Abstract
Manure from swine treated with antimicrobials as feed additives is a major source for the expansion of the antibiotic resistance gene (ARG) reservoir in the environment. Vermicomposting via housefly larvae (Musca domestica) can be efficiently used to treat manure and regenerate biofertilizer, but few studies have investigated its effect on ARG attenuation. Here, we tracked the abundances of 9 ARGs and the composition and structure of the bacterial communities in manure samples across 6 days of full-scale manure vermicomposting. On day 6, the abundances of genes encoding tetracycline resistance [tet(M), tet(O), tet(Q), and tet(W)] were reduced (P < 0.05), while those of genes encoding sulfonamide resistance (sul1 and sul2) were increased (P < 0.05) when normalized to 16S rRNA. The abundances of tetracycline resistance genes were correlated (P < 0.05) with the changing concentrations of tetracyclines in the manure. The overall diversity and richness of the bacteria significantly decreased during vermicomposting, accompanied by a 100 times increase in the relative abundance of Flavobacteriaceae spp. Variations in the abundances of ARGs were correlated with the changing microbial community structure and the relative abundances of the family Ruminococcaceae, class Bacilli, or phylum Proteobacteria. Vermicomposting, as a waste management practice, can reduce the overall abundance of ARGs. More research is warranted to assess the use of this waste management practice as a measure to attenuate the dissemination of antimicrobial residues and ARGs from livestock production before vermicompost can be safely used as biofertilizer in agroecosystems.
INTRODUCTION
Antimicrobials have been widely used at therapeutic levels for microbial infection control and at subtherapeutic levels for growth promotion as veterinary feed additives, administered in water or parenterally in animal husbandry (1). Antimicrobials can select for antibiotic resistance genes (ARGs) in host-associated bacteria from concentrated animal feeding operations and in the environment where antimicrobial-containing feces are excreted or applied to fields (2). Through horizontal gene transfer mechanisms triggered by mobile genetic elements, such as conjugative plasmids, integrons, and transposons (3, 4), different microbes can share their genetic information, potentially causing the transfer and dissemination of ARGs from feces-associated bacteria to indigenous environmental microbes. For instance, an intI1 gene encoding a site-specific integrase responsible for integration is closely related to the transfer of sulfonamide resistance genes among bacteria (5). Specifically, swine manure increases the resistant gene reservoir in the environment, as most applied veterinary antimicrobials are poorly absorbed by livestock (6). These antimicrobials are largely excreted in feces and then dispersed to soil when the manure is used as fertilizer. Unmanaged waste disposal is commonly regarded as one of the major sources for the environmental expansion of ARGs (3); therefore, it is extremely important that waste treatment and disposal technologies to reduce antimicrobials and the associated ARGs are developed prior to the applications of manure as a fertilizer to farmlands.
Vermicomposting is a biological process that involves the oxidation and stabilization of organic wastes through combined larvae and bacterial activities (7, 8). In China, vermicomposting with housefly larvae (Musca domestica) has reached full-scale operations in developed regions (7, 8), primarily to cope with the volume of swine manure generated by the swine industries. Compared with the traditional composting practice without larvae, vermicomposting is much more beneficial. Within 5 to 7 days, a typical cycle of larva vermicomposting can reduce the mass and moisture content of manure by 70 to 80%, reduce odor and the abundance of fecal coliform bacteria, and provide a dehydrated compost fertilizer (a reduction rate of 106 ± 17 kg m−3 day−1) ready for land applications (9); this process can also produce a fresh larva yield of 95 to 120 kg m−3 fresh raw manure (7). Recent studies have focused on composting techniques to retain the nutritional values of manure for soil fertilization but have not determined whether such techniques can reduce antimicrobials or associated resistance genes.
Previously, we demonstrated that the total mass of 8 antimicrobials was significantly attenuated by vermicomposting, and the profiling of the microbial community structure showed an increase in the relative abundance of the Proteobacteria phylum (10). In the current study, we furthered this investigation by quantifying the concentration dynamics of 17 antimicrobials (4 tetracyclines [ACs], 8 sulfonamides [SAs], 4 quinolones [QLs], and 1 macrolide) according to the antimicrobials administered in a finishing swine farm (see Materials and Methods). Although the clinical use of these antimicrobials in human medicines has been limited in recent years in China, these antibacterial agents are still widely used to add to feed or treat bacterial infections in livestock industries because of their low price (especially tetracyclines) (4, 11–13). We also determined the relative abundances of 9 ARGs conferring resistance to these antimicrobials over the course of 6 days of vermicomposting. These ARGs included four tet genes involved in ribosomal protection [tet(M), tet(O), tet(Q), and tet(W)], two sul genes (sul1 and sul2), and three qnr genes (qnrA, qnrB, and qnrS). These genes were selected based on preliminary investigations showing their appearance in fresh manure in Chinese concentrated feeding operations (2, 4, 12, 14). Particularly, within 40 classes of tet genes studied in the environment, the tet genes involved in ribosomal protection proteins have shown greater relative abundance than the tet genes involved in efflux pumps in swine feedlots (15, 16); therefore, these tet genes were selectively investigated in manure samples under vermicomposting practice. In addition, we investigated the changing microbial community structure and ecological succession. The objectives of this study were to determine (i) whether larval vermicomposting attenuates antimicrobials and ARGs in swine manure, (ii) how bacterial community structures in manure are changed after vermicomposting, and (iii) what are the underlying mechanisms of ARG dynamics from the aspects of the changing physicochemical environment, antimicrobials, and bacterial communities.
MATERIALS AND METHODS
Study sites and housefly larva vermicomposting approach.
A full-scale swine manure vermicomposting system with housefly larvae (Musca domestica) was established by the Hangzhou TianYuan Agriculture Development Company in 2008, located in Xiaoshan District, Hangzhou, Zhejiang Province, China (30°49′47.02″N, 120°39′22.12″E). The company receives unprocessed fresh manure daily from a neighboring finishing swine farm (155,000 finishing hogs), with approximately 35,000 tons of fresh manure (moisture content of 80 to 85% in total weight) processed annually. Tetracyclines were used for in-feed antimicrobials, while sulfonamides and quinolones were used for therapy in this finishing swine farm. Beta-lactams were also used occasionally. The housefly larva vermicomposting processes consisted of several stages in series: (i) seed pupation and eclosion, (ii) housefly oviposition, (iii) larval inoculation in unprocessed manure and vermicomposting, (iv) separation of larval residues, and (v) seed sifting and breeding (9). The unprocessed manure, with a surface density of 11.0 kg m−2, was evenly spread on the surface of cement block pools (7.2 × 2.5 × 0.2 m) in greenhouse-assisted larva vermireactors prior to larva inoculation (10). The thickness of the unprocessed manure was approximately 5 cm, and juvenile larvae (12 h old) were applied to the surface of the manure with an average population density of 580,000 m−2. Twelve hours after the larval inoculation, unprocessed fresh manure was added daily at a rate of 5.56 (day 1), 15.6 (day 2), 15.6 (day 3), and 5.56 (day 4) kg m−2. These additions increased the vermicomposting layer from 5 cm to 8 to 10 cm in depth. This semicontinuous feeding mode was shown to be an efficient technique for larval bioconversion (7, 9). The life cycle of vermicomposting with inoculated larvae is 5 (summer) to 10 (winter) days, depending on the ambient temperature. After vermicomposting, more than half of the total weight of the added manure was reduced, and the associated total antimicrobial mass was also significantly attenuated (10). The mass reduction was primarily due to moisture removal and organic biodegradation by larval metabolism and growth. Detailed information about these larva vermireactors, housefly oviposition, and larva residue separation processes has been reported previously (7, 9, 10).
Field sampling and physicochemical measurements.
A summer strategy typical of a 6-day treatment cycle was chosen in this study, given that the summer strategy was used during the majority of time (more than 9 months) for manure processing. The manure residues on the 1st, 3rd, 5th, and 6th days of vermicomposting (treatments designated T-1, T-3, T-5, and T-6, respectively) were sampled in May 2013. In order to compare the samples for the larva-treated vermicomposting and those without larva treatment, the unprocessed raw manure (C-0) was stored in plastic buckets at ambient temperatures (21 to 28°C) mimicking traditional unmanaged composting without larvae, and samples were collected from plastic buckets on the 3rd and 6th days of traditional composting (C-3 and C-6, respectively). Unprocessed fresh manure was also added into the plastic buckets daily at the same rate as that for the samples under vermicomposting. Therefore, traditional composting was identical to vermicomposting with the sole exception that these samples did not contain larva additions. Three samples (C-0, C-3, and C-6) served as controls for the comparison of ARG abundance dynamics against those for the vermicomposting treatment. T-6 represented the final vermicompost. Three replicates of each sample from both the control and treatment collection points were randomly sampled from three different cement block pools. Details of the sampling protocols have been reported previously (10). The pH and temperature of the interior of the manure stockpiles during vermicomposting and/or control composting were recorded on site. Samples were then placed in a portable freezer container within 2 h of collection for transportation and subsequently stored at −20°C, pending physicochemical examination. The moisture contents in samples collected from the unprocessed manure and vermicompost were determined after drying at 105°C for 24 h, while the organic matter contents were determined by calculating the difference between the samples before and after heating at 550°C for 4 h. Total nitrogen was analyzed using the Kjeldahl method (17). Another portion of the samples was freeze-dried using the Lab-1-80 freeze dryer (Biocool Co. Limited, Beijing, China) at −80°C and vacuum at 20 Pa. The freeze-dried samples were sieved through a 0.15-mm mesh before microbial analysis.
