Supplemental Digital Content is available in the text
Abstract
Tumor tissues are often absent or insufficient for testing epidermal growth factor receptor (EGFR) mutations to guide EGFR tyrosine kinase inhibitors (TKIs) treatment of patients with nonsmall cell lung cancer (NSCLC).
We conducted this systematic review and meta-analysis to assess whether blood can be used as a substitute for tumor tissue in detecting EGFR mutations.
MEDLINE, EMBASE, and the Cochrane Library were searched for studies that provided data to estimate the accuracy of blood testing against tissue testing in NSCLC patients and/or those directly compared the efficacy of EGFR TKIs in EGFR mutant and wild-type patients according to sources of specimens.
Sensitivity, specificity, and concordance rate were used as measures of the accuracy. Risk ratio (RR) for objective response and hazard ratio (HR) for progression-free survival (PFS) and overall survival (OS) were used as measures for treatment efficacy. We combined the effects by using the fixed-effects model unless there was evidence of heterogeneity, in which case a random-effects mode was used.
This systematic review included 25 studies with 2605 patients. The pooled overall sensitivity, specificity, and concordance rate were 0.61, 0.90, and 0.79, respectively. Serum showed lower sensitivity (0.56 vs 0.65) but higher specificity (0.95 vs 0.85) and higher concordance (0.86 vs 0.74) than plasma. EGFR mutations (exon 19 or 21) in blood were significantly associated with objective response (RR: 4.08; 95% confidence interval [CI] 2.48–6.70), PFS (HR: 0.72; 95% CI 0.64–0.80), and OS (HR: 0.71; 95% CI 0.50–0.99). Importantly, the association of the mutations with the 3 clinical outcomes for serum was similar to that for tumor tissue and higher than that for plasma.
Blood, in particular serum, is a good substitute when tumor tissue is absent or insufficient for testing EGFR mutations to guide EGFR TKIs treatment in patients with NSCLC. EGFR mutation positivity in blood could be used to recommend EGFR TKIs treatment, but the absence of blood positivity should not necessarily be construed with confirmed negativity.
INTRODUCTION
Lung cancer is a leading cause of cancer-related deaths worldwide and some 85% of lung cancer patients were having nonsmall cell lung cancer (NSCLC).1,2 Gefitinib and erlotinib, 2 tyrosine kinase inhibitors (TKIs) that are targeted at epidermal growth factor receptor (EGFR), are widely recommended for advanced NSCLC but only some 10% of patients respond to the treatment.3–5 Clinical trials have shown that patients with mutations in the kinase domain of the EGFR gene are much more likely to respond to EGFR TKIs treatment than EGFR wild-type patients.6,7 Testing EGFR mutations is now a common practice in selecting patients for EGFR TKIs treatment.
However, some two-third of NSCLC patients8 are already at an advanced stage at the time of diagnosis for which surgical operation is normally not recommended. Biopsy is thus required to obtain tumor tissues for testing EGFR mutations.9 Biopsies can fail in 10% to 50% of patients to obtain sufficient tumor tissues for EGFR mutation analysis.10 Even in well-organized clinical trials, more than half of the patients did not have sufficient tumor tissues for the testing.11 Surrogate biological samples for EGFR mutation testing have been investigated.
The level of circulating DNA in blood has been found to be higher in lung cancer patients than cancer-free patients.12,13 Most of the excess circulating DNA is believed to be released from dying lung cancer cells at primary and/or metastatic sites.13 Therefore, blood is a potential substitute for tumor tissues to provide a noninvasive, easily accessible, and repeatedly measurable source of genotypic information that may predict response and prognosis after treatment. EGFR mutations have been detected in plasma DNA14,15 and serum DNA16,17 and some consistency in EGFR mutation status is observed between blood and tumor tissues.14–17 As a result, EGFR mutations detected in blood may be a good predictor of response to EGFR TKIs treatment.14,17–20
We conducted this study to identify and summarize the current best research evidence to evaluate the accuracy of EGFR mutations status in blood against that in tumor tissues as the reference and to compare the power of EGFR mutations in blood and in tumor tissues in predicting clinical outcomes of EGFR TKIs treatment in patients with NSCLC.
MATERIALS AND METHODS
Data Sources and Search Strategy
We conducted a computerized literature search of the Cochrane Library, PubMed, and EMBASE from their inception to June 2013, with different combinations of the following keywords: “non-small cell lung cancer,” “epidermal growth factor receptor,” “mutation,” “plasma,” and “serum.” In addition, we searched the abstracts database of the American Society of Clinical Oncology using the previously mentioned terms. We subsequently manually searched the bibliographies of included studies and recent narrative reviews for additional studies. No language restrictions were applied. We considered both published and unpublished studies for inclusion, including those only published in abstracts.
We included all studies that provided enough raw data to create the 2 × 2 diagnostic tables for EGFR mutation status in tumor tissue specimens and blood samples in NSCLC patients and/or those that directly compared the clinical outcomes of EGFR TKIs in EGFR mutant and wild-type patients according to sources of specimens. Two reviewers independently reviewed the titles, abstracts, and full texts of all citations that were likely to meet the predefined selection criteria. The discrepancies were resolved by discussion or by consulting with a third reviewer. For duplicate publications, we only included the most recent and complete version of studies.
