Abstract
It was shown recently that individual cells of an isogenic Saccharomyces cerevisiae population show variability in acetic acid tolerance, and this variability affects the quantitative manifestation of the trait at the population level. In the current study, we investigated whether cell-to-cell variability in acetic acid tolerance could be explained by the observed differences in the cytosolic pHs of individual cells immediately before exposure to the acid. Results obtained with cells of the strain CEN.PK113-7D in synthetic medium containing 96 mM acetic acid (pH 4.5) showed a direct correlation between the initial cytosolic pH and the cytosolic pH drop after exposure to the acid. Moreover, only cells with a low initial cytosolic pH, which experienced a less severe drop in cytosolic pH, were able to proliferate. A similar correlation between initial cytosolic pH and cytosolic pH drop was also observed in the more acid-tolerant strain MUCL 11987-9. Interestingly, a fraction of cells in the MUCL 11987-9 population showed initial cytosolic pH values below the minimal cytosolic pH detected in cells of the strain CEN.PK113-7D; consequently, these cells experienced less severe drops in cytosolic pH. Although this might explain in part the difference between the two strains with regard to the number of cells that resumed proliferation, it was observed that all cells from strain MUCL 11987-9 were able to proliferate, independently of their initial cytosolic pH. Therefore, other factors must also be involved in the greater ability of MUCL 11987-9 cells to endure strong drops in cytosolic pH.
INTRODUCTION
The study of microbial acetic acid tolerance is relevant in different fields of applied microbiology. Acetic acid, like other weak acids, such as sorbic acid and lactic acid, traditionally has been used as a preservative agent in food and beverages, where it prevents microbial spoilage by arresting the growth of yeasts and other fungi (1). However, certain strains of the species Zygosaccharomyces bailii and Saccharomyces cerevisiae still grow in the presence of relatively highly weak acid concentrations (2, 3), and, therefore, it is crucial to understand the underlying tolerance mechanisms in order to avoid food spoilage more effectively. More recently, understanding acetic acid tolerance of the platform yeast S. cerevisiae became important in the field of industrial biotechnology once hydrolysates of lignocellulosic biomass were considered renewable feedstock for microbial fermentations (4). Notably, the acetic acid concentrations in those hydrolysates can reach up to 133 mM (8 g liter−1) (5–7), at which the acid becomes a strong inhibitor of microbial growth and fermentation, especially at the low medium pH values typically used in industrial batch fermentations. Therefore, an understanding of the molecular mechanisms underlying S. cerevisiae tolerance to acetic acid is important for the generation of robust industrial strains that are able to ferment lignocellulosic hydrolysates efficiently.
The inhibitory effect of acetic acid is associated predominantly with its undissociated form, which can diffuse across the plasma membranes of cells mainly by simple diffusion (8). Once inside the cytoplasm, acetic acid (pKa = 4.76) dissociates into a proton and its counterion, resulting in a decrease in intracellular pH and an accumulation of acetate anions. The yeast S. cerevisiae has developed several mechanisms by which it can counteract the harmful effects that acetic acid exerts on the cells. In general, adaptation to acetic acid has been associated with the abilities to recover intracellular pH (3, 9–11), to inhibit further uptake of acetic acid (12), to activate multidrug transporters to pump out acetate anions (3, 13), and to adjust the membrane lipid profile (14). Among these mechanisms, recovery of intracellular pH is thought to be of predominant importance in the responses of S. cerevisiae to acetic acid (9). In fact, exposure of cells to acetic acid has been shown to increase the activities of plasma membrane and vacuolar H+-ATPases, which pump protons out of the cytosol (3, 11, 13, 15). Another indication for the importance of pH homeostasis in weak acid tolerance is given by two studies that investigated interspecies diversity with regard to short-term changes in intracellular pH upon exposure to weak acid. It has been suggested that the higher tolerance of the species Z. bailii and Candida krusei compared to that of S. cerevisiae is a consequence of their ability to preserve physiological pH better after shifting to acid-containing medium (16, 17).
Although S. cerevisiae has an innate tolerance to acetic acid, moderate to high concentrations have been shown to affect the cell's physiology negatively (18, 19). A frequently reported effect is significant prolongation of the latency phase in the presence of inhibitory acetic acid concentrations (20–23). This effect was demonstrated recently to be attributable to the fact that only a relatively small fraction of cells in the entire population are able to resume proliferation in the presence of acetic acid (20). The size of this fraction was shown to decrease with increasing acetic acid concentrations, in a strain-dependent manner. The occurrence of such a fraction was observed previously in populations of S. cerevisiae cells exposed to other weak acids (24).
The fact that single cells of a genetically uniform population show different levels of stress tolerance is not a novel observation (25–27). In fact, phenotypic cell-to-cell heterogeneity is assumed to be a strategy for microbial populations to survive unanticipated environmental changes. Although research regarding the molecular basis of cell-to-cell heterogeneity is still in its infancy, a few studies provide the first insights into the possible mechanisms involved (25, 27–29). With regard to weak acid tolerance, it has been suggested that cell-to-cell heterogeneity could be explained by the variations in cytosolic pH (pHc) among individual cells at the moment when the cells are exposed to the acid (24, 30). It is assumed that only cells with pHc values around neutrality start cell division in the presence of the acid (24). More recently, a study of Z. bailii proposed that tolerance to weak acids is due to a subpopulation of cells showing low pHc values of around 5.5 (30). As the uptake of weak acids appears to involve mainly a simple mechanism based on diffusion of the protonated form, lower pHc values would allow the outside/inside equilibrium to be reached at lower intracellular concentrations of acid, thus reducing the accumulation of anions and the amounts of protons that have to be removed in order to restore the pHc. However, no experimental evidence based on correlating pHc kinetics with the cell's ability to proliferate in the presence of acid has been provided so far.
In the current study, we recorded pHc changes of individual cells in S. cerevisiae populations during a shift from nonstress to acetic acid stress conditions, and we correlated both the initial pHc and the maximal pHc drop with the cell's ability to proliferate in the presence of the acid. Data are provided for two different S. cerevisiae strains which significantly differed in acetic acid tolerance.
MATERIALS AND METHODS
Medium composition.
