Abstract
MiR-143/145 is down-regulated in cervical cancer, which may serve as a tumor suppressor by targeting KRAS and Ras-responsive element-binding protein (RREB1). Activated KRAS leads to down-regulation of miR-143/145 transcription in a RREB1-dependent manner, establishing a miR-143/145-KRAS-RREB1 feedback loop. A polymorphism rs4705343C/T in the promoter of miR-143/145 might influence the binding of TATA-binding protein. We hypothesized that the miR-143/145 rs4705343 and KRAS rs712 may be related to the occurrence of cervical squamous cell carcinoma (CSCC). In this study, we genotyped the 2 polymorphisms in 415 patients with CSCC and 504 controls using polymerase chain reaction–restriction fragment length polymorphism. The promoter activities were measured by the Dual-Luciferase Reporter Assay System. We found that the rs4705343TC genotype was associated with an increased risk of CSCC (adjusted odds ratio [OR] = 1.37; 95% confidence interval [CI], 1.05–1.80). The significantly increased association was also observed in a dominant genetic model (adjusted OR = 1.32; 95% CI, 1.01–1.72). Combined analysis showed that individuals carrying the genotypes of rs4705343 TC/CC and rs712GT/TT had a 1.47-fold increased risk of CSCC (adjusted OR = 1.47; 95% CI, 1.01–2.15). By using multifactor dimensionality reduction software method, we identified a significant interaction between the miR-143/145 rs4705343 and KRAS rs712. Dual-Luciferase Reporter Assay showed that the luciferase activity was significantly lower in cells transfected with the rs4705343C allele than that of the rs4705343T allele. These findings indicate that miR-143/145 rs4705343 and KRAS rs712 may contribute to the etiology of CSCC in Chinese women.
INTRODUCTION
Cervical cancer, arising from the cervix, is the fourth most common cancer among women. In 2012, it accounted for about 528,000 cases and 266,000 deaths worldwide, and 80% of the cases occurred in developing countries.1,2 Epidemiologic studies have identified that human papillomavirus (HPV) infection is involved in the development of cervical cancer; some HPV-infected individuals, however, do not develop the disease, indicating that HPV infection is a prerequisite but insufficient cause of cervical cancer.3–6 To date, several other risk factors have been identified, such as tobacco smoking, reproductive habits, long-term use of oral contraceptives, as well as genetic factors.7–10
MicroRNAs are a group of small noncoding RNAs of ∼22 nucleotides in length that can regulate RNA degradation and translational suppression by binding to target genes.11,12 Accumulating evidence has shown that miRNAs play key roles in the control of cell proliferation, differentiation, apoptosis, and tumorigenesis.13 Abnormal expression of miRNAs is detected in many tumors, acting as oncogenes or tumor suppressors.14,15 Wang et al16 reported that miR-143/145 is down-regulated in cervical cancer, and over-expression of miR-143/145 in HeLa cells can suppress cell growth. The significant down-regulation of miR-143/145 in cervical cancer was demonstrated by several other independent groups.17–21 Moreover, the down-regulation of miR-143 was related to tumor size, lymph node metastasis, HPV16 infection, and poor prognosis in cervical cancer.17,20 These findings indicate that miR-143/145 may serve as a target for the therapy of cervical cancer.
Regarding the mechanism of miR-143/145 in tumorigenesis, it has been found that miR-143/145 suppresses cell growth by targeting KRAS and Ras-responsive element-binding protein (RREB1).22–26KRAS is an oncogene, with 6.3% to 17.5% activating mutations detected in cervical cancer, indicating that KRAS is an essential event in the development of cervical carcinogenesis.27–29 After activation, KRAS can lead to down-regulation of miR-143/145 transcription in an RREB1-dependent manner, establishing a miR-143/145-KRAS-RREB1 feedback loop (Figure 1A).24,25 Genetic variant in the 3′ untranslated region (3′ UTR) of expressed gene or promoter of miRNA can modulate its expression and confer disease risk.30–32 One example is that a variant rs712 in the KRAS 3′ UTR with a function of regulation KRAS expression by disrupting complementary sites of let-7 and miR-181. The rs712∗G allele generated ∼15% derepression of luciferase activity compared with the rs712∗T allele in HeLa cell.32 Another example is a rs4705343C/T polymorphism in the promoter of miR-143/145 (400 bp upstream from the transcription start site).31 In silico analysis predicted that the rs4705343T allele but not the rs4705343C can bind to the TATA-binding protein (TBP) (http://www.gene-regulation.com/pub/databases.html) (Figure 1B).
FIGURE 1.
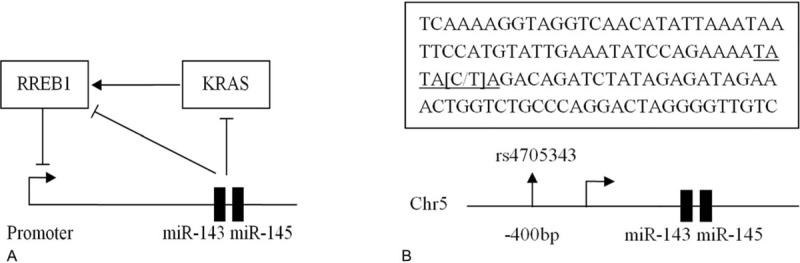
A: miR-143/145-KRAS-RREB1 feedback loop. B: Bioinformatics analysis predicted a TATA-binding protein (TBP) binding site in the promoter region of miR-143/145 (underlined sequence). If the polymorphic location is T, it can bind to the transcriptional factor TBP; otherwise, if the polymorphic location is C, it cannot bind to the TBP.