Antimicrobial concentration analyses.
The methods established by Huang et al. (18) were used to simultaneously extract four classes of antimicrobials in our samples (C-0, T-1, T-3, T-5, T-6, C-3, and C-6), including 4 tetracyclines, 8 sulfonamides, 4 quinolones, and 1 macrolide. The extracted aliquot (10 μl) was injected into a Kromasil C18 column (Akzo Nobel, Bohuslän, Sweden) loaded with a mobile phase for the chromatographic separation of the antimicrobials followed by the analysis under the positive electrospray ionization mode. The gradient elution and two optimized operating parameters (i.e., the declustering potential and collision energy for each antimicrobial analyzed) were used to achieve the needed resolution and method sensitivity. The internal standard calibration method was used to quantify the analytes (18), and the method detection limit was determined to be 0.5 to 14.1 μg kg−1 for manure samples. The detailed protocols for the antimicrobial analyses can be found in our previous study (10).
Real-time quantitative PCR for the ARGs, intI1 gene, and 16S rRNA.
Total DNA was extracted from 0.1 g of the freeze-dried samples (C-0, T-1, T-3, T-5, T-6, C-3, and C-6) using a PowerSoil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA, USA), following the manufacturer's protocol. The quality and the concentrations of the extracted DNA were checked and determined using 2.5% agarose gel electrophoresis and a spectrophotometer (NanoDrop ND-2000c; Thermo, Waltham, MA, USA). The extracted DNA was also purified using the inhibitor removal technology from the PowerSoil DNA isolation kit.
Real-time quantitative PCR (qPCR) was used to quantify nine ARGs, one intI1 gene, and total 16S rRNA. Each 20-μl qPCR aliquot contained 10 μl of Bestar real-time PCR master mix SYBR green (DBI, Germany), 0.5 μl of each 10 μM forward and reverse primer, 8.0 μl of sterile, DNA-free water, and 1.0 μl of DNA sample as the template. The reaction was run using a StepOnePlus real-time PCR system (Applied Biosystems, Foster City, CA, USA). The running protocols consisted of 95°C for 2 min followed by 40 cycles of 95°C for 10 s; the annealing temperatures were 55°C for 34 s and 72°C for 30 s. The fluorescence acquisition temperature was set at 72°C. The real-time qPCR specificity was confirmed using a melting curve analysis with temperature ramping from 60 to 95°C (0.5°C per read, 30-s hold) and agarose gel electrophoresis. The primers used for the amplification of the tet genes (19), sul genes (20), qnr genes (14), intI1 gene (21), and 16S rRNA (22) with the annealing temperature for each individual gene have been previously reported.
Total DNA extracted from the raw manure was analyzed for the presence of ARGs, the intI1 gene, and 16S rRNA using conventional PCR and agarose gel electrophoresis. After the PCR amplification, the gel slices from an agarose gel containing the target genes were excised and purified using the AxyPrep DNA gel extraction kit (Axygen; Corning Inc., Corning, NY, USA). The purified PCR products were ligated into a pMD 19-T simple vector (TaKaRa, Dalian, China) and then cloned into Escherichia coli DH5α (TaKaRa, Dalian, China). Positive clones with target gene inserts were confirmed by conventional PCR and BLAST alignment (http://www.ncbi.nlm.nih.gov/blast/) after sequencing. The plasmids carrying the target genes were extracted from these clones using an AxyPrep plasmid miniprep kit (Axygen) and quantitatively analyzed by a spectrophotometer (NanoDrop ND-2000c) and then used as the standards for real-time qPCR. Each standard curve was made using six points (three replicates for each point) from 10-fold serial dilutions of a known copy number of the plasmid, the concentration of which ranged from 100 to 10−5 ng of DNA per reaction for 16S rRNA and the intI1 gene, from 10−1 to 10−6 for the tet and sul genes, and from 10−2 to 10−7 for the qnr genes. The qPCR amplification efficiency and R2 values of standard curves for these target genes were 86.3 to 113% and >0.985, respectively. From the threshold cycle (CT) values linked to the standard curves, the target gene copies of the ARGs and intI1 gene were first calculated in absolute values (normalized to a dry weight basis). In order to eliminate the variance due to the differences in the background bacterial abundances as a result of varied DNA yields among different samples, we further presented our results in relative values: relative number of ARG gene copies = absolute number of ARG gene copies/absolute number of gene copies of 16S rRNA.
If the relative gene copies of ARGs were reduced after vermicomposting, it indicated ARG attenuation in a given sample. We chose 16S rRNA to normalize the ARGs and intI1 gene because the absolute gene copies of 16S rRNA have been commonly used in environmental samples as a proxy for cell number or bacterial biomass. Therefore, if the relative gene copy of one ARG is 0.50 in a fresh manure sample when normalized to the 16S rRNA copy number, this single gene would be found in nearly one in every second bacterium, assuming a single copy of each gene in a single genome.
All qPCRs were run in triplicate for each DNA sample. The detection limits for target genes were calculated from the following assumptions: (i) ≥102 copies per reaction, (ii) 0.1 g dry weight for DNA extraction, and (iii) 1.5 × 1012 copies of 16S rRNA per g dry weight (the average concentration for all samples). According to these assumptions, the detection limits for the ARGs and intI1 gene were estimated to be 8.0 × 104 copies per g dry weight or 5.0 × 10−8 copies normalized to the 16S rRNA copy number.
16S rRNA amplicon pyrosequencing and data processing.
Based on the results of the ARG variations and preliminary physicochemical properties among the samples studied, the extracted DNA samples including C-0, T-3, and T-6 with three replicates of each were further analyzed by sequencing 16S rRNA to reveal their microbial community profiling under a 6-day vermicomposting practice. A total of 9 PCR amplicon libraries were constructed (triplicates of each sample) with purified DNA as the template for the nine samples. The V3 to V6 regions of the bacterial 16S rRNA were amplified using the primer set 341F/1073R (CCTACGGGAGGCAGCAG/ACGAGCTGACGACARCCATG). The PCR products were run on a Roche FLX 454 sequencer (Roche Diagnostics Corporation, Branford, CT, USA). Details regarding the pyrosequencing analysis of the amplicon have been previously reported (10).
Quality filtering, demultiplexing, and chimera identification of the raw sequence data from the nine amplicon libraries were performed using the mothur 454 standard operating procedure (23). In brief, sequences with an average quality score of <35 over a 50-bp sliding window that had one or more ambiguous nucleotides, contained homopolymer over 8 bases, or had at least one mismatch to the barcode and two mismatches to the primer were removed. Chimeras were identified using the UCHIME algorithm (24). The preprocessed sequences were aligned to the SILVA-compatible alignment database. After filtering and screening, the high-quality sequences were binned into operational taxonomic units (OTUs) using a 97% identity threshold with the most abundant sequence selected as a representative sequence for that OTU. Taxonomic assignments of sequences to the genus level were made using data from the Ribosomal Database Project (RDP) (25). The OTU-based bacterial richness (observed numbers of OTUs and Chao1), diversity (invsimpson index), and sampling coverage (Good's coverage) were calculated by randomly selecting different sequence sizes from 1 to 5,000 with a bootstep of 200. The relative abundances (percentages) of bacterial memberships within each sample at the family level were compared among samples. Reads in each OTU were normalized by calculating the percentage of reads in each OTU for each sample multiplied by the means of the total reads in all samples studied to correct for differences in sequencing depths.
Statistical analyses.
One-way repeated-measures analysis of variance (ANOVA), followed by Student-Newman-Keuls (S-N-K) tests for pairwise comparison, was used to determine dynamic changes in the relative abundances of ARGs and antimicrobial concentrations in manure across the 6-day vermicomposting practices using the software package SPSS 16.0. Samples within the same day of two contrasting composting practices (i.e., C-3 versus T-3 and C-6 versus T-6) were also compared using Student's t test. Based on the initial results from the detrended correspondence analysis (DCA), a linear model distance-based redundancy analysis (RDA) was performed with the software CANOCO 4.5, using a Bray-Curtis distance measure with Monte Carlo permutation tests (4,999 permutations) to find the main physicochemical factors (organic matter, nitrogen, pH, temperature, and moisture) that determined the variations in ARGs among the samples. Before the DCA or RDA, all environmental variables were centered and standardized. The linear correlations between (i) the relative abundances of ARGs and detected antimicrobial concentrations and (ii) the relative abundances of ARGs and derived OTUs were analyzed by calculating all possible Spearman's rank correlation coefficients at a statistically significant level (P < 0.05) to show their relationship to the detected antimicrobials or microbial taxonomic structures or both.
