Abstract
Animal manures and municipal biosolids recycled onto crop production land carry antibiotic-resistant bacteria that can influence the antibiotic resistome of agricultural soils, but little is known about the contribution of bacteriophage to the dissemination of antibiotic resistance genes (ARGs) in this context. In this work, we quantified a set of ARGs in the bacterial and bacteriophage fractions of agricultural soil by quantitative PCR. All tested ARGs were present in both the bacterial and phage fractions. We demonstrate that fertilization of soil with dairy manure or human biosolids increases ARG abundance in the bacterial fraction but not the bacteriophage fraction and further show that pretreatment of dairy manure can impact ARG abundance in the bacterial fraction. Finally, we show that purified bacteriophage can confer increased antibiotic resistance to soil bacteria when combined with selective pressure. The results indicate that soilborne bacteriophage represents a substantial reservoir of antibiotic resistance and that bacteriophage could play a significant role in the horizontal transfer of resistance genes in the context of an agricultural soil microbiome. Overall, our work reinforces the advisability of composting or digesting fecal material prior to field application and suggests that application of some antibiotics at subclinical concentrations can promote bacteriophage-mediated horizontal transfer of ARGs in agricultural soil microbiomes.
INTRODUCTION
Overuse of antibiotics has contributed to the spread of antibiotic resistance due to the release of antibiotics, antibiotic-resistant bacteria, and antibiotic resistance genes (ARGs) into the environment (1). This phenomenon is mediated by horizontal gene transfer (HGT) of mobile genetic elements—such as plasmids, transposons, and integrons—between bacterial cells through conjugation and viral transduction (2–4).
Transduction has been shown to occur in environmental matrices, including freshwater and wastewater; moreover, bacterial 16S ribosomal RNA sequences have been observed in the viral fraction of wastewater, confirming the ability of bacteriophage to carry bacterial genes (5, 6). In fact, while much attention has been paid to conjugation, more recent work has additionally implicated bacteriophage as a major vehicle for horizontal gene transfer and recombination (7–10).
Using high-throughput sequencing of murine fecal phage populations, Modi et al. (11) demonstrated that antibiotic treatment leads to the enrichment of genes conferring resistance to the administered drug as well as to unrelated antibiotics in the phage genome. Furthermore, bacteriophage from drug-treated mice provided cultured naive microbiota with increased levels of resistance to the corresponding drug. Overall, this work concluded that antibiotic residues potentiate the transduction-mediated dissemination of ARGs.
Antibiotic resistance genes have been found in the bacteriophage DNA fraction of various environmental matrices, such as activated sludge (12), urban sewage and river water (13), and wastewater effluents from hospitals and wastewater treatment plants (14). The aforementioned studies indicate that bacteriophage represents a reservoir of ARGs across a broad selection of environments. However, it remains to be determined if bacteriophage has such a role in agricultural soils.
In the present study, a set of ARGs previously detected in manured soils was quantified in bacteria and in bacteriophage recovered from agricultural soil using quantitative PCR (qPCR) (15–17). The impact of soil amendment either with dairy manure or with biosolids on the abundance and distribution of ARGs in bacteria and bacteriophage was determined. In the case of dairy manure, the effect of various preapplication treatments on the composition of ARGs in the bacterial and bacteriophage fractions was determined. Finally, we tested whether bacteriophage enriched from biosolids conferred increased antibiotic resistance to soilborne bacteria when combined with selective pressure.
MATERIALS AND METHODS
Field operations and soil sampling.
The soil samples used in the experiments described here were obtained from field experiments undertaken during the 2014 growing season on the Agriculture and Agri-Food Canada research farm in London, Ontario, Canada (42.984°N, 81.248°W). The field installations and methods were described in detail by Marti et al. (17). Briefly, the soil is a silt loam (gray-brown luvisol) with the following properties: a pH of 7.5; a cation exchange capacity of 13.2; a sand, silt, and clay composition of 18%, 67%, and 15%, respectively; and an organic matter content of 3.4%. Manure for application in the spring of 2014 was obtained from dairy farms in the London area, and biosolids that were aerobically digested and dewatered were obtained from the municipality of Tilsonburg, Ontario, Canada. The dairy manures were variously untreated (raw), anaerobically digested, mechanically dewatered, or composted. The dewatered and composted manures were applied at a rate of 5 dry metric tons/ha, and the raw and the digested manures were applied at a rate of 80,000 liters/ha.