Based on this background, we hypothesized that the miR-143/145 rs4705343 and KRAS rs712 polymorphisms may be associated with the risk of cervical squamous cell carcinoma (CSCC). To test this hypothesis, we performed an association analysis of 415 CSCC cases and 504 controls in Chinese women.
MATERIALS AND METHODS
Study Subjects
A hospital-based case–control study was performed to assess the association of miR-143/145 rs4705343 and KRAS rs712 with risk of CSCC. The study protocol was approved by the Institutional Review Board of the West China Second University Hospital, and written informed consent was obtained from all participants. Four hundred fifteen newly diagnosed CSCC patients were consecutively enrolled from the West China Second University Hospital of Sichuan University and the People's Hospital of Leshan between January 2010 and July 2013 with a response rate of 93.9% (415/442). All cases had histological confirmation of the diagnosis. The mean age of the case group was 44.2 ± 8.7 years. Clinical information was collected from medical record, including gravidity, parity, clinical stages, differentiated status, and lymph node metastasis status. Five hundred four individuals who came to the hospital for physical examination during the same period as the cases were recruited as the controls. The response rate of the controls was 90.3% (504/558). Exclusion criteria of the controls included acute and chronic cervicitis, endometritis, myoma of uterus, or a self-reported family history of cancer. The mean age of the controls was 43.4 ± 9.2 years. The controls were frequency matched to the cases based on age and residence area. All the subjects were genetically unrelated Han Chinese women.
DNA Extraction
Approximately, 2 mL venous blood sample was taken from each subject before surgery treatment. Genomic DNA was isolated using an extraction kit (Bioteke, Beijing, China) according to the manufacturer's instructions. The purity and concentration of each DNA sample were determined by Nucleic Acid/Protein Analyzer (DU730, Beckman Coulter, Inc., Brea, CA). The ratio of OD260/OD280 is 1.7 to 1.9, and the concentration of each sample is >20 ng/μL.
Genotyping
The miR-143/145 rs4705343 and KRAS rs712 polymorphisms were genotyped by polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP). The primer sequences, PCR reaction conditions, and restriction enzymes (New England BioLabs, Beverly, MA) were described in our previous work.31,33–35 Briefly, PCR amplification was performed in a final volume of 10 μL containing 50 ng genomic DNA, 20 pmol of each primer, and 5 μL 2× PCR mix (Bioteke). After amplification, PCR products were digested with AccI for the miR-143/145 rs4705343 and TaqI for the KRAS rs712. The digestion products were separated on polyacrylamide gels and stained with 1.5 g/L argent nitrate. The rs4705343C allele yielded 2 fragments of 121 and 26 bp, and the rs4705343T allele yielded a single fragment of 147 bp. The rs712G allele produced 2 fragments of 300 and 25 bp, and the rs712T allele was visualized as a single fragment of 325 bp. For quality control, the PCR–RFLP analyses were carried out by 2 staffs independently with a blindness of the subjects’ case–control status and positive and negative controls were included in each 96-well plate. Of the samples, 10% were randomly selected to verify the genotyping results by direct sequencing (Invitrogen, Shanghai, China) (Figure 2), and the results were completely identical with those of PCR–RFLP.
FIGURE 2.
Deoxyribonucleic acid sequencing of the miR-143/145 rs4705343 and KRAS rs712 polymorphisms.
Construction of Reporter Plasmids
The promoter region of miR-143/145 (from −2786 to +45) containing rs4705343T was synthesized by PCR. The primers were 5′-AGTGGTACCGCCGTGGAGAGTG GAATAGA-3′ (forward) and 5′-GTGAAGCTTCCAACTGACCAGAGATGCAG-3′ (reverse), including the KpnI and HindIII restriction sites (underlined bases). After restriction enzyme digestion, the fragment was cloned into the pGL3-basic vector (Promega, Madison, WI). Using the rs4705343T plasmid as template, rs4705343C construct was created using a QuickChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA). The mutagenic oligonucleotide primers were 5′-GAAATATCCAGAAAATATACAGACAGATCTATAGAGATA-3′ (forward), and 5′-TATCTCTATAGATCTGTCTGTATATTTTCTGGATATTTC-3′ (reverse). After construction of rs4705343T and rs4705343C plasmids, the orientation and integrity of the inserts were confirmed by direct sequencing (Invitrogen). The generated reporter vectors were named pGL3-rs4705343T and pGL3-rs4705343C, respectively.