S. cerevisiae cells were routinely maintained on solid yeast extract-peptone-dextrose (YPD) medium containing 10 g liter−1 yeast extract, 20 g liter−1 peptone, 20 g liter−1 glucose, and 15 g liter−1 agar. Throughout this study, all acetic acid tolerance assays were performed in synthetic medium containing 5 g liter−1 (NH4)2SO4, 3 g liter−1 KH2PO4, 0.5 g liter−1 MgSO4·7H2O, 20 g liter−1 glucose, and appropriate amounts of trace elements and vitamins, as described by Verduyn et al. (31). Acetic acid was added to the medium at the concentrations indicated in Results, and the pH was adjusted to 4.5 with 5 M KOH. In order to prepare solid medium, 20 g liter−1 agar was added.
Strain construction.
All S. cerevisiae strains used in this study are listed in Table 1. The URA3 gene was disrupted in strains CEN.PK113-7D and MUCL 11987-9 by using a loxP-KanMX-loxP cassette conferring resistance to the antibiotic G418 (32). The disruption cassette was obtained by PCR amplification of pUG6 template DNA using the primers ura3_KanMX_FW and ura3_KanMX_RV (Table 2) (32); each of the primers contained a sequence homologous to that of pUG6 at its 3′ end and a sequence homologous to the integration site at the chromosomal URA3 locus at its 5′ end. The cassette was subsequently purified from the PCR mixture using a PCR purification kit (Qiagen, The Netherlands) and was used for transformation of CEN.PK113-7D and MUCL 11987-9. Transformations were carried out according to the lithium acetate method described by Gietz et al. (33), and selection of transformants was performed on solid YPD medium containing 100 mg liter−1 G418. Correct disruption of the URA3 gene was verified by PCR using the primers KanMX_verif_FW and ura3_verif_RV (Table 2).
TABLE 1.
Strains used in this study
| Strain | Description | Reference |
|---|---|---|
| CEN.PK113-7D | Prototrophic S. cerevisiae laboratory strain | 43 |
| MUCL 11987-9 | Haploid segregant from diploid strain MUCL 11987 with high acetic acid tolerance (Belgian Coordinated Collections of Microorganisms) (20), showing acetic acid tolerance similar to that of its parent strain | This study |
| CEN.PK113-7D ura3Δ | ura3Δ::loxP-KanMX-loxP | This study |
| MUCL 11987-9 ura3Δ | ura3Δ::loxP-KanMX-loxP | This study |
| CEN.PK113-7D (pHluorin) | CEN.PK113-7D ura3Δ with plasmid pYES-PACT1-pHluorin | This study |
| MUCL 11987-9 (pHluorin) | MUCL 11987-9 ura3Δ with plasmid pYES-PACT1-pHluorin | This study |
TABLE 2.
Primers used in this study
| Name | Sequence (5′ to 3′) |
|---|---|
| ura3_KanMX_FW | ACAAAATCTTTGTCGCTCTTCGCAATGTCAACAGTACCCTTAGTAGCATAGGCCACTAGTGGATCTG |
| ura3_KanMX_RV | GTATACAGAATAGCAGAATGGGCAGACATTACGAATGCACACGGTCAGCTGAAGCTTCGTACGC |
| KanMX_verif_FW | TGCATGGTTACTCACCACTG |
| ura3_verif_RV | AACCTTCATCTCTTCCACCC |
Strains CEN.PK113-7D ura3Δ and MUCL 11987-9 ura3Δ were subsequently transformed with plasmid pYES-PACT1-pHluorin (URA3), which contains the pHluorin (a pH-sensitive variant of the green fluorescent protein) open reading frame, under the control of the constitutive S. cerevisiae ACT1 actin gene promoter, and the URA3 gene as a selectable marker (34, 35). The plasmid pYES-PACT1-pHluorin (URA3) was kindly provided by Gertien J. Smits (University of Amsterdam, The Netherlands). Selection of transformants was performed on solid synthetic medium, and expression of the pHluorin gene in the selected transformants was checked by fluorescence microscopy.
Acetic acid tolerance assay.
Cells from a single colony on a plate were used to inoculate 4 ml of synthetic medium in a glass tube, and the cells were subsequently cultivated overnight in an orbital shaker at 200 rpm and 30°C. An aliquot of the overnight culture was transferred to 4 ml of fresh synthetic medium to obtain an optical density at 600 nm (OD600) of 0.2. This culture was cultivated under the same conditions as for the overnight culture for approximately 6 to 7 h, until the mid-exponential phase was reached (OD600 between 1.0 and 1.5). An amount of cells from the latter culture required to obtain an OD600 of 0.2 in 5 ml was harvested by centrifugation at 800 × g for 7 min, and the cells were subsequently resuspended in 5 ml of synthetic medium without acetic acid and without glucose. This culture was further serially diluted in the same medium to obtain 10-fold dilution steps over the range of 10−1 to 10−4. An aliquot of 250 μl from each dilution was spread on solid synthetic medium either with or without acetic acid, and cells were subsequently incubated in a static incubator at 30°C. Cells spread on medium without acetic acid were incubated for 2 days, while cells spread on medium with acetic acid were incubated for 2 to 5 days according to the concentration used. Plates containing 50 to 150 CFU were used to determine the numbers of cells that were able to proliferate under each condition.
Single-cell static fluorescence microscopy.
Strains CEN.PK113-7D ura3Δ and MUCL 11987-9 ura3Δ containing the plasmid pYES-PACT1-pHluorin (URA3) were grown under the same conditions as in the acetic acid tolerance assay, with the exception that the cells harvested in mid-exponential phase were resuspended in liquid synthetic medium containing the acetic acid concentration indicated in Results (with adjustment of the OD600 to 0.2). One milliliter of the resulting culture was then transferred to a 35-mm imaging dish with a hydrophilic adhesive bottom (Ibidi, Germany). After the cells had settled (10 min), the fluorescence intensity of individual S. cerevisiae cells was recorded using a Keyence fluorescence microscope (BZ-9000 series Generation II [Biorevo]) with 410-nm and 480-nm filters. In particular, images were obtained at ×40 magnification after exposure times of 0.005 s in bright field and 0.25 s at excitation wavelengths of 410 nm and 480 nm. Under these conditions, the autofluorescence of strains CEN.PK113-7D and MUCL 11987-9 was negligible. The fluorescence images obtained at both excitation wavelengths were subsequently analyzed using ImageJ software (36) in order to quantify fluorescence intensities.
Microfluidics-based time-lapse fluorescence microscopy.