Soil cores (2 cm wide, 15 cm deep) were taken immediately after manure or biosolid application, as well as at 7 and 30 days postapplication (days 0, 7, and 30, respectively), in order to assess any temporal impact on ARG composition. Six cores were sampled from each of three replicated plots using a T sampler rinsed with 70% ethanol between samplings. Cores were bulked into a labeled Ziploc bag, mixed by hand until they were homogeneous, and transported to the laboratory in a cooler with cool packs. Thus, three independent soil samples (each representing the average for 6 cores from a given plot) from control, dairy manure, and biosolids treatments were analyzed at each sampling time.
Extraction of bacterial and phage DNA fractions from soil.
Twenty-five-gram portions of soil were mixed with 25 ml of sterile SM buffer (100 mM NaCl, 8 mM MgSO4 · 7H2O, 50 mM Tris-HCl, pH 7.5) in a 50-ml Falcon tube and thoroughly mixed by vortexing for 1 h. The resulting slurry was centrifuged at 2,500 × g for 4 min to pellet the soil, and the supernatant was passed through a 0.22-μm-pore-size Durapore membrane filter (Millipore) under vacuum. The filter was rinsed with 10 ml of SM buffer. Bacteria were retained on the filter, whereas bacteriophage passed through and was in the filtrate.
The membrane was transferred to a 15-ml Falcon tube and incubated with 1 ml GITC (5 M guanidine isothiocyanate, 100 mM EDTA, 0.5% N-lauroylsarcosine) buffer for 1 h at 75°C. DNA was extracted using a modification of the Qiagen DNeasy blood and tissue kit procedure. Briefly, 0.5 ml of buffer AL was added to each tube before vortexing for 1 min, adding 0.5 ml 95% ethanol, vortexing again, and loading the contents of the tubes onto the provided spin columns in 700-μl increments. The columns were then washed once with 500 μl of buffer AW1 and twice with 700 μl of buffer AW2. The columns were centrifuged for 2 min at maximum speed to remove residual wash buffer, before they were eluted twice with 50 μl buffer AE. The resulting 100 μl of DNA, now enriched for bacterial DNA, was further purified and concentrated by ethanol precipitation, using a 1:10 volume of 3 M sodium acetate, 3 volumes of 95% ethanol, and RNA-grade glycogen (Life Technologies) as a coprecipitate. The pellet was resuspended in 12 μl of 1× TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA), quantified with a NanoDrop ND1000 microspectrophotometer (NanoDrop Technologies, Wilmington, DE), and diluted to a final concentration of 1 ng/μl using deionized water. The DNA extract was aliquoted and stored at −20°C.
The soil filtrate was filtered once more using a 0.22-μm-pore-size Sterivex syringe filter. The phage in the filtrate was concentrated using Amicon Ultra-15 filtration devices (30-kDa-molecular-mass cutoff) per the manufacturer's instructions. The resulting 400 to 600 μl was further enriched for bacteriophage by ultracentrifugation on CsCl density gradients of 1.7, 1.5, and 1.35 g/ml as described in reference 18. The resulting 1 ml of fluid containing enriched phage was desalted using Amicon Ultra-0.5 filtration devices (30-kDa-molecular-mass cutoff) per the manufacturer's instructions. The 30 μl of concentrate was adjusted to 1 ml with sterile SM buffer and stored at 4°C. To ensure the removal of nonphage DNA prior to lysis of the phage particles, 0.5 ml of enriched bacteriophage was treated with 10 U of Turbo DNase (Life Technologies) for 1 h at 37°C and then for 5 min at 65°C. To isolate bacteriophage DNA, 0.4 mg of proteinase K (final concentration, 0.7 mg/ml) was incubated with the DNase-treated bacteriophage preparation for 1 h at 37°C to digest the capsid proteins. The resulting lysate was extracted twice with phenol-chloroform-isoamyl alcohol, and the DNA was further purified and concentrated by ethanol precipitation as described above.