Quality Assessment and Data Extraction
Two reviewers independently extracted data using a predesigned data abstraction form and critiqued the quality of the studies, with a third reviewer contacted in case of disagreement. We constructed 2 × 2 tables that contained the number of true positives (EGFR mutations detected in both tumor tissue and blood samples), true negatives (wild-type EGFR detected in both tumor tissue and blood samples), false positives (EGFR mutations detected in blood samples but not in tumor tissue samples), and false negatives (wild-type EGFR detected in blood samples but not in tumor tissue samples). In addition, the following data were extracted from each study: study characteristics (such as first author's name, year of publication, study design, and number of patients enrolled), patients’ characteristics (such as mean or median of age, percentage of male, smoking status, and histology), EGFR mutation testing methods, clinical outcomes of EGFR TKIs treatment stratified by EGFR mutation status (such as objective response, progression-free survival [PFS], and overall survival [OS]) and items necessary to assess study quality. We assessed the methodological quality of studies with the Quality Assessment of Diagnostic Accuracy Studies-2 (QUADAS-2) tool.21
Statistical Analysis
We used the EGFR mutation status in tumor tissue samples as reference standard, and calculated sensitivity, specificity, and concordance rate for each study according to the 2 × 2 tables mentioned in the previous subsection. The association between the status of EGFR mutations and clinical outcomes of EGFR TKIs treatment was measured by risk ratio (RR) for objective response and hazard ratio (HR) for PFS and OS. We combined the EGFR mutation rate, sensitivity, specificity, concordance rate, RR, and HR by using the fixed-effects model22 unless there was evidence of heterogeneity, in which case a random-effects model23 was used. In the meta-analysis of EGFR mutation rate of all exons, we only included the data of direct sequencing, the most commonly used mutation test method, when ≥2 mutation test methods were used in the same original trials. Heterogeneity was explored by the Q test with degree of freedom equal to the number of analyzed studies minus one.24 A P value of ≤0.10 in the Q test is deemed statistically significant and suggests presence of heterogeneity across studies. We performed subgroup analyses to detect the source of heterogeneity according to ethnicity, sex, age, histology, tumor stages, smoking history, type of blood samples, and EGFR mutation testing methods, wherever deemed appropriate. We performed funnel plots to assess the possible presence of publication bias and Egger test to assess the symmetry of the funnel plot.25 We employed a 2-tailed significance level of 0.05 for all the statistical tests except for the assessment of heterogeneity. Data analyses were carried out with STATA Version 13 (STATA Corporation, College Station, TX), Review Manager (RevMan 5.2.9) and MetaAnalyst Version Beta 3.13 (Tufts MedicalCenter, Boston, MA). The study was reported according to the guidelines provided by the Preferred Reporting Items for Systematic Reviews and Meta-Analyses report.26
RESULTS
Study Characteristics
A total of 6261 citations were retrieved from the Cochrane Library, MEDLINE, EMBASE, and other resources. After excluding duplicates and screening of titles and abstracts, 353 citations were left for full-text screening, among which 328 were excluded, leaving 25 studies14–20,27–44 including 2605 patients who were eligible for inclusion. The details for study selection are indicated in Figure 1.
FIGURE 1.
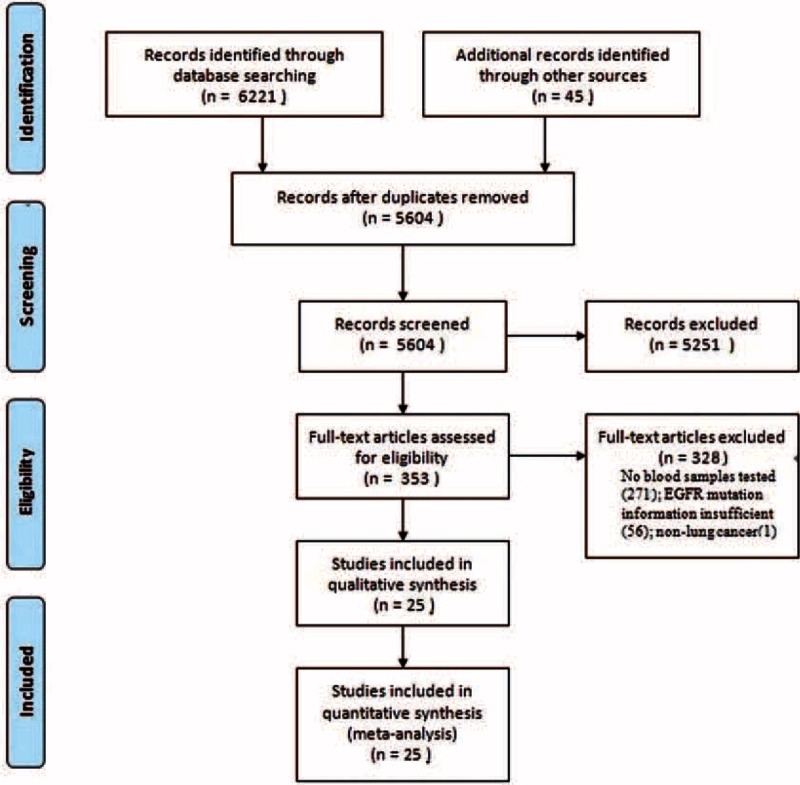
Flowchart of study selection.