The strains containing plasmid pYES-PACT1-pHluorin (URA3) were cultivated and harvested under the same conditions as described for the acetic acid tolerance assay, with the exception that the mid-exponential-phase cells were resuspended in liquid synthetic medium without acetic acid (with adjustment of the OD600 to 0.2). A 300-μl aliquot of this culture was subsequently transferred to a CellASIC ONIX Y04C microfluidic yeast plate (height of 3.5 to 4.5 μm; Merck Millipore). The plate was then connected to the ONIX microfluidic platform (CellASIC EV262 system) and placed in an inverted fluorescence microscope (Nikon Eclipse TE2000-E) with 410-nm and 480-nm filters. Before the microfluidic chamber was perfused with the cell suspension, the capillaries and chamber were washed with fresh synthetic medium without acetic acid at 3 lb/in2 for 1 min. The cells were then injected into the chamber at 6 lb/in2 for 20 s. Once an appropriate number of cells were trapped in the chamber, untrapped cells were washed out at 5 lb/in2 for 5 min. The medium was shifted to synthetic medium containing acetic acid starting at 1 lb/in2 for 2 min (to replace the medium in the chamber quickly), and then the flow rate was adjusted to 37.5 μl h−1 until the end of the experiment. Changes in fluorescence intensities of individual cells under these conditions were recorded by taking images at ×40 magnification after excitation at 410 nm and 480 nm (exposure time of 90 ms), using MetaMorph (Molecular Devices) and ImageJ software. Images were acquired just before the shift (time zero) and at different time points after the shift (2, 5, 15, and 30 min and then every 30 min for a maximum of 12 h). Images acquired before the shift (time zero) were used to determine the initial budding status (budding versus nonbudding) of each individual cell. Additionally, bright-field images were taken at each time point in order to monitor cell proliferation.
pHluorin calibration of single cells.
The ratio between the fluorescence intensities recorded at 410 nm and 480 nm (referred to as the 410/480 ratio) has been shown to be correlated with the pHc within a defined range of values (34). In order to convert the 410/480 ratios into the corresponding pHc values, a calibration curve was generated as described by Orij et al. (35), with the exception that the pHc values in the current study were obtained by analyzing individual cells instead of population average values. Briefly, cells of strain CEN.PK113-7D ura3Δ containing plasmid pYES-PACT1-pHluorin (URA3) were grown to mid-exponential phase. A number of cells sufficient to yield an OD600 of 0.2 in 1 ml were collected by centrifugation at 800 × g for 5 min, and the cells were subsequently resuspended in 350 μl of phosphate-buffered saline (PBS) solution containing 100 μg ml−1 digitonin (AppliChem, Germany), which permeabilizes cell membranes without affecting mitochondrial function. After 10 min of incubation at room temperature, the cells were harvested by centrifugation at 800 × g for 5 min and resuspended in 1 ml of citrate/phosphate buffers with different pH values (ranging from 5.0 to 8.0). With permeabilization of the cells, the pHc was equilibrated to the extracellular pH, yielding cells with known pHc values. Fluorescence intensities of 22 individual cells per pH value tested were quantified from images taken by fluorescence microscopy, using both the Keyence and Nikon inverted fluorescence microscopes at the aforementioned excitation wavelengths. The intensities were then used to calculate the 410/480 ratios. Mean values and standard deviations were obtained from three biological replicates. Based on the two calibration curves obtained (see Fig. S1 in the supplemental material), the following equations were derived for calculation of pHc values from 410/480 ratios: pHc = −14,054(410/480 ratio)4 + 65,042(410/480 ratio)3 − 110,780(410/480 ratio)2 + 84,448(410/480 ratio) − 17,808 (R2 = 0.9918) for the Keyence fluorescence microscope and pHc = 3.2059(410/480 ratio)3 − 10.821(410/480 ratio)2 + 14.837(410/480 ratio) − 0.2759 (R2 = 0.9951) for the Nikon inverted fluorescence microscope. As pHc values lower than 5 have been shown to result in loss of fluorescence of the pHluorin protein (37) and pHc values higher than 8 are less accurate, as deduced from the calibration curves, only cells with pHc values between 5.0 and 8.0 were considered in the current study.
RESULTS
Cell-to-cell heterogeneity in acetic acid tolerance.
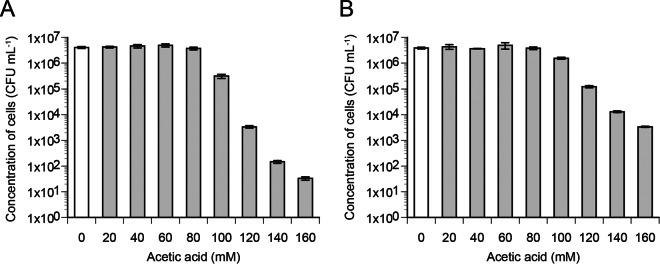
In a previous study, it was shown that only a fraction of cells in an S. cerevisiae population resumed proliferation upon exposure to acetic acid (20). This effect was observed in several strains that were exposed to a relatively high acetic acid concentration (157 mM acetic acid at pH 4.5). As a first step in the current study, the effects of different acetic acid concentrations on the size of the subpopulation of proliferating cells were analyzed. Cells of the strain CEN.PK113-7D were therefore spread on solid synthetic medium containing acetic acid at concentrations ranging from 20 to 160 mM. This quantification method was demonstrated previously to represent properly the fraction of cells able to resume proliferation in liquid acetic acid-containing medium (20). Data showed that all cells (4 × 106 CFU ml−1) were able to proliferate in the presence of the acid at concentrations of up to 80 mM (Fig. 1A); however, we confirmed the findings that only a very small fraction of cells from the entire population (0.001 to 10%) were able to proliferate at concentrations of ≥100 mM and the size of this fraction decreased with increasing acetic acid concentrations.
FIG 1.
Effects of increasing acetic acid concentrations on the fraction of proliferating cells. Cells cultivated in synthetic medium to the mid-exponential phase were dispersed on solid synthetic medium without or with increasing concentrations of acetic acid at pH 4.5. The concentration of cells able to resume proliferation under each condition was determined by counting CFU after incubation for 2 days for plates without acetic acid (white bars) or 2 to 5 days for plates with acetic acid (gray bars). Mean values and standard deviations for CEN.PK113-7D (A) and MUCL 11987-9 (B) were obtained from three biological replicates.