Cell Lines and Real-Time PCR
HeLa, A549, and HepG2 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum at 37°C in an atmosphere of 5% CO2. The cell lines were identified by short tandem repeat (STR) sequencing, and the relative expression of miR-143/145 in the cell lines was analyzed by real-time PCR. Total RNA was extracted using TriPure Isolation Reagent (Roche, Indianapolis, IN). The concentration was quantified using Nucleic Acid/Protein Analyzer (DU730, Beckman Coulter, Inc.). cDNA for miR-143/145 was synthesized with specific miRNA primers from Ribobio Co. Ltd (Guangzhou, China). The quantification of miR-143/145 was performed using QuantiFast SYBR Green PCR Kit (Qiagen, Hilden, Germany). The threshold cycle (Ct) is defined as number of cycles which reaches the fixed fluorescence threshold. U6 snRNA was used as an internal control and relative expression of miR-143/145 was calculated using the 2−ΔΔCt method.36
Transient Transfections and Luciferase Assays
For transfection, HeLa, A549, and HepG2 cells were seeded onto 24-well plates at a density of 1 × 105 cells/well. After an overnight incubation, each well was transiently transfected with 2 μg pGL3-basic vector DNA or pGL3-rs4705343T or pGL3-rs4705343C using X-tremeGENE HP reagent (Roche). As an internal control, 40 ng pRL-TK vector DNA (Promega) was co-transfected. At 48 h after transfection, the luciferase activities were measured by the Dual-Luciferase Reporter Assay System (Promega) following the manufacturer's protocol. Each experiment was independently done in triplicate. Relative luciferase activity was calculated as the ratio of firefly/Renilla luciferase activity.
Statistical Analysis
Age distribution between the cases and controls was evaluated using the Student t test. Hardy–Weinberg equilibrium was tested using a goodness-of-fit χ2 test to determine whether the observed genotype frequencies were in agreement with expected values. The associations between the miR-143/145 rs4705343 and KRAS rs712 and the presence of CSCC were estimated using the χ2 test. Odds ratios (OR) and their 95% confidence interval (CI) were calculated using a logistic regression model adjusting for age. The levels of luciferase reporter gene expression were examined using the Student t test between different constructs. All statistical analyses were done using the SPSS software program version 13.0 (SPSS Science, Chicago, IL). Rs4705343–rs712 interaction analysis was performed using multifactor dimensionality reduction (MDR) software (version 3.0.2), which is freely available at the website www.epistasis.org.37,38 All tests were 2 sided, and P < 0.05 was considered significant.
RESULTS
Characteristics of Study Subjects
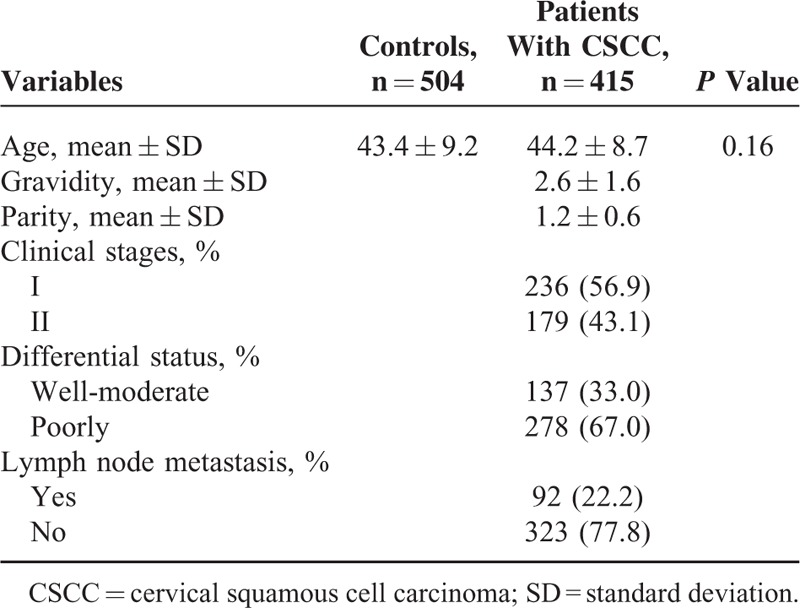
The characteristics of patients with CSCC and controls are summarized in Table 1. No significant difference was observed in the distribution of age between the 2 groups (P = 0.16). The mean gravidity is 2.6 ± 1.6 and the mean parity is 1.2 ± 0.6 in cases. Of the 415 patients with CSCC, 236 (56.9%) were stage I, and 179 (43.1%) were stage II. One hundred thirty-seven (33.0%) were well-moderate differentiated carcinoma, and 278 (67.0%) were poorly differentiated carcinoma. Ninety two (22.2%) were carcinoma with lymph node metastasis, and 323 (77.8%) were carcinoma without lymph node metastasis.
TABLE 1.