Each time that the above-described procedures were carried out, a series of conventional PCRs was performed in order to assess the quality of bacteriophage isolation. The presence of bacterial rrnS DNA (encoding the 16S rRNA) was assessed using the primers described in Table 1. PCRs were carried out with the following cycle conditions: 1 cycle at 95°C for 10 min followed by 30 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 1 min. GoTaq Flexi reagents (Promega) used in the reaction mixtures, and the reaction mixtures were composed of 1× Green GoTaq Flexi buffer; 1.5 mM MgCl2; 0.2 mM (each) dATP, dGTP, dCTP, and dTTP; 0.4 μM each primer; 1 U of GoTaq DNA polymerase; and deionized water to 25 μl. The reactions were carried out at the following stages of the extraction procedure: for bacterial DNA, after the full procedure, including ethanol precipitation; for bacteriophage particles, after Turbo DNase treatment but before proteinase K lysis; and for bacteriophage DNA, after the full procedure, including ethanol precipitation. The ideal result was a positive signal from the bacterial DNA preparations and a negative result from bacteriophage particle preparations, indicating the absence of bacterial DNA outside the bacteriophage particles. Thus, any bacterial DNA detected in the enriched bacteriophage DNA fraction must be due to a legitimate uptake of that DNA into the phage particle during the viral replication cycle. PCR products were analyzed by agarose gel electrophoresis; a representative gel is shown in Fig. S1 in the supplemental material. In all cases, no bacterial DNA (above the levels seen for a no-template negative control) was detected for either intact phage particles or purified phage DNA. To ensure that the lack of amplification from bacteriophage particles or bacteriophage DNA was not simply due to the presence of PCR inhibitors, these reactions were also performed in the presence of the same DNA used for the positive control, with positive results being obtained (see Fig. S1 in the supplemental material).
TABLE 1.
Primers and probes used for conventional or quantitative PCR in this study
| Primer specificity and primer namea | Sequence (5′ to 3′)b | Product size (bp) | Annealing temp (°C) | Final primer concn (nM) | Target | Reference |
|---|---|---|---|---|---|---|
| Universal bacteria (conventional PCR) | ||||||
| GM5-F | CCTACGGGAGGCAGCAG | 550 | 55 | 400 | rrnS gene | 19 |
| 907-R | CCGTCAATTCCTTTGAGTTT | 400 | ||||
| Universal bacteria | ||||||
| BACT1369F | CGGTGAATACGTTCYCGG | 123 | 59 | 300 | rrnS gene | 20 |
| PROK1492R | GGWTACCTTGTTACGACTT | 300 | ||||
| TM1389F | HEX-CTTGTACACACCGCCCGTC-BHQI | 300 | ||||
| strA | ||||||
| strA-F | TATGGTTGTTTGCCATGGTG | 149 | 62 | 400 | Streptomycin | 15 |
| strA-R | TTCTCTTCGGCGTTAGCAAT | 400 | Phosphotransferase A | |||
| strB | ||||||
| strB-F | ATCGCTTTGCAGCTTTGTTT | 143 | 61 | 300 | Streptomycin | 21 |
| strB-R | ATGATGCAGATCGCCATGTA | 300 | Phosphotransferase B | |||
| strB-P | HEX-ATGCCTCGGAACTGCGT-BHQI | 200 | ||||
| sul1 | ||||||
| sul1-F | GACTGCAGGCTGGTGGTTAT | 105 | 64 | 200 | Sulfamethazine resistance gene 1 | 16 |
| sul1-R | GAAGAACCGCACAATCTCGT | 200 | ||||
| aadA | ||||||
| aadA-F | CAGCGCAATGACATTCTTGC | 293 | 63 | 200 | Aminoglycoside adenyltransferase A | 21 |
| aadA-R | GTCGGCAGCGACA(C/T)CCTTCG | 200 | ||||
| aadA-P | HEX-TGGTAGGTCCAGCGGCGGAG-BHQI | 300 | ||||
| blaOXA-20 | ||||||
| blaOXA-20-F | TGATGATTGTCGAAGCCAAA | 100 | 60 | 400 | Beta-lactamase, class D (oxacillinase) | 22 |
| blaOXA-20-R | GCCTGTAGGCCACTCTACCC | 400 |
F, forward primer; R, reverse primer; P, probe.
HEX, 2′,4′,5′,7′-tetrachloro-6-carboxy-4,7-dichlorofluorescein succinimidyl ester; BHQI, black hole quencher 1.
Quantification of gene target copies.
