Supplemental Digital Content is available in the text
Abstract
As an immunotoxin, diphtheria toxin has been widely used in gene therapy and gene function assays for its roles in protein synthesis inhibition, and the aim of our study is to set up a nonintegrating lentiviral system for specific expression of diphtheria toxin A (DTA) used in cancer gene therapy.
Here, we established a lentiviral system that could coordinately express fluorescent protein and DTA driven by the cytomegalovirus (CMV) promoter, which is convenient for us to precisely trace the expression of DTA and monitor the process of lentivirus packaging. To achieve safer cancer therapy, we replaced the CMV promoter with the Survivin promoter, a specific promoter that is dramatically activated in cancer tissues and cells, but not in normal tissues and cells, and that will impose greater therapeutic potential because a significant expression difference occurred between these 2 groups. Meanwhile, we obtained integrase-deficient lentivirus (IDLV) after packaging with the integrase mutant, which expresses defective integrase RRK262263264AAH, to minimize the side effects that derived from the insertional mutagenesis of the host genome.
Our results suggest that the IDLV system that we generated possesses therapeutic potential in cancers in vitro and in vivo.
INTRODUCTION
Survivin, one member of the inhibitor of the apoptosis protein family, is expressed in multiple types of cancers but is undetectable in normal tissues.1–3 A series of studies has revealed that this protein could serve as a biomarker for the diagnosis and prognosis of multiple cancers, including breast cancer,4,5 esophageal cancer,6,7 endometrial cancer,8 salivary gland cancer,9 nonsmall-cell lung cancer and small-cell lung cancer,10,11 gastric cancer,12 and epithelial ovarian cancer.13 Primary studies have revealed that Survivin correlates with apoptosis inhibition, while recent studies demonstrated that Survivin is also involved in multiple positive progress of malignant processes and cancer progression, including tumorigenesis,14 invasion and metastasis,15,16 and drug resistance,17–20 and is also closely correlated with the clinical stage of cancer, lymph node and distant metastasis. Furthermore, Survivin may also serve as a signal mediator because it can be regulated by multiple biomolecules in biological processes.21–25 Thus, Survivin has been proposed as an effective target for cancer therapy alone or combination with other therapeutic strategies.26–31 High expression level of Survivin is always detected in multiple cancers since its promoter is specifically and highly activated in cancers, thus Survivin promoter is a valuable tool to drive specific expression of target genes, especially for toxic genes, in cancer cells for gene therapy.2,32–37
Immunotoxins, including diphtheria toxin (DT), have been widely used for cancer therapy.38,39 Previous studies revealed that cancer cells are more sensitive to DT than are normal cells, and DT could inactivate elongation factor 2 (EF-2) by adenosine diphosphate ribosylation and inhibit protein translation, thereby triggering apoptosis and achieving cancer therapy through the application of DT with specific promoters40–46 or a fused protein,47,48 which can selectively target and kill cancer cells. However, the limitation of these studies, including the difficulties in translocation of target proteins or genes into cancer cells and the efficacy or safety of these tools used for translocation, always occurred. Although lentiviruses have been extensively used in genetic manipulation due to their powerful ability to deliver target sequences, they also harbor multiple advantages, including greater transgene capacity and the transduction of genes into the nucleus of nondividing cells mediated by the preintegration complex (PIC), low immunogenicity, and the persistent expression of target genes for their insertion within the genomic DNA of host cells. However, many concerns have also arisen regarding their insertion, and integrase-deficient lentivirus (IDLV) represents a promising tool for human gene therapy, which has been well reviewed in our previous studies.49
In this study, we achieved the coordinate expression of fluorescent protein and diphtheria toxin A (DTA) through lentiviral vectors, and these expression cassettes are driven by the cytomegalovirus (CMV) or Survivin promoter, thereby achieving the specific and efficient expression of the toxic gene in cancer cells but not normal cells. Additionally, we obtained IDLV expressing DTA in modified HEK293T cells, which could inhibit cell proliferation through protein synthesis inhibition in vitro. Meanwhile, we also demonstrated that these IDLVs could inhibit the tumor growth in immune-deficient nude mice.
MATERIALS AND METHODS
Cell Culture
L-O2, HFL-1, TE-1, Eca109, A549, MDA-MB-231, SK-BR-3, ZR-7530, HeLa, and HEK293T cells were grown in Dulbecco's Modified Eagle medium (DMEM) plus 10% heat-inactivated fetal bovine serum containing penicillin and streptomycin (pen/strep). To establish the modified stable cell line HEK293T[mEF-2(G717R)], we added 1.5 μg/mL puromycin to the culture medium.
Clinical Sample Collection
All of the human normal and tumor samples were obtained from patients of Fuzhou General Hospital under a standard protocol that was approved by the Fuzhou General Hospital Ethics Committee. Total RNA was extracted from fresh samples using TRIzol, and cDNA synthesis was performed with the ReverTra Ace qPCR RT Kit (TOYOBO, Osaka, Japan, Cat No. FSQ-301); meanwhile, total protein was also extracted after lysing with Ripa lysis buffer.
Western Blots
The total protein of cells or tissues was harvested and transferred to a PVDF membrane for western blotting. The human Survivin protein level was detected using polyclonal rabbit anti-survivin antibody (Abcam, Cambridge, USA, Cat No. ab24479), and human β-actin was used as an internal control using a mouse anti-β-actin monoclonal antibody (Beyotime, Nantong, China, Cat No. AA128). The corresponding Horse Radish Peroxidase (HRP)-conjugated goat anti-rabbit IgG (Abcam, Cat No. ab136817) and HRP-conjugated goat anti-mouse IgG (ZSGB-Bio, Cat No. ZB-2305) served as secondary antibodies to detect the abundance of these proteins.
Quantitative PCR
The RNA of cells or tissues was extracted, and cDNA was produced using cDNA synthesis kit. Real-time PCRs were performed following the manual of the BIO-RAD Mini Thermal Cycler and the SYBR Green qPCR Mix Reagent (TOYOBO, Cat No. QPS-201). The primers that were used for all of the target genes are described in Table S1 http://links.lww.com/MD/A359.
Target Sequences Clone, Vectors Construction, and Luciferase Reporter Assays
All of the target sequences were obtained using Pfu polymerase (Transgene, Beijing, China, Cat No. M0254L) and specific primers (Tables S2 and S3 http://links.lww.com/MD/A359). The primers in Table S2 http://links.lww.com/MD/A359 were used to amplify the Survivin promoters, and pGL3-Surp269-Luciferase and pGL3-Surp1430-luciferase were, respectively, obtained after the insertion of these promoter fragments into a pGL3-Basic vector that was linearized with the same restricted enzymes. Lentiviral vectors for coordinately expressing fluorescent protein and DTA were constructed, and these expression cassettes were driven by the CMV or Survivin-1430 promoters (Surp1430). The open-reading frame (ORF) of DTA was amplified from pBSDT-AII plasmids (kindly provided by Prof Yoh Wada at Osaka University, Japan), and other target sequences were derived from our previous study.50 The primers that were used for lentiviral vector construction are described in Table S3 http://links.lww.com/MD/A359, and the steps for cloning and construction are summarized in Figure S1 http://links.lww.com/MD/A359.