Characteristics of Study Subjects
Association of the MiR-143/145 rs4705343 and KRAS rs712 With Risk of CSCC
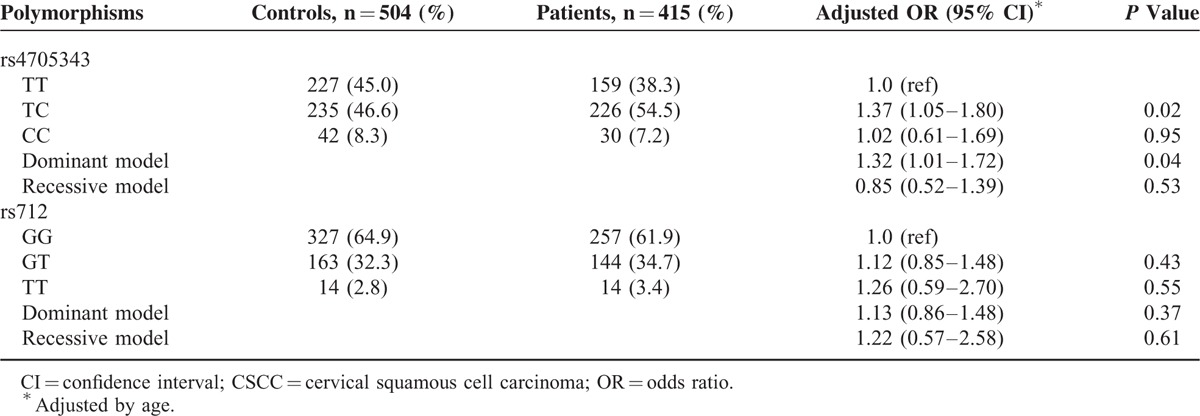
The genotype frequencies of the miR-143/145 rs4705343 and KRAS rs712 in cases and controls are presented in Table 2. The observed genotype distributions of the 2 polymorphisms in the controls showed no evidence of deviation from Hardy–Weinberg equilibrium (P = 0.08 and 0.23 for miR-143/145 rs4705343 and KRAS rs712, respectively). The frequency of the miR-143/145 rs4705343TC genotype was higher among the cases (54.5%) than among the controls (46.6%), and the difference was statistically significant (adjusted OR = 1.37; 95% CI, 1.05–1.80). Similarly, the significantly increased frequency was also observed in a dominant genetic model (adjusted OR = 1.32; 95% CI, 1.01–1.72). However, no evidence of association was found between the KRAS rs712 polymorphism and the risk of CSCC. After stratification analysis according to clinical stages, differential status, and lymph node metastasis, no significant association was observed between the 2 polymorphisms and the features of CSCC (data not shown). Further association analysis of the 2 polymorphisms with prognosis was not available due to lack of survival data in this study.
TABLE 2.
Genotype Frequencies of the MiR-143/145 rs4705343 and KRAS rs712 Between Patients With CSCC and Controls
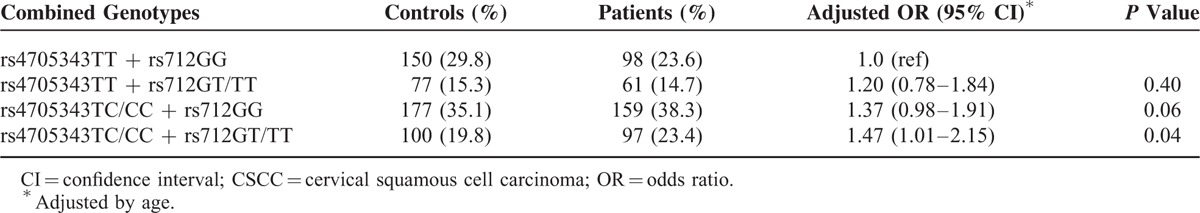
The combined effects of the miR-143/145 rs4705343 and KRAS rs712 on CSCC risk are shown in Table 3. Carriers with the genotypes of rs4705343 TC/CC and rs712GT/TT had a 1.47-fold increased risk of CSCC compared with those with the genotypes of rs4705343TT and rs712GG (adjusted OR = 1.47; 95% CI, 1.01–2.15).
TABLE 3.
Combined Effects of the MiR-143/145 rs4705343 and KRAS rs712 on CSCC Risk
We then did interaction analysis between the miR-143/145 rs4705343 and KRAS rs712 using MDR approach. We found that a genetic interaction between the miR-143/145 rs4705343 and KRAS rs712 conferred susceptibility to CSCC, with the testing accuracy of 0.5386, cross-validation consistency of 10/10, and P value of 0.001.
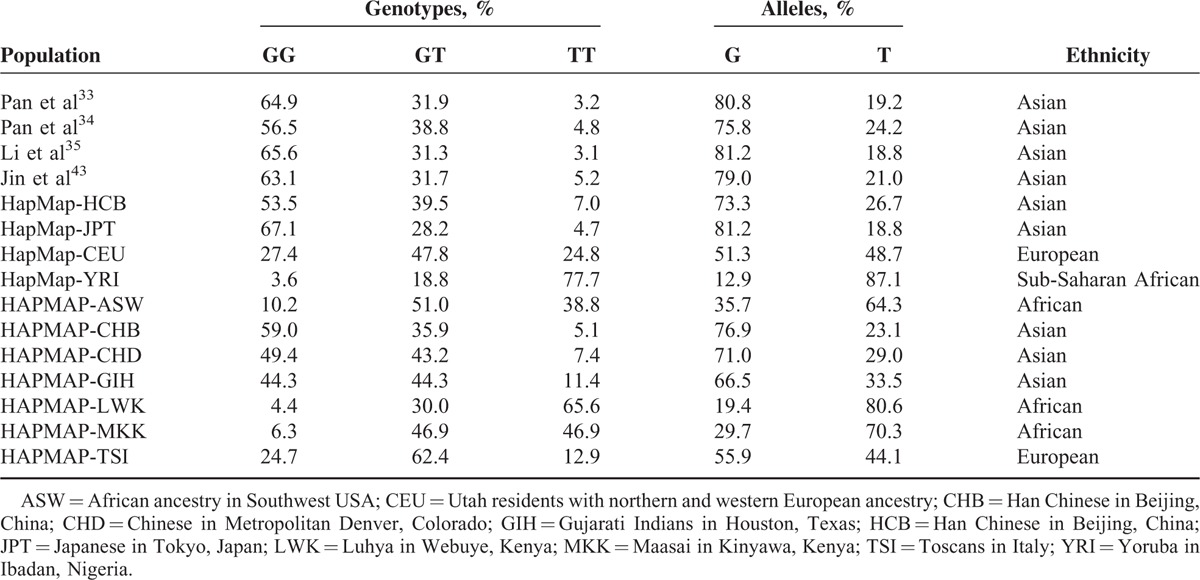
In view of the important role of the miR-143/145 rs4705343 and KRAS rs712 in the onset of CSCC, it is valuable to review the frequencies of the 2 polymorphisms in different populations. Due to lack of available data of the miR-143/145 rs4705343, only the distribution of the KRAS rs712 in different populations is listed in Table 4.
