified in patients with LN and SLE compared with healthy controls, highlighting a likely important role of rare ATG5 variants in Chinese SLE patients.
INTRODUCTION
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with a strong genetic component.1 To date, >50 loci showed robust association with SLE have been identified by hypothesis-free genome-wide association studies using common tagging single nucleotide polymorphisms (SNPs) with minor allele frequency (MAF) >5%,2 significantly broadening our views about genetic pathogenesis of SLE. However, all these variants can explain no >15% of genetic risk in SLE. For the unexplained heritability, a further check for the rare variants may be helpful.
Autophagy is a phylogenetically ancient mechanism by which the cell can remove the long-lived proteins and damaged organelles through lysosomal degradation. Recently, genetic studies reported that polymorphisms in various autophagy-related genes, including ATG5, ATG7, IRGM, DRAM1, CDKA1B, APOL1, and MTMR3, were associated with SLE, suggesting the possible role of autophagy in SLE.3–7ATG5 is an important gene in the initiation of autophagosome formation.8 Several large-scale replication studies have repeatedly associated common variants within or near ATG5 with susceptibility to SLE.6,9 The functional studies targeting ATG5 consistently demonstrated its pivotal role in autoimmune diseases. Elevated ATG5 expression has been observed in a mouse model of autoimmune demyelination, as well as in blood and brain tissues from patients with multiple sclerosis.10 Besides, the ATG5 rs573775, T allele was reported to have effects on SLE susceptibility, cytokine production, and disease features.3,11 Also in our previous study,6 the risk alleles within PRDM1-ATG5 region correlated with high ATG5 instead of PRDM1 messenger RNA expression in B cells, and higher ATG5 expression was observed in B cells from SLE patients compared with normal controls. Thus, ATG5 was suggested to be a strong candidate gene, which may play an important role in SLE.
However, the SLE-associated variants of ATG5 were observed in different populations, and moreover, no significant associations between variants within ATG5 gene region and SLE were observed in several Chinese populations,6,12 whereas, the rare variants of ATG5 have been reported to be associated with several complex diseases, including prostate cancers, gastrointestinal cancers, and Parkinson disease.13–15 So, it is of high interest to further check the genetic role of ATG5 in SLE, especially rare variants of ATG5 in the Chinese population. Lupus nephritis (LN), a major manifestation and fatal target-organ damage of SLE, is one of the strongest indicators of poor prognosis and possibly a kind of extreme phenotype. Thus, the present study was to further explore the genetic role of ATG5 in a Chinese population by detecting the association of both common and rare variants of ATG5 with LN.
materials and methods
Patients and Controls
To replicate the association of common variants of ATG5 with LN, 500 LN patients (31.9 ± 11.2 years, 423 women) and 500 healthy blood donors (40.0 ± 8.6 years, 140 women), who were of Han Chinese in Beijing (CHB) origin, were enrolled in the study. Furthermore, to detect the association of rare variants of ATG5 with LN, exon sequencing was performed in 90 LN patients (26.4 ± 10.8 years, 57 women), 30 SLE patients (33.7 ± 10.0 years, 29 women), and 60 healthy blood donors (34.6 ± 10.3 years, 48 women).
The SLE patients met the revised SLE criteria of the American College of Rheumatology in this study.16 All the LN patients were confirmed by renal biopsy using light microscopy, immunofluorescence, and electron microscopy, and the SLE patients without renal damage were defined as the none of proteinuria, abnormal urinary sediment, or renal biopsy evidence of nephropathy. The study was approved by the medical ethics committee of Peking University First Hospital, and all patients gave written informed consents.
SNP Selection and Genotyping
Seven SNPs designed in immunochip aiming for fine-mapping of immune-related genes in autoimmune diseases, including rs78200552, rs144506815, rs9386514, rs9373839, rs78937934, rs6906688, and rs510432, were genotyped by an Illumina Solexa HiSeq 2000 platform (Shenzhen Huada Gene Research Institute, ShenZhen, China). Of note, rs9373839 was reported to be one of the top associated variants with systemic sclerosis. Four SNPs, which were reported to be associated with SLE in whites,3,9 including rs2245214, rs4945747, rs573775, and rs2757133, were also selected and genotyped by TaqMan allele discrimination assays (Applied Biosystems, Foster City, CA). Thus, a total of 11 common tagging SNPs were included in the present study. Among them, rs9386541, rs9373839, and rs4945747 were in the same block, whereas rs2757133 and rs573775 were in the same block. The genotyping data were verified by direct sequencing in selected samples with concordance of 100%.