Nucleotide sequence accession number.
The raw sequences have been deposited into the DDBJ Sequence Read Archive (DRA) database under accession number DRA001286.
RESULTS
Antibiotics in raw manure and after vermicomposting.
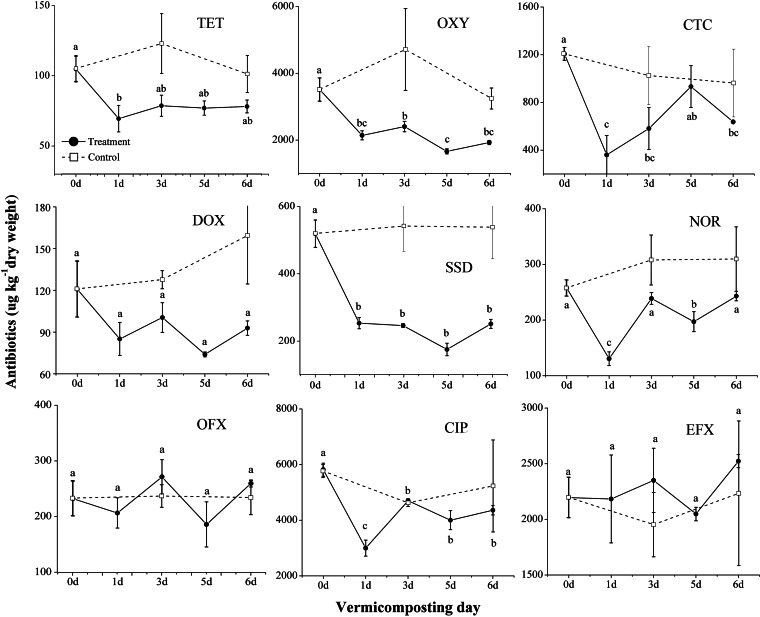
Of the 17 antimicrobials examined, 9 of them, which included sulfadiazine, tetracyclines (tetracycline, oxytetracycline, chlortetracycline, and doxycycline), and quinolones (norfloxacin, ofloxacin, ciprofloxacin, and enrofloxacin), were above the detection limit in unprocessed raw manure samples. Of the tetracyclines, oxytetracycline and chlortetracycline dominated, with average concentrations of 3,511 and 1,207 μg kg−1, respectively, which were an order of magnitude greater than those of doxycycline (121 μg kg−1) and tetracycline (105 μg kg−1) (Fig. 1). The quinolones were dominated by ciprofloxacin and enrofloxacin (5,819 and 2,196 μg kg−1, respectively), which were an order of magnitude greater than norfloxacin (258 μg kg−1) and ofloxacin (232 μg kg−1). The ciprofloxacin concentration showed the highest concentration of all antimicrobials studied.
FIG 1.
Quantification of antimicrobials (micrograms per kilogram dry weight) in unprocessed fresh manure (control samples, C-0), manure during 6-day traditional composting (control samples, C-3 and C-6), and manure during 6-day housefly larvae (Musca domestica) vermicomposting (larva-treated samples, T-1, T-3, T-5, and T-6). The error bars are the standard variations for triplicate samples. The different letters above the error bars indicate significant differences among samples including C-0, T-1, T-3, T-5, and T-6 at P < 0.05 levels using the S-N-K test combined with one-way repeated-measures ANOVA. TET, tetracycline; OXY, oxytetracycline; CTC, chlortetracycline; DOX, doxycycline; SSD, sulfadiazine; NOR, norfloxacin; OFX, ofloxacin; CIP, ciprofloxacin; EFX, enrofloxacin.
Under vermicomposting, the concentrations of chlortetracycline, sulfadiazine, norfloxacin, and ciprofloxacin were reduced significantly after 1 day (P < 0.05) by 70.2%, 51.1%, 49.3%, and 48.4%, respectively, compared to those on C-0 (Fig. 1). However, the concentrations of chlortetracycline, norfloxacin, and ciprofloxacin rebounded on day 2, most likely due to the daily reintroduction of unprocessed manure under the semicontinuous feeding mode. After 6 days of vermicomposting, the concentrations of 5 of the 9 antimicrobials, including tetracycline, oxytetracycline, chlortetracycline, sulfonamide, and ciprofloxacin, were reduced (P < 0.05), while the concentrations of doxycycline, norfloxacin, ofloxacin, and enrofloxacin remained unchanged (Fig. 1), comparing C-0 with T-6. There were no significant changes across all antimicrobials studied among the control samples (C-0, C-3, and C-6, control lines). In a comparison of C-6 with T-6 (the two contrasting composting practices) within the same day, the concentrations of many of the antimicrobials were also reduced (P < 0.05).
Antibiotic resistance genes in raw manure and after vermicomposting.
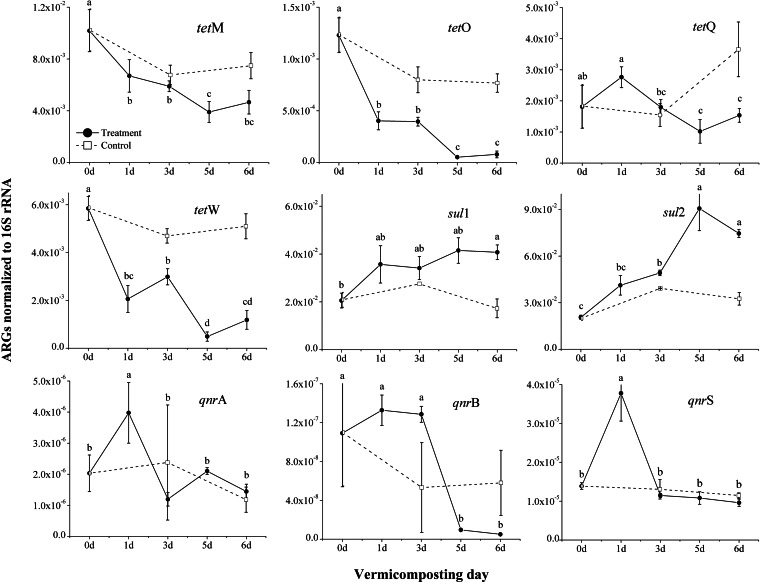
Of the 9 ARGs, the sul genes dominated in the unprocessed raw manure samples (Fig. 2), with average sul1 and sul2 gene copy abundances of 2.06 × 10−2 and 2.07 × 10−2 (normalized to 16S rRNA), respectively. Of the tet genes, tet(M) was the most abundant at 1.02 × 10−2 gene copies, which was an order of magnitude greater than the abundances of the other tet genes. The qnr genes were the least abundant, ranging from 1.39 × 10−5 (qnrS) to 1.09 × 10−7 (qnrB) gene copies, with the latter being only slightly higher than the detection limit. When gene copies were normalized to the dry weight of samples (see Fig. S1 in the supplemental material), the patterns were similar.
FIG 2.
Gene copies of antibiotic resistance genes (ARGs) normalized to 16S rRNA. The error bars are the standard variations for triplicate samples. The tetracycline-related ARGs include tet(M), tet(O), tet(Q), and tet(W), the sulfonamide-related resistance genes include sul1 and sul2, and the quinolone-related ARGs include qnrA, qnrB, and qnrS. Refer to the legend to Fig. 1 for sample designations.
Over the course of vermicomposting, the gene copies of all tet genes [except for tet(Q)] decreased after 1 day and then continued to decrease (P < 0.01) by nearly an order of magnitude by day 6 (Fig. 2). Compared to the initial levels (C-0), the final concentrations in T-6 were 54.2%, 93.5%, 38.5%, and 79.7% lower for tet(M), tet(O), tet(Q), and tet(W), respectively. When T-6 and C-6 (the two contrasting composting practices) were compared within the same day, the differences in these tet genes were also significant. When data were normalized to the dry basis of the manure samples, the gene copies of tet(O) and tet(W) had significantly decreased, while those of tet(M) and tet(Q) remained unchanged, comparing T-6 to C-0 or C-6 (see Fig. S1 in the supplemental material). The gene copies of sul1 (normalized to 16S rRNA) increased after 1 day and then remained constant, while sul2 peaked in abundance on day 5 and remained significantly (P < 0.05) higher until day 6, compared to all control samples (C-0, C-3, and C-6) (Fig. 2). Both qnrA and qnrS significantly (P < 0.05) increased in abundance on day 1 and then rapidly returned to the pretreatment levels (C-0) over the course of vermicomposting (normalized to 16S rRNA in Fig. 2 and to dry weight in Fig. S1 in the supplemental material). There were also no significant differences between C-6 and T-6 for qnrA and qnrS. The gene copy abundance of qnrB was higher in T-3 than in C-3; however, the final value was below the level of detection after 6 days of vermicomposting (T-6) compared with that in C-0 or C-6 (Fig. 2; see also Fig. S1 in the supplemental material).