TABLE 4.
Distribution of the KRAS rs712 in Different Populations
Relative Expression of MiR-143/145 in Cell Lines
To detect the expression of miR-143/145 in HeLa, A549, and HepG2 cell lines, real-time PCR was performed. Relative expression of miR-143/145 in cell lines is presented in Figure 3.
FIGURE 3.
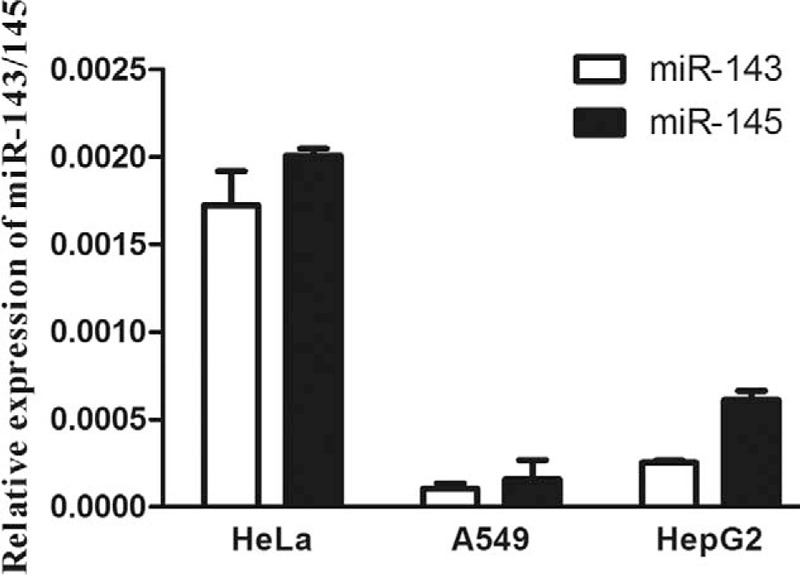
Expression of miR-143/145 in HeLa, A549, and HepG2 cell lines. Real-time polymerized chain reaction was performed to detect miR-143/145 expression. Relative expression was normalized using U6 snRNA. Data were presented as mean ± standard deviation.
Dual Luciferase Reporter Assay of the MiR-143/145 rs4705343 Polymorphism
To determine whether the miR-143/145 rs4705343 polymorphism could influence activity of the promoter, the length of 2841 bp in the promoter region of miR-143/145 was cloned into the pGL3-basic vector. After transient transfection in HeLa, A549, and HepG2 cell lines, the promoter strength was measured by Dual-Luciferase Reporter Assay System. As shown in Figure 4, the luciferase activity of pGL3-rs4705343C was significantly lower than that of pGL3-rs4705343T. In HeLa cell, the rs4705343C allele decreased transcriptional efficiency by 2.1-fold compared with the rs4705343T allele (P = 0.0009). Similar results were obtained in A549 and HepG2 cells (A549: 1.7-fold decrease and P = 0.0006; HepG2: 1.9-fold decrease and P = 0.001, respectively).
FIGURE 4.
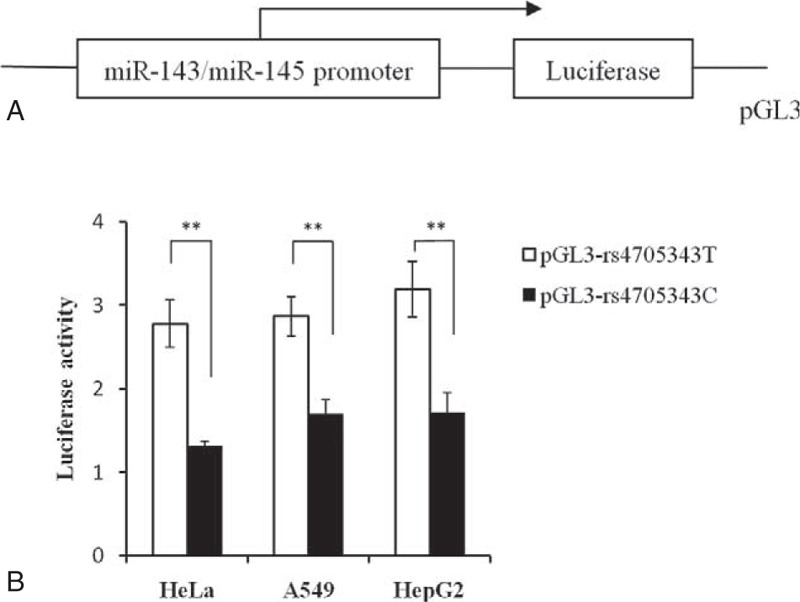
Effect of the miR-143/145 rs4705343 polymorphism on the transcriptional activity. A: Schematic representation of the promoter of miR-143/145 into pGL3-basic plasmid. B: The pGL3-rs4705343C and pGL3-rs4705343T constructs were transiently transfected into HeLa, A549, and HepG2 cells. The relative luciferase activity was normalized with the internal control of Renilla luciferase activity. Each assay was performed in triplicate. Data were presented as mean ± standard deviation. ∗∗P < 0.01.