Sequencing and Analysis
All 8 exons, including all coding exons and splice sites of ATG5, were sequenced using genomic DNA. Polymerase chain reaction (PCR) primers were designed using Primer3 based on the genomic sequence of human ATG5 gene (GeneBank access number, NC_0000016.11). The PCR product sequencing was performed by Beijing Genomics Institute based on Sanger sequencing method. The sequencing results were analyzed by Chromas 15.0 (http://technelysium.com.au/?page id=13) and compared with wild type of ATG5 gene reference sequences (http://blast.ncbi.nlm.nih.gov/). The PCR products were sequenced forward and backward, and repeated twice. Variants were considered to be common (previously reported with MAF >1%), rare (previously reported with MAF <1%), or novel (not previously reported in 1000 Genome (1000G) project or the Single Nucleotide Polymorphism Database). When a novel variant was identified, another pair of primers was designed to repeat the sequencing to make sure the novel variant was real.
Genotype data of 103 CHB controls were downloaded from 1000G project (http://www.ensembl.org/Homo_sapiens/Gene/Variation_Gene/Table?db=core;g=ENSG00000057663;r=6:106184476-106325820). Using these data and our sequencing data, association analysis and statistical analysis were performed.
Bioinformation Mechanism Prediction
Functional significance of the common or rare variants was annotated by the Encyclopedia of DNA Elements project data or related open databases. The variant regulatory effects, such as whether they were transcriptional factor binding sites, were searched by the Variant Effect Predictor (http://asia.ensembl.org/info/docs/tools/vep/index.html) and the Genomatix Database (http://www.genomatix.de/cgi-bin/tools/tools.pl). The RegRNA Database (http://regrna.mbc.nctu.edu.tw/html/tutorial.html) was used to identify the homologs of regulatory RNA motifs and elements. Moreover, to further predict whether genetic variants were micro RNA (miRNA) synthetic and target sites, the miRBase Database (http://www.mirbase.org/) and the miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) were searched.
Statistical Analysis
The genotype frequencies of SNPs were tested for Hardy–Weinberg equilibrium separately in patients and controls. Associations between disease and SNPs were analyzed by χ2 tests, and Fisher exact test was used when necessary. Results of the measurement data for clinical information were expressed as mean ± SD, and t tests were used to analyze the difference between patients carrying rare variants or not. The same procedure was applied to subgroups stratified according to sex and pathologic classes of LN patients. Statistical analysis was performed with SPSS16.0 software (SPSS Inc, Chicago, IL). A 2-tailed P value of <0.05 was considered statistically significant.
RESULTS
Association Analysis of the Tagging Common SNPs Within ATG5
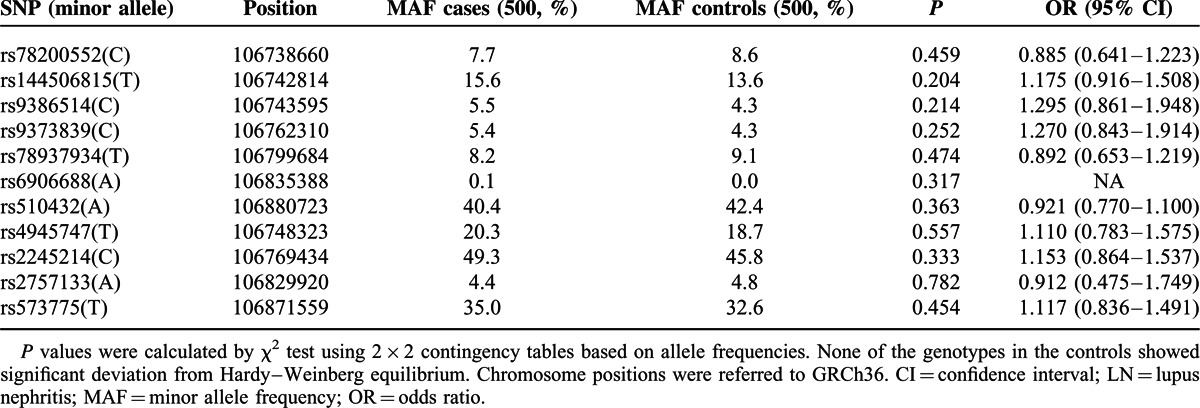
A total of 11 tagging SNPs within ATG5 were genotyped in 500 LN patients and 500 healthy controls. Deviation from Hardy–Weinberg equilibrium was not observed for any of the SNPs in the patients or controls (P > 0.05). At allele level, results of the case–control association analyses were shown in Table 1, and none of the SNPs were observed to be associated with LN significantly (P > 0.05). Sex ratio and pathologic classes of LN patients were 2 important confounding factors. In the current study, the female ratio of the patients was 84.6% (423/500), and percentages of class I, II, III, IV, V, and VI pathology of the 500 LN patients were 0%, 7.5%, 22.2%, 53.9%, 15.9%, and 0.4%, respectively. To investigate the possible association of the common SNPs with different sex and pathologic classes of LN patients, we further stratified the patients into subgroups. However, no association was observed between the LN subsets and any of the 11 SNPs investigated (P values ranged from 0.07 to 0.83).