Altered bacterial community structure during vermicomposting.
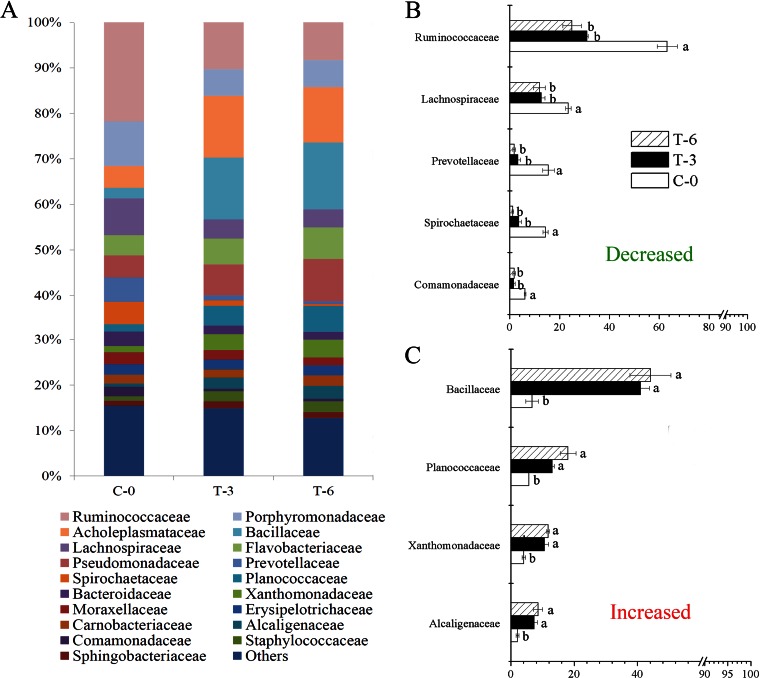
A total of 140,139 sequences were detected from all nine samples with the 341F/1073R primer set. After preprocessing, a total of 61,496 high-quality sequences (4,349 unique sequences) with an average length of 204 bp for each sequence were obtained. The majority of the sequences detected (21,778 sequences corresponding to 35.4% of the total) belonged to Bacteroidetes. The second largest group was Firmicutes (20,906 sequences, 34.0%), which was followed by Proteobacteria (13,847 sequences, 22.5%). After clustering, these sequences produced 2,456 OTUs in total. Further analysis of these data demonstrated a significant (P < 0.001) reduction in species richness and alpha diversity (number of OTUs, Chao1, and Invsimpson) when C-0 was compared with T-3 and T-6. The Good's coverage value in C-0 was 0.87 on average, which was significantly (P < 0.001) lower than those in T-3 (0.95) and T-6 (0.96). All samples had Good's coverage values that were >0.85, which is indicative of high coverage. When annotated to the family level, the relative abundances of the taxonomic families, which included Ruminococcaceae, Lachnospiraceae, Prevotellaceae, Spirochaetaceae, and Comamonadaceae, decreased (P < 0.05) after vermicomposting, while Bacillaceae, Planococcaceae, Xanthomonadaceae, and Alcaligenaceae increased (Fig. 3). Ruminococcaceae was the most dominant taxon in fresh manure and was decreased by 94.1% during vermicomposting. Of the top 100 most abundant OTUs (Table 1), those belonging to Xanthomonadaceae, Alcaligenaceae, and Pseudomonadaceae all increased significantly in relative abundance. Bacillaceae also increased in relative abundance, while the OTUs from the families Lachnospiraceae, Carnobacteriaceae, and Streptococcaceae significantly decreased in relative abundance. A single Flavobacteriaceae OTU showed a 100-fold increase in relative abundance, which was closely related to the genus Riemerella, a typical pathogen in the family Flavobacteriaceae. Despite the fact that such an increase may affect the evenness of a bacterial community, the overall diversity as reflected by the Invsimpson index was still significantly higher in the raw manure than in samples after larva treatment.
FIG 3.
Relative abundances of bacterial taxa at the family level based on 16S rRNA amplicon sequences. (A) Percentages of total sequence reads between three groups, i.e., C-0, T-3, and T-6, are compared. Each group has three replicates. The significantly (P < 0.05) decreased (B) or increased (C) taxa are represented. The error bars are the standard variations for triplicate samples. The different letters above the error bars indicate significant differences at P < 0.05 levels using the S-N-K test combined with one-way repeated-measures ANOVA.
TABLE 1.
The most abundant and significantly changed OTUs (top 100) labeled with taxonomic assignmentsa
| OTU no. | Phylum | Family (no. of OTUs) | No. of sequences in C-0 | No. of sequences in T-6 |
|---|---|---|---|---|
| 1 | Bacteroidetes | Flavobacteriaceae (100) | 11.6 ± 3.38 | 1,155 ± 327 |
| 2 | Proteobacteria | Xanthomonadaceae (100) | 84.0 ± 44.1 | 383 ± 105 |
| 5 | Proteobacteria | Unclassified (100) | 11.0 ± 10.0 | 495 ± 407 |
| 6 | Bacteroidetes | Bacteroidaceae (100) | 303 ± 68.2 | 22.3 ± 11.6 |
| 9 | Firmicutes | Lachnospiraceae (100) | 226 ± 37 | 51.3 ± 21.2 |
| 12 | Firmicutes | Clostridiales family XI incertae sedis (100) | 56.7 ± 5.5 | 64.8 ± 23.2 |
| 15 | Firmicutes | Unclassified (100) | 10.0 ± 2.08 | 85.6 ± 37.7 |
| 16 | Tenericutes | Acholeplasmataceae (100) | 5.00 ± 2.52 | 63.1 ± 30.6 |
| 17 | Firmicutes | Clostridiales family XI incertae sedis (64) | 4.67 ± 1.86 | 59.2 ± 24.7 |
| 18 | Firmicutes | Bacillaceae (100) | 3.33 ± 0.33 | 124 ± 66.8 |
| 19 | Firmicutes | Bacillaceae (100) | 11.7 ± 10.2 | 50.9 ± 17.9 |
| 20 | Firmicutes | Bacillaceae (63) | 7.67 ± 3.71 | 97.6 ± 39.3 |
| 23 | Firmicutes | Unclassified (100) | 0.00 ± 0.00 | 67.2 ± 45.5 |
| 24 | Firmicutes | Carnobacteriaceae (100) | 109.7 ± 36.2 | 22.7 ± 15.2 |
| 25 | Firmicutes | Unclassified (100) | 0 ± 0 | 57.4 ± 31.3 |
| 27 | Firmicutes | Planococcaceae (100) | 16.0 ± 5.13 | 69.8 ± 24.0 |
| 28 | Firmicutes | Lachnospiraceae (100) | 113 ± 13 | 6.46 ± 5.90 |
| 32 | Bacteroidetes | Bacteroidaceae (100) | 109 ± 53 | 0.58 ± 0.58 |
| 33 | Bacteroidetes | Unclassified (100) | 0.67 ± 0.33 | 40.3 ± 14.4 |
| 35 | Firmicutes | Clostridiaceae (100) | 45.2 ± 18.3 | 2.36 ± 1.08 |
| 38 | Firmicutes | Unclassified (100) | 1.33 ± 0.67 | 26.2 ± 11.5 |
| 39 | Bacteroidetes | Unclassified (100) | 56.7 ± 17.8 | 11.5 ± 4.57 |
| 40 | Proteobacteria | Alcaligenaceae (100) | 0.33 ± 0.33 | 57.4 ± 6.32 |
| 44 | Bacteroidetes | Unclassified (100) | 73.7 ± 26.3 | 0.29 ± 0.29 |
| 47 | Firmicutes | Streptococcaceae (100) | 60.7 ± 15.4 | 4.56 ± 1.65 |
| 48 | Proteobacteria | Pseudomonadaceae (100) | 1.33 ± 0.88 | 48.7 ± 25.5 |
| 54 | Bacteroidetes | Unclassified (100) | 52.0 ± 16.09 | 2.61 ± 2.61 |
| 55 | Proteobacteria | Xanthomonadaceae (100) | 0.00 ± 0.00 | 23.6 ± 15.7 |
| 57 | Firmicutes | Unclassified (54) | 1.00 ± 0.58 | 14.4 ± 7.32 |
| 58 | Proteobacteria | Unclassified (97) | 1.67 ± 1.67 | 32.1 ± 21.3 |
| 60 | Firmicutes | Bacillaceae (100) | 0.00 ± 0.00 | 20.1 ± 16.5 |
| 61 | Firmicutes | Lachnospiraceae (100) | 49.7 ± 15.4 | 0.29 ± 0.29 |
| 63 | Fibrobacteres | Fibrobacteraceae (100) | 43.0 ± 6.0 | 0.00 ± 0.00 |
| 66 | Bacteroidetes | Flavobacteriaceae (100) | 34.7 ± 5.8 | 4.77 ± 1.92 |
| 68 | Proteobacteria | Moraxellaceae (100) | 39.7 ± 14.9 | 0.58 ± 0.58 |
| 69 | Firmicutes | Ruminococcaceae (100) | 37.1 ± 7.79 | 0.29 ± 0.29 |
| 72 | Bacteroidetes | Unclassified (100) | 40.3 ± 16.9 | 0.29 ± 0.29 |
| 78 | Proteobacteria | Pseudomonadaceae (100) | 0.00 ± 0.00 | 25.3 ± 17.7 |
| 80 | Bacteroidetes | Unclassified (100) | 34.3 ± 14.2 | 0.29 ± 0.29 |
| 81 | Firmicutes | Ruminococcaceae (100) | 48.2 ± 16.7 | 0.00 ± 0.00 |
| 82 | Proteobacteria | Unclassified (100) | 1.00 ± 1.00 | 21.6 ± 16.3 |
| 85 | Unclassified | Unclassified (100) | 0.67 ± 0.33 | 12.0 ± 3.11 |
| 86 | Tenericutes | Acholeplasmataceae (100) | 1.67 ± 1.67 | 10.67 ± 2.31 |
| 88 | Tenericutes | Acholeplasmataceae (100) | 0.67 ± 0.67 | 6.51 ± 6.08 |
| 89 | Bacteroidetes | Bacteroidaceae (100) | 29.0 ± 4.9 | 1.16 ± 1.16 |
| 96 | Tenericutes | Acholeplasmataceae (100) | 1.00 ± 1.00 | 14.9 ± 11.2 |
| 97 | Firmicutes | Ruminococcaceae (100) | 27.0 ± 10.2 | 0.66 ± 0.34 |
| 100 | Firmicutes | Unclassified (100) | 26.7 ± 6.5 | 0.58 ± 0.58 |
The relative abundance in each OTU indicated by the number of detected sequences between unprocessed manure (C-0) and the residues after the 6-day vermicomposting treatment (T-6) is compared (n = 3). The data are expressed as means ± SD. The significantly decreased abundance in each OTU is in bold.