DISCUSSION
In this study, we investigated for the first time the association of the miR-143/145 rs4705343 and KRAS rs712 polymorphisms with CSCC risk in Chinese women. We found that the rs4705343TC and TC/CC genotypes were associated with increased risks of CSCC. Combined analysis showed that individuals carrying the genotypes of rs4705343 TC/CC and rs712GT/TT had a 1.47-fold increased risk of CSCC. MDR analysis revealed that a significant interaction between the miR-143/145 rs4705343 and KRAS rs712 was observed. Dual-Luciferase Reporter Assay showed that the luciferase activity was significantly lower in cells transfected with the rs4705343C allele than that of the rs4705343T allele. These findings indicate that miR-143/145 rs4705343 and KRAS rs712 may contribute to the etiology of CSCC.
Emerging evidence has shown that the expression of miR-143/145 is down-regulated in cervical cancer, and the down-regulation is involved in not only clinical features but also the outcome of cervical cancer, suggesting that miR-143/145 may serve as tumor suppressors in the pathogenesis of cervical cancer.17–21 Recently, we discovered a functional polymorphism (rs4705343C/T) and the rs4705343C/T polymorphism may contribute to the susceptibility of colorectal cancer.31 In this study, we analyzed the genotype distribution of the rs4705343 polymorphism in CSCC cases and controls, and found that the rs4705343TC and TC/CC genotypes had a 1.37- and 1.32-fold increased risk of CSCC, respectively. The effect of the rs4705343 polymorphism on increased CSCC risk may be explained by allele-specific nucleotide variation. The rs4705343T allele might possess the binding site of TBP and result in stronger promoter activity compared with the counterpart rs4705343C allele.
KRAS, an intracellular signal transducer, plays a key role in the context of various malignancies, including cervical cancer. Somatic KRAS mutations were frequently detected in cervical cancer,27–29,39–41 and the mutations were linked to worse outcome in patients treated by radiotherapy.41 Previously, a functional polymorphism rs712 in the KRAS 3′ UTR was discovered, and the rs712 polymorphism was not only associated with the risk of colorectal cancer,33 gastric cancer,35 and oral squamous cell carcinoma42 but also correlated to clinical features, such as tumor size, clinical stages, and distant metastasis.33,43 However, no significant association of the rs712 polymorphism with the risk of nasopharyngeal carcinoma was observed.34 In this study, we failed to find any association of the rs712 polymorphism with the presence of CSCC. As is known, different cancer types have different susceptibility loci, and HPV infection is a major risk factor for both nasopharyngeal carcinoma44,45 and CSCC.3–6 The negative result of the rs712 polymorphism with nasopharyngeal carcinoma and CSCC may be caused by HPV infection. Additionally, the sample size in this study is moderate. We cannot exclude the possibility that the negative result is due to insufficient statistical power. Further association study is of great value to confirm the results.
Regarding the positive result of the rs4705343-rs712 combined analysis and interaction analysis, it may be attributable to the feed-forward regulatory circuit of miR-143/145-KRAS-RREB1 (Figure 1A).24,25 Loss of miR-143/145 expression is frequently observed in KRAS mutant tumors, which is related to a good progression-free survival.24,46 Over-expression of miR-143/145 reduces KRAS and RREB1 expression. Conversely, activated KRAS represses transcription of the miR-143/145 cluster through the direct action of RREB1 at the miR-143/145 promoter. Therefore, our results of the combined analysis and interaction analysis seem to be biologically plausible.
We have realized some limitations in the hospital-based case–control study. Firstly, the controls were not selected from the general population, and we cannot rule out the possibility of selection bias. By matching on age, ethnicity, and residence area, the potential bias might be minimized. Secondly, birth control, HPV status, and survival data were not available in this study, which prevented our further analysis of the association between the 2 polymorphisms and the clinical information. Thirdly, all the subjects were Chinese, and thus the results cannot be directly applicable to other ethnic groups. To confirm the role of the miR-143/145 rs4705343 polymorphism in CSCC risk requires additional studies with larger sample size in different populations. Although the P values in the study are borderline, the present study provides first evidence that the miR-143/145 rs4705343 polymorphism is involved in the development of CSCC. Notably, the miR-143/145 rs4705343 and KRAS rs712 may jointly contribute to the risk of CSCC in Chinese women, which improves our understanding of CSCC carcinogenesis.