TABLE 1.
Allelic association of the tagging SNPs in ATG5 with LN
Association Analysis of the Sequencing Variants of ATG5
All 8 exons, including essential splice sites were sequenced by Sanger technology and 13 variants were identified. Among them, 5 were common SNPs previously reported in CHB population, including rs510432, rs138203657, rs1624701, rs41292420, and rs77859116, whereas 8 were rare or novel (see the Sequencing and Analysis section for definitions). For the 5 common SNPs, they were found in patients and controls with similar frequencies (P > 0.05) (supplementary Table 1, http://links.lww.com/MD/A286).
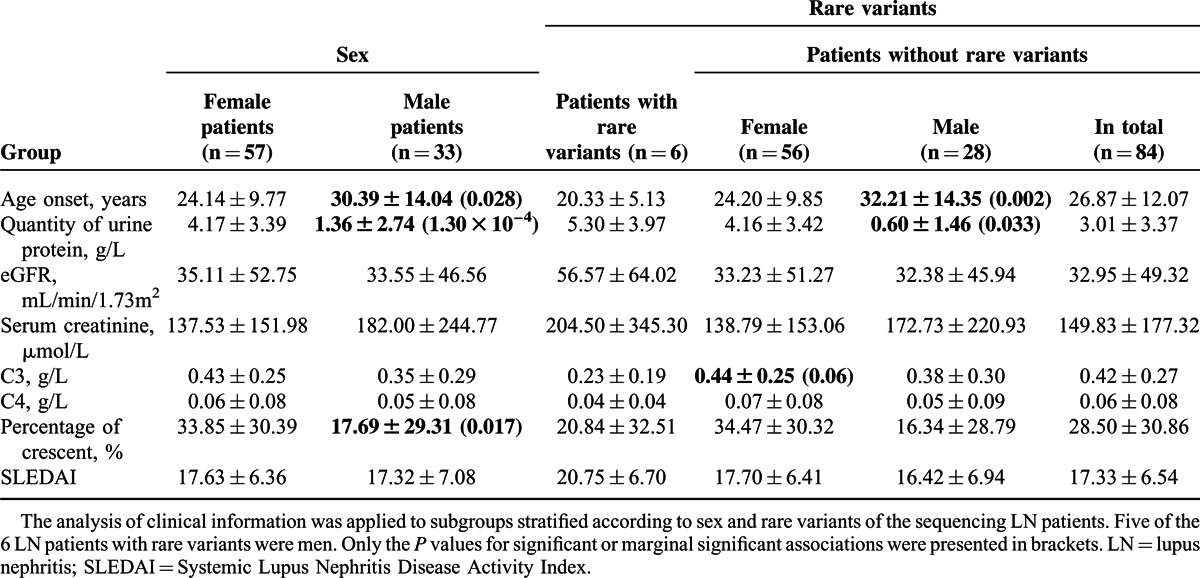
Among the 8 rare or novel variants, none was reported in 1000G database or rare variant database except 106773567C>T (only reported in a Puerto Rico population with MAF 0.5%), indicating they were novel variants (supplementary Fig. 1, http://links.lww.com/MD/A286). Five heterozygous variants, including 106773751C>T, 106773700G>T, 106773530T>G, 106773567C>T, and 106696229T>C, were identified in 1 LN patient each, and 2 variants, including 106740994G>A and 106740756->TTAT, were observed in 1 SLE patient each. Although 106696268G>T was identified in 1 LN patient and 1 healthy control, the ratio of rare variants in LN patients, SLE patients, and reference controls (the 60 healthy controls in the present study and 103 CHB controls in 1000G database) were 6/90 (6.7%), 2/30(6.7%) and 1/163 (0.6%) with calculated odds ratio 11.6 between LN or SLE patients and controls (P < 0.05) (Table 2). Moreover, among