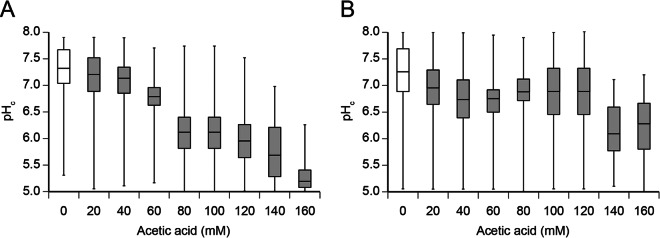
It is known that unstressed, exponentially growing, S. cerevisiae cells exhibit significant heterogeneity with regard to their pHc values (24). In addition, it is known that exposure of such cells to acetic acid results in significant immediate decreases in their pHc values (19). Here, we studied the kinetics of pHc changes at the single-cell level. For this purpose, a pH-sensitive variant of the green fluorescent protein (i.e., pHluorin) was used (34). First, the strain CEN.PK113-7D ura3Δ was transformed with plasmid pYES-PACT1-pHluorin (35), after which the effects of increasing acetic acid concentrations on pHc distribution were analyzed. Figure 2A shows the pHc values of individual cells before (no acetic acid) and 10 min after exposure of the cells to medium containing different acid concentrations. The results obtained with the nonstressed cells confirmed the previously documented cell-to-cell heterogeneity with regard to pHc values. As a control, cells from mid-exponential-phase cultures were not subjected to prior centrifugation but were used directly for cytosolic pH determinations. Cell-to-cell heterogeneity was also observed, thus excluding potential artifacts caused by the conditions used here to prepare the cells for measurements.
FIG 2.
Shifts in the pHc distribution upon exposure to different concentrations of acetic acid. Cells expressing pHluorin were cultivated in synthetic medium to the mid-exponential phase, after which they were transferred to identical media either without (white box) or with (gray boxes) different concentrations of acetic acid at pH 4.5. After 10 min, the pHc values of at least 182 individual cells for each condition were determined as described in Materials and Methods. The pHc distributions of CEN.PK113-7D (A) and MUCL 11987-9 (B) are represented by box-and-whisker plots. Boxes indicate median values and 25th and 75th percentiles; whiskers indicate where minimum and maximum values were measured.
The data presented in Fig. 2 also showed that the pHc distributions for cells cultivated in the presence of acetic acid concentrations of up to 40 mM were not markedly different from the pHc distributions for cells cultivated in the absence of acetic acid (unstressed cells). However, any further increase in the acetic acid concentration shifted the pHc median toward lower values (Fig. 2A).
S. cerevisiae cells with low initial pHc values show less severe drops in pHc when shifted to acetic acid stress.
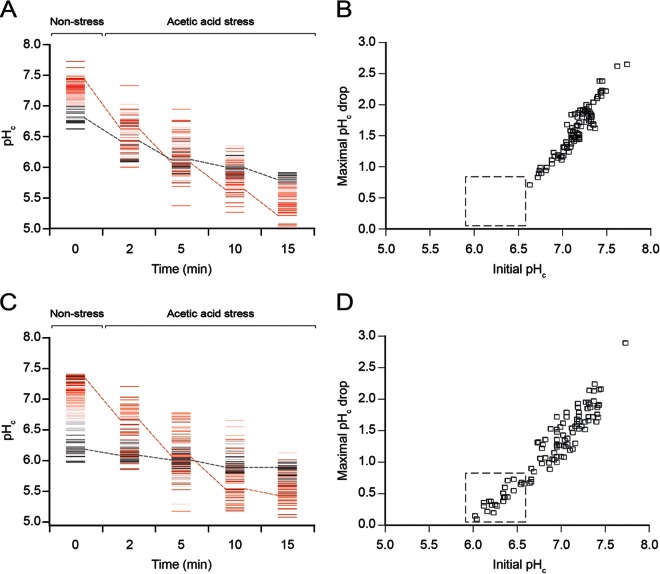
To investigate whether there was a correlation between the pHc values under nonstress conditions (referred to as initial pHc) and the magnitude of the pHc drop after the shift to acetic acid stress, cells of the strain CEN.PK113-7D were cultivated in synthetic medium without acetic acid to the mid-exponential phase and then were shifted to synthetic medium containing 120 mM acetic acid (pH 4.5). This concentration was chosen because it was the highest at which the pHc values of most cells after the shift were still in the measurable range for pHluorin. The pHc values of 125 individual cells were recorded just before and 2, 5, 10, and 15 min after the shift. Our data showed that the cells with the highest initial pHc values tended to have the lowest pHc values after the first 15 min in the presence of the acid (Fig. 3A). In contrast, cells with the lowest initial pHc values showed the highest pHc values after exposure to the acid. At time points later than 15 min, the distribution of the pHc values shifted back to higher values, which suggests that most cells showed their maximal pHc drop (calculated by subtracting the lowest detected pHc from the initial pHc) within the first 15 min (see Fig. S2 in the supplemental material).
FIG 3.
Effects of the pHc before the addition of acetic acid on the magnitude of the maximal pHc drop after exposure to the acid. The pHc values of 125 individual cells per strain were recorded during a shift from synthetic medium without acetic acid to synthetic medium with 120 mM acetic acid at pH 4.5. (A and C) In detail, data are shown for CEN.PK113-7D (A) and MUCL 11987-9 (C) cells just before the shift in medium (time zero) and at different time points during the first 15 min after the shift. The first time point at which pHc was measured under acetic acid stress was 2 min, as this was the time needed to replace the original medium completely with medium containing acetic acid. Data are shown only for cells that maintained fluorescence during the entire experiment and whose pHc values stayed within the range of 5 to 8 and therefore could be precisely determined (80.8% for each strain). Each line in panels A and C represents an individual cell, and colors were used to differentiate between cells with different pHc values under nonstress conditions (referred to as the initial pHc). The means of pHc values for the 10% of cells with the lowest and highest pHc values under nonstress conditions (time zero) were calculated over time and connected by black and red dashed lines, respectively. (B and D) For cells represented in panels A and C, the maximal drop in pHc within the first 15 min after exposure to the acid was calculated and plotted against the cell's initial pHc; data are shown for CEN.PK113-7D (B) and MUCL 11987-9 (D) cells. The dashed boxes denote the major differences between the two populations, as only the tolerant strain MUCL 11987-9 showed initial pHc values <6.6 and experienced minimal acid-induced pHc drops.