Footnotes
Abbreviations: 3′ UTR = 3′ untranslated region, ASW = African ancestry in Southwest USA, CEU = Utah residents with northern and western European ancestry, CHB = Han Chinese in Beijing, China, CHD = Chinese in Metropolitan Denver, Colorado, CI = confidence interval, CSCC = cervical squamous cell carcinoma, Ct = threshold cycle, GIH = Gujarati Indians in Houston, Texas, HCB = Han Chinese in Beijing, China, HPV = human papillomavirus, JPT = Japanese in Tokyo, Japan, LWK = Luhya in Webuye, Kenya, MDR = multifactor dimensionality reduction, MKK = Maasai in Kinyawa, Kenya, OR = odds ratio, PCR–RFLP = polymerase chain reaction–restriction fragment length polymorphism, RREB1 = Ras-responsive element-binding protein, STR = short tandem repeat, TBP = TATA-binding protein, TSI = Toscans in Italy, YRI = Yoruba in Ibadan, Nigeria.
YL and RS have contributed equally to this work.
This work was supported by the National Natural Science Foundation of China (no. 81302149), Distinguished Young Scientist of Sichuan University (no. 2013SCU04A38), the Science and Technology Pillar Program of Sichuan Province (no. 2014KJT059–2014SZ0001), and the PhD Programs Foundation of Ministry of Education of China (no. 20130181120011).
The authors have no funding and conflicts of interest to disclose.
REFERENCES
- 1.Malleter M, Jacquot C, Moreau D, et al. A novel large regulator RNA, B2, partially overlaps the HEF1/NEDD9/Cas-L gene. Int J Mol Med 2010; 25:897–903. [DOI] [PubMed] [Google Scholar]
- 2.Kent A. HPV vaccination and testing. Rev Obstet Gynecol 2010; 3:33–34. [PMC free article] [PubMed] [Google Scholar]
- 3.Herrington CS. Do HPV-negative cervical carcinomas exist?—revisited. J Pathol 1999; 189:1–3. [DOI] [PubMed] [Google Scholar]
- 4.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999; 189:12–19. [DOI] [PubMed] [Google Scholar]
- 5.Schiffman M, Castle PE, Jeronimo J, et al. Human papillomavirus and cervical cancer. Lancet 2007; 370:890–907. [DOI] [PubMed] [Google Scholar]
- 6.Dunne EF, Park IU. HPV and HPV-associated diseases. Infect Dis Clin N Am 2013; 27:765–778. [DOI] [PubMed] [Google Scholar]
- 7.Appleby P, Beral V, et al. International Collaboration of Epidemiological Studies of Cervical C. Carcinoma of the cervix and tobacco smoking: collaborative reanalysis of individual data on 13,541 women with carcinoma of the cervix and 23,017 women without carcinoma of the cervix from 23 epidemiological studies. Int J Cancer 2006; 118:1481–1495. [DOI] [PubMed] [Google Scholar]
- 8.International Collaboration of Epidemiological Studies of Cervical C. Cervical carcinoma and reproductive factors: collaborative reanalysis of individual data on 16,563 women with cervical carcinoma and 33,542 women without cervical carcinoma from 25 epidemiological studies. Int J Cancer 2006; 119:1108–1124. [DOI] [PubMed] [Google Scholar]
- 9.Appleby P, Beral V, Berrington de Gonzalez A, et al. Cervical cancer and hormonal contraceptives: collaborative reanalysis of individual data for 16,573 women with cervical cancer and 35,509 women without cervical cancer from 24 epidemiological studies. Lancet 2007; 370:1609–1621. [DOI] [PubMed] [Google Scholar]
- 10.Gao LB, Pan XM, Li LJ, et al. Null genotypes of GSTM1 and GSTT1 contribute to risk of cervical neoplasia: an evidence-based meta-analysis. PLoS ONE 2011; 6:e20157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281–297. [DOI] [PubMed] [Google Scholar]
- 12.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009; 136:215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowland JB, Hother C, Gronbaek K. MicroRNAs and cancer. APMIS 2007; 115:1090–1106. [DOI] [PubMed] [Google Scholar]
- 14.Nieters A, Yuan JM, Sun CL, et al. Effect of cytokine genotypes on the hepatitis B virus-hepatocellular carcinoma association. Cancer 2005; 103:740–748. [DOI] [PubMed] [Google Scholar]
- 15.Voorhoeve PM, Le Sage C, Schrier M, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell 2006; 124:1169–1181. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Tang S, Le SY, et al. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 2008; 3:e2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Y, Ma C, Zhang W, et al. Down regulation of miR-143 is related with tumor size, lymph node metastasis and HPV16 infection in cervical squamous cancer. Diagn Pathol 2014; 9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lui WO, Pourmand N, Patterson BK, et al. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 2007; 67:6031–6043. [DOI] [PubMed] [Google Scholar]
- 19.Liu L, Yu X, Guo X, et al. miR-143 is downregulated in cervical cancer and promotes apoptosis and inhibits tumor formation by targeting Bcl-2. Mol Med Rep 2012; 5:753–760. [DOI] [PubMed] [Google Scholar]
- 20.Huang L, Lin JX, Yu YH, et al. Downregulation of six microRNAs is associated with advanced stage, lymph node metastasis and poor prognosis in small cell carcinoma of the cervix. PLoS ONE 2012; 7:e33762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deftereos G, Corrie SR, Feng Q, et al. Expression of miR-21 and miR-143 in cervical specimens ranging from histologically normal through to invasive cervical cancer. PLoS ONE 2011; 6:e28423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Guo X, Zhang H, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene 2009; 28:1385–1392. [DOI] [PubMed] [Google Scholar]