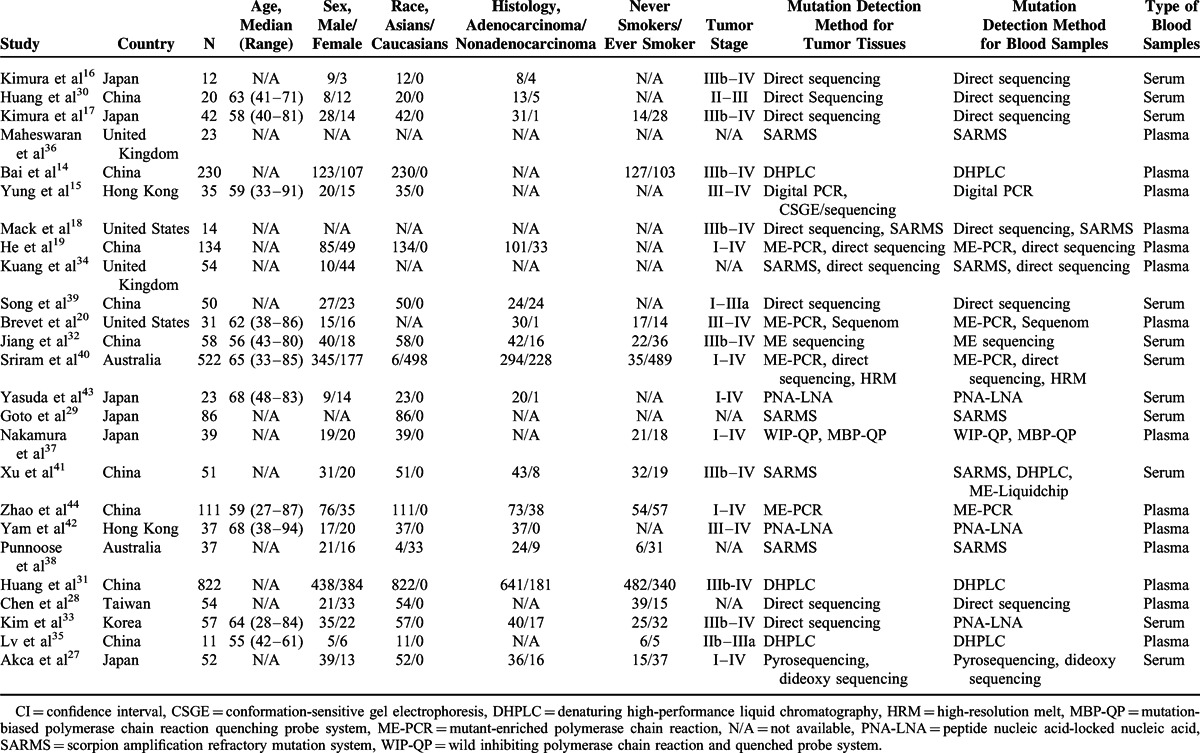
Of all included studies, 19 studies14–17,19,27–33,35,37,39,41–44 were carried out mainly in Asians and 618,20,34,36,38,40 in Caucasians. The participants in 12 included studies14–18,20,31–33,35,41,42 were in stages III–IV and 6 studies19,27,37,40,43,44 in I–IV. Thirteen studies14,15,18–20,28,31,35–38,42,44 tested EGFR mutations in plasma and 12 studies16,17,27,29,30,32–34,39–41,43 in serum. Direct sequencing and scorpion amplification refractory mutation system (SARMS) were the main methods used for mutation test, which were adopted in 816–19,28,30,39,40 and 718,29,34–36,38,41 studies, respectively. The detailed baseline characteristics are presented in Table 1.
TABLE 1.
Characteristics of Included Studies
Overall, the methodological quality of included studies is moderate. In terms of patient selection, 11 studies14,17,30–34,38,39,41,44 enrolled a consecutive or random sample of patients, 14 studies14,16,18,29–31,33,35,36,38,41–44 avoided a case–control design, and inappropriate exclusion was avoided in 3 studies.34,38,44 As for the index test, most of the included studies did not clearly report whether the interpretation of index test results was blinded and if a threshold was prespecified. As for a reference standard, only 2 studies34,44 clearly stated that the reference standard results were interpreted without knowledge of the results of the index test. In terms of flow and timing, the patients of all studies received the same reference standard, but 15 studies14–16,18,19,29,34–36,38,40–44 did not include all the study population in the analysis. The quality assessment results are indicated in Supplemental Digital Content—Table 1, http://links.lww.com/MD/A260.
Characteristics of EGFR Mutations in Blood Samples
The number of studies contributed to the meta-analysis of the EGFR mutation rate in blood samples ranged from 3 to 25 (Table 2). The pooled rate of blood EGFR mutations of all exons was 35.0% (95% CI 26.0%–44.0%), with 27.20% (95% CI 16.0%–38.3%) of the mutations in exon 19, 18.8% (95% CI 10.4%–27.2%) in exon 21, 3.9% (95% CI 0.0%–9.2%) in exon 18, and 2.9% (95% CI 0.0%–6.4%) in exon 20.
TABLE 2.
Pooled Rates of EGFR Mutation in Blood Samples
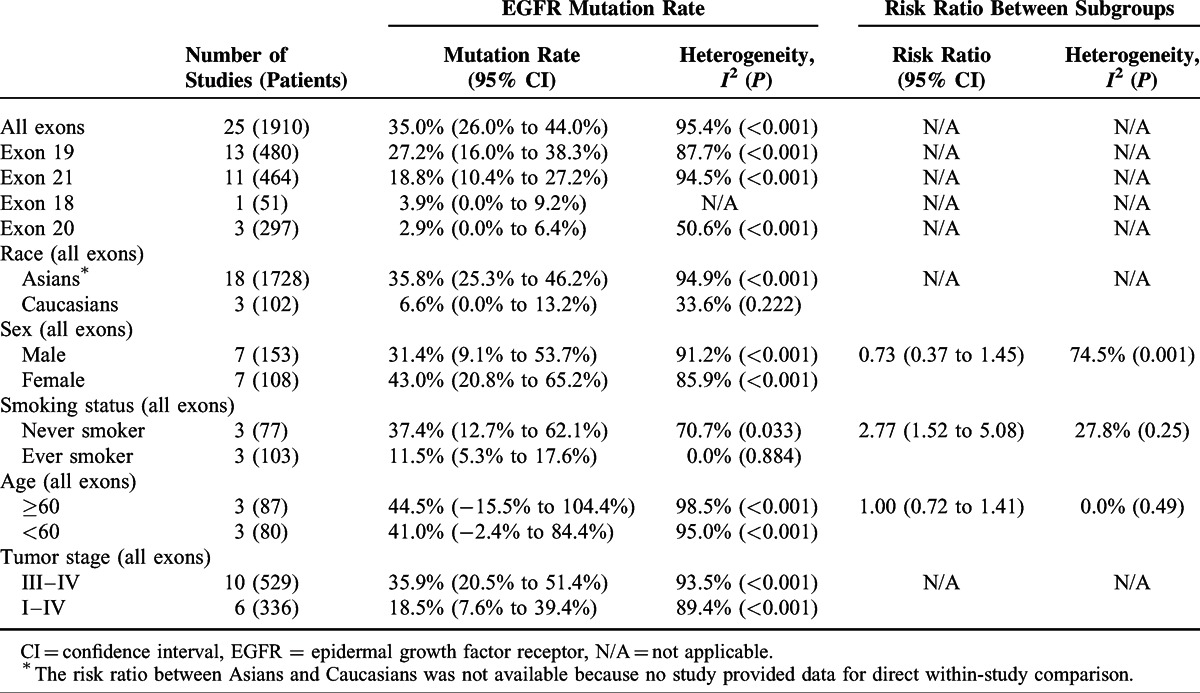
The EGFR mutation rates of all exons were pooled separately according to ethnicity, sex, smoking status, and age. The mutation rate was 35.8% (95% CI 25.3%–46.2%) in the Asian ethnicity and 6.6% (95% CI 0.0%–13.2%) in the Caucasians ethnicity. The mutation rate in females (43.0%; 95% CI 20.8%–65.2%) was higher than that in males (31.4%; 95% CI 9.1%–53.7%) but the difference was not statistically significant (RR: 0.73; 95% CI 0.37%–1.45%). Smoking status was significantly associated with EGFR mutations in blood, with a higher mutation rate in never smokers than in ever smokers (37.4% vs 11.50%; RR: 2.77; 95% CI 1.52–5.08). No difference was shown in the mutation rate between those aged ≥60 and those aged <60 (44.5% vs 41.0%; RR: 1.00; 95% CI 0.72–1.41). Subgroup analysis by tumor stage (stages I–II vs stages III–IV) was not done due to insufficient data for patients with stages I–II NSCLC. The pooled mutation rate in patients with stages III–IV NSCLC was 35.9%.