all of the patients, the LN patients carrying the rare variants tended to have severe clinical manifestations (Table 3). Thus, further analysis of the clinical information in the subset LN patients with rare variants or not were taken (Table 4). As for the relatively higher male ratio of the sequencing LN patients, we first compared the clinical information between male and female LN patients. In this study, patients carrying the rare variants tended to be predominantly male (male ratio 83.3% vs 33.3%, P < 0.05). However, it seemed to be interesting that the patients carrying the rare variants tended to be earlier onset, having heavier proteinuria, and with worse renal function, higher Systemic Lupus Erythematosus Disease Activity Index Scores, and lower levels of C3 and C4. The same procedure was applied to subgroups stratified according to the sex of LN patients without rare variants, and similar results were observed, highlighting a likely important role of rare variants. However, due to the relatively small sample size, only age onset, quantity of urine protein, and C3 level of the LN patients carrying rare variants showed significant or marginally significant difference from the controls, suggesting the necessity to validate the association of these rare variants and LN in a larger cohort.
TABLE 2.
Association analysis of the rare or novel sequencing variants of ATG5
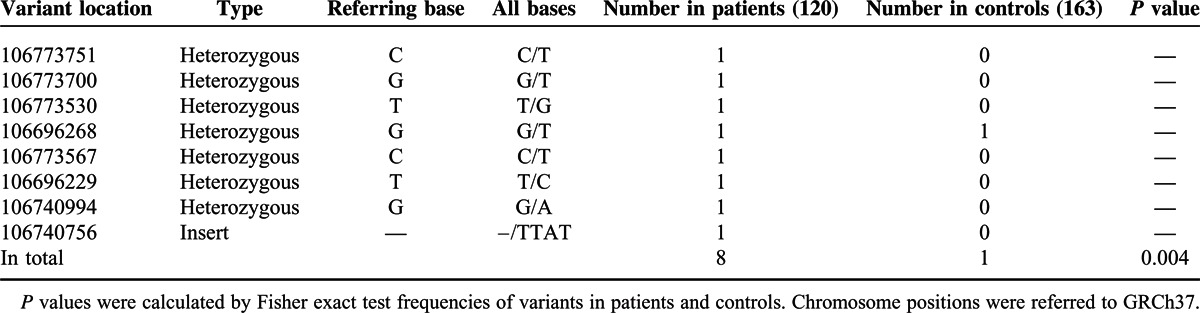
TABLE 3.
Clinical characters of patients carrying the rare variants
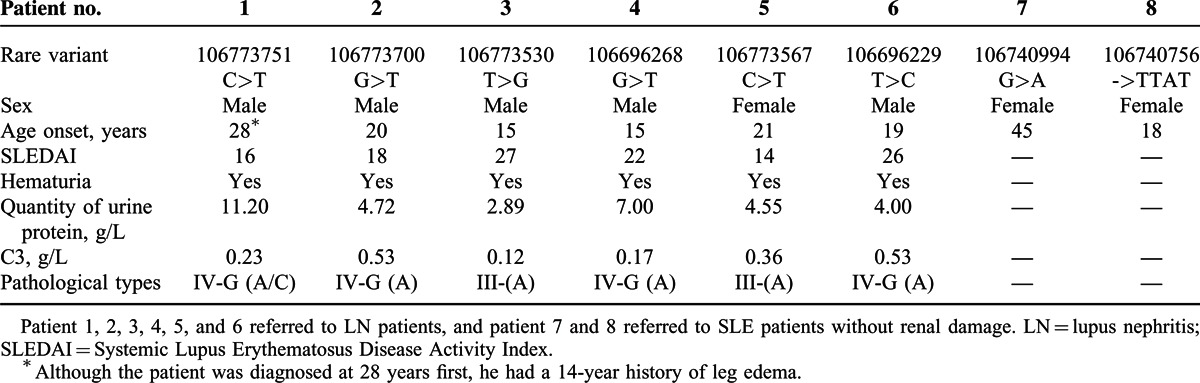
TABLE 4.
