Abstract
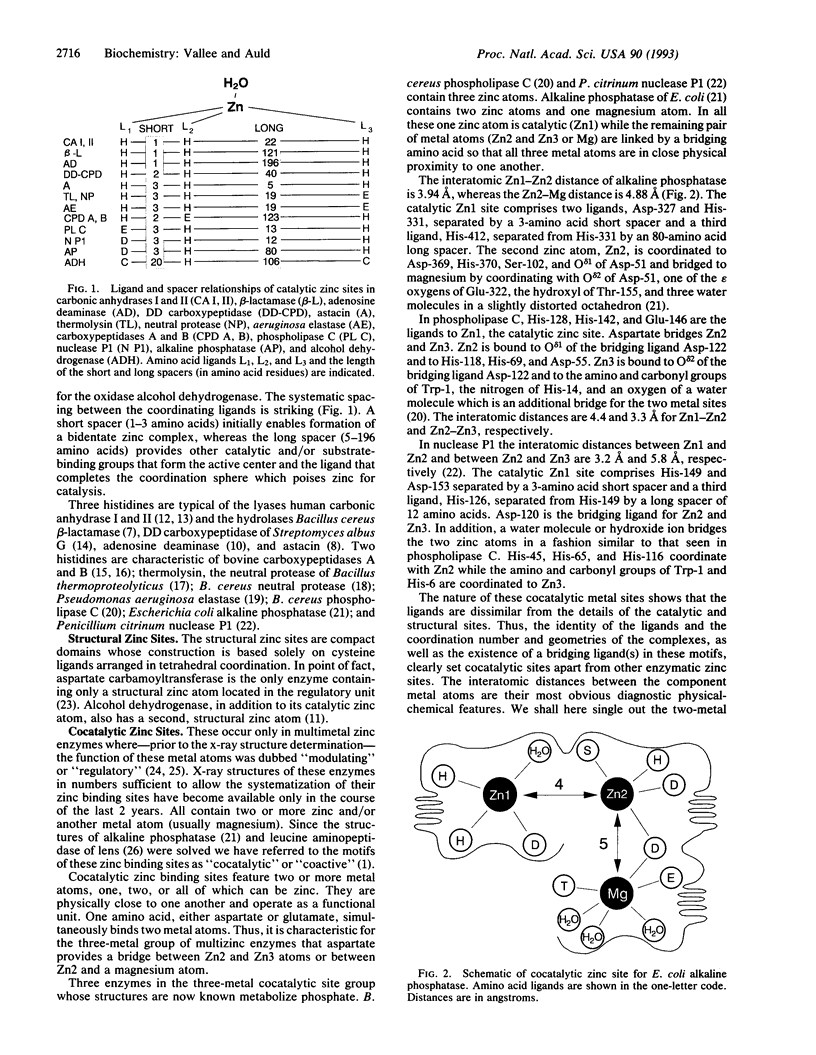
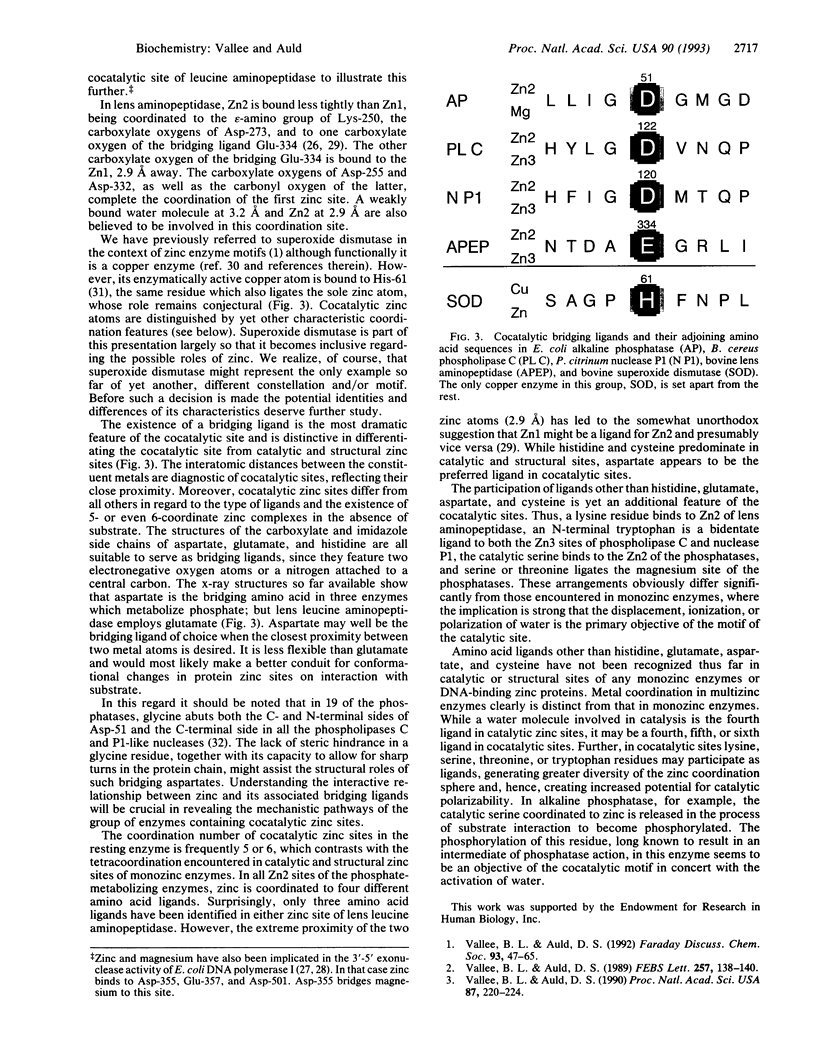
Cocatalytic zinc binding sites are characteristic of enzyme molecules which contain two or more zinc and/or other metal atoms. In each site an aspartate, glutamate, or histidine residue simultaneously binds to two zinc atoms or a zinc and a different metal atom. In the resultant amino acid bridge, two of the cocatalytic metal atoms bind to the same amino acid. Consequently the participating metal atoms are in close proximity and function as a catalytic unit, typical of this motif. In these functional units aspartate seems to be preferred over glutamate. Serine, threonine, tryptophan, and lysine residues are encountered as zinc ligands, although they have not so far been identified as ligands in monozinc enzymes or DNA-binding zinc proteins. The resultant coordination spheres and their mechanistic implications raise interesting questions for further study.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bannister J. V., Bannister W. H., Rotilio G. Aspects of the structure, function, and applications of superoxide dismutase. CRC Crit Rev Biochem. 1987;22(2):111–180. doi: 10.3109/10409238709083738. [DOI] [PubMed] [Google Scholar]
- Beese L. S., Steitz T. A. Structural basis for the 3'-5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991 Jan;10(1):25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bode W., Gomis-Rüth F. X., Huber R., Zwilling R., Stöcker W. Structure of astacin and implications for activation of astacins and zinc-ligation of collagenases. Nature. 1992 Jul 9;358(6382):164–167. doi: 10.1038/358164a0. [DOI] [PubMed] [Google Scholar]
- Burley S. K., David P. R., Lipscomb W. N. Leucine aminopeptidase: bestatin inhibition and a model for enzyme-catalyzed peptide hydrolysis. Proc Natl Acad Sci U S A. 1991 Aug 15;88(16):6916–6920. doi: 10.1073/pnas.88.16.6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S. K., David P. R., Sweet R. M., Taylor A., Lipscomb W. N. Structure determination and refinement of bovine lens leucine aminopeptidase and its complex with bestatin. J Mol Biol. 1992 Mar 5;224(1):113–140. doi: 10.1016/0022-2836(92)90580-d. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dideberg O., Charlier P., Dive G., Joris B., Frère J. M., Ghuysen J. M. Structure of a Zn2+-containing D-alanyl-D-alanine-cleaving carboxypeptidase at 2.5 A resolution. Nature. 1982 Sep 30;299(5882):469–470. doi: 10.1038/299469a0. [DOI] [PubMed] [Google Scholar]