Consistency of EGFR Mutations in Tumor Tissue and Blood
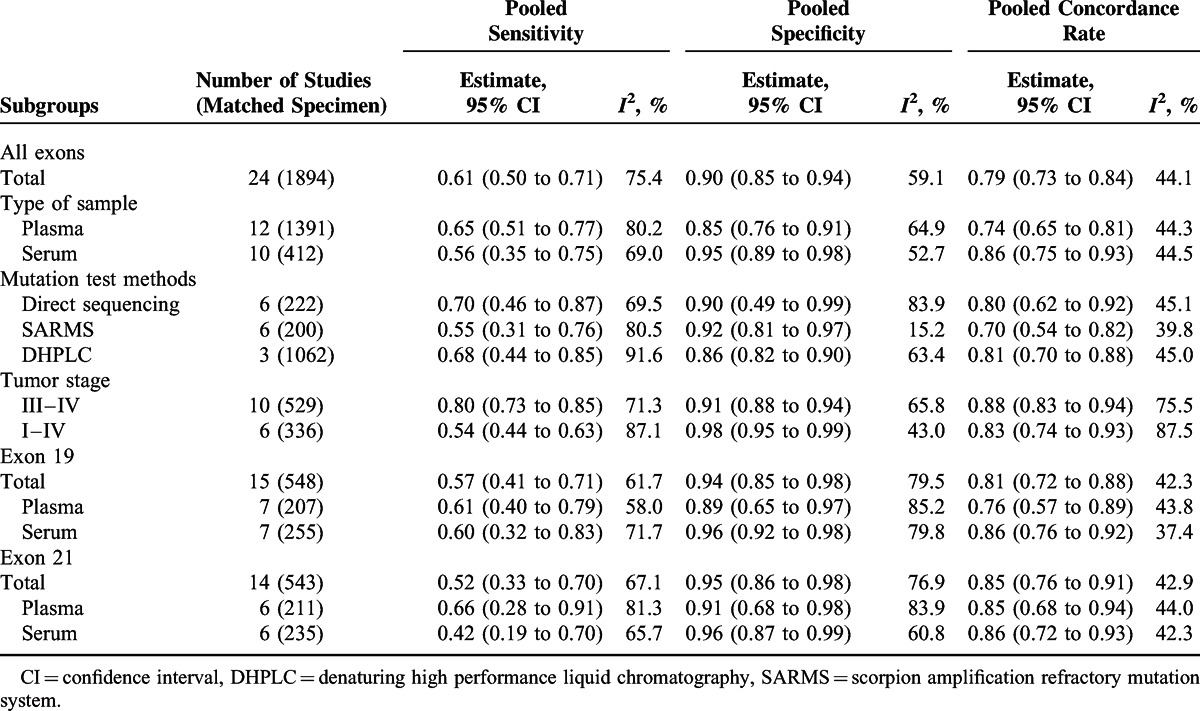
Table 3 presents the meta-analysis of diagnostic tests for EGFR mutation in blood using EGFR mutation in tumor tissues as the gold standard. Overall, the sensitivity, specificity, and concordance rate were 0.61 (95% CI 0.50–0.71), 0.90 (95% CI 0.85–0.94), and 0.79 (95% CI 0.73–0.84), respectively. There was no major difference in the subgroup analysis for mutations in exons 19 and 21 when compared with the mutations of all exons. As for the subgroup analysis according to the type of sample, the serum was generally associated with lower sensitivity but higher specificity and concordance when compared with plasma. As for mutation test methods, the direct sequencing showed the highest sensitivity (0.70; 95% CI 0.46–0.87) and SARMS showed the highest specificity (0.92; 95% CI 0.81–0.97). Subgroup analysis by tumor stage was not done due to insufficient data for patients with stages I–II NSCLC. The pooled sensitivity, specificity, and concordance rate in patients with stages III–IV NSCLC were 0.80 (95% CI 0.73–0.85), 0.91 (95% CI 0.88–0.94), and 0.88 (95% CI 0.83–0.94), respectively. Of note, the meta-analyses of diagnostic test were generally associated with high heterogeneity.
TABLE 3.
Accuracy and Agreement of EGFR Mutations in Blood Against Mutation Status in Tumor Tissue as the Reference
Association of EGFR Mutations and Response to EGFR TKIs Treatment
Five studies16–18,30,41 including 185 patients contributed to the meta-analysis of EGFR mutations (exons 19 and 21) and response to EGFR TKIs treatment. Meta-analysis demonstrated that the association between objective response to TKIs treatment and EGFR mutations were significant in both blood and tumor tissue, but the association was slightly stronger in tumor tissue (RR: 4.49; 95% CI 2.34–8.62) than in blood (RR: 4.08; 95% CI 2.48–6.70), though the subgroup difference was not significant (P = 0.82) (Figure 2). The Egger test (P = 0.628) and funnel plot suggested that there was no publication bias (Supplemental Digital Content—Figure 1, http://links.lww.com/MD/A260).