Pooled analysis of associations with the rare variants of ATG5 in the subsets of sequencing LN patients
Bioinformatics Analysis of the Sequencing Rare Variants
Among the 8 rare or novel variants, 4 variants (106696229 T>C, 106696268G>T, 106740756->TTAT, and 106740994G>A) located in the intron region, 2 (106773530T>G and 106773567C>T) in the 5′UTC region and 2 (106773700G>T and 106773751C>T) in the promoter region. By searching Variant Effect Predictor, although the variants were not missense mutations, they have several types of features, including transcript, regulatory feature, and motif feature. Moreover, by searching Genomatix, a variant in promoter region, 106773700G>T, was identified to be the binding site of several transcription factors, including Kruppel-like transcription factors (KLFSs), GLI zinc finger (GLIF), and the family member of heat shock proteins (HEATs). In addition, by searching RegRNA Databases, the rare variants may play a role in SLE through regulation of RNA transcription, splicing, and miRNA binding. Among the involved miRNAs, the association of miRNA-155 with ATG3 was also identified in miRTarBase (supplementary Table 2, http://links.lww.com/MD/A286).
DISCUSSION
Recent studies showed that SNPs, rs2245214 and rs573775, in ATG5 were associated with SLE susceptibility in white populations instead of Asian populations.3,6,9,12 As ATG5 plays an important role in immune responses, a deep exploration of genetic variants of ATG5 in SLE is of importance to get a better understanding of its genetic role in SLE. Identifying rare variants by targeted gene sequencing is an efficient approach to identify gene mutations, including low frequency and novel variants for complex diseases.17 In the present study, we tested 11 tagging SNPs within ATG5 in LN patients. Furthermore, exon sequencing of ATG5 was performed in LN patients, SLE patients, and healthy controls. By sequencing, 5 common SNPs and 8 rare variants were detected. The results showed that no significant association was found with the 11 tagging and the 5 sequencing common SNPs in ATG5, whereas the elevated frequencies of rare variants of ATG5 in LN patients and SLE patients were detected. Moreover, LN patients carrying the rare variants tended to have more severe clinical manifestations.
As many previous genetic studies about ATG5 reported that the rare variants of ATG5 were associated with complex diseases, such as prostate cancer, stomach cancer, colon cancer, and Parkinson disease,13–15 our results suggested the potential role of rare variants of ATG5 in LN and SLE patients in the present population. Although the rare variants did not change the amino acids, bioinformation mechanism prediction indicated their regulatory effects on transcription. One rare variant in the promoter region was identified to be the binding site of several transcriptional factors, including KLFS, GLIF, and HEAT. Moreover, by searching the RegRNA Database, the rare variants may be involved in the pathogenesis of LN by regulating RNA transcription, RNA splicing, and miRNA binding. Among the binding miRNAs, miRNA143 and miRNA155 have been reported to be associated with the regulation of cell proliferation, apoptosis, and autophagy.18,19 Mutation of ATG5 in kidney epithelium resulted in tubulointerstitial disease, loss of organ function, and death,20 and all ATG5-null mice died within 24 hours of birth,8 suggesting the vital role of ATG5 for life. Our data may be consistent with the data from ATG5 knockout mice that noncoding variants rather than missense mutations may be involved in SLE susceptibility without lethal effect at birth. As we also observed that patients with ATG5 mutations tended to be younger in age onset, future studies targeting SLE patients with family history or patients died in early age would be of importance.
However, in this study, except for age onset, quantity of urine protein, and C3 level, no significant difference of the clinical features, including serum creatinine, and pathological classes, existed in LN patients carrying rare variants and controls, which may be mainly due to the limitation of our relatively small sample size. In the future, the validation of the association of these rare variants and LN in a larger cohort is necessary. Besides, as both SLE and LN were still a complex phenotype with obscure pathogenesis, extreme phenotype was still difficult to be clearly defined. Thus, identifying the rare variants in extreme-phenotype LN patients may be helpful. Furthermore, as only the exons and flanking regions of ATG5 were sequenced in our study, the noncoding variants emerging as important contributors to genetic disease but missed by exon capture, are still needed to be widely investigated.