The primers and probes (Sigma-Aldrich, Toronto, ON, Canada) used in the present study are summarized in Table 1. The resistance genes quantified were strA, strB, sul1, aadA, and blaOXA-20, which encode resistance to streptomycin (strA and strB), sulfamethazine (sul1), aminoglycosides (aadA), and β-lactams (blaOXA-20). Note that the amount of blaOXA-20 in dairy manure-amended soils was consistently below the limit of detection, and therefore, blaOXA-20 is not discussed. As a control for total bacterial DNA abundance among the various samples, rrnS (encoding the 16S rRNA) was also quantified. PCR amplification was performed using a Bio-Rad CFX96 real-time PCR instrument with Bio-Rad CFX Manager software, version 3.0. The reactions were performed with the Brilliant II quantitative PCR (qPCR) master mix (Agilent, Toronto, ON, Canada) for the TaqMan PCR and the Brilliant II SYBR green qPCR master mix (Agilent) for the SYBR green PCR. Two microliters of template DNA (corresponding to 2 ng of either bacteriophage or bacterial DNA) was added, and deionized water was used to reach a final volume of 25 μl. Negative controls without template DNA were run in triplicate. Each reaction was run in triplicate with the following cycle conditions: 1 cycle at 95°C for 10 min followed by 40 cycles of 95°C for 15 s, the annealing temperature indicated in Table 1 for 35 s, and 72°C for 1 min. For the SYBR green assay, a melting curve step was added in order to check the purity of the PCR product. This step consisted of a ramp of the temperature from 65 to 95°C at an increment of 0.5°C and a hold for 5 s for each step. The plasmids used to generate standard curves were created as described by Marti et al. (16). Standard curves were prepared with a 10-fold serial dilution of the known concentration of the plasmid solution for each marker, in order to have final concentrations of the plasmid ranging from 107 to 10° copies per microliter. The identities of the quantified gene targets were ensured on the basis of hybridization when using TaqMan chemistry or melting behavior when using SYBR green. Gene abundance data are presented as the number of gene copies (GC) per nanogram of bacterial or phage DNA included in the reaction mixture.
Transduction of soil bacteria by biosolids-derived phage.
Total bacteriophage was extracted and enriched from biosolids (three independent replicates) essentially as described above for soil, except that the initial centrifugation was increased to 15 min at 10,000 × g and the filtration steps were repeated two to three times to ensure the adequate removal of bacteria. Phage was stored at 4°C in SM buffer. In order to ensure that the aforementioned phage purification procedure, while stringent, yielded an adequate number of infection-competent phage particles, the titer of coliphage within the resulting bacteriophage enrichments was estimated using the single-agar-layer method essentially as described in reference 23. Escherichia coli K-12 (grown in tryptone-yeast extract-glucose broth) was used as the host, and tryptone-yeast extract-glucose agar was the medium. Control plates that received the enriched bacteriophage preparation but no strain K-12 bacteria had no visible colonies, corroborating the conclusion from the rrnS-specific PCR that the phage preparations were free of bacterial contamination. The average titer of coliphage was determined to be about 300 PFU/ml in our enrichments. For comparison, Colomer-Lluch et al. (13) reported coliphage titers of about 104 PFU/ml and 102 PFU/ml in urban sewage and river water, respectively; Ashelford et al. (24) reported about 107 phage per gram of soil associated with plant roots in sugar beet fields. While our coliphage titer was low by comparison to the titers in the aforementioned studies, it should be noted that these were assessments of the bacteriophage titers in environmental matrices that had not undergone phage purification procedures and that the titer of K-12-infecting coliphage reported here is likely a vast underestimate of the total number of infection-competent bacteriophage recovered.
We chose to assess the impact of biosolids-derived phage on soilborne microbiota without the complication of soil particles, which might adsorb to antibiotics or bacteriophage and thus interfere with the effective free concentration of drugs or phage particles. Accordingly, total bacteria were extracted from soil in an aqueous context as follows: 20 g of soil was mixed with 20 ml of SM buffer in a 50-ml Falcon tube by vortexing for 1 h. The resulting slurry was centrifuged at 2,000 × g for 2 min to remove most soil and large particulates, and the supernatant was removed and placed into two 15-ml Falcon tubes (5 ml per tube). One tube served as a no-phage control and received 500 μl of SM buffer. The other tube was amended with 500 μl of biosolids-derived phage; this was performed for three independent biological isolates of phage and soil-derived supernatant. After 1 h of incubation at 30°C, ampicillin, cefoxitin, or sulfamethazine was added to various final concentrations (see Table 2). The resulting transduction reaction mixtures were incubated for 3 days at 30°C, serially diluted, and plated on either selective or control Chromocult agar. Thus, we chose to focus on resistant versus total coliform bacteria as a subset of the total population of potential bacterial transductants generated in this assay. Control plates lacked antibiotics (in order to count the total number of coliforms), while selective plates contained ampicillin (32 μg/ml), cefoxitin (32 μg/ml), or sulfamethazine (512 μg/ml) in order to count the number of resistant coliform bacteria. Drug concentrations were based on breakpoints (i.e., MICs) reported by the Canadian Integrated Program for Antimicrobial Resistance Surveillance (25). The frequency of resistant coliforms was calculated by dividing the resistant coliform count (in numbers of CFU per milliliter) by the total coliform count (also in numbers of CFU per milliliter) counted in the absence of antibiotics. Phage-only controls were also included and yielded no bacterial growth. In all cases, the Chromocult plates were incubated at 37°C overnight. Note that these 3 antibiotics were chosen on the basis of an initial qualitative screen of 10 antibiotics (see Fig. S2 in the supplemental material). These 10 antibiotics were chosen to represent several major classes (e.g., penicillins, aminoglycosides, macrolides, cephalosporins, carbapenems, quinolones, sulfonamides).