FIGURE 2.
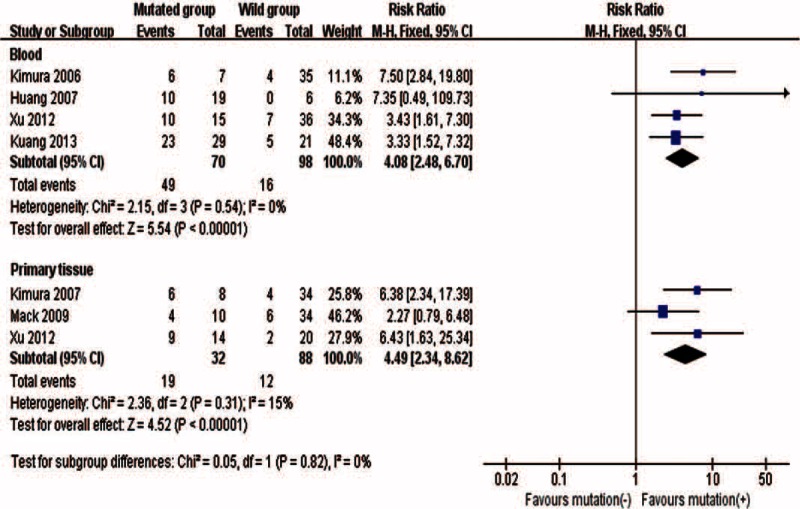
Association between objective response to TKIs treatment and EGFR mutations in blood or tumor tissue samples.
Association of EGFR Mutations and Survival
Eight studies14,16–19,28,30,38,41 with a total of 564 patients contributed to the meta-analysis of EGFR mutations (exons 19 and 21) and survival. The pooled HR for PFS was larger in blood (HR: 0.72; 95% CI 0.64–0.80) than in tumor tissue (HR: 0.57; 95% CI 0.39–0.85) (Figure 3). Although no significant subgroup difference was identified (P = 0.20), the EGFR mutation in serum (HR: 0.58; 95% CI 0.4–0.83) tended to have better predictive value than that in plasma (HR: 0.73; 95% CI 0.65–0.83).
FIGURE 3.
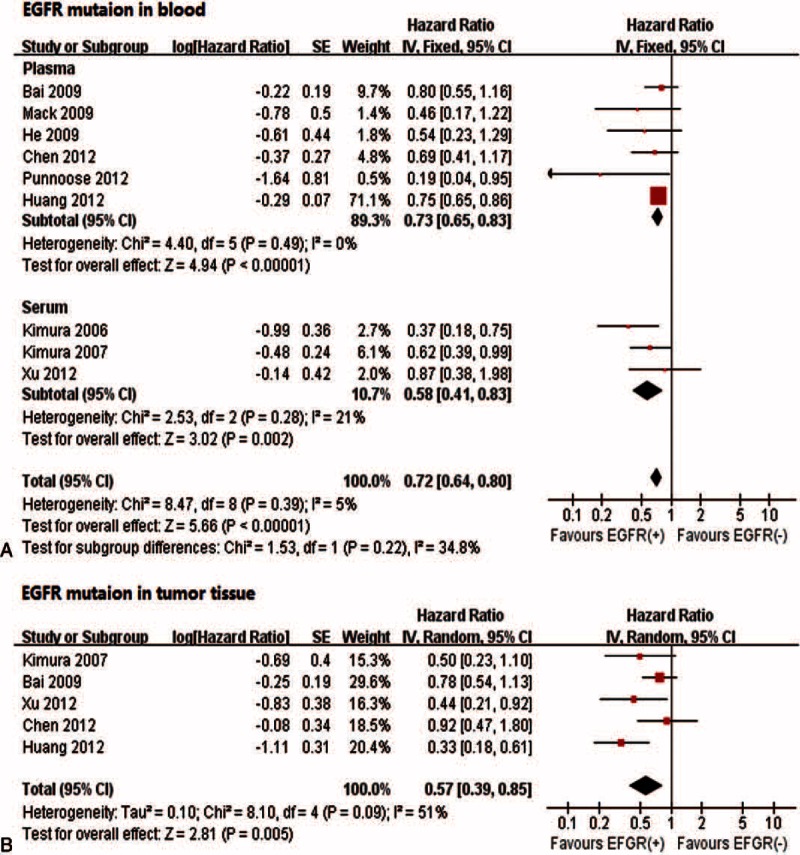
Associations between progression-free survival and EGFR mutations in blood (A) and tumor tissue samples (B). EGFR = epidermal growth factor receptor.
Likewise, the pooled HR for OS was 0.71 (95% CI 0.50–0.99) for EGFR mutations in blood and 0.54 (95% CI 0.38–0.76) in tumor tissue (Figure 4). The subgroup analysis indicated that the HR was significant in serum (HR: 0.59; 95% CI 0.39–0.91), but not in plasma (HR: 0.95; 95% CI 0.54–1.67). No significant subgroup difference was seen (P = 0.19).
FIGURE 4.