The luciferase reporter activities were determined using the Dual-Luciferase Reporter Assay System (Promega, Madison, USA, Cat No. E1910). For all of the luciferase assays, pGL3 and pRL-TK were co-transfected into cells, and the Renilla luciferase activities were determined to calibrate the transfection efficiency. The calibrated value for internal control was used to normalize all of the other values to obtain the normalized relative luciferase units representing the activities of the corresponding promoters.
EF-2 and Integrase Direct Mutation
To produce DTA-expressing lentivirus in HEK293T cells, a modified HEK293T cell line that could express mutated elongation factor 2 (mEF-2) should be established. The specific primers for human EF-2 clone and mutagenesis are described in Table S4 http://links.lww.com/MD/A359. The PCR products of EF-2 were extracted and then digested with EcoRI and BamHI enzymes; following the digestion, pCDH-CMV-EF-2-EF1-puro was obtained after the insertion of digested products of EF-2 into pCDH-CMV-MCS-EF1-puro, and the mutagenesis of human EF-2 was subsequently performed by the site-directed mutagenesis of codon 717(G717R, GGA to CGA) using a Site-Directed Mutagenesis Kit (Stratagene, San Diego, USA).
To generate mutations at specific sites in integrase, an AgeI-integrase-BspT1 fragment was obtained by high-fidelity PCR with a template pMDLg/pRRE vector and the specific primers in Table S4 http://links.lww.com/MD/A359, and AgeI-integrase-BspT1 was inserted into the pMD-18T simple vector (TaKaRa, Dalian, China, Cat No. D103A). The sequence of integrase was confirmed by enzyme digestion and sequencing. The pMD-18T simple vector containing the integrase ORF served as the template, and mutagenesis was performed with the Site-Directed Mutagenesis Kit (Stratagene) and the specific primers in Table S4 http://links.lww.com/MD/A359. Some single bacteria were selected, and plasmids were isolated for identification by enzyme digestion and sequencing. After the identification of mutations, enzyme digestion and ligation was performed for the construction of pMDLg/pRRE (RRK262263264AAH), a packaging vector containing mutated sites at RRK262263264.
Lentivirus Production, Precipitation, and Titration
All of the lentiviruses were generated in HEK293T[mEF-2(G717R)] after transient transfection with the PEI reagent (Sigma, Saint Louis, USA, Cat No. 40872-7) and plasmids (Figure S3 http://links.lww.com/MD/A359). Lentivirus-containing medium was collected every 24 h for 3 days, and cellular debris was cleared by low-speed centrifugation and passage through a 0.45-μm filter (Millipore, Billerica, USA, Cat No. SLHV033RB). The collected medium was concentrated with the PEG-it™ virus precipitation solution (SBI, Mountain View, USA, Cat No. LV810A-1), and pellets containing viral particles were re-suspended with DMEM. Lentiviral titers was measured with the QuickTiter™ Lentivirus Quantitation Kit (CELL BIOLABS, San Diego, USA, Cat No. VPK-108-HIV-P24).
Xenograft Studies and Lentivirus Injection
Viable ZR7530 cells were harvested and re-suspended in DMEM and then injected subcutaneously into 4- to 6-week-old female nude mice. The animal experiments were performed according to the ethics rules that were approved by the Fuzhou General Hospital Ethics Committee. Among these nude mice, 5 nude mice per group were randomly divided for receiving different treatments. The tumor sizes were measured every 5 days using a vernier calliper after injection of concentrated lentivirus, and the tumor volumes were analyzed using the formula 0.52 × width2 × length. The tumors were dissected and separated from the bodies at the time of sacrifice, which was 60 days post-tumor implantation.
Statistical Analysis
The data represent at least 3 independent experiments using cells or extracts from a minimum of 3 separate isolations. The differences between the groups were compared using an analysis of variance for repeated measures. All of the statistical analyses were performed with the software GraphPad. Prism. v5.0, and the error bars indicate the standard deviation.
RESULTS
Increased Level of Survivin Protein and mRNA in Cancer Tissues and Cells
We tested the endogenous expression level of Survivin protein and mRNA in several human clinical samples, including normal rectum tissue, esophageal squamous cell carcinoma adjacent tissue, esophageal squamous cell carcinoma tissue, gastric cancer adjacent tissue, gastric cancer tissue, breast cancer tissue, rectal cancer adjacent tissue, and rectal cancer tissue. As shown in Figure 1A, we did not detect the expression of Survivin protein in normal rectum tissue, and a very weak level of Survivin protein was found in esophageal squamous cell carcinoma adjacent tissue; however, Survivin protein was easily detectable in the other tissues. Moreover, the most distinct difference in protein level occurred between the normal rectum group and the gastric cancer adjacent tissue group, and the gastric cancer adjacent tissue had a 288-fold increase compared to that of the normal tissue. Even the rectal cancer tissue, the group with the lowest expression of Survivin protein, had a 7.6-fold increase compared to that of the normal group. The Survivin transcript was also evaluated through quantitative real-time PCR with specific primers that are described in Table S1 http://links.lww.com/MD/A359. The levels of Survivin mRNA transcript were also quite lower in normal rectum tissue and esophageal squamous cell carcinoma adjacent tissue than in the other tissues that were used in this study, especially in breast cancer tissue, which had a >7000-fold increase compared to that of normal rectum tissue (Figure 1B).
FIGURE 1.
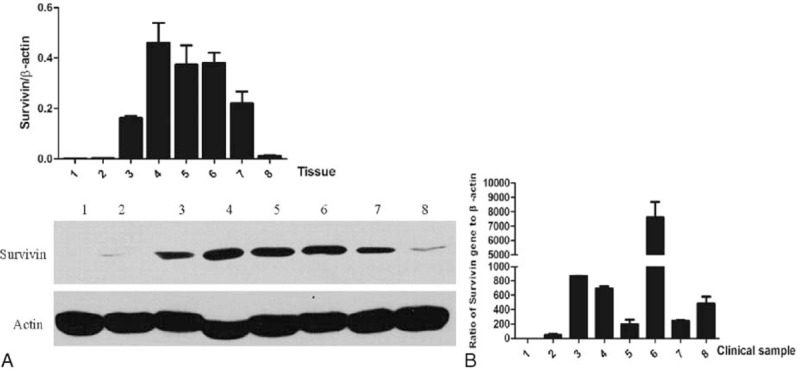
Survivin protein and mRNA levels are dramatically increased in cancer tissues. 1, normal rectum tissue; 2, esophageal squamous cell carcinoma adjacent tissue; 3, esophageal squamous cell carcinoma tissue; 4, gastric cancer adjacent tissue; 5, gastric cancer tissue; 6, breast cancer tissue; 7, rectal cancer adjacent tissue; 8, rectal cancer tissue. (A) Comparison of the protein levels in normal and cancer tissues by western blotting. The protein levels of Survivin are normalized with the β-actin protein level. The analysis and histogram of the protein level are described above the protein bands; (B) Analysis of the mRNA levels in normal and cancer tissues by quantitative polymerized chain reaction. The mRNA levels of Survivin are normalized with the β-actin mRNA level.