Taken together, this study indicates that interrogation of rare variants will enable identification of potential genetic variants that can refine current pathogenesis models for the disease. The study provided further genetic data for ATG5 variants in Chinese SLE patients, highlighting a likely important role of rare variants.
Acknowledgments
We appreciate the work of all of our colleagues, and we are grateful for the participation of all of the patients and healthy control subjects.
Footnotes
Abbreviations: CHB = Han Chinese in Beijing, ENCODE = the Encyclopedia of DNA Elements project, GLIF = GLI zinc finger, HEAT = heat shock protein, KLFS = Kruppel-like transcription factor, LN = lupus nephritis, MAF = minor allele frequency, miRNA = micro RNA, PCR = polymerase chain reaction, SLE = systemic lupus erythematosus, SNP = single nucleotide polymorphism, SSc = systemic sclerosis, 1000 Genome = 1000G.
This work was supported by grants from the National Natural Science Foundation of China (81200524), the Foundation of Ministry of Education of China (20120001120008), Beijing Natural Science Foundation (7152148), the Major State Basic Research Development Program of China (973 program, 2012CB517700), the Research Fund of Beijing Municipal Science and Technology for the Outstanding PhD Program (20121000110), and the Natural Science Fund of China to the Innovation Research Group (81021004). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH, et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum 2005; 52:1138–1147. [DOI] [PubMed] [Google Scholar]
- 2.Rullo OJ, Tsao BP. Recent insights into the genetic basis of systemic lupus erythematosus. Ann Rheum Dis 2013; 72 (suppl 2):56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harley JB, Alarcon-Riquelme ME, Criswell LA, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 2008; 40:204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W, Tang H, Zhang Y, et al. Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians. Am J Hum Genet 2013; 92:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman BI, Langefeld CD, Andringa KK, et al. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol 2014; 66:390–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou XJ, Lu XL, Lv JC, et al. Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population. Ann Rheum Dis 2011; 70:1330–1337. [DOI] [PubMed] [Google Scholar]
- 7.Zhou XJ, Zhang H. Autophagy in immunity: implications in etiology of autoimmune/auto-inflammatory diseases. Autophagy 2012; 8:1286–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuma A, Hatano M, Matsui M, et al. The role of autophagy during the early neonatal starvation period. Nature 2004; 432:1032–1036. [DOI] [PubMed] [Google Scholar]
- 9.Gateva V, Sandling JK, Hom G, et al. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 2009; 41:1228–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alirezaei M, Fox HS, Flynn CT, et al. Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis. Autophagy 2009; 5:152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.López P, Alonso-Pérez E, Rodríguez-Carrio J, et al. Influence of Atg5 mutation in SLE depends on functional IL-10 genotype. PLoS One 2013; 8:e78756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han JW, Zheng HF, Cui Y, et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009; 41:1234–1237. [DOI] [PubMed] [Google Scholar]
- 13.Kim MS, Song SY, Lee JY, et al. Expressional and mutational analyses of ATG5 gene in prostate cancers. APMIS 2011; 119:802–807. [DOI] [PubMed] [Google Scholar]
- 14.An CH, Kim MS, Yoo NJ, et al. Mutational and expressional analyses of ATG5, an autophagy-related gene, in gastrointestinal cancers. Pathol Res Pract 2011; 207:433–437. [DOI] [PubMed] [Google Scholar]
- 15.Kang MR, Kim MS, Oh JE, et al. Frame shift mutations of autophagy-related genes ATG2B, ATG5, ATG9B and ATG12 in gastric and colorectal cancers with micro satellite instability. J Pathol 2009; 217:702–706. [DOI] [PubMed] [Google Scholar]
- 16.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 1982; 25:1271–1277. [DOI] [PubMed] [Google Scholar]
- 17.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature 2009; 461:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen JZ, Zhang YY, Fu HY, et al. Over expression of microRNA-143 inhibits growth and induces apoptosis in human leukemia cells. Oncol Rep 2014; 31:2035–2042. [DOI] [PubMed] [Google Scholar]
- 19.Holla S, Kurowska-Stolarska M, Bayry J, et al. Selective inhibition of IFNG-induced autophagy by Mir155- and Mir31-responsive WNT5A and SHH signaling. Autophagy 2014; 10:311–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kawakami T, Gomez IG, Ren S, et al. Deficient autophagy results in mitochondrial dysfunction and FSGF. J Am Soc Nephrol 2015; 26:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]