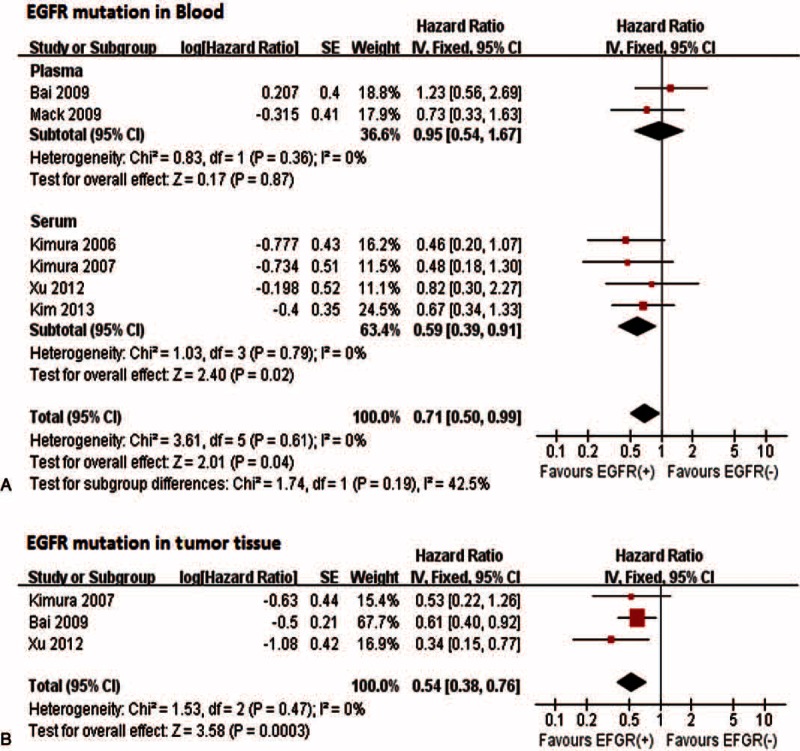
Associations between overall survival and EGFR mutations in blood (A) or tumor tissue samples (B). EGFR = epidermal growth factor receptor.
The risk of publication bias was not assessed in the meta-analyses in tumor tissues as the number of included studies was ≤5. In terms of EFGR mutations in blood, the funnel plot for PFS showed some asymmetry (Egger test: P = 0.04) (Supplemental Digital Content—Figure 2, http://links.lww.com/MD/A260) but the funnel plots for OS did not (Egger test: P = 0.69) (Supplemental Digital Content—Figure 3, http://links.lww.com/MD/A260).
DISCUSSION
Testing EGFR mutations in tumor tissue plays an important role in selecting patients for EGFR TKIs treatment,45,46 but in many patients, tumor tissues are either lacking or insufficient for mutations analysis. Blood may be a potential substitute for tumor tissues for this purpose. The present study reviewed 25 studies including 2605 patients and provided the most comprehensive analysis about the relation of EGFR mutations in blood to those in tumor tissues, and the value of EGFR mutations in blood in predicting the clinical outcomes of EGFR TKIs treatment.
We found a high overall concordance rate in EGFR mutation status between blood and tumor tissues, although the sensitivity can be further improved. Importantly, the high specificity of the blood testing against tissue analysis will guarantee high positive predictive values and avoid unnecessary treatment in those who have a false-positive result and will not respond to the treatment. This will be particularly true in populations that have a relatively high EGFR mutation rate. For example, based on the overall sensitivity of 0.64 and specificity of 0.89 and assuming a mutation rate of 10%, 30%, and 60% representing various populations, the positive predictive value will be 40%, 72%, and 90%, respectively. Interestingly, testing EGFR mutations in serum seems to have a higher specificity than in plasma, although the differences are not statistically significant.
Given the high positive predictive values, it is anticipated that those who are blood test positive would respond similarly or be slightly less well to EGFR TKIs treatment than those who are tissue test positive. Indeed, the difference in response to the treatment between EGFR mutation-positive and negative patients is very similar when either blood or tumor tissues are used for the testing. As PFS and OS are concerned, this difference is also evident when blood testing is used but slightly lower than that with tumor tissue testing.
For similar reasoning, we would also expect plasma test-positive patients benefit less than serum test-positive patients, which is confirmed with empirical data in our study. Importantly, the response to EGFR TKIs treatment in serum test-positive patients relative to that in serum-negative patients is almost as large as that observed when mutation status is determined based on tissue testing. This seems to give strong support for serum as a good surrogate for tumor tissue for EGFR mutation testing. Having said this, it appears that more future research is required to confirm the superiority of serum over plasma in this context.
In using EGFR mutation status to guide EGFR TKIs treatment, it is important to achieve a high positive predictive value so that this expensive treatment that also has severe side effects will not be used in those are false positive and will not benefit from the treatment. It is known that the positive predictive value is also determined by the prevalence of the condition (which is EGFR mutation in our case) in addition to the sensitivity and specificity of the testing method. This means that the higher the prevalence of the mutation, the higher the positive predictive value, the more useful blood testing would be.
It has been shown that the prevalence of EGFR mutations is higher in females, nonsmokers, adenocarcinoma, and Asians.47,48 These associations were also reflected in the EGFR mutation rate in blood in our study although the data here are limited to a few small studies relevant to this project and biased by the false positives. This is important information for determining the applications of the blood mutation testing. We thus suggest blood testing, in particular serum testing, to be used in females, nonsmokers, adenocarcinoma, and Asians in whom the positive predictive value could reach >90% as they usually have a mutation rate >50%.
Our study also suggested that the EGFR mutation test methods may have different sensitivity and specificity. The sensitivity by direct sequencing, the current gold standard for testing EGFR mutations,49 was the highest in our study. Different from the work by Ellison et al,50 we found that SARMS had a relatively lower sensitivity than other methods. Further studies are needed as the sample size is generally small in the studies included. The difference in the accuracy of the methods might be partially a result of the hidden cutoff used in different methods.
Limitations of this meta-analysis primarily arise from the quality of original studies reviewed. Owing to insufficient reporting of the original studies, the assessment of many QUADAS items could only be judged as unclear, which suggests that there may be a risk of bias in patient selection and interpretation of reference and index test. Regarding the estimate of the efficacy of treatment, the bias would affect both the blood test and tissue test groups, and these may cancel each other out. Second, the sample sizes in most included studies were small and some important subgroup differences may truly exist but cannot be confirmed. Third, like many other meta-analyses of single rates, our study showed large statistical heterogeneity that could not be explained. We employed the random-effects model in combing the result. Last, the level of circulating DNA could be related to tumor volume that largely depends on tumor stage, so tumor stage could be a potential factor that may influence the results. Owing to insufficient data for patients with stages I–II NSCLC, we were unable to perform subgroup analysis according to tumor stage. However, we pooled the mutation rate, sensitivity, specificity, and concordance rate in studies including stages III–IV and I–IV NSCLC, separately. As expected, the pooled sensitivity and concordance rate in studies including stages III–IV NSCLC were relatively higher than those in studies including all stages NSCLC. This suggests that blood EGFR mutation testing is particularly suitable for patients in advanced NSCLC, though further investigation is still needed.