- 23.Pekow J, Meckel K, Dougherty U, et al. Tumor suppressors miRNA-143 and miR-145 and predicted target proteins API5, ERK5, KRAS, and IRS-1 are differentially expressed in proximal and distal colon. Am J Physiol 2015; 308:G179–G187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent OA, Chivukula RR, Mullendore M, et al. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev 2010; 24:2754–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent OA, McCall MN, Cornish TC, et al. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res 2014; 42:7528–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kent OA, Fox-Talbot K, Halushka MK. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene 2013; 32:2576–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pappa KI, Choleza M, Markaki S, et al. Consistent absence of BRAF mutations in cervical and endometrial cancer despite KRAS mutation status. Gynecol Oncol 2006; 100:596–600. [DOI] [PubMed] [Google Scholar]
- 28.Kang S, Kim HS, Seo SS, et al. Inverse correlation between RASSF1A hypermethylation, KRAS and BRAF mutations in cervical adenocarcinoma. Gynecol Oncol 2007; 105:662–666. [DOI] [PubMed] [Google Scholar]
- 29.Wright AA, Howitt BE, Myers AP, et al. Oncogenic mutations in cervical cancer: genomic differences between adenocarcinomas and squamous cell carcinomas of the cervix. Cancer 2013; 119:3776–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo X, Yang W, Ye DQ, et al. A functional variant in microRNA-146a promoter modulates its expression and confers disease risk for systemic lupus erythematosus. PLoS Genet 2011; 7:e1002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li L, Pan X, Li Z, et al. Association between polymorphisms in the promoter region of miR-143/145 and risk of colorectal cancer. Hum Immunol 2013; 74:993–997. [DOI] [PubMed] [Google Scholar]
- 32.Kim M, Chen X, Chin LJ, et al. Extensive sequence variation in the 3′ untranslated region of the KRAS gene in lung and ovarian cancer cases. Cell Cycle 2014; 13:1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan XM, Sun RF, Li ZH, et al. A let-7 KRAS rs712 polymorphism increases colorectal cancer risk. Tumour Biol 2014; 35:831–835. [DOI] [PubMed] [Google Scholar]
- 34.Pan XM, Jia J, Guo XM, et al. Lack of association between let-7 binding site polymorphism rs712 and risk of nasopharyngeal carcinoma. Fam Cancer 2014; 13:93–97. [DOI] [PubMed] [Google Scholar]
- 35.Li ZH, Pan XM, Han BW, et al. A let-7 binding site polymorphism rs712 in the KRAS 3′ UTR is associated with an increased risk of gastric cancer. Tumour Biol 2013; 34:3159–3163. [DOI] [PubMed] [Google Scholar]
- 36.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 37.Hahn LW, Ritchie MD, Moore JH. Multifactor dimensionality reduction software for detecting gene–gene and gene–environment interactions. Bioinformatics 2003; 19:376–382. [DOI] [PubMed] [Google Scholar]
- 38.Moore JH, Andrews PC. Epistasis analysis using multifactor dimensionality reduction. Method Mol Biol 2015; 1253:301–314. [DOI] [PubMed] [Google Scholar]
- 39.Mouron SA, Abba MC, Guerci A, et al. Association between activated K-ras and c-erbB-2 oncogenes with “high-risk” and “low-risk” human papilloma virus types in preinvasive cervical lesions. Mut Res 2000; 469:127–134. [DOI] [PubMed] [Google Scholar]
- 40.Stenzel A, Semczuk A, Rozynskal K, et al. Low-risk” and “high-risk” HPV-infection and K-ras gene point mutations in human cervical cancer: a study of 31 cases. Pathol Res Pract 2001; 197:597–603. [PubMed] [Google Scholar]
- 41.Wegman P, Ahlin C, Sorbe B. Genetic alterations in the K-Ras gene influence the prognosis in patients with cervical cancer treated by radiotherapy. Int J Gynecol Cancer 2011; 21:86–91. [DOI] [PubMed] [Google Scholar]
- 42.Wang WY, Chien YC, Wong YK, et al. Effects of KRAS mutation and polymorphism on the risk and prognosis of oral squamous cell carcinoma. Head Neck 2012; 34:663–666. [DOI] [PubMed] [Google Scholar]
- 43.Jin H, Liang Y, Wang X, et al. Association between a functional polymorphism rs712 within let-7-binding site and risk of papillary thyroid cancer. Med Oncol 2014; 31:221. [DOI] [PubMed] [Google Scholar]
- 44.Atighechi S, Baghdadabad MRA, Mirvakili SA, et al. Human papilloma virus and nasopharyngeal carcinoma: pathology, prognosis, recurrence and mortality of the disease. Exp Oncol 2014; 36:215–216. [PubMed] [Google Scholar]
- 45.Robinson M, Suh YE, Paleri V, et al. Oncogenic human papillomavirus-associated nasopharyngeal carcinoma: an observational study of correlation with ethnicity, histological subtype and outcome in a UK population. Infect Agents Cancer 2013; 8:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mekenkamp LJ, Tol J, Dijkstra JR, et al. Beyond KRAS mutation status: influence of KRAS copy number status and microRNAs on clinical outcome to cetuximab in metastatic colorectal cancer patients. BMC Cancer 2012; 12:292. [DOI] [PMC free article] [PubMed] [Google Scholar]