In addition to the collection of clinical samples, we also examined the expression level of Survivin mRNA and protein in normal and cancer cells, including 2 normal cell lines L-O2 and HFL-1; 2 lines of esophageal cancer cells TE-1 and Eca109; the lung cancer cell line A549; 3 lines of breast cancer cells MDA-MB-231, SK-BR-3, and ZR7530; and the cervical cancer cell line HeLa. As shown in Figure 2A, no or a weak expression of Survivin protein was found in normal cells, whereas a more abundant level of Survivin protein occurred in cancer cells compared to normal cells. The greatest level of Survivin occurred in SK-BR-3 cells, and the lowest level occurred in A549, the difference between these 2 groups was 24.1-fold. On average, cancer cells showed a 46.1-fold increase in the Survivin protein level. In addition, the Survivin transcript was also analyzed through quantitative real-time PCR with the same primers as described above. The levels of the Survivin transcript in cancer cells were higher than in normal cells, and the most significant difference occurred between HFL-1 and ZR7530 cells, with a 5.8-fold increase in ZR7530 cells. On average, cancer cells showed a 1.8-fold increase in the Survivin mRNA level compared to those of normal cells (Figure 2B).
FIGURE 2.
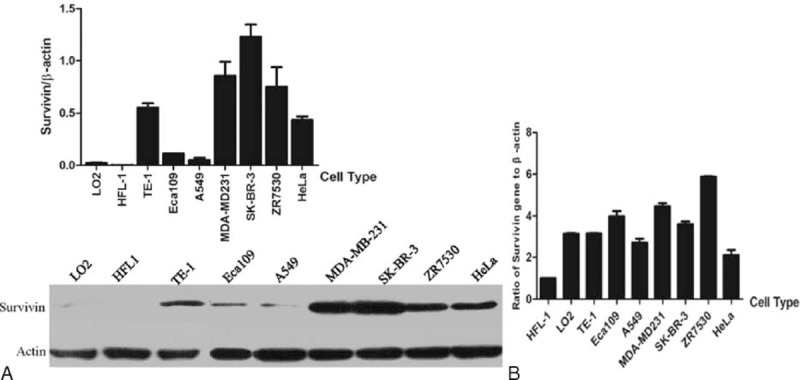
Survivin protein and mRNA levels are dramatically increased in cancer cells. (A) Comparison of the protein levels in normal and cancer cells by western blotting. The protein levels of Survivin are normalized with the β-actin protein level. The analysis and histogram of the protein level are described above the protein bands; (B) Analysis of mRNA levels in normal and cancer cells by quantitative polymerized chain reaction. The mRNA levels of Survivin are normalized with the β-actin mRNA level.
Survivin-1430 Promoter Has a Stronger Activity Than Survivin-269 Promoter and Is Dramatically Elevated in Cancer Cells
To better use the Survivin promoter in cancer gene therapy, 2 pairs of specific primers were designed to clone Surp1430 and Survivin-269 promoters (Surp269), respectively. As shown in Figure 3A, we obtained these target bands of the Survivin promoter with high-fidelity PCR, and we also obtained the corresponding pGL3 vectors as driven by Surp1430 or Surp269 (Figure 3B). Moreover, the activities of these 2 promoters in different cancer cells, including HeLa, Eca109, and ZR-7530, were analyzed after the co-transfection of pGL3 and pRL-TK, and we found that Surp1430 has a stronger activity than does Surp269 in all of these cells (Figure 3C).
FIGURE 3.
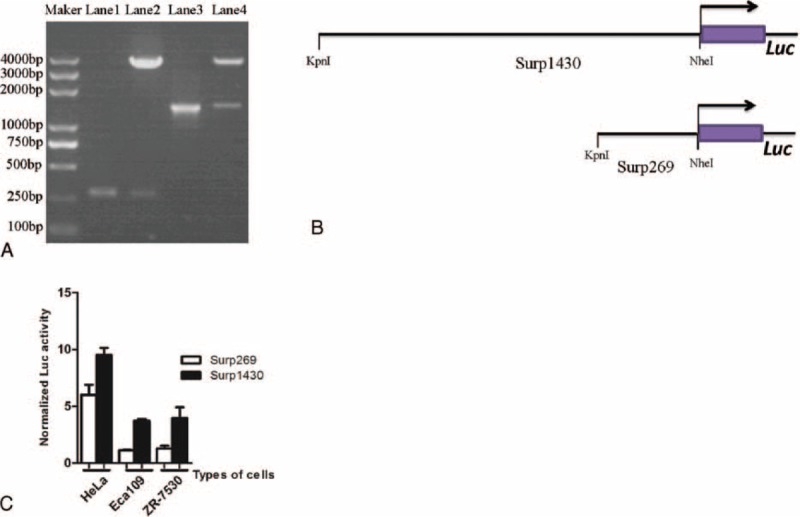
Clone different sizes of the Survivin promoter and compare their activities in cancer cells. (A) Clone of Survivin promoters and construction of pGL3 reporter vectors as driven by the Survivin promoter. Lane 1 is the PCR product of Surp269, lane 2 is the product of pGL3-Surp269-Luciferase digested with the KpnI and NheI enzymes, lane 3 is the PCR product of Surp1430, and lane 4 is the product of pGL3-Surp1430-Luciferase digested with KpnI and NheI enzymes; (B) Diagram of the Survivin promoter-Luc construct with the firefly luciferase gene under the control of Surp1430 or Surp269; (C) Comparison of the Surp1430 and Surp269 activities in different cancer cells. PCR = polymerized chain reaction.
Additionally, the activities of the CMV promoter and Surp1430 in multiple normal and cancer cells were compared through luciferase reporter assays, and the luciferase activities as driven by Surp1430 were tested after the co-transfection of pGL3-Surp1430 and pRL-TK. As shown in Figure 4, we found that the activity of Surp1430 is very weak in normal cells compared to that in cancer cells; however, the activity of Surp1430 dramatically increased in multiple cancer cells. The most significant difference occurred between HFL-1 and SK-BR-3, with up to a 55.6-fold difference between these 2 groups. Compared to the CMV promoter, a universal promoter that is always used to drive the ubiquitous expression of target genes, the activities of the Surp1430 promoter were slightly lower than those of the CMV promoter in tested cells. Therefore, we conclude that the Survivin promoter is a good driver for the expression of toxic genes, and Surp1430 is a better candidate than Surp269 in cancer gene therapy.
FIGURE 4.