It has to be mentioned that the exact time points at which the maximal pHc drops were reached in individual cells were different. Taking this into account, we plotted the maximal pHc drop for each individual cell against its initial pHc, and we observed a strong correlation (r = 0.94) between the two parameters (Fig. 3B), which clearly showed that, the higher the pHc was before exposure to acetic acid, the larger the pHc drop was after exposure to the acid. It should be noted that, in contrast to Fig. 2, Fig. 3 shows only cells that kept their pHc values within the range of the calibration curve (between pH 5 and pH 8) for the entire experiment (representing 80.8% of the total number of cells). The fractions of cells that had to be omitted from the graphical representation in Fig. 3 due to pHc drops to values below 5 or a total loss of fluorescence were the same for the two strains.
The pHc under nonstress conditions determines the ability of an individual cell to resume proliferation in the presence of acetic acid.
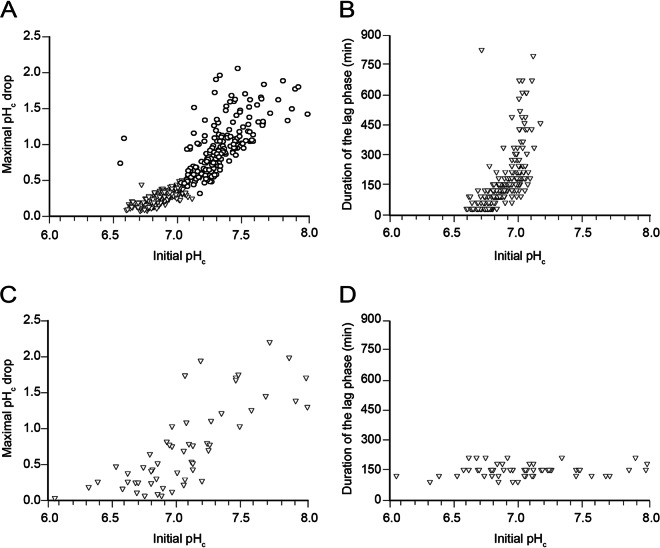
As described above, the pHc of an individual cell before exposure to acetic acid determined the magnitude of the pHc drop after exposure to the acid (Fig. 3A and B). As it is generally assumed that the ability to retain pHc is an important factor in acetic acid tolerance, we investigated whether the initial pHc of a cell (and thus the magnitude of the pHc drop) determined the cell's ability to resume proliferation. The changes in pHc and the ability to proliferate were monitored in individual CEN.PK113-7D cells for a long period after the shift from synthetic medium without acetic acid to acetic acid-containing medium, using a microfluidics device. This experiment was performed at a concentration of 96 mM acetic acid (pH 4.5), at which approximately 60% of the cells were able to proliferate within the time span of the experiment. In agreement with the data obtained at 120 mM (Fig. 3B), the initial pHc also was correlated with the maximal drop in pHc (Fig. 4A). This effect was observed for both subpopulations of proliferating and nonproliferating cells, with correlation coefficients of 0.84 and 0.53, respectively. In addition, the cells that were able to proliferate were the ones that had initial pHc values in the lower range and thus experienced the lowest pHc drop after exposure to the acid (Fig. 4A). In contrast, the cells that were not able to proliferate were the ones that had the highest initial pHc values and consequently experienced the most dramatic drop in pHc.
FIG 4.
Effects of initial pHc values for individual S. cerevisiae cells on the cells' ability to proliferate in the presence of acetic acid. CEN.PK113-7D (A and B) or MUCL 11987-9 (C and D) cells were cultivated in synthetic medium to the mid-exponential phase, after which the medium was replaced with identical medium containing 96 mM acetic acid at pH 4.5. The pHc values for 512 (CEN.PK113-7D) or 55 (MUCL 11987-9) individual cells were determined just before and at different time points after the shift in medium. For all cells, the time required to resume proliferation in the presence of the acid was determined by monitoring bud formation over a time period of 15 h. Cells that did not resume proliferation within this time period were considered to be nonproliferating cells. (A and C) For each cell in the proliferating (319 cells) (▽) and nonproliferating (193 cells) (○) subpopulations, the maximal pHc drop was calculated and plotted against the initial pHc. (B and D) For each proliferating cell, the time required to resume proliferation was plotted against the initial pHc. Data were collected from four (CEN.PK113-7D) or two (MUCL 11987-9) independent experiments.
The subpopulation of proliferating cells was also studied with regard to the time required to resume proliferation (referred to as the lag phase). Bright-field images were obtained before and at different time points up to 12 h after the shift to medium containing acetic acid. The images were used to determine the time required (lag phase) until bud formation (for initially nonbudded cells) or until an increase in bud size was observed (for cells already budding at the beginning of the experiment). For each cell, the duration of the lag phase was plotted against the initial pHc (Fig. 4B), which revealed a direct correlation between the two parameters (r = 0.66). This correlation suggests that, the higher the initial pHc is, the longer it takes for a cell to resume proliferation. A similar conclusion could be drawn when the duration of the lag phase was plotted against the maximal pHc drop after exposure to acetic acid (see Fig. S3 in the supplemental material).
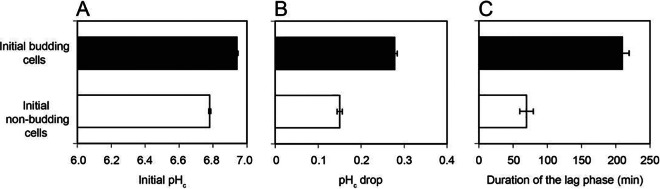
Our data collected from four biological replicates also showed that cells that were budding at time zero (before the addition of acetic acid) had significantly higher initial pHc values, experienced larger pHc drops, and consequently required longer periods to resume proliferation upon acetic acid stress than did cells that were in a nonbudding state immediately before the shift to the acid (Fig. 5).
FIG 5.
Correlations of budding status before the addition of acetic acid with the initial pHc (A), the severity of the pHc drop (B), and the duration of the lag phase (C) for individual CEN.PK113-7D cells after exposure to acetic acid. In total, 512 individual CEN.PK113-7D cells (four biological replicates) were monitored in time-lapse experiments during a shift to 96 mM acetic acid at pH 4.5, over a time period of 12 h. Of the total number of cells, 319 resumed proliferation after exposure to acetic acid; for those cells, the budding status, the initial pHc, the pHc drop (calculated as the difference between the initial pHc and the pHc after 2 min of exposure to acetic acid), and the time required to resume proliferation (individual lag phases) were determined. Individual lag phases were defined by monitoring bud formation (for initially nonbudding cells) or the increase in bud size (for cells budding at time zero). Data are shown as the means ± standard errors of the means for 187 budding and 132 nonbudding cells.