- Fujishima A., Honda K. Electrochemical photolysis of water at a semiconductor electrode. Nature. 1972 Jul 7;238(5358):37–38. doi: 10.1038/238037a0. [DOI] [PubMed] [Google Scholar]
- Honzatko R. B., Crawford J. L., Monaco H. L., Ladner J. E., Ewards B. F., Evans D. R., Warren S. G., Wiley D. C., Ladner R. C., Lipscomb W. N. Crystal and molecular structures of native and CTP-liganded aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1982 Sep 15;160(2):219–263. doi: 10.1016/0022-2836(82)90175-9. [DOI] [PubMed] [Google Scholar]
- Hough E., Hansen L. K., Birknes B., Jynge K., Hansen S., Hordvik A., Little C., Dodson E., Derewenda Z. High-resolution (1.5 A) crystal structure of phospholipase C from Bacillus cereus. Nature. 1989 Mar 23;338(6213):357–360. doi: 10.1038/338357a0. [DOI] [PubMed] [Google Scholar]
- Kannan K. K., Notstrand B., Fridborg K., Lövgren S., Ohlsson A., Petef M. Crystal structure of human erythrocyte carbonic anhydrase B. Three-dimensional structure at a nominal 2.2-A resolution. Proc Natl Acad Sci U S A. 1975 Jan;72(1):51–55. doi: 10.1073/pnas.72.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E. E., Wyckoff H. W. Reaction mechanism of alkaline phosphatase based on crystal structures. Two-metal ion catalysis. J Mol Biol. 1991 Mar 20;218(2):449–464. doi: 10.1016/0022-2836(91)90724-k. [DOI] [PubMed] [Google Scholar]
- Liljas A., Kannan K. K., Bergstén P. C., Waara I., Fridborg K., Strandberg B., Carlbom U., Järup L., Lövgren S., Petef M. Crystal structure of human carbonic anhydrase C. Nat New Biol. 1972 Feb 2;235(57):131–137. doi: 10.1038/newbio235131a0. [DOI] [PubMed] [Google Scholar]
- Pauptit R. A., Karlsson R., Picot D., Jenkins J. A., Niklaus-Reimer A. S., Jansonius J. N. Crystal structure of neutral protease from Bacillus cereus refined at 3.0 A resolution and comparison with the homologous but more thermostable enzyme thermolysin. J Mol Biol. 1988 Feb 5;199(3):525–537. doi: 10.1016/0022-2836(88)90623-7. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A., Lipscomb W. N. Carboxypeptidase A: a protein and an enzyme. Adv Protein Chem. 1971;25:1–78. doi: 10.1016/s0065-3233(08)60278-8. [DOI] [PubMed] [Google Scholar]
- Schmid M. F., Herriott J. R. Structure of carboxypeptidase B at 2-8 A resolution. J Mol Biol. 1976 May 5;103(1):175–190. doi: 10.1016/0022-2836(76)90058-9. [DOI] [PubMed] [Google Scholar]
- Sharff A. J., Wilson D. K., Chang Z., Quiocho F. A. Refined 2.5 A structure of murine adenosine deaminase at pH 6.0. J Mol Biol. 1992 Aug 20;226(4):917–921. doi: 10.1016/0022-2836(92)91040-v. [DOI] [PubMed] [Google Scholar]