TABLE 2.
Abundance of ampicillin-, cefoxitin-, and sulfamethazine-resistant coliforms recovered from soil suspensions following 3 days of incubation in presence or absence of bacteriophage enriched from biosolidsa
| Drug concnb during incubation | Bacteriophage supplementation | Drug-resistant coliform count/total coliform count |
||
|---|---|---|---|---|
| Ampicillin | Cefoxitin | Sulfamethazine | ||
| 0 | − | 0.000337 ± 0.000052 | 0.00724 ± 0.00917 | 0.245 ± 0.145 |
| + | 0.00101 ± 0.00099 | 0.00335 ± 0.00273 | 0.129 ± 0.037 | |
| 1:100 | − | 0.00143 ± 0.00142 | 0.00315 ± 0.00044 | 0.00326 ± 0.00291 |
| + | 0.00252 ± 0.01480 | 0.00168 ± 0.00067* | 0.0207 ± 0.0132* | |
| 1:10 | − | 0.411 ± 0.323 | 0.0512 ± 0.0239 | 0.0342 ± 0.0459 |
| + | 0.116 ± 0.031 | 0.192 ± 0.060* | 0.243 ± 0.098* | |
Suspensions were incubated without the addition of antibiotics or with one of the antibiotics at either a 1:100 or a 1:10 clinical breakpoint concentration. Data represent the mean ± standard deviation (n = 3) of the count of resistant coliforms (in numbers of CFU per milliliter) normalized to the total coliform count (in numbers of CFU per milliliter). *, statistically significant differences between phage treatment and no phage treatment.
1:100 and 1:10, final concentrations 100-fold and 10-fold, respectively, below the breakpoint concentrations of the indicated antibiotics (ampicillin, 32 μg/ml; cefoxitin, 32 μg/ml; sulfamethazine, 512 μg/ml).
Calculations and statistics.
Each condition was analyzed in triplicate. In the qPCR experiments, three technical replicates were performed for each of three independent biological isolates for each sample. In the transduction experiment, we report the averages from three independent biological replicates. Statistically significant treatment effects were determined using an unpaired t test without assuming equal variance. Data were analyzed using SigmaPlot software, version 12.5 (Systat Software Inc.). The significance level was set at a P value of 0.05.
RESULTS
Impact of manure application on abundance and distribution of antibiotic resistance genes in bacteriophage and bacterial fractions.
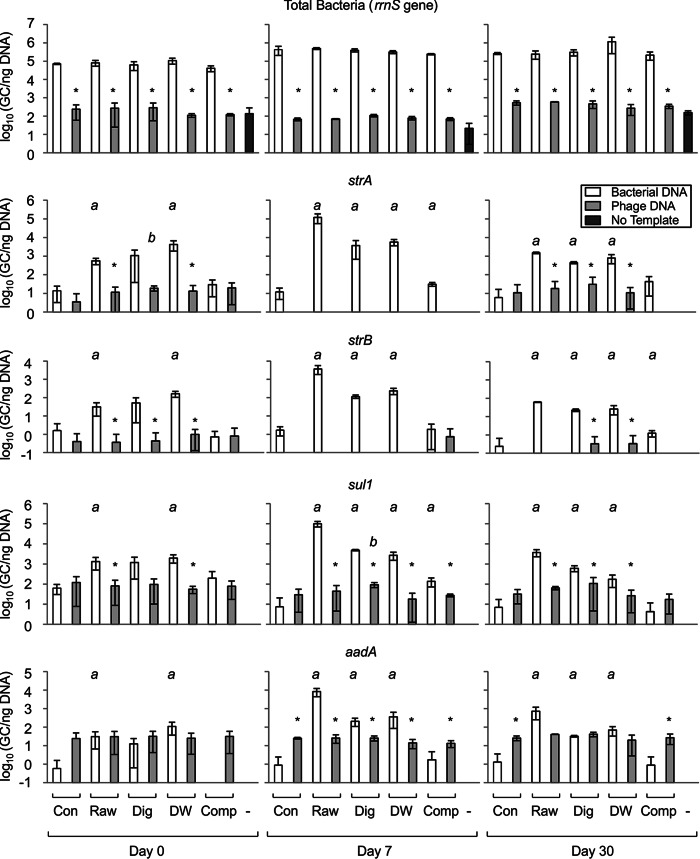
The abundance of rrnS in the bacterial fraction did not vary with manure application (Fig. 1, top). Soil carried about 105 gene copies (GC) per ng of template DNA at day 0 and about 106 GC/ng DNA 7 and 30 days following manure application. The increase in rrnS copy numbers at days 7 and 30 relative to the numbers at day 0 was statistically significant for most of the corresponding treatments, namely, the control, raw, digested, dewatered, and composted treatments for day 0 versus day 7 and the control, digested, and composted treatments for day 0 versus day 30. For simplicity, the results of these statistical analyses are not indicated in Fig. 1. Overall, there was a time-dependent increase in bacterial abundance following manure application. The absence of any effect of manure preapplication treatment on rrnS abundance indicates that the overall abundance of bacteria was unaffected by these treatments. Thus, any changes in ARG abundance seen between manure treatments would not simply reflect a gross variation in the overall bacterial abundance.