Correlating ARG abundance with environmental factors, antimicrobial concentrations, microbial community structure, and the intI1 gene.
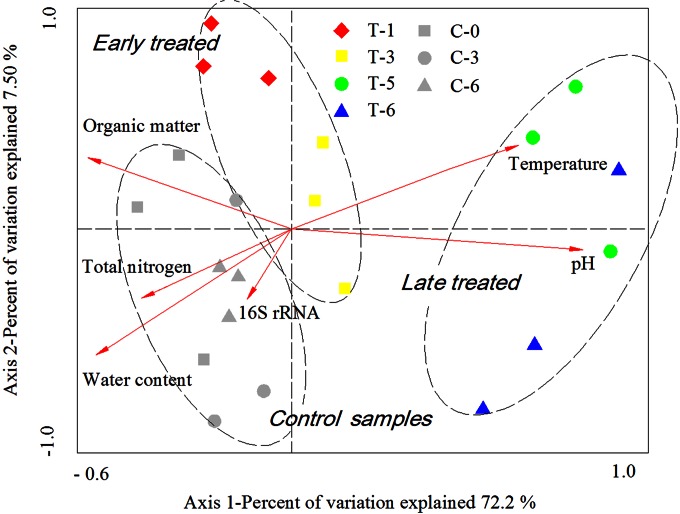
ARG abundances were clearly separated between the control samples (C-0, C-3, and C-6), early-treated samples (T-1 and T-3), and later-treated samples (T-5 and T-6) (Fig. 4). Vermicomposting significantly reduced the organic matter, total nitrogen, and water content compared to those in the control samples. The pH increased significantly from 6.68 (C-0) to 8.46 (T-6), while the temperature showed marked changes with a peak at ∼47°C on day 5 (T-5) (see Table S1 in the supplemental material). There were no significant changes in the physicochemical properties among the control samples over the period of 6 days of storage. The RDA analysis shows that the ARG dynamics through vermicomposting were positively correlated (P < 0.05) with the changes in the water content and organic matter in the manure samples, indicating that the overall ARG abundance was high when the water content and organic matter were also high. Negative correlations were found between ARG dynamics and changes in temperature and pH (P < 0.01) (Fig. 4).
FIG 4.
Distance-based redundancy analysis (RDA) comparing antibiotic resistance genes (ARGs) (symbols) and environmental variables (arrows) using the Bray-Curtis distance measure. The percentage of variation explained by each axis is shown, and the relationship is significant (P = 0.004).
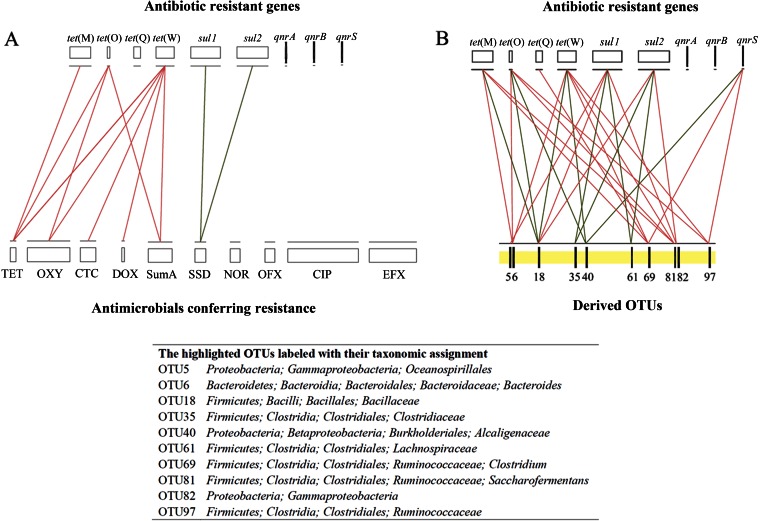
Among 981 pairwise correlations, 17 and 169 were significant (P < 0.05) for ARGs versus antimicrobials and for ARGs versus OTUs, respectively (Fig. 5). The tet genes were strongly correlated with the tetracycline concentrations and OTUs, while the qnr genes were only weakly correlated with the quinolone concentrations (Fig. 5A). Certain specific OTUs significantly affected changes in the ARG abundance (Fig. 5B). Several Ruminococcaceae OTUs, which were associated with Clostridiales, maintained significantly positive correlations (P < 0.05) with the abundance of the tet genes. Approximately 95% of OTUs were negatively correlated (P < 0.05) with the sul genes (Fig. 5B), except for two OTUs from the Proteobacteria phylum and one OTU from the Bacilli, which showed strong positive (P < 0.01; Spearman's r of >0.7) correlations with the sul gene abundance.
FIG 5.
Network analyses as inferred by pairwise Spearman correlations between antibiotic resistance genes and antimicrobials (A) and antibiotic resistance genes and normalized OTU abundances (B). The normalized reads in each OTU are calculated by the percentage of reads in each OTU for each sample multiplied by the means of the total reads in all samples studied. In order to filter the data for reduced network complexity, the connections (red lines for positive and green lines for negative) shown here stand only for strong (Spearman's r of >0.7) and remarkably significant (P < 0.01) correlations. The most abundant and significantly changed top 100 OTUs were used for network analysis (see Table 1 for details), and the top 10 with the most correlations with antibiotic resistance genes are highlighted and labeled with their taxonomic assignments. The background values of the ARG abundances and antimicrobial concentrations detected in fresh manure are illustrated by the lengths of the bars. SumA, the total abundance of four tetracyclines.
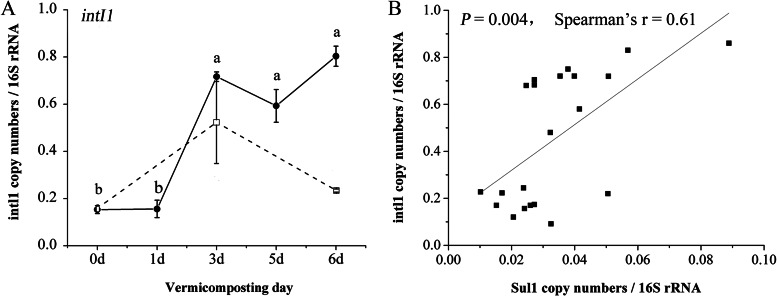
The intI1 gene increased (P < 0.05) after 3 days of vermicomposting and then remained constantly higher in T-6 (day 6) than in C-0 (Fig. 6). In comparison with the samples under traditional composting (C-6), the intI1 gene in T-6 increased by 2.68-fold. The relative abundance of sul1 was remarkably related to that of the intI1 gene, as reported by Spearman correlations.
FIG 6.