- Smith R. F., Smith T. F. Automatic generation of primary sequence patterns from sets of related protein sequences. Proc Natl Acad Sci U S A. 1990 Jan;87(1):118–122. doi: 10.1073/pnas.87.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton B. J., Artymiuk P. J., Cordero-Borboa A. E., Little C., Phillips D. C., Waley S. G. An X-ray-crystallographic study of beta-lactamase II from Bacillus cereus at 0.35 nm resolution. Biochem J. 1987 Nov 15;248(1):181–188. doi: 10.1042/bj2480181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tainer J. A., Getzoff E. D., Beem K. M., Richardson J. S., Richardson D. C. Determination and analysis of the 2 A-structure of copper, zinc superoxide dismutase. J Mol Biol. 1982 Sep 15;160(2):181–217. doi: 10.1016/0022-2836(82)90174-7. [DOI] [PubMed] [Google Scholar]
- Thayer M. M., Flaherty K. M., McKay D. B. Three-dimensional structure of the elastase of Pseudomonas aeruginosa at 1.5-A resolution. J Biol Chem. 1991 Feb 15;266(5):2864–2871. doi: 10.2210/pdb1ezm/pdb. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Active-site zinc ligands and activated H2O of zinc enzymes. Proc Natl Acad Sci U S A. 1990 Jan;87(1):220–224. doi: 10.1073/pnas.87.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Functional zinc-binding motifs in enzymes and DNA-binding proteins. Faraday Discuss. 1992;(93):47–65. doi: 10.1039/fd9929300047. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Short and long spacer sequences and other structural features of zinc binding sites in zinc enzymes. FEBS Lett. 1989 Oct 23;257(1):138–140. doi: 10.1016/0014-5793(89)81805-8. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Auld D. S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990 Jun 19;29(24):5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- Vallee B. L., Coleman J. E., Auld D. S. Zinc fingers, zinc clusters, and zinc twists in DNA-binding protein domains. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):999–1003. doi: 10.1073/pnas.88.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volbeda A., Lahm A., Sakiyama F., Suck D. Crystal structure of Penicillium citrinum P1 nuclease at 2.8 A resolution. EMBO J. 1991 Jul;10(7):1607–1618. doi: 10.1002/j.1460-2075.1991.tb07683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. K., Rudolph F. B., Quiocho F. A. Atomic structure of adenosine deaminase complexed with a transition-state analog: understanding catalysis and immunodeficiency mutations. Science. 1991 May 31;252(5010):1278–1284. doi: 10.1126/science.1925539. [DOI] [PubMed] [Google Scholar]