FIG 1.
Abundance of gene targets in the bacterial and bacteriophage DNA fractions of soil amended with dairy manure. Bar graphs represent the number of gene copies per nanogram of template DNA, as measured by qPCR. The gene targets quantified are indicated above the corresponding graphs. As indicated at the bottom, soil either did not receive manure (control [Con]) or was amended with raw (Raw) dairy manure or manure that had been digested (Dig), dewatered (DW), or composted (Comp) prior to application. Soil from these plots was sampled immediately after application, as well as 7 and 30 days later (days 0, 7, and 30, respectively). A no-template control (−) was also included in the qPCRs. Data are presented as the mean and standard deviation from three biological replicates. *, a statistically significant difference between the bacterial and corresponding phage DNA fractions. A statistically significant difference between a given manure treatment and the corresponding control not receiving manure is indicated by the letter a or b for the bacterial or phage fraction, respectively.
As expected, the rrnS gene target was far less abundant in the bacteriophage DNA fraction for all treatments, where it was present at a level of about 102 GC/ng DNA (Fig. 1, top). Importantly, this level was not significantly different from the background level of rrnS measured in a no-template control. This means that any ARGs observed in the phage fraction do not simply reflect the rampant acquisition of bacterial genes overall, since the rrnS gene is very rarely acquired by bacteriophage in our soil samples. As with the bacterial fraction, the rrnS copy number in the bacteriophage DNA fraction was consistent among all treatments, indicating little or no impact of manure application or pretreatment of manure on the overall bacterial DNA abundance in bacteriophage.
A no-template control was also evaluated for each of the ARG primer/probe sets, and no gene targets were detected, indicating low background levels for each of the ARGs. Within the bacterial fraction, each of the detected ARGs was significantly more abundant in soil receiving raw manure than in control soil not receiving manure (Fig. 1). For instance, the level of sul1 increased from about 101 GC/ng DNA in control soil to about 105 GC/ng DNA in soil amended with raw manure. Furthermore, ARG abundance was significantly higher for soils amended with digested and dewatered manure than for control soil, indicating that these treatments are ineffective at mediating the dissemination of ARGs by manure application. In contrast, composting of manure prior to field application did not increase the abundance of ARG targets above that in soils not receiving manure, indicating the efficacy of this practice. The ARG abundance in the bacterial fraction transiently increased on day 7 relative to that on day 0 and then decreased by day 30. This suggests that there may have been an initial proliferation of ARG-harboring bacteria in the days following manure application, followed by their decline by day 30.
In soils not receiving manure, each ARG quantified in the bacterial fraction was also detected in the phage fraction (Fig. 1). Moreover, ARG abundance was not significantly lower in the phage fraction than in the corresponding bacterial fraction in soils not receiving manure, with the exception of the abundances of strA (not detected on day 7) and strB (not detected on day 7 or 30). Strikingly, aadA was significantly more abundant in the phage fraction than in the bacterial fraction on days 7 and 30 (Fig. 1, bottom). In stark contrast to the findings for the bacterial fraction, ARG abundance in the phage fraction did not generally respond significantly to manure treatment or time (Fig. 1). Instead, the phage fraction appeared to harbor a constant reservoir of ARGs at a background abundance roughly matching that seen in the bacterial fraction in the absence of manure. This abundance did not respond to amendment with manure, regardless of preapplication treatment.
Impact of biosolids application on abundance and distribution of antibiotic resistance genes in bacteriophage and bacterial fractions.
The abundance of rrnS was between 104 and 105 GC/ng DNA in untreated soil and did not increase significantly following application of aerobically digested biosolids (Fig. 2). In fact, there was a significant decrease on days 0 and 7 relative to that in untreated soil. Thus, any increase in ARG abundance in treated versus control soil would not simply reflect an increase in bacterial DNA abundance.