In conclusion, blood, in particular serum, is a good substitute when tumor tissue is absent or insufficient for testing EGFR mutations to guide EGFR TKIs treatment in patients with NSCLC. EGFR mutation positivity in blood could be used to recommend EGFR TKIs treatment, but the absence of blood positivity should not necessarily be construed with confirmed negativity. Further research is necessary to further improve the sensitivity and identify the best testing methods for testing EGFR mutations in blood.
Footnotes
Abbreviations: CI = confidence interval, EGFR = epidermal growth factor receptor, HR = hazard ratio, NSCLC = nonsmall cell lung cancer, OS = overall survival, PFS = progression-free survival, QUADAS-2 = Quality Assessment of Diagnostic Accuracy Studies-2, RR = risk ratio, TKIs = tyrosine kinase inhibitors.
CM and JQY contributed equally to this study and should be considered as co-first authors.
This study was partly supported by the Health and Medical Research Fund from Food and Health Bureau of Hong Kong (Project no: 11120971).
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, et al. Global cancer statistics, 2002. CA Cancer J Clin 2005; 55:74–108. [DOI] [PubMed] [Google Scholar]
- 2.Brambilla E, Travis WD, Colby TV, et al. The new World Health Organization classification of lung tumours. Eur Respir J 2001; 18:1059–1068. [DOI] [PubMed] [Google Scholar]
- 3.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial). J Clin Oncol 2003; 21:2237–2246. [DOI] [PubMed] [Google Scholar]
- 4.Jonker DJ, O’Callaghan CJ, Karapetis CS, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med 2007; 357:2040–2048. [DOI] [PubMed] [Google Scholar]
- 5.Kris MG, Natale RB, Herbst RS, et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. J Am Med Assoc 2003; 290:2149–2158. [DOI] [PubMed] [Google Scholar]
- 6.Shepherd FA, Rodrigues Pereira J, Ciuleanu T, et al. Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 2005; 353:123–132. [DOI] [PubMed] [Google Scholar]
- 7.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. Lancet Oncol 2010; 11:121–128. [DOI] [PubMed] [Google Scholar]
- 8.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin 2009; 59:225–249. [DOI] [PubMed] [Google Scholar]
- 9.Shih JY, Gow CH, Yu CJ, et al. Epidermal growth factor receptor mutations in needle biopsy/aspiration samples predict response to gefitinib therapy and survival of patients with advanced nonsmall cell lung cancer. Int J Cancer 2006; 118:963–969. [DOI] [PubMed] [Google Scholar]
- 10.Coghlin CL, Smith LJ, Bakar S, et al. Quantitative analysis of tumor in bronchial biopsy specimens. J Thorac Oncol 2010; 5:448–452. [DOI] [PubMed] [Google Scholar]
- 11.Costa DB, Kobayashi S, Tenen DG, et al. Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non-small cell lung cancers. Lung Cancer 2007; 58:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruhn N, Beinert T, Oehm C, et al. Detection of microsatellite alterations in the DNA isolated from tumor cells and from plasma DNA of patients with lung cancer. Ann N Y Acad Sci 2000; 906:72–82. [DOI] [PubMed] [Google Scholar]
- 13.Sozzi G, Conte D, Leon M, et al. Quantification of free circulating DNA as a diagnostic marker in lung cancer. J Clin Oncol 2003; 21:3902–3908. [DOI] [PubMed] [Google Scholar]
- 14.Bai H, Mao L, Wang HS, et al. Epidermal growth factor receptor mutations in plasma DNA samples predict tumor response in Chinese patients with stages IIIB to IV non-small-cell lung cancer. J Clin Oncol 2009; 27:2653–2659. [DOI] [PubMed] [Google Scholar]
- 15.Yung TK, Chan KC, Mok TS, et al. Single-molecule detection of epidermal growth factor receptor mutations in plasma by microfluidics digital PCR in non-small cell lung cancer patients. Clin Cancer Res 2009; 15:2076–2084. [DOI] [PubMed] [Google Scholar]
- 16.Kimura H, Kasahara K, Shibata K, et al. EGFR mutation of tumor and serum in gefitinib-treated patients with chemotherapy-naive non-small cell lung cancer. J Thorac Oncol 2006; 1:260–267. [DOI] [PubMed] [Google Scholar]
- 17.Kimura H, Suminoe M, Kasahara K, et al. Evaluation of epidermal growth factor receptor mutation status in serum DNA as a predictor of response to gefitinib (IRESSA). Br J Cancer 2007; 97:778–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mack PC, Holland WS, Burich RA, et al. EGFR mutations detected in plasma are associated with patient outcomes in erlotinib plus docetaxel-treated non-small cell lung cancer. J Thorac Oncol 2009; 4:1466–1472. [DOI] [PubMed] [Google Scholar]
- 19.He C, Liu M, Zhou C, et al. Detection of epidermal growth factor receptor mutations in plasma by mutant-enriched PCR assay for prediction of the response to gefitinib in patients with non-small-cell lung cancer. Int J Cancer 2009; 125:2393–2399. [DOI] [PubMed] [Google Scholar]
- 20.Brevet M, Johnson ML, Azzoli CG, et al. Detection of EGFR mutations in plasma DNA from lung cancer patients by mass spectrometry genotyping is predictive of tumor EGFR status and response to EGFR inhibitors. Lung Cancer 2011; 73:96–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 2011; 155:529–536. [DOI] [PubMed] [Google Scholar]
- 22.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 1959; 22:719–748. [PubMed] [Google Scholar]
- 23.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials 1986; 7:177–188. [DOI] [PubMed] [Google Scholar]
- 24.WG C. The combination of estimates from different experiments. Biometrics 1954; 10:101–129. [Google Scholar]