(A) The gene copies of intI1 normalized to 16S rRNA. The error bars are the standard variations from triplicate samples. Refer to Fig. 1 for sample designations. (B) The pairwise Spearman correlations between the gene copies of intI1 and the gene copies of sul1.
DISCUSSION
We have previously shown that housefly larva vermicomposting significantly attenuates the total mass of antimicrobials and increases the relative abundance of Proteobacteria (10). Here, we furthered this research by coexamining the abundance of ARGs and the concentration of antimicrobials and expanded our analysis of the microbial community composition. We found positive correlations between the concentration of tetracyclines and the abundance of tetracycline resistance genes, which also showed significant correlations with the relative abundance of the Clostridiales taxa (especially with the family Ruminococcaceae). Genes coding for resistance to sulfonamides increased in abundance in the vermicompost after vermicomposting, despite a lack of increase in the concentration of sulfonamides; this was correlated with increased relative abundances of two proteobacterial taxa and one Bacilli taxon.
Schematic of various ARGs responding to vermicomposting.
Previous reports have shown that the tet gene abundance is positively correlated with the tetracycline concentration (12), suggesting that tetracycline residues in swine manure may exert direct selection pressure on tetracycline resistance in microorganisms. The values for the tet gene abundances obtained in this study were similar to those reported in other livestock samples. The tet gene [tet(M), tet(O), tet(Q), and tet(W)] copies normalized to 16S rRNA ranged from 10−2 to 10−3 in raw manure with the highest values found for the tet(M) gene, which is comparable to values for swine feedlots (10−2 to 10−5) (12), waste lagoons at cattle feedlots (10−2 to 10−6) (26), and swine lagoons (10−1 to 10−4) (27, 28); yet, these values are much lower than those observed in natural river sediments (10−7 to 10−8) (29). Such a relatively high abundance implies that the swine manure may become a crucial tet-resistant source of exposure in natural environments, and vermicomposting can significantly reduce the abundance of tet genes involved in ribosomal protection proteins. Other classes of tet genes that acted as efflux pumps [i.e., tet(A), tet(B), tet(C), tet(D), and tet(G)] were also frequently detected in the environment (12), especially in populations of Escherichia coli (30). Traditional composting without larvae has been reported to significantly reduce these tet genes (31).
Although the concentrations of quinolones detected were the highest among all antimicrobials studied, the number of associated qnr gene copies (10−5 to 10−7) normalized to 16S rRNA was near or even lower than the values found in natural surface sediments of Famosa Slough (10−4 to 10−5) (14). The selection pressure exerted by quinolones on qnr genes was extremely weak in manure samples, as demonstrated by our network analysis. A previous study suggested that qnr genes only produce low-level resistance (32), which was consistent with our findings. Also similar to the results of this study, Rutgersson et al. (33) did not find qnr gene enrichment in fecal samples from quinolone-contaminated villages; the high concentrations of quinolones only select for the low abundances of qnr genes in the surrounding soil, sediment, and water samples. Quinolones exhibit concentration-dependent effects, and high concentrations may increase the extent of bacterial destruction (34). When quinolones were used at high concentrations as veterinary feed additives, many strains harboring qnr genes did not survive in swine gut or manure with quinolone concentrations exceeding the MIC (35); however, the residual concentrations of ciprofloxacin and enrofloxacin after 6 days of vermicomposting were greater than 4 μg g−1 and 2 μg g−1, respectively, which are above the resistance breakpoint for these antimicrobials (36). Therefore, the potential risk of their selection for the resistance in microbiota is still a concern if this vermicompost is applied to agricultural land for a long duration. More importantly, since a primary mechanism for quinolone resistance is derived from gene mutations in a susceptible target (32), the presence of quinolones at high concentrations may facilitate the selection of mutations to various extents. In addition to qnr genes, these other mutated genes in manure samples under vermicomposting practice need further identification.
Among all of the ARGs studied, sul1 and sul2 had the highest background abundances of genes (10−2) in unprocessed manure; such values were also similar with those reported in manure samples collected from other pig feedlots across China (4). After vermicomposting, sul1 and sul2 gene copies were one to two orders of magnitude greater than those for tet genes and three to six orders of magnitude greater than those for qnr genes. Moreover, the relative abundance of the sul genes (normalized to 16S rRNA) after vermicomposting was increased, suggesting that these genes may be more frequently detected in a single genome of each bacterium through gene mutation or possibly there was preferred growth of sul-harboring bacteria in the background of indigenous microbes in larva-treated samples. We also found a strong correlation between the abundances of sul1 genes and one integron (intI1), suggesting the potential of horizontal gene transfer. Cell-to-cell contact (transconjugation) experiments have also demonstrated that sul2 is transmissible between different bacteria exposed to adverse situations (37, 38). Coenrichment of multiple genes may be a result of the aggregation of various resistance loci on a single mobile genetic element (3, 38). It is therefore possible that the observed enrichment of these sul genes may be due to their location on mobile genetic elements harboring genes conferring resistance to other antimicrobials. After vermicomposting, sulfonamides were below the detection limit, while sul genes were highly abundant.
Niche alternations and microbial succession responsible for changed ARGs.
Similar to other antimicrobials studied previously (10), ARG abundance was also significantly correlated with environmental conditions (moisture, temperature, total nitrogen, and pH). Compared to results for the unprocessed manure, vermicomposting significantly reduced organic matter, total nitrogen, and water content, which was accompanied by significantly elevated pH and much fluctuation in temperature (10). Previous studies have found a significantly negative relationship between the tet genes and pH (12); however, the underlying mechanisms of such a relationship remained unknown. Reduced tet gene abundance has been found to be correlated with increasing temperature (22 to 55°C) during the anaerobic digestion of wastewater solids (39). The temperature in our vermicompost peaked at ∼47°C on days 3 to 5 and then gradually returned to the pretreatment level on day 6.
Besides the increased relative abundance of the Proteobacteria phylum (10), the relative abundance of a single taxon of Flavobacteriaceae spp. was also significantly increased through vermicomposting. Taxa in the family Flavobacteriaceae are common bacterial intracellular symbionts of various insects, including cockroaches, termites, ladybird beetles, ticks, and wasps (40), and their related strains are highly resistant to many antimicrobial agents. Whether the increase in Flavobacteriaceae spp. following vermicomposting was the result of shedding of intestinal intracellular symbionts through larval gut transit warrants more genetic evidence from larval gut microbiome analyses. However, there was evidence showing growth promotion of the family Flavobacteriaceae in decomposed (nonenriched) substrates through earthworm processing (41), which was consistent with our findings. These r-selected bacteria are known to degrade many complex organic compounds. Most genera assigned to the family Flavobacteriaceae are aerobic or microaerophilic, which might explain why they were absent in the anaerobic and water-saturated unprocessed manure but became abundant immediately following vermicomposting with the enhanced aeration by the larvae (40). Through larval gut, the excreted casts and feces may greatly modify the manure microbiota; this may also have caused the reduced abundance of OTUs from the obligate anaerobic family Ruminococcaceae, which was also correlated with the attenuation of the tet genes.
Shifts in the bacterial community under antimicrobial stresses might provide new insight into the taxa which are favored by antimicrobial selection pressures. A study using in-feed antimicrobials in pigs demonstrated increased proteobacterial abundance by 1 to 11% in the swine intestinal microbiome (42). Another study observed increased members of Proteobacteria in the human gut microbiome following treatment with broad-spectrum antimicrobials used for their therapeutic effect against a wide range of bacterial species (43). In oxytetracycline- and penicillin G-contaminated rivers, the deeply rooting classes Deltaproteobacteria and Epsilonproteobacteria were also highly represented (44). These findings demonstrated a potentially close linkage between antimicrobials and increased Proteobacteria. Certain taxa belonging to Proteobacteria might be favored by application of antimicrobials when they act as the preferred hosts for harboring multiple ARGs selected by these antimicrobials. Additionally in our network analyses, Clostridiales showed the most correlations with the ARGs studied. These low-GC Gram-positive bacteria have also been demonstrated to be closely associated with antibiotic-containing environments (44). From a literature review, however, we found that until now no straightforward evidence suggested reduced diversity of bacterial communities under antimicrobial stresses (42, 44, 45). In contrast, antimicrobial treatment leads to a highly connected phage-bacterial network for gene exchange (46). Such community-based exploration implied that the decreased bacterial diversity in residues after larva treatments is responsible for the overall attenuated ARGs by impairing their preexisting host-ARG genetic relationships. Despite the decreased diversity of the bacterial community, the sul genes were intriguingly represented with higher abundance in larva-treated samples. Taxa in Bacilli showed positive correlations with the increases in these sul genes. Previously, class Bacilli were found to dominate in the human and livestock gut after antimicrobial treatment, as well as in antibiotic-containing downstream rivers (44). Some species in Bacilli can excrete cellulase and polyphenoloxidase, promoting compost maturity (47), and function well even when the surrounding temperature is much higher (48). These properties enable them to survive thermophilic periods and become effective recolonizers in treated vermicompost. There was also evidence demonstrating that Bacillus are the preferred hosts for the sul genes. Based on culture-dependent approaches, sulfonamide-resistant bacteria isolated from agriculture sites contaminated by sulfonamides incorporated mainly Bacillus genera (49). Similarly, Bacillus has been found to be the most prevalent genus for all sulfonamide-resistant bacteria isolated from manure samples (50). In another study, a phylogenetic analysis further showed that there were possibilities for the genetic transfer of the sul1 gene from intestinal Enterococcus spp. to indigenous Bacillus spp. (51). Sulfonamide-resistant bacteria may be selectively distributed in certain bacterial groups, including even potential human pathogens. In clinical research, sul1 has been frequently detected in the proteobacterium Pseudomonas aeruginosa as indicated by the taxonomic distribution of molecular data from the Comprehensive Antibiotic Resistance Database (52). Such a common Gram-negative, aerobic bacterium is an opportunistic human pathogen, which might easily acquire resistance by the horizontal gene transfer of antibiotic resistance determinants (53) and thus increase the potential risk for sul1 dissemination during microbial succession when raw manure become more aerobic or microaerophilic after larva treatment.