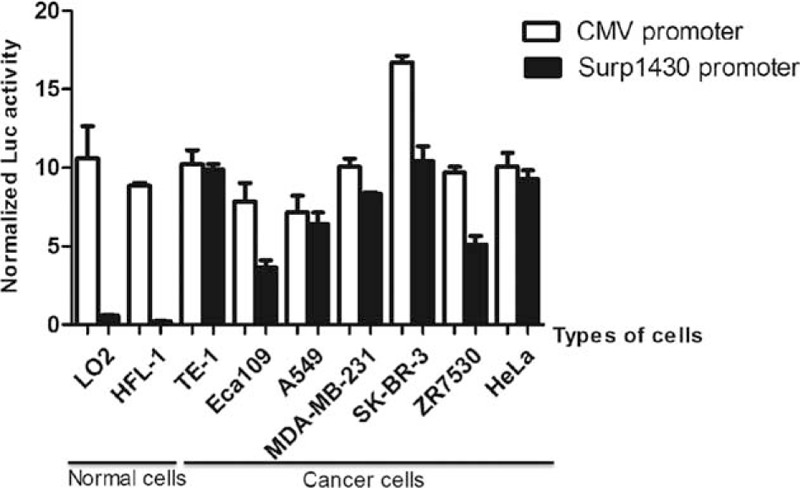
In vitro analyses of the Surp1430 and CMV promoter activities through transient transfection. The transcriptional activities of the Surp1430 and CMV promoters were measured in normal and multiple cancer cells. CMV = cytomegalovirus.
Specifically Killing Cancer Cells and Achieving Minimal Adverse Effects After Transfection With DTA-Expressing Lentiviral Vector Driven by Surp1430
Because the Survivin promoter is specifically activated in cancer cells, we designed and constructed 2 groups of lentiviral vectors, which could coordinately express fluorescent protein and DTA mediated by foot and mouth disease viruses (FMDV) 2A, to test their capabilities of killing cancer cells. Among these lentiviral vectors, coordinate expression cassettes in 1 group are driven by the CMV promoter, while coordinate expression cassettes in another group are driven by Surp1430 (Figure S2 http://links.lww.com/MD/A359). After the transfection of a different amount of DTA-expressing lentiviral vectors into normal and cancer cells, we counted the numbers of viable cells in each treated well, calculated their inhibitory effect on cell proliferation, and found that normal cells are not sensitive to DTA-expressing lentiviral vectors as driven by Surp1430; however, cancer cells differentially responded to DTA and eventually resulted in a cell survival reduction from 42% to 92%. Furthermore, 0.1 μg of DTA-expressing lentiviral vector is enough to kill cancer cells (Figure 5A). Subsequently, we detected the RNA level of DTA in multiple cells after transfection with 0.1 μg of DTA-expressing lentiviral vectors (Figure 5B). Concluded from Figure 5A and 5B, the result suggested that these cells differentially respond to the toxicity of DTA.
FIGURE 5.
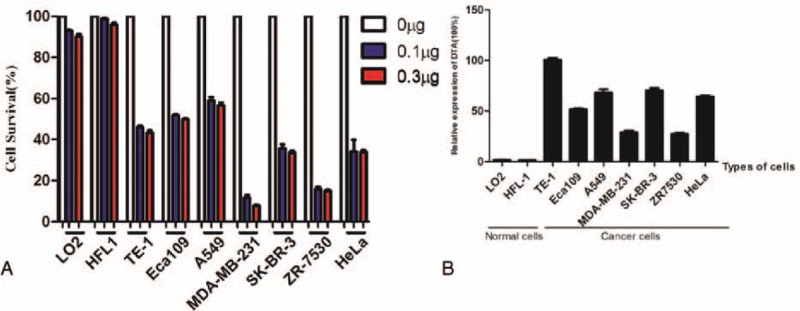
In vitro assays of killing effect on cancer cells with DTA-expressing lentiviral vectors driven by Surp1430 promoters. (A) Lentiviral vectors comprising the coordinate expression of fluorescent protein and DTA as driven by Surp1430 specifically kill cancer cells in vitro. Cell proliferation was inhibited and cell counts decreased after the transient transfection of DTA-expressing lentiviral vectors as driven by Surp1430 in cancer cells. The 0-μg group represents the co-transfection of 0.3 μg of pPRIME-CMV-GFP-FF3, 0.2 μg of pGL3-control and 25 ng of pRL-TK into cells; the 0.1-μg group represents the co-transfection of 0.1 μg of pPRIME-Surp1430-GFP-2A-DTA-FF3, 0.2 μg of pPRIME-CMV-GFP-FF3, 0.2 μg of pGL3-control and 25 ng of pRL-TK into cells; and the 0.3-μg group represents the co-transfection of 0.3 μg of pPRIME-Surp1430-GFP-2A-DTA-FF3, 0.2 μg of pGL3-control and 25 ng of pRL-TK into cells; (B) Quantifying the RNA level of DTA in multiple cells after transfection with DTA-expressing lentiviral vectors. CMV = cytomegalovirus, DTA = diphtheria toxin A.
Production of DTA-Expressing IDLV in HEK293T[mEF-2(G717R)]
Based on the result of the cellular survival assays described above, novel lentiviruses, which could not only simultaneously express fluorescent protein and DTA but also avoid the insertion of its genome into host cells, were suggested for production for an in vivo assay. However, wild-type HEK293T could also be killed by DTA due to its role in protein synthesis inhibition; thus, it is not feasible to produce DTA-expressing lentivirus without some modifications. For DTA-expressing lentivirus production, we cloned the EF-2 gene and obtained pCDH-CMV-EF-2-EF1-Puro after vector construction. In addition, we induced a direct point mutation at the G717 residue of EF-2 with specific primers, thereby achieving the overexpression of mEF-2 in wild-type HEK293T after the establishment of a stable cell line named HEK293T[mEF-2(G717R)], which was infected with mEF-2-expressing lentivirus and selected with drugs. Therefore, HEK293T[mEF-2(G717R)] could resist the inhibitive effect of protein synthesis that derived from DTA expression due to its overexpression of mEF-2.
After the co-transfection of the lentiviral vector with pMDLg/pRRE (RRK262263264AAH), pRSV-Rev and VSVG into HEK293T[mEF-2(G717R)], we obtained all of the IDLVs. Among these IDLVs, pPRIME-CMV-GFP-FF3 and pPRIME-Surp1430-GFP-FF3 served as controls, while pPRIME-CMV-GFP-2A-DTA-FF3 served as a positive control because its coordinate expression cassette was driven by the CMV promoter. As shown in Figure 6, compared to pPRIME-CMV-GFP-FF3, a weaker fluorescence occurred after the transfection of pPRIME-CMV-GFP-2A-DTA-FF3 into HEK293T[mEF-2(G717R)]; however, the absence of fluorescence was found after the transfection of pPRIME-CMV-GFP-2A-DTA-FF3 into wild-type HEK293T (Figure S4 http://links.lww.com/MD/A359). Additionally, very weak fluorescence was observed after the transfection of pPRIME-Surp1430-GFP-FF3 into HEK293T. Moreover, almost no fluorescence was observed after the transfection of pPRIME-Surp1430-GFP-2A-DTA-FF3 into HEK293T[mEF-2(G717R)], indicating that HEK293T[mEF-2(G717R)] was successfully obtained and the DTA we used is effective for protein synthesis inhibition.