Cells of an acetic acid-tolerant S. cerevisiae strain can better endure strong drops in pHc.
In order to address whether S. cerevisiae strain-to-strain diversity with regard to acetic acid tolerance can be explained by differences in the distributions of initial pHc values, the strain MUCL 11987 was introduced in the current study. This strain was shown previously to have significantly greater acetic acid tolerance than strain CEN.PK113-7D (20). First, a haploid segregant of MUCL 11987 (referred to as MUCL 11987-9) with acetic acid tolerance similar to that of its parent was isolated. This segregant was then characterized in more detail with regard to the fraction of cells able to resume proliferation in the presence of different acetic acid concentrations. At up to 80 mM, all cells of MUCL 11987-9 were able to proliferate (Fig. 1B), similar to findings observed previously for CEN.PK113-7D under the same conditions (Fig. 1A). At concentrations of ≥100 mM, however, the fraction of proliferating cells was always higher for MUCL 11987-9 than for CEN.PK113-7D, thus confirming the acetic acid-tolerant phenotype of strain MUCL 11987-9. With regard to the pHc distribution, our data showed that this parameter was not significantly affected at acetic acid concentrations of up to 120 mM (Fig. 2B). In comparison, the pHc distribution of CEN.PK113-7D was already strongly shifted down at 80 mM (Fig. 2A).
In order to study the dependence of the pHc drop on the initial pHc, the pHc values of individual MUCL 11987-9 cells were monitored during a shift from medium without acetic acid to medium with acetic acid, using the same conditions as described above. Similar to findings observed for CEN.PK113-7D, cells of strain MUCL 11987-9 with relatively low pHc values in the unstressed population were better able to retain their initial pHc values upon exposure to 120 mM acetic acid, in contrast to cells with higher initial pHc values (Fig. 3C). Remarkably, the strain MUCL 11987-9 showed a fraction of cells with initial pHc values below ∼6.6 (Fig. 3D); such a fraction of cells was not observed in strain CEN.PK113-7D (Fig. 3C). Accordingly, this fraction experienced only a small drop in pHc (<0.7 units) upon exposure to the acid (Fig. 3D).
Strain MUCL 11987-9 was analyzed using the microfluidics device under exactly the same conditions as used for CEN.PK113-7D (96 mM acetic acid at pH 4.5). Under these conditions, all cells of strain MUCL 11987-9 were able to resume proliferation. Interestingly, all cells showed similarly short lag phases (90 to 210 min), even though the initial pHc values of the respective cells and the pHc drops differed remarkably (Fig. 4C and D). In fact, even cells suffering large pHc drops (∼2 units) were able to resume proliferation, which suggests that MUCL 11987-9 has a better capacity to recover from the pHc drop than does CEN.PK113-7D.
DISCUSSION
In this study, we showed that cell-to-cell heterogeneity in acetic acid tolerance could be at least partly explained by the variations in pHc values of individual cells at the moment immediately before acetic acid exposure. However, the acetic acid tolerance of an S. cerevisiae cell is also affected by genetic factors, as became obvious when S. cerevisiae strains differing in acetic acid tolerance were studied.
The first part of the current work focused on the cell-to-cell heterogeneity in acetic acid tolerance in the well-studied prototrophic laboratory strain CEN.PK113-7D. We clearly demonstrated that the initial pHc of individual S. cerevisiae cells determined the magnitude of the pHc drop after exposure to acetic acid. Indeed, cells with low initial pHc values (ranging from 6.6 to 7.1) experienced less severe drops in pHc than did cells with high initial pHc values (ranging from 7.1 to 7.9).
Based on our data, it is tempting to assume that the correlation between the initial pHc and the magnitude of the pHc drop can be explained simply by the fact that the equilibrium between undissociated acetic acid and internal acetate is dependent on the pHc. In fact, a lower initial pHc results in a smaller proportion of acetic acid that dissociates inside the cell, thereby avoiding excessive acid accumulation, as proposed previously by Stratford et al. (30). Those authors proposed a correlation between the pHc and the intracellular accumulation of acetic acid at the average population level for Z. bailii. However, the possibility that the difference in the pHc drops observed for cells with low versus high initial pHc values could also be attributed to other intrinsic physiological factors cannot be excluded, since it was shown previously that pHc acts as an intracellular signal in yeast (38). For instance, H+-ATPase pumps might be more active in cells with lower initial pH values, thus contributing to fast removal of newly produced cytosolic protons. In fact, it has been demonstrated that Pma1, the major yeast H+-ATPase, is activated when intracellular pH decreases (39). Further studies are necessary to evaluate these hypotheses.
Valli et al. (40) demonstrated that S. cerevisiae mutants with high intracellular pH are those that accumulate the most lactic acid (when expressing heterologous lactate dehydrogenase). Obviously, the high pH was used as an indicator for improved lactic acid tolerance. This result is not in contradiction to our findings. The context of the study by Valli et al. (40) is very different from that of the current one. In particular, it has to be considered that the cells were continuously producing lactic acid intracellularly and that mutations that led to improved tolerance were caused by an increased ability to maintain physiological pH even though lactic acid accumulated. In the current study, nonstressed cells were challenged by sudden exposure to extracellular acetic acid.
As only a fraction of CEN.PK113-7D cells (approximately 60%) were shown to contribute to growth of the culture in the presence of 96 mM acetic acid (pH 4.5), we also studied whether the initial pHc determined the cell's ability to resume proliferation. Indeed, only cells with low initial pHc values were able to recover pHc to neutral values and resume proliferation in the presence of the acid. Moreover, the time periods required for individual CEN.PK113-7D cells to resume proliferation (lag phases) varied among the cells and were correlated with both their initial pHc values and their budding status before the addition of acetic acid. The latest observation is congruent with the fact that nonbudding cells are considered more stress resistant (41).
In contrast to the proliferating CEN.PK113-7D cells, cells with high initial pHc values experienced severe drops in pHc and were unable to resume proliferation. Our previous study showed that these nonproliferating cells do not die immediately upon exposure to acetic acid and that they stay viable for a relatively long period of time (20). A possible explanation for the fact that cells with large drops in pHc cannot resume proliferation is that these cells are unable to restore their pHc to neutral values, which might be caused by a deficiency in ATP. In fact, ATP drives the activity of plasma membrane and vacuolar H+-ATPases, which pump protons out of the cytosol in order to restore the pHc after weak acid stress (24). However, a recent study based on measuring average intracellular ATP levels in S. cerevisiae populations suggests that ATP depletion alone is not the only cause of growth inhibition (determined by optical density measurements) upon acetic acid stress (42).