Implications of vermicomposting for antibiotic resistance gene management.
The Chinese agricultural industry uses 97 million kg of antimicrobials annually in livestock production, accounting for 46% of the total volume used every year (1, 54), with swine manure identified as a major source of antimicrobial pollution in the environment (55). It is possible to use a traditional composting practice without larvae such that antimicrobials and ARGs in manure samples would be reduced to a certain degree after 2 to 3 months of treatment (13); however, within the comparable 6 days (for instance, T-6 versus C-6), vermicomposting is much more beneficial than the traditional practice. A total of 600 million tons of swine manure are produced in China annually (4); such a large amount is striking, but if the manure is treated faster, more can be treated overall. Vermicomposting is a time-based improvement, with an added advantage of controlling ARGs more efficiently than traditional composting practice. While only a single sampling site was analyzed during the current study, the categories of antimicrobials administered to the swine were similar to those reported elsewhere (4, 18). Therefore, the selection pressure on ARGs exerted by these antimicrobials and the fate of the ARGs through vermicomposting technology is likely comparable to those of other systems.
One potential risk for vermicomposting is the possibility of the acquisition of ARGs by mature flies, since the housefly gut is a favorable environment for the transfer of ARG-containing strains from their food to their bodies (56). We assumed that such a risk is not of significant importance. When harvested, >95% of housefly larvae will be dehydrated and dried. Another portion of the larvae (<5%) will develop into mature flies, which will mate and spawn to give birth to the new larvae for the next batch of vermicomposting. Being restricted to the breeding room during oviposition, these mature flies will not live by artificially affecting supplies of water and feeding as they become senile with a weak capacity for oviposition. Consequently, the mature flies that escape, if any, would rarely have the potential to cause any danger to their surroundings. The sustained abundance of sulfonamide resistance genes requires attention, especially if treated vermicompost is to be used as an agricultural biofertilizer. Furthering our understanding of how vermicomposting management practices can be augmented to reduce antimicrobial residues, and ARGs requires a more detailed investigation of the potential host range and genetic transferability of various ARGs and ultimately of the possible consequences on environmental health.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (grant 41373074), the National Ministry of Science and Technology (grant 2013GB23600658), and the Zhejiang Science and Technology Innovation Program (grant 2013C33001) and in part by the U.S. Department of Energy under contract DE-AC02-06CH11357.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01367-15.
REFERENCES
- 1.Hvistendahl M. 2012. China takes aim at rampant antibiotic resistance. Science 336:795–795. doi: 10.1126/science.336.6083.795. [DOI] [PubMed] [Google Scholar]
- 2.Pruden A, Pei RT, Storteboom H, Carlson KH. 2006. Antibiotic resistance genes as emerging contaminants: studies in northern Colorado. Environ Sci Technol 40:7445–7450. doi: 10.1021/es060413l. [DOI] [PubMed] [Google Scholar]
- 3.Su JQ, Wei B, Xu CY, Qiao M, Zhu YG. 2014. Functional metagenomic characterization of antibiotic resistance genes in agricultural soils from China. Environ Int 65:9–15. doi: 10.1016/j.envint.2013.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Zhu YG, Johnson TA, Su JQ, Qiao M, Guo GX, Stedtfeld RD, Hashsham SA, Tiedje JM. 2013. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc Natl Acad Sci U S A 110:3435–3440. doi: 10.1073/pnas.1222743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaze WH, Zhang LH, Abdouslam NA, Hawkey PM, Calvo-Bado L, Royle J, Brown H, Davis S, Kay P, Boxall ABA, Wellington EMH. 2011. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 5:1253–1261. doi: 10.1038/ismej.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alcock RE, Sweetman A, Jones KC. 1999. Assessment of organic contaminant fate in waste water treatment plants. I. Selected compounds and physicochemical properties. Chemosphere 38:2247–2262. doi: 10.1016/S0045-6535(98)00444-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang H, Zhang ZJ, Czapar GF, Winkler MKH, Zheng JG. 2013. A full-scale house fly (Diptera: Muscidae) larvae bioconversion system for value-added swine manure reduction. Waste Manage Res 31:223–231. doi: 10.1177/0734242X12469431. [DOI] [PubMed] [Google Scholar]
- 8.Čičková H, Pastor B, Kozanek M, Martinez-Sanchez A, Rojo S, Takac P. 2012. Biodegradation of pig manure by the housefly, Musca domestica: a viable ecological strategy for pig manure management. PLoS One 7:e32798. doi: 10.1371/journal.pone.0032798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang ZJ, Wang H, Zhu J, Suneethi S, Zheng JG. 2012. Swine manure vermicomposting via housefly larvae (Musca domestica): the dynamics of biochemical and microbial features. Bioresour Technol 118:563–571. doi: 10.1016/j.biortech.2012.05.048. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z, Shen J, Wang H, Liu M, Wu L, Ping F, He Q, Li H, Zheng C, Xu X. 2014. Attenuation of veterinary antibiotics in full-scale vermicomposting of swine manure via the housefly larvae (Musca domestica). Sci Rep 4:6844. doi: 10.1038/srep06844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen H, Zhang MM. 2013. Occurrence and removal of antibiotic resistance genes in municipal wastewater and rural domestic sewage treatment systems in eastern China. Environ Int 55:9–14. doi: 10.1016/j.envint.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 12.Wu N, Qiao M, Zhang B, Cheng WD, Zhu YG. 2010. Abundance and diversity of tetracycline resistance genes in soils adjacent to representative swine feedlots in China. Environ Sci Technol 44:6933–6939. doi: 10.1021/es1007802. [DOI] [PubMed] [Google Scholar]
- 13.Dolliver H, Gupta S, Noll S. 2008. Antibiotic degradation during manure composting. J Environ Qual 37:1245–1253. doi: 10.2134/jeq2007.0399. [DOI] [PubMed] [Google Scholar]
- 14.Cummings DE, Archer KF, Arriola DJ, Baker PA, Faucett KG, Laroya JB, Pfeil KL, Ryan CR, Ryan KRU, Zuill DE. 2011. Broad dissemination of plasmid-mediated quinolone resistance genes in sediments of two urban coastal wetlands. Environ Sci Technol 45:447–454. doi: 10.1021/es1029206. [DOI] [PubMed] [Google Scholar]
- 15.Jindal A, Kocherginskaya S, Mehboob A, Robert M, Mackie RI, Raskin L, Zilles JL. 2006. Antimicrobial use and resistance in swine waste treatment systems. Appl Environ Microbiol 72:7813–7820. doi: 10.1128/AEM.01087-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koike S, Krapac IG, Oliver HD, Yannarell AC, Chee-Sanford JC, Aminov RI, Mackie RI. 2007. Monitoring and source tracking of tetracycline resistance genes in lagoons and groundwater adjacent to swine production facilities over a 3-year period. Appl Environ Microbiol 73:4813–4823. doi: 10.1128/AEM.00665-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bao S. 2000. Agro-chemical analysis of soil. China Agricultural Press, Beijing, China. [Google Scholar]
- 18.Huang YJ, Cheng MM, Li WH, Wu LH, Chen YS, Luo YM, Christie P, Zhang HB. 2013. Simultaneous extraction of four classes of antibiotics in soil, manure and sewage sludge and analysis by liquid chromatography-tandem mass spectrometry with the isotope-labelled internal standard method. Anal Methods 5:3721–3731. doi: 10.1039/c3ay40220g. [DOI] [Google Scholar]