FIG 2.
Abundance of gene targets in the bacterial and bacteriophage DNA fractions of soil amended with aerobically digested biosolids. Bar graphs represent the number of gene copies per nanogram of template DNA, as measured by qPCR. The gene targets quantified are indicated above the corresponding graphs. As indicated at the bottom of each graph, soil was untreated (control [Con]; sampled immediately before application) or received biosolids and was sampled immediately after application (day 0) or at 7 and 30 days postapplication. A no-template control (−) was also included in the qPCRs. Data are presented as the mean and standard deviation from three replicates. *, a statistically significant difference between the corresponding bacterial and phage DNA fractions. A statistically significant difference between a given biosolids-treated time point and the corresponding untreated control is indicated by the letter a or b for the bacterial or phage fraction, respectively.
Following biosolids application, the gene targets strA, strB, and sul1 were more abundant in the bacterial fraction of treated soil than it that of control soil (Fig. 2). The abundance of these gene targets declined thereafter through day 30. In contrast, aadA and blaOXA-20 were not enriched by biosolids application. Furthermore, other than aadA on day 30, both aadA and blaOXA-20 otherwise remained at comparable levels in the treated and control soils through the period of investigation.
Every gene target was detected in the bacteriophage fraction in control soils, with the exception of strA, which was undetected in both the bacterial and bacteriophage fractions (Fig. 2). Only sul1 was significantly less abundant in the bacteriophage fraction than in the bacterial fraction for untreated soil. ARG abundance in the bacteriophage fraction did not respond significantly to biosolids treatment or time for most samples, with the exception of sul1 abundance. On day 0, the abundance of this gene target increased slightly but significantly in treated soil relative to that in control soil (Fig. 2). As was observed with dairy manure, the phage fraction appears to harbor a constant reservoir of ARGs at the background abundance, and the abundance in the phage fraction had little or no response to amendment with biosolids.
Taken together, these results suggest that soilborne bacteriophage represents a reservoir of antibiotic resistance genes. Amendment of soil with dairy manure or biosolids had no significant effect on the abundance of gene targets in the bacteriophage fraction but increased the abundance in the bacterial fraction.
Potential for transduction.
Aqueous suspensions of soil bacteria were incubated for 3 days with or without bacteriophage enriched from biosolids and with or without supplementation with antibiotics (Table 2). Note that we chose to quantify coliform bacteria as a representative subset of bacteria within the total soilborne microbiome. In the absence of added antibiotics, there was no effect of bacteriophage supplementation on the abundance of viable antibiotic-resistant coliform bacteria (Table 2). However, after 3 days in the presence of 1/10 the breakpoint concentration of cefoxitin, the abundance of cefoxitin-resistant coliforms was 3.7-fold higher in the presence than in the absence of bacteriophage (Table 2). Likewise, phage conferred a 6.3-fold increase in the abundance of sulfamethazine-resistant coliforms when incubated in the presence of 1/100 of the sulfamethazine breakpoint concentration and a 7.1-fold increase in the presence of 1/10 of its breakpoint concentration (Table 2). Phage appeared to confer a slight decrease (less than 2-fold) in the abundance of cefoxitin-resistant coliforms when incubated in the presence of 1/100 of the cefoxitin breakpoint concentration. On the other hand, the abundance of ampicillin-resistant coliforms did not vary with any treatment. Taken together, these results suggest that bacteriophage from biosolids increased the abundance of coliform bacteria resistant to sulfamethazine or cefoxitin in the presence but not the absence of each antibiotic.
DISCUSSION
Distribution of antibiotic resistance genes in bacterial and bacteriophage fractions of agricultural soils.
Several studies have detected ARGs in the bacteriophage metagenome (or phageome) of a wide variety of environmental matrices, including activated sludge (9, 12), urban sewage and river water (13), and various wastewater effluents from hospitals and wastewater treatment plants (14, 26). When specific ARGs are detected by real-time PCR in the bacteriophage populations of the above-described environments, their levels are only about 10-fold lower than those in the corresponding bacterial fractions, on average (reviewed in reference 2). The present study further demonstrates that the phageome of agricultural soil harbors ARGs. The ARGs tend to be present in similar numbers in the corresponding bacterial and phage DNA fractions from agricultural soils, at least in the absence of manure or biosolids application. ARG copy abundance in the bacterial fraction rose sharply in response to dairy manure application and, in the case of strA, strB, and sul1, also rose in response to biosolids application. In the case of manure, some preapplication treatment options, specifically, composting, attenuated the enrichment effect. As measured by rrnS gene target copy abundance, there was no stimulation of total bacterial populations with the addition of manure, regardless of manure treatment. In contrast, the abundance of ARG target copies did respond to manure application. It is likely that the ARGs originated from bacteria that were introduced by application of dairy manure and biosolids, especially given that composting of the manure attenuated this enrichment effect.