FIGURE 6.
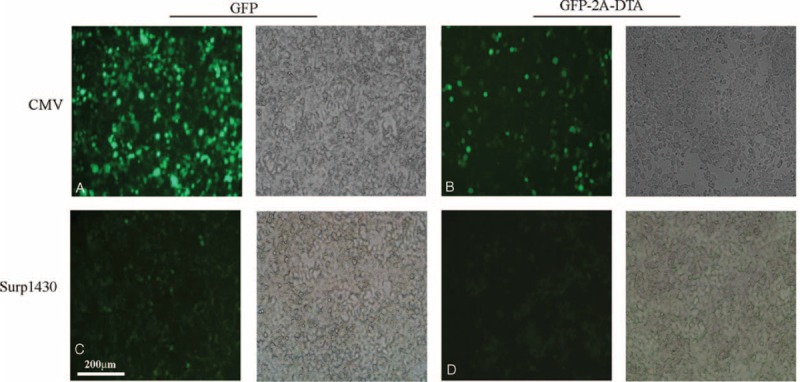
Production of IDLVs after the co-transfection of lentiviral plasmids and packaging plasmids into HEK293T[mEF-2(G717R)] cells. (A) Production of pPRIME-CMV-GFP-FF3 IDLVs in cells; (B) Production of pPRIME-CMV-GFP-2A-DTA-FF3 IDLVs in cells; (C) Production of pPRIME-Surp1430-GFP-FF3 IDLVs in cells; (D) Production of pPRIME-Surp1430-GFP-2A-DTA-FF3 IDLVs in cells. CMV = cytomegalovirus, IDLV = integrase-deficient lentivirus.
Eradication of Breast Cancer Cells in Nude Mice With DTA-Expressing IDLV
To validate the effect of DTA-expressing IDLV in cancer gene therapy, we established xenografts with ZR7530 in nude mice; these nude mice were randomly assigned into 4 different groups, and every group had 5 nude mice that were used in the experiment (n = 5). When tumors were produced in these nude mice and when the volume of these tumors grew to approximately 35 mm3, we injected concentrated DTA-expressing IDLVs into different groups; the first group received the pPRIME-CMV-GFP-FF3, the second group received the pPRIME-Surp1430-GFP-FF3, the third group received the pPRIME-CMV-GFP-2A-DTA-FF3, and the fourth group received the pPRIME-Surp1430-GFP-2A-DTA-FF3. Among these IDLVs, both pPRIME-CMV-GFP-FF3 and pPRIME-Surp1430-GFP-FF3 served as negative controls, while pPRIME-CMV-GFP-2A-DTA-FF3 served as a positive control because the coordinate expression cassette was controlled by the CMV promoter, a ubiquitous promoter that is always used for the overexpression of target genes in cells and tissues.
As shown in Figure 7, after the repeated administration of IDLVs containing 4 × 106 TU every time at 3 different time points (30, 40, and 50 days after implantation), we found that neither of the negative controls had any effect on the eradication of cancer cells because the tumors receiving these IDLVs still grew rapidly after IDLV injection. However, the tumors were significantly repressed after the injection of DTA-expressing IDLVs compared to the negative controls (CMV-GFP vs CMV-DTA-2A-GFP, P = 0.0154; Survivin-GFP vs Survivin-DTA-2A-GFP, P = 0.0165; P < 0.05), and no significant difference occurred between DTA-expressing IDLVs that was driven by CMV or Surp1430 promoters(CMV-DTA-2A-GFP vs Survivin-DTA-2A-GFP, P = 0.3445, P > 0.05), suggesting that DTA-expressing IDLVs are feasible for cancer gene therapy. Although we observed tumor repression in the tested groups after repeated injection of DTA-expressing IDLVs, the tumors could not be completely eradicated, and a very small mass of tumors survived until mouse dissection. As expected, we found no potential toxicity on the whole body weight after dissection in the nude mice those were injected with pPRIME-CMV-GFP-FF3 or pPRIME-Surp1430-GFP-FF3, and no significant difference occurred between these 2 groups (P = 0.8119, P > 0.05); in addition, no adverse effect on whole body weight of nude mice injected with pPRIME-Surp1430-GFP-2A-DTA-FF3 when compared with pPRIME-CMV-GFP-FF3 (P = 0.3453, P > 0.05). However, obvious adverse effect was found after injection with pPRIME-CMV-GFP-2A-FF3 (P < 0.0001).
FIGURE 7.
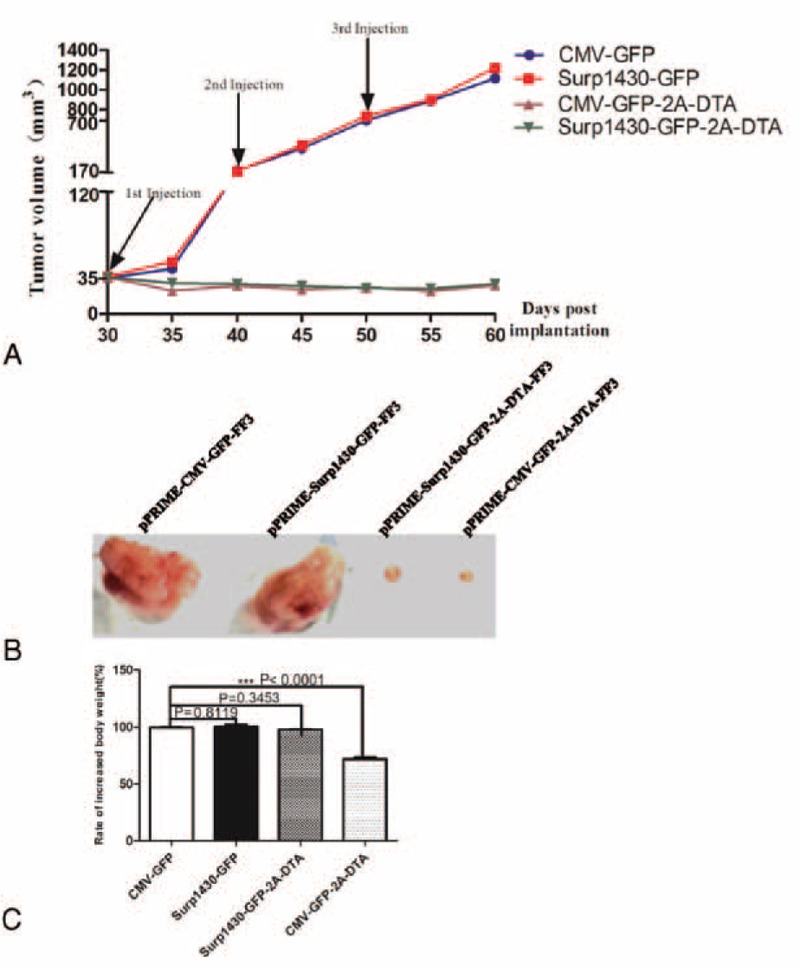
The effect of the intratumoral injection of DTA-expressing IDLVs on subcutaneous breast tumor growth in nude mice (n = 5). (A) The tumor growth curve after the repeated injection of pPRIME-CMV-GFP-FF3, pPRIME-Surp1430-GFP-FF3 IDLVs, pPRIME-CMV-GFP-2A-DTA-FF3, and pPRIME-Surp1430-GFP-FF3 IDLVs; (B) The dissection of tumors from nude mice after multiple injection of IDLVs, the injected IDLVs are described above the tumors; (C) The rate of increased body weight between mice after injection with IDLVs. Before the initial injection, the whole body weight of mice was recorded, the value of body weight was also recorded after tumors dissection, and the increased value was calculated, then the effect of IDLVs on body weight change was evaluated when compared with group injected with pPRIME-CMV-GFP-FF3. DTA = diphtheria toxin A; IDLV = integrase-deficient lentivirus. CMV, cytomegalovirus.