- 19.Aminov RI, Garrigues-Jeanjean N, Mackie RI. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl Environ Microbiol 67:22–32. doi: 10.1128/AEM.67.1.22-32.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo Y, Mao DQ, Rysz M, Zhou DX, Zhang HJ, Xu L, Alvarez PJJ. 2010. Trends in antibiotic resistance genes occurrence in the Haihe River, China. Environ Sci Technol 44:7220–7225. doi: 10.1021/es100233w. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein C, Lee MD, Sanchez S, Hudson C, Phillips B, Register B, Grady M, Liebert C, Summers AO, White DG, Maurer JJ. 2001. Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob Agents Chemother 45:723–726. doi: 10.1128/AAC.45.3.723-726.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fierer N, Jackson JA, Vilgalys R, Jackson RB. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl Environ Microbiol 71:4117–4120. doi: 10.1128/AEM.71.7.4117-4120.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schloss PD, Gevers D, Westcott SL. 2011. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS One 6:e27310. doi: 10.1371/journal.pone.0027310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knapp CW, Dolfing J, Ehlert PAI, Graham DW. 2010. Evidence of increasing antibiotic resistance gene abundances in archived soils since 1940. Environ Sci Technol 44:580–587. doi: 10.1021/es901221x. [DOI] [PubMed] [Google Scholar]
- 27.Marshall BM, Levy SB. 2011. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev 24:718−733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng W, Chen H, Su C, Yan S. 2013. Abundance and persistence of antibiotic resistance genes in livestock farms: a comprehensive investigation in eastern China. Environ Int 61:1–7. doi: 10.1016/j.envint.2013.08.023. [DOI] [PubMed] [Google Scholar]
- 29.Pei RT, Kim SC, Carlson KH, Pruden A. 2006. Effect of river landscape on the sediment concentrations of antibiotics and corresponding antibiotic resistance genes (ARG). Water Res 40:2427–2435. doi: 10.1016/j.watres.2006.04.017. [DOI] [PubMed] [Google Scholar]
- 30.Bryan A, Shapir N, Sadowsky MJ. 2004. Frequency and distribution of tetracycline resistance genes in genetically diverse, nonselected, and nonclinical Escherichia coli strains, isolated from diverse human and animal sources. Appl Environ Microbiol 70:2503–2507. doi: 10.1128/AEM.70.4.2503-2507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma R, Larney FJ, Chen J, Yanke LJ, Morrison M, Topp E, McAllister TA, Yu ZT. 2009. Selected antimicrobial resistance during composting of manure from cattle administered sub-therapeutic antimicrobials. J Environ Qual 38:567–575. doi: 10.2134/jeq2007.0638. [DOI] [PubMed] [Google Scholar]
- 32.Jacoby GA. 2005. Mechanisms of resistance to quinolones. Clin Infect Dis 41:S120–S126. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- 33.Rutgersson C, Fick J, Marathe N, Kristiansson E, Janzon A, Angelin M, Johansson A, Shouche Y, Flach CF, Larsson DGJ. 2014. Fluoroquinolones and qnr genes in sediment, water, soil, and human fecal flora in an environment polluted by manufacturing discharges. Environ Sci Technol 48:7825–7832. doi: 10.1021/es501452a. [DOI] [PubMed] [Google Scholar]
- 34.Wright DH, Brown GH, Peterson ML, Rotschafer JC. 2000. Application of fluoroquinolone pharmacodynamics. J Antimicrob Chemother 46:669–683. doi: 10.1093/jac/46.5.669. [DOI] [PubMed] [Google Scholar]
- 35.Duggirala A, Joseph J, Sharma S, Nutheti R, Garg P, Das T. 2007. Activity of newer fluoroquinolones against Gram-positive and Gram-negative bacteria isolated from ocular infections: an in vitro comparison. Indian J Ophthalmol 55:15–19. doi: 10.4103/0301-4738.29489. [DOI] [PubMed] [Google Scholar]
- 36.Aarestrup FM, Wiuff C, Molbak K, Threlfall EJ. 2003. Is it time to change fluoroquinolone breakpoints for Salmonella spp.? Antimicrob Agents Chemother 47:827–829. doi: 10.1128/AAC.47.2.827-829.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Enne VI, Livermore DM, Stephens P, Hall LMC. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325–1328. doi: 10.1016/S0140-6736(00)04519-0. [DOI] [PubMed] [Google Scholar]
- 38.Sköld O. 2000. Sulfonamide resistance: mechanisms and trends. Drug Resist Updat 3:155–160. doi: 10.1054/drup.2000.0146. [DOI] [PubMed] [Google Scholar]
- 39.Diehl DL, LaPara TM. 2010. Effect of temperature on the fate of genes encoding tetracycline resistance and the integrase of class 1 integrons within anaerobic and aerobic digesters treating municipal wastewater solids. Environ Sci Technol 44:9128–9133. doi: 10.1021/es102765a. [DOI] [PubMed] [Google Scholar]
- 40.Bernardet JF, Nakagawa Y, Holmes B, Subcommittee on the Taxonomy of Flavobacterium and Cytophaga-like Bacteria of the International Committee on Systematics of Prokaryotes . 2002. Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070. doi: 10.1099/ijs.0.02136-0. [DOI] [PubMed] [Google Scholar]
- 41.Bernard L, Chapuis-Lardy L, Razafimbelo T, Razafindrakoto M, Pablo AL, Legname E, Poulain J, Bruls T, O'Donohue M, Brauman A, Chotte JL, Blanchart E. 2012. Endogeic earthworms shape bacterial functional communities and affect organic matter mineralization in a tropical soil. ISME J 6:213–222. doi: 10.1038/ismej.2011.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Looft T, Johnson TA, Allen HK, Bayles DO, Alt DP, Stedtfeld RD, Sul WJ, Stedtfeld TM, Chai BL, Cole JR, Hashsham SA, Tiedje JM, Stanton TB. 2012. In-feed antibiotic effects on the swine intestinal microbiome. Proc Natl Acad Sci U S A 109:1691–1696. doi: 10.1073/pnas.1120238109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rea MC, Dobson A, O'Sullivan O, Crispie F, Fouhy F, Cotter PD, Shanahan F, Kiely B, Hill C, Ross RP. 2011. Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci U S A 108:4639–4644. doi: 10.1073/pnas.1001224107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li D, Qi R, Yang M, Zhang Y, Yu T. 2011. Bacterial community characteristics under long-term antibiotic selection pressures. Water Res 45:6063–6073. doi: 10.1016/j.watres.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Rooks MG, Veiga P, Wardwell-Scott LH, Tickle T, Segata N, Michaud M, Gallini CA, Beal C, van Hylckama-Vlieg JET, Ballal SA, Morgan XC, Glickman JN, Gevers D, Huttenhower C, Garrett WS. 2014. Gut microbiome composition and function in experimental colitis during active disease and treatment-induced remission. ISME J 8:1403–1417. doi: 10.1038/ismej.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Modi SR, Lee HH, Spina CS, Collins JJ. 2013. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499:219−222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayende L, Wilhelmi BS, Pletschke BI. 2006. Cellulases (CMCases) and polyphenol oxidases from thermophilic Bacillus spp. isolated from compost. Soil Biol Biochem 38:2963–2966. doi: 10.1016/j.soilbio.2006.03.019. [DOI] [Google Scholar]
- 48.Strom PF. 1985. Effect of temperature on bacterial species-diversity in thermophilic solid-waste composting. Appl Environ Microbiol 50:899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phuong Hoa PT, Nonaka L, Hung Viet P, Suzuki S. 2008. Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci Total Environ 405:377–384. doi: 10.1016/j.scitotenv.2008.06.023. [DOI] [PubMed] [Google Scholar]
- 50.Wang N, Yang XH, Jiao SJ, Zhang J, Ye BP, Gao SX. 2014. Sulfonamide-resistant bacteria and their resistance genes in soils fertilized with manures from Jiangsu Province, Southeastern China. PLoS One 9:e112626. doi: 10.1371/journal.pone.0112626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao PP, Mao DQ, Luo Y, Wang LM, Xu BJ, Xu L. 2012. Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res 46:2355–2364. doi: 10.1016/j.watres.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 52.McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O'Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang WL, Yan M, Yu T, Wright GD. 2013. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother 57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu X, Kulasekara BR, Lory S. 2009. Role of horizontal gene transfer in the evolution of Pseudomonas aeruginosa virulence. Genome Dyn 6:126–139. doi: 10.1159/000235767. [DOI] [PubMed] [Google Scholar]
- 54.Liu JL, Wong MH. 2013. Pharmaceuticals and personal care products (PPCPs): a review on environmental contamination in China. Environ Int 59:208–224. doi: 10.1016/j.envint.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 55.Martinez JL. 2009. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 157:2893–2902. doi: 10.1016/j.envpol.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 56.Petridis M, Bagdasarian M, Waldor MK, Walker E. 2006. Horizontal transfer of shiga toxin and antibiotic resistance genes among Escherichia coli strains in house fly (Diptera: Muscidae) gut. J Med Entomol 43:288–295. doi: 10.1093/jmedent/43.2.288. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.