As our data obtained from strain CEN.PK113-7D showed that only cells with relatively low initial pHc values were able to resume proliferation, the question arose as to whether a population of cells from a strain with higher acetic acid tolerance would contain a larger fraction of cells with relatively low initial pHc values. The analysis of the highly tolerant strain MUCL 11987-9 showed that the distribution of initial pHc values was indeed broader than for CEN.PK113-7D. Most importantly, a fraction of cells exhibited lower initial pHc values than did the reference strain. These data might explain why a larger fraction of cells in the MUCL 11987-9 population experienced less dramatic pHc drops, which might facilitate their proliferation in the presence of acetic acid. All cells from MUCL 11987-9 were able to proliferate in the presence of 96 mM acetic acid (pH 4.5), and this was even independent of the initial pHc of the cells. Therefore, the larger fraction of cells with lower initial pHc values in strain MUCL 11987-9 cannot be the only parameter that explains the difference in the acetic acid tolerances of the two strains. Obviously, the cells of strain MUCL 11987-9 have a genetically determined greater capability to recover from severe pHc drops. Our results emphasize the relevance of studying weak acid tolerance at the single-cell level, as well as the population level, and can serve as a starting point for developing industrially useful strains that are tolerant to weak acids.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant PIM2010EEI-00610 (ERA-NET IB, MINECO, Spain), an Ajut 2014SGR-4 award, and an Institució Catalana de Recerca i Estudis Avançats Academia 2009 award to J.A. and the ERA-NET Scheme of the 6th EU Framework Program (INTACT; German Federal Ministry of Education and Research project 0315933 [to E.N.]). M.F.-N. received a personal stipend from Colciencias (Colombia).
We thank Johan Thevelein (KU Leuven, Belgium) and Jack Pronk (TU Delft, The Netherlands) for kindly providing us with the strains MUCL 11987 and CEN.PK113-7D, respectively. We are also grateful to Gertien J. Smits (University of Amsterdam, The Netherlands) for providing us with the plasmid pYES-PACT1-pHluorin (URA3). We thank Nuria Barba, Inka Schönebeck, Martina Carrillo, Solvejg Sevecke, and Meneka Ruvi Rupasinghe for technical support and Mathias Klein for fruitful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02313-15.
REFERENCES
- 1.Moon NJ. 1983. Inhibition of the growth of acid tolerant yeasts by acetate, lactate and propionate and their synergistic mixtures. J Appl Bacteriol 55:453–460. doi: 10.1111/j.1365-2672.1983.tb01685.x. [DOI] [Google Scholar]
- 2.Mira NP, Teixeira MC. 2013. Microbial mechanisms of tolerance to weak acid stress. Front Microbiol 4:416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piper P, Calderon CO, Hatzixanthis K, Mollapour M. 2001. Weak acid adaptation: the stress response that confers yeasts with resistance to organic acid food preservatives. Microbiology 147:2635–2642. doi: 10.1099/00221287-147-10-2635. [DOI] [PubMed] [Google Scholar]
- 4.Almario MP, Reyes LH, Kao KC. 2013. Evolutionary engineering of Saccharomyces cerevisiae for enhanced tolerance to hydrolysates of lignocellulosic biomass. Biotechnol Bioeng 110:2616–2623. doi: 10.1002/bit.24938. [DOI] [PubMed] [Google Scholar]
- 5.Chandel AK, Silverio da Silva S, Singh OV. 2011. Detoxification of lignocellulosic hydrolysates for improved bioethanol production, p 225–246. In Dos Santos Bernardes MA. (ed), Biofuel production: recent developments and prospects. InTech, Rijeka, Croatia: http://cdn.intechopen.com/pdfs-wm/20063.pdf. [Google Scholar]
- 6.Demeke MM, Dietz H, Li Y, Foulquie-Moreno MR, Mutturi S, Deprez S, Den AT, Bonini BM, Liden G, Dumortier F, Verplaetse A, Boles E, Thevelein JM. 2013. Development of a d-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol Biofuels 6:89. doi: 10.1186/1754-6834-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zha Y, Muilwijk B, Coulier L, Punt PJ. 2012. Inhibitory compounds in lignocellulosic biomass hydrolysates during hydrolysate fermentation processes. J Bioprocess Biotech 2:112. doi: 10.4172/2155-9821.1000112. [DOI] [Google Scholar]
- 8.Casal M, Cardoso H, Leao C. 1996. Mechanisms regulating the transport of acetic acid in Saccharomyces cerevisiae. Microbiology 142:1385–1390. [DOI] [PubMed] [Google Scholar]
- 9.Giannattasio S, Guaragnella N, Zdralevic M, Marra E. 2013. Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Front Microbiol 4:33. doi: 10.3389/fmicb.2013.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mira NP, Palma M, Guerreiro JF, Sa-Correia I. 2010. Genome-wide identification of Saccharomyces cerevisiae genes required for tolerance to acetic acid. Microb Cell Fact 9:79. doi: 10.1186/1475-2859-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bidani A, Heming TA. 1995. Kinetic analysis of cytosolic pH regulation in alveolar macrophages: V-ATPase-mediated responses to a weak acid. Am J Physiol 269:L20–L29. [DOI] [PubMed] [Google Scholar]
- 12.Mollapour M, Piper PW. 2007. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol 27:6446–6456. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mira NP, Teixeira MC, Sa-Correia I. 2010. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: a genome-wide view. OMICS 14:525–540. doi: 10.1089/omi.2010.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindberg L, Santos AX, Riezman H, Olsson L, Bettiga M. 2013. Lipidomic profiling of Saccharomyces cerevisiae and Zygosaccharomyces bailii reveals critical changes in lipid composition in response to acetic acid stress. PLoS One 8:e73936. doi: 10.1371/journal.pone.0073936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carmelo V, Santos H, Sa-Correia I. 1997. Effect of extracellular acidification on the activity of plasma membrane ATPase and on the cytosolic and vacuolar pH of Saccharomyces cerevisiae. Biochim Biophys Acta 1325:63–70. doi: 10.1016/S0005-2736(96)00245-3. [DOI] [PubMed] [Google Scholar]