DISCUSSION
In addition to Survivin protein, recent studies found that receptor-tyrosine-kinase-like orphan receptor 1, a cell surface receptor tyrosine kinase expressed in chronic lymphocytic leukemia, could also be served as a marker to identify malignant cells from normal cells,51 and it has also been identified as a potential therapeutic target for the treatment of leukemia and lung adenocarcinoma,52–54 thus both of them may be useful for cancer gene therapy. To achieve the specific expression of DTA in cancer cells and to avoid the side effect on normal cells and tissues, we obtained 2 Survivin promoters and then compared their activities in different types of cancer cells. Luciferase reporter assays in our study showed that Surp1430 possesses stronger activities in cancer cells than Surp269, suggesting that different nucleotides between Surp1430 and Surp269 are helpful for increasing the activities of Surp1430, and finally we used Surp1430 to drive the strong and specific expression of DTA in cancer cells. Compared to the CMV promoter, the activity of Surp1430 is much lower than that of the CMV promoter in normal cells; however, the activity of Surp1430 is comparable to that of the CMV promoter in cancer cells, thereby guaranteeing the abundant and specific expression of DTA in cancer cells.
Additionally, to monitor the process of lentivirus packaging and to trace the activating sites of DTA, we linked fluorescent protein and DTA with FMDV 2A sequence, which has been validated in previous studies for their advantages in bicistronic expression.55–58 Namely, we constructed a coordinate expression cassette, fluorescent protein-2A-DTA, to achieve the expression of fluorescent protein and to avoid the inhibition of fluorescent protein synthesis, and the expression of fluorescent protein can be used to determine the effect of lentivirus packaging. As shown in Figure S4 http://links.lww.com/MD/A359, we found no fluorescence in wild-type HEK293T cells, suggesting that fluorescent protein synthesis was totally inhibited in spite of these expression cassettes being driven by the CMV promoter. In contrast, distinct fluorescence was observed in HEK293T[mEF-2(G717R)] cells even though they were also driven by the CMV promoter, indicating that HEK293T[mEF-2(G717R)] cells can effectively avoid the toxicity of DTA. However, the activity of Surp1430 is very weak in HEK293T cells because the fluorescence that is driven by Surp1430 is much weaker than that is driven by CMV promoter. In contrast, almost no fluorescence was observed after the transfection of pPRIME-Surp1430-GFP-2A-DTA-FF3 into HEK293T[mEF-2(G717R)] cells. Therefore, DTA could effectively inhibit the expression of fluorescent protein in unmodified cells, and the HEK293T[mEF-2(G717R)] cells that were established in our study provided an effective pool for DTA-expressing lentivirus packaging.
Although lentiviruses have some advantages compared to other viruses, including the persistent expression of exogenous genes, weak immunogenicity, greater transgene capacity, and convenient production, there are safety concerns regarding insertional mutagenesis; therefore, it is necessary to modify lentiviruses to relieve the risk of insertional mutagenesis while leaving the attractive properties of lentiviruses. IDLVs could solve these problems and have been widely used in human gene therapy.49 As discussed in our previous study, many key amino acid residues in integrase have been modified for the generation of IDLVs, and these IDLVs have been used to infect mouse or human cells in vitro and in vivo. Among these mutants, the RRK262263264AAH mutant is effective in the long-term expression of exogenous genes in nondividing cells and in low residual integrase activities.59 Because a single molecule of DT is enough to kill a cancer cell as long as it is included in cell, the transient expression level of DTA as mediated by IDLVs is a good candidate for cancer gene therapy. Moreover, the application of IDLV also enhances the safety of cancer gene therapy. Therefore, we obtained the RRK262263264AAH mutant after direct mutation and then used pMDLg/pRRE (RRK262263264AAH), which expresses the RRK262263264AAH mutant, to perform lentivirus packaging. These DTA-expressing IDLVs not only possess the property of integrase deficiency but also coordinately express fluorescent protein and DTA driven by the CMV or Survivin promoter, respectively. Moreover, in vitro and in vivo assays both demonstrated the potential and safety of killing cancer cells after using our system, suggesting that our system represents a promising tool in cancer gene therapy.
Although we noted a very small mass of tumors in nude mice after the injection of DTA-expressing IDLVs, their killing effect on xenografts tumors is significant, and their failure in the penetration of IDLVs across tumor masses may be responsible for the survival of these tumors. Moreover, we also found that no obvious pathogenic effects on whole body weight of nude mice after the injection of DTA-expressing IDLVs as driven by the tumor-specific promoter. These results are similar to those of previous studies,40,46 but we used IDLVs, a safer system, to achieve a safer cancer therapy. However, since Survivin protein overexpression was also detected in megakaryocyte and erythrocytes, and it is also up-regulated by β-catenin,60 which is activated during erythropoiesis process.61 Therefore, a long-term observation for side effect on hematopoiesis and large-scale experiments using our Survivin promoter-driven IDLV system should be performed to validate the safety of our system before medical trials.
CONCLUSIONS
In conclusion, we demonstrated that the Survivin promoter is specifically and highly activated in cancer cells and tissues; however, its activity was repressed in normal cells and tissues. Moreover, we compared and used Surp1430 to drive the bicistronic expression cassette, including fluorescent protein-2A-DTA in a lentiviral vector. Meanwhile, we successfully obtained a modified HEK293T, HEK293T[mEF-2(G717R)], for use in DTA-expressing IDLVs production. In vitro and in vivo assays both showed that our DTA-expressing IDLV system can specifically kill a variety of cancer cells with low side effects, thereby providing a promising tool for targeted gene therapy for cancers.
Footnotes
Abbreviations: DMEM = Dulbecco's Modified Eagle medium, DT = diphtheria toxin, DTA = diphtheria toxin A, EF-2 = elongation factor 2, FBS = fetal bovine serum, FMDV = foot and mouth disease viruses, IDLV = integrase-deficient lentivirus, mEF-2 = mutated elongation factor 2, ORF = open-reading frame, PIC = preintegration complex, RLUs = relative luciferase units, Surp1430 = Survivin-1430 promoters, Surp269 = Survivin-269 promoters.
Baoshun Lin, Anding Gao, Rui Zhang, and Hongyu Ma have contributed equally to this work.