- 25.Soeken K, Sripusanapan A. Assessing publication bias in meta-analysis. Nurs Res 2003; 52:57–60. [DOI] [PubMed] [Google Scholar]
- 26.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. Ann Intern Med 2009; 151:W65–94. [DOI] [PubMed] [Google Scholar]
- 27.Akca H, Demiray A, Yaren A, et al. Utility of serum DNA and pyrosequencing for the detection of EGFR mutations in non-small cell lung cancer. Cancer Genet 2013; 206:73–80. [DOI] [PubMed] [Google Scholar]
- 28.Chen YM, Fan WC, Tseng PC, et al. Plasma epidermal growth factor receptor mutation analysis and possible clinical applications in pulmonary adenocarcinoma patients treated with erlotinib. Oncol Lett 2012; 3:713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goto K, Ichinose Y, Ohe Y, et al. Epidermal growth factor receptor mutation status in circulating free DNA in serum: from IPASS, a phase III study of gefitinib or carboplatin/paclitaxel in non-small cell lung cancer. J Thorac Oncol 2012; 7:115–121. [DOI] [PubMed] [Google Scholar]
- 30.Huang JS, Dong QG, Xu KL, et al. Epidermal growth factor receptor mutation in serum circulating DNA and selective targeting therapy against lung cancer. Tumor 2007; 27:968–972. [Google Scholar]
- 31.Huang Z, Wang Z, Bai H, et al. The detection of EGFR mutation status in plasma is reproducible and can dynamically predict the efficacy of EGFR-TKI. Thoracic Cancer 2012; 3:334–340. [DOI] [PubMed] [Google Scholar]
- 32.Jiang B, Liu F, Yang L, et al. Serum detection of epidermal growth factor receptor gene mutations using mutant-enriched sequencing in Chinese patients with advanced non-small cell lung cancer. J Int Med Res 2011; 39:1392–1401. [DOI] [PubMed] [Google Scholar]
- 33.Kim S, Sung J, Jo U, et al. Can mutations of EGFR and KRAS in serum be predictive and prognostic markers in patients with advanced non-small cell lung cancer (NSCLC)? Med Oncol 2013; 30:328. [DOI] [PubMed] [Google Scholar]
- 34.Kuang Y, Rogers A, Yeap BY, et al. Noninvasive detection of EGFR T790 M in gefitinib or erlotinib resistant non-small cell lung cancer. Clin Cancer Res 2009; 15:2630–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lv C, Ma Y, Feng Q, et al. A pilot study: sequential gemcitabine/cisplatin and icotinib as induction therapy for stage IIB to IIIA non-small-cell lung adenocarcinoma. World J Surg Oncol 2013; 11:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maheswaran S, Sequist LV, Nagrath S, et al. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med 2008; 359:366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura T, Sueoka-Aragane N, Iwanaga K, et al. Application of a highly sensitive detection system for epidermal growth factor receptor mutations in plasma DNA. J Thorac Oncol 2012; 7:1369–1381. [DOI] [PubMed] [Google Scholar]
- 38.Punnoose EA, Atwal S, Liu W, et al. Evaluation of circulating tumor cells and circulating tumor DNA in non-small cell lung cancer: association with clinical endpoints in a phase II clinical trial of pertuzumab and erlotinib. Clin Cancer Res 2012; 18:2391–2401. [DOI] [PubMed] [Google Scholar]
- 39.Song G, Ren J, Zhang L, et al. Low correspondence of EGFR mutations in tumor tissue and paired serum of non-small-cell lung cancer patients. Chin J Cancer Res 2010; 22:27–31. [Google Scholar]
- 40.Sriram KB, Tan ME, Savarimuthu SM, et al. Screening for activating EGFR mutations in surgically resected nonsmall cell lung cancer. Eur Respir J 2011; 38:903–910. [DOI] [PubMed] [Google Scholar]
- 41.Xu F, Wu J, Xue C, et al. Comparison of different methods for detecting epidermal growth factor receptor mutations in peripheral blood and tumor tissue of non-small cell lung cancer as a predictor of response to gefitinib. Onco Targets Ther 2012; 5:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yam I, Lam DC, Chan K, et al. EGFR array: uses in the detection of plasma EGFR mutations in non-small cell lung cancer patients. J Thorac Oncol 2012; 7:1131–1140. [DOI] [PubMed] [Google Scholar]
- 43.Yasuda H, Soejima K, Nakayama S, et al. Bronchoscopic microsampling is a useful complementary diagnostic tool for detecting lung cancer. Lung Cancer 2011; 72:32–38. [DOI] [PubMed] [Google Scholar]
- 44.Zhao X, Han RB, Zhao J, et al. Comparison of epidermal growth factor receptor mutation statuses in tissue and plasma in stage I-IV non-small cell lung cancer patients. Respiration 2012; 85:119–125. [DOI] [PubMed] [Google Scholar]
- 45.Bentel JM, Thomas MA, Sadowska A, et al. From theory to practice: EGFR mutation testing in current practice and roadblocks to its implementation. J OncoPathol 2013; 1:93–102. [Google Scholar]
- 46.Sequist LV, Heist RS, Shaw AT, et al. Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann Oncol 2011; 22:2616–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med 2009; 361:958–967. [DOI] [PubMed] [Google Scholar]
- 48.Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007; 7:169–181. [DOI] [PubMed] [Google Scholar]
- 49.Pirker R, Herth FJ, Kerr KM, et al. Consensus for EGFR mutation testing in non-small cell lung cancer: results from a European workshop. J Thorac Oncol 2010; 5:1706–1713. [DOI] [PubMed] [Google Scholar]
- 50.Ellison G, Donald E, McWalter G, et al. A comparison of ARMS and DNA sequencing for mutation analysis in clinical biopsy samples. J Exp Clin Cancer Res 2010; 29:132. [DOI] [PMC free article] [PubMed] [Google Scholar]