- 16.Halm M, Hornbaek T, Arneborg N, Sefa-Dedeh S, Jespersen L. 2004. Lactic acid tolerance determined by measurement of intracellular pH of single cells of Candida krusei and Saccharomyces cerevisiae isolated from fermented maize dough. Int J Food Microbiol 94:97–103. doi: 10.1016/j.ijfoodmicro.2003.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Arneborg N, Jespersen L, Jakobsen M. 2000. Individual cells of Saccharomyces cerevisiae and Zygosaccharomyces bailii exhibit different short-term intracellular pH responses to acetic acid. Arch Microbiol 174:125–128. doi: 10.1007/s002030000185. [DOI] [PubMed] [Google Scholar]
- 18.Ullah A, Orij R, Brul S, Smits GJ. 2012. Quantitative analysis of the modes of growth inhibition by weak organic acids in Saccharomyces cerevisiae. Appl Environ Microbiol 78:8377–8387. doi: 10.1128/AEM.02126-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guldfeldt LU, Arneborg N. 1998. Measurement of the effects of acetic acid and extracellular pH on intracellular pH of nonfermenting, individual Saccharomyces cerevisiae cells by fluorescence microscopy. Appl Environ Microbiol 64:530–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swinnen S, Fernandez-Nino M, Gonzalez-Ramos D, van Maris AJ, Nevoigt E. 2014. The fraction of cells that resume growth after acetic acid addition is a strain-dependent parameter of acetic acid tolerance in Saccharomyces cerevisiae. FEMS Yeast Res 14:642–653. doi: 10.1111/1567-1364.12151. [DOI] [PubMed] [Google Scholar]
- 21.Lambert RJ, Stratford M. 1999. Weak-acid preservatives: modelling microbial inhibition and response. J Appl Microbiol 86:157–164. doi: 10.1046/j.1365-2672.1999.00646.x. [DOI] [PubMed] [Google Scholar]
- 22.Pampulha ME, Loureiro-Dias MC. 2000. Energetics of the effect of acetic acid on growth of Saccharomyces cerevisiae. FEMS Microbiol Lett 184:69–72. doi: 10.1111/j.1574-6968.2000.tb08992.x. [DOI] [PubMed] [Google Scholar]
- 23.Narendranath NV, Thomas KC, Ingledew WM. 2001. Effects of acetic acid and lactic acid on the growth of Saccharomyces cerevisiae in a minimal medium. J Ind Microbiol Biotechnol 26:171–177. doi: 10.1038/sj.jim.7000090. [DOI] [PubMed] [Google Scholar]
- 24.Viegas CA, Almeida PF, Cavaco M, Sa-Correia I. 1998. The H+-ATPase in the plasma membrane of Saccharomyces cerevisiae is activated during growth latency in octanoic acid-supplemented medium accompanying the decrease in intracellular pH and cell viability. Appl Environ Microbiol 64:779–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy SF, Ziv N, Siegal ML. 2012. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol 10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaern M, Elston TC, Blake WJ, Collins JJ. 2005. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet 6:451–464. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 27.Stratford M, Steels H, Nebe-von-Caron G, Avery SV, Novodvorska M, Archer DB. 2014. Population heterogeneity and dynamics in starter culture and lag phase adaptation of the spoilage yeast Zygosaccharomyces bailii to weak acid preservatives. Int J Food Microbiol 181:40–47. doi: 10.1016/j.ijfoodmicro.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avery SV. 2006. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 4:577–587. doi: 10.1038/nrmicro1460. [DOI] [PubMed] [Google Scholar]
- 29.Holland SL, Reader T, Dyer PS, Avery SV. 2014. Phenotypic heterogeneity is a selected trait in natural yeast populations subject to environmental stress. Environ Microbiol 16:1729–1740. doi: 10.1111/1462-2920.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stratford M, Steels H, Nebe-von-Caron G, Novodvorska M, Hayer K, Archer DB. 2013. Extreme resistance to weak-acid preservatives in the spoilage yeast Zygosaccharomyces bailii. Int J Food Microbiol 166:126–134. doi: 10.1016/j.ijfoodmicro.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verduyn C, Postma E, Scheffers WA, Van Dijken JP. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 32.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gietz RD, Schiestl RH, Willems AR, Woods RA. 1995. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 34.Miesenbock G, De Angelis DA, Rothman JE. 1998. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394:192–195. doi: 10.1038/28190. [DOI] [PubMed] [Google Scholar]
- 35.Orij R, Postmus J, Ter Beek A, Brul S, Smits GJ. 2009. In vivo measurement of cytosolic and mitochondrial pH using a pH-sensitive GFP derivative in Saccharomyces cerevisiae reveals a relation between intracellular pH and growth. Microbiology 155:268–278. doi: 10.1099/mic.0.022038-0. [DOI] [PubMed] [Google Scholar]
- 36.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zdraljevic S, Wagner D, Cheng K, Ruohonen L, Jantti J, Penttila M, Resnekov O, Pesce CG. 2013. Single-cell measurements of enzyme levels as a predictive tool for cellular fates during organic acid production. Appl Environ Microbiol 79:7569–7582. doi: 10.1128/AEM.01749-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Orij R, Brul S, Smits GJ. 2011. Intracellular pH is a tightly controlled signal in yeast. Biochim Biophys Acta 1810:933–944. doi: 10.1016/j.bbagen.2011.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Cyert MS, Philpott CC. 2013. Regulation of cation balance in Saccharomyces cerevisiae. Genetics 193:677–713. doi: 10.1534/genetics.112.147207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valli M, Sauer M, Branduardi P, Borth N, Porro D, Mattanovich D. 2006. Improvement of lactic acid production in Saccharomyces cerevisiae by cell sorting for high intracellular pH. Appl Environ Microbiol 72:5492–5499. doi: 10.1128/AEM.00683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plesset J, Ludwig JR, Cox BS, McLaughlin CS. 1987. Effect of cell cycle position on thermotolerance in Saccharomyces cerevisiae. J Bacteriol 169:779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ullah A, Chandrasekaran G, Brul S, Smits GJ. 2013. Yeast adaptation to weak acids prevents futile energy expenditure. Front Microbiol 4:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin MLF, Heijnen JJ, Hoare M, Lange HC, Madden EA, Niederberger P, Nielsen J, Parrou JL, Petit T, Porro D, Reuss M, van Riel N, Rizzi M, Steensma HY, Verrips CT, Vindelov J, Pronk JT. 2000. An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Technol 26:706–714. doi: 10.1016/S0141-0229(00)00162-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.