This work was supported by the National High Technology Research and Development Program of China (863 Program, No. 2014AA020541, to KL), the National Science Foundation of China (No. 81302068, to KL), the program for the Top Young Innovative Talents of Fujian province ( to KL) and the China Postdoctoral Science Foundation (No. 2013M542580, to KL).
The authors have no conflicts of interest to disclose.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website (www.md-journal.com).
REFERENCES
- 1.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 1997; 3:917–921. [DOI] [PubMed] [Google Scholar]
- 2.Bao R, Connolly DC, Murphy M, et al. Activation of cancer-specific gene expression by the survivin promoter. J Natl Cancer Inst 2002; 94:522–528. [DOI] [PubMed] [Google Scholar]
- 3.Chen JS, Liu JC, Shen L, et al. Cancer-specific activation of the survivin promoter and its potential use in gene therapy. Cancer Gene Ther 2004; 11:740–747. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka K, Iwamoto S, Gon G, et al. Expression of survivin and its relationship to loss of apoptosis in breast carcinomas. Clin Cancer Res 2000; 6:127–134. [PubMed] [Google Scholar]
- 5.Zhang M, Zhang X, Zhao S, et al. Prognostic value of survivin and EGFR protein expression in triple-negative breast cancer (TNBC) patients. Target Oncol 2014; 9:349–357. [DOI] [PubMed] [Google Scholar]
- 6.Kato J, Kuwabara Y, Mitani M, et al. Expression of survivin in esophageal cancer: correlation with the prognosis and response to chemotherapy. Int J Cancer 2001; 95:92–95. [DOI] [PubMed] [Google Scholar]
- 7.Malhotra U, Zaidi AH, Kosovec JE, et al. Prognostic value and targeted inhibition of survivin expression in esophageal adenocarcinoma and cancer-adjacent squamous epithelium. PLoS ONE 2013; 8:e78343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunner A, Riss P, Heinze G. Brustmann H. pHH3 and survivin are co-expressed in high-risk endometrial cancer and are prognostic relevant. Br J Cancer 2012; 107:84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ettl T, Stiegler C, Zeitler K, et al. EGFR, HER2, survivin, and loss of pSTAT3 characterize high-grade malignancy in salivary gland cancer with impact on prognosis. Hum Pathol 2012; 43:921–931. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Zhu J, Liu DY, et al. Over-expression of survivin and VEGF in small-cell lung cancer may predict the poorer prognosis. Med Oncol 2014; 31:775. [DOI] [PubMed] [Google Scholar]
- 11.Xu P, Xu XL, Huang Q, et al. CIP2A with survivin protein expressions in human non-small-cell lung cancer correlates with prognosis. Med Oncol 2012; 29:1643–1647. [DOI] [PubMed] [Google Scholar]
- 12.Meng JR, Tang HZ, Zhou KZ, et al. TFF3 and survivin expressions associate with a lower survival rate in gastric cancer. Clin Exp Med 2013; 13:297–303. [DOI] [PubMed] [Google Scholar]
- 13.Chen L, Liang L, Yan X, et al. Survivin status affects prognosis and chemosensitivity in epithelial ovarian cancer. Int J Gynecol Cancer 2013; 23:256–263. [DOI] [PubMed] [Google Scholar]
- 14.Min L, Ji Y, Bakiri L, et al. Liver cancer initiation is controlled by AP-1 through SIRT6-dependent inhibition of survivin. Nat Cell Biol 2012; 14:1203–1211. [DOI] [PubMed] [Google Scholar]
- 15.Chu XY, Chen LB, Wang JH, et al. Overexpression of survivin is correlated with increased invasion and metastasis of colorectal cancer. J Surg Oncol 2012; 105:520–528. [DOI] [PubMed] [Google Scholar]
- 16.Hori M, Miki T, Okamoto M, et al. The detergent-soluble cytoplasmic pool of survivin suppresses anoikis and its expression is associated with metastatic disease of human colon cancer. PLoS ONE 2013; 8:e55710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okamoto K, Okamoto I, Hatashita E, et al. Overcoming erlotinib resistance in EGFR mutation-positive non-small cell lung cancer cells by targeting survivin. Mol Cancer Ther 2012; 11:204–213. [DOI] [PubMed] [Google Scholar]
- 18.Lechler P, Renkawitz T, Campean V, et al. The antiapoptotic gene survivin is highly expressed in human chondrosarcoma and promotes drug resistance in chondrosarcoma cells in vitro. BMC Cancer 2011; 11:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun XP, Dong X, Lin L, et al. Up-regulation of survivin by AKT and hypoxia-inducible factor 1alpha contributes to cisplatin resistance in gastric cancer. FEBS J 2014; 281:115–128. [DOI] [PubMed] [Google Scholar]
- 20.Zhang M, Latham DE, Delaney MA, et al. Survivin mediates resistance to antiandrogen therapy in prostate cancer. Oncogene 2005; 24:2474–2482. [DOI] [PubMed] [Google Scholar]
- 21.Hoffman WH, Biade S, Zilfou JT, et al. Transcriptional repression of the anti-apoptotic survivin gene by wild type p53. J Biol Chem 2002; 277:3247–3257. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Yan Y, Ji W, et al. OCT4 positively regulates survivin expression to promote cancer cell proliferation and leads to poor prognosis in esophageal squamous cell carcinoma. PLoS ONE 2012; 7:e49693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoon MJ, Park SS, Kang YJ, et al. Aurora B confers cancer cell resistance to TRAIL-induced apoptosis via phosphorylation of survivin. Carcinogenesis 2012; 33:492–500. [DOI] [PubMed] [Google Scholar]
- 24.Ye Q, Cai W, Zheng Y, et al. ERK and AKT signaling cooperate to translationally regulate survivin expression for metastatic progression of colorectal cancer. Oncogene 2014; 33:1828–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen S, Li X, Lu D, et al. SOX2 regulates apoptosis through MAP4K4-survivin signaling pathway in human lung cancer cells. Carcinogenesis 2014; 35:613–623. [DOI] [PubMed] [Google Scholar]
- 26.Altieri DC. Validating survivin as a cancer therapeutic target. Nat Rev Cancer 2003; 3:46–54. [DOI] [PubMed] [Google Scholar]
- 27.Altieri DC. Targeting survivin in cancer. Cancer Lett 2013; 332:225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu Q, Li W, Hu X, et al. Synergistic treatment of ovarian cancer by co-delivery of survivin shRNA and paclitaxel via supramolecular micellar assembly. Biomaterials 2012; 33:6580–6591. [DOI] [PubMed] [Google Scholar]
- 29.Kumar B, Yadav A, Lang JC, et al. YM155 reverses cisplatin resistance in head and neck cancer by decreasing cytoplasmic survivin levels. Mol Cancer Ther 2012; 11:1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tolcher AW, Quinn DI, Ferrari A, et al. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann Oncol 2012; 23:968–973. [DOI] [PubMed] [Google Scholar]
- 31.Kunze D, Erdmann K, Froehner M, et al. Enhanced inhibition of bladder cancer cell growth by simultaneous knockdown of antiapoptotic Bcl-xL and survivin in combination with chemotherapy. Int J Mol Sci 2013; 14:12297–12312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen H. Exploiting the intron-splicing mechanism of insect cells to produce viral vectors harboring toxic genes for suicide gene therapy. Mol Ther Nucleic Acids 2012; 1:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R, Wang X, Li B, et al. Tumor-specific adenovirus-mediated PUMA gene transfer using the survivin promoter enhances radiosensitivity of breast cancer cells in vitro and in vivo. Breast Cancer Res Treat 2009; 117:45–54. [DOI] [PubMed] [Google Scholar]
- 34.Garg H, Salcedo R, Trinchieri G, et al. Improved nonviral cancer suicide gene therapy using survivin promoter-driven mutant Bax. Cancer Gene Ther 2010; 17:155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C, Sun B, An N, et al. Inhibitory effect of survivin promoter-regulated oncolytic adenovirus carrying P53 gene against gallbladder cancer. Mol Oncol 2011; 5:545–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z, Zhou X, Li J, et al. Suppression of hepatoma tumor growth by systemic administration of the phytotoxin gelonin driven by the survivin promoter. Neoplasma 2013; 60:469–479. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, Ji W, Hu H, et al. Survivin promoter-regulated oncolytic adenovirus with Hsp70 gene exerts effective antitumor efficacy in gastric cancer immunotherapy. Oncotarget 2014; 5:150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastan I, Hassan R, FitzGerald DJ, et al. Immunotoxin treatment of cancer. Annu Rev Med 2007; 58:221–237. [DOI] [PubMed] [Google Scholar]
- 39.Li Z, Yu T, Zhao P, et al. Immunotoxins and cancer therapy. Cell Mol Immunol 2005; 2:106–112. [PubMed] [Google Scholar]
- 40.Zheng JY, Chen D, Chan J, et al. Regression of prostate cancer xenografts by a lentiviral vector specifically expressing diphtheria toxin A. Cancer Gene Ther 2003; 10:764–770. [DOI] [PubMed] [Google Scholar]
- 41.Peng W, Verbitsky A, Bao Y, et al. Regulated expression of diphtheria toxin in prostate cancer cells. Mol Ther 2002; 6:537–545. [DOI] [PubMed] [Google Scholar]
- 42.Showalter SL, Huang YH, Witkiewicz A, et al. Nanoparticulate delivery of diphtheria toxin DNA effectively kills mesothelin expressing pancreatic cancer cells. Cancer Biol Ther 2008; 7:1584–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mizrahi A, Czerniak A, Levy T, et al. Development of targeted therapy for ovarian cancer mediated by a plasmid expressing diphtheria toxin under the control of H19 regulatory sequences. J Transl Med 2009; 7:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amit D, Hochberg A. Development of targeted therapy for bladder cancer mediated by a double promoter plasmid expressing diphtheria toxin under the control of H19 and IGF2-P4 regulatory sequences. J Transl Med 2010; 8:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fogar P, Navaglia F, Basso D, et al. Heat-induced transcription of diphtheria toxin A or its variants, CRM176 and CRM197: implications for pancreatic cancer gene therapy. Cancer Gene Ther 2010; 17:58–68. [DOI] [PubMed] [Google Scholar]
- 46.Hine CM, Seluanov A, Gorbunova V. Rad51 promoter-targeted gene therapy is effective for in vivo visualization and treatment of cancer. Mol Ther 2012; 20:347–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thorburn J, Frankel AE, Thorburn A. Apoptosis by leukemia cell-targeted diphtheria toxin occurs via receptor-independent activation of Fas-associated death domain protein. Clin Cancer Res 2003; 9:861–865. [PubMed] [Google Scholar]
- 48.Yang X, Kessler E, Su LJ, et al. Diphtheria toxin-epidermal growth factor fusion protein DAB389EGF for the treatment of bladder cancer. Clin Cancer Res 2013; 19:148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu KC, Lin BS, Gao AD, et al. Integrase-deficient lentivirus: opportunities and challenges for human gene therapy. Curr Gene Ther 2014; 14:352–364. [DOI] [PubMed] [Google Scholar]
- 50.Liu K, Wang H, Long Y, et al. Coordinate lentiviral expression of Cre recombinase and RFP/EGFP mediated by FMDV 2A and analysis of Cre activity. J Cell Biochem 2012; 113:2909–2919. [DOI] [PubMed] [Google Scholar]
- 51.Daneshmanesh AH, Mikaelsson E, Jeddi-Tehrani M, et al. Ror1, a cell surface receptor tyrosine kinase is expressed in chronic lymphocytic leukemia and may serve as a putative target for therapy. Int J Cancer 2008; 123:1190–1195. [DOI] [PubMed] [Google Scholar]
- 52.Mani R, Mao Y, Frissora FW, et al. Tumor antigen ROR1 targeted drug delivery mediated selective leukemic but not normal B-cell cytotoxicity in chronic lymphocytic leukemia. Leukemia 2015; 29:346–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mani R, Chiang CL, Frissora FW, et al. ROR1-targeted delivery of OSU-2S, a nonimmunosuppressive FTY720 derivative, exerts potent cytotoxicity in mantle-cell lymphoma in vitro and in vivo. Exp Hematol 2015; doi: 10.1016/j.exphem.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu Y, Yang H, Chen T, et al. Silencing of receptor tyrosine kinase ROR1 inhibits tumor-cell proliferation via PI3K/AKT/mTOR signaling pathway in lung adenocarcinoma. PLoS ONE 2015; 10:e0127092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chinnasamy D, Milsom MD, Shaffer J, et al. Multicistronic lentiviral vectors containing the FMDV 2A cleavage factor demonstrate robust expression of encoded genes at limiting MOI. Virol J 2006; 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang J, Qian JJ, Yi S, et al. Stable antibody expression at therapeutic levels using the 2A peptide. Nat Biotechnol 2005; 23:584–590. [DOI] [PubMed] [Google Scholar]
- 57.Fang J, Yi S, Simmons A, et al. An antibody delivery system for regulated expression of therapeutic levels of monoclonal antibodies in vivo. Mol Ther 2007; 15:1153–1159. [DOI] [PubMed] [Google Scholar]
- 58.Sommer CA, Stadtfeld M, Murphy GJ, et al. Induced pluripotent stem cell generation using a single lentiviral stem cell cassette. Stem Cells 2009; 27:543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Philippe S, Sarkis C, Barkats M, et al. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc Natl Acad Sci USA 2006; 103:17684–17689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCrann DJ, Yezefski T, Nguyen HG, et al. Survivin overexpression alone does not alter megakaryocyte ploidy nor interfere with erythroid/megakaryocytic lineage development in transgenic mice. Blood 2008; 111:4092–4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang CL, Chen SS, Lee SJ, et al. Lysophosphatidic acid induces erythropoiesis through activating lysophosphatidic acid receptor 3. Stem Cells 2011; 29:1763–1773. [DOI] [PubMed] [Google Scholar]