In contrast to the responsiveness of the ARG target abundance in the bacterial fraction, ARG levels were virtually identical in the phageome, regardless of treatment or the time of sampling. It is unclear why ARG levels in the phageome did not respond to manure application or pretreatment in parallel to the bacteriome.
Here, we used rrnS (the 16S rRNA gene) as an indicator of overall bacterial abundance; rrnS is also useful as a representative gene for measurement of overall bacterial gene acquisition by bacteriophage (6). Since rrnS is ubiquitous in bacterial species, it should represent a baseline level of bacterial gene acquisition by phage. Notably, rrnS levels in the phage fraction of all soils sampled in this study were lower than those in the corresponding bacterial fraction by several orders of magnitude (indeed, in most cases, rrnS levels in the phage fraction were not even above the background rrnS levels, defined in control qPCRs lacking a template). This suggests that bacterial gene uptake by phage is a relatively rare event, in agreement with previous estimates of about one transduction event per every 108 phage infections (27).
Potential transduction of soil coliform bacteria by biosolids-derived phage.
We chose to focus on coliform bacteria as a subset of the total population of potential bacterial transductants generated in our experimental setup. This had the advantage of allowing us to quantify a relatively homogeneous set of bacterial species; the counting of total viable bacteria from a complex matrix like soil is cumbersome, and we would have no idea if increases in the population of viable bacteria were due to the transduction of the various species or due to some shift in the complex dynamics of interspecies competition due to the perturbations of both antibiotic selection and increased phage infective burden. By focusing on coliforms, we thus took a more reductionist approach. A major drawback is that we likely grossly underestimated the true frequencies of transduction in agricultural soil, as well as the true extent of the influence of subclinical selective pressure. Nevertheless, our results serve as a proof of concept that subclinical concentrations of some antibiotics can potentiate the phage-mediated horizontal transfer of resistance genes into potential human pathogens in the context of an agricultural soil microbiome.
We assume that the enrichment of antibiotic-resistant coliforms in the presence of bacteriophage is due to transduction, although we use this term broadly, as the mechanism of gene transfer and whether it is generalized or specialized (4) have not been proven. A few lines of evidence support the hypothesis that transduction was at play. First, the effect occurred specifically when enriched bacteriophage was incubated with soil-derived bacteria. Second, PCR analyses of the purified bacteriophage preparations yielded no evidence of bacterial DNA, and phage-only plating controls gave no indication of bacterial growth, indicating that the bacteriophage enrichments were not contaminated with bacterial DNA or viable coliform bacteria. The increased resistance in the presence of enriched bacteriophage must therefore be specific to the phage; the resistance genes may have originated from bacteria in the biosolids from which the bacteriophage was enriched, or the bacteriophage may be promoting the transfer of resistance genes within the soil bacterial community.
Transduction is known to be feasible in soil (28) and is undoubtedly a widespread phenomenon in many environments. For instance, retail chicken meat carries a number of phage capable of transferring antimicrobial resistance; of 243 phage randomly isolated from chicken meat, about 25% was able to transduce into E. coli resistance to 1 or more of the 35 antimicrobials tested (29). Based on factors like phage versus bacterial abundance, the number of phage in a transduction-competent state, and the physical conditions of various environments, Muniesa et al. (8) concluded that phage-mediated horizontal transfer between intestinal bacteria or between intestinal and indigenous bacteria in extraintestinal environments was probable. Moreover, bacteriophage isolated from antibiotic-treated mouse feces can confer antibiotic resistance to cultured murine intestinal microbiota (11). In-feed antibiotics induce prophage in swine fecal microbiomes (30), and horizontal gene transfer (including transduction) is subject to selective pressure (31). For instance, a study examining the incidence of HGT of ARGs in thousands of microbial genomes found that the genes involved in HGT events are 25-fold more likely to become fixed in human-associated bacteria than in diverse environmental isolates, due to selective pressure (32). Combined with our observation of probable transduction only in the presence of selective pressure, it is tempting to speculate that transduction is generally potentiated by antibiotic selection.
It is unclear why ampicillin selection did not confer the transduction of ampicillin resistance and why cefoxitin selection (at 1/100 of the breakpoint concentration) caused a slight but significant decrease in cefoxitin resistance in the presence versus absence of phage. In the case of the latter, the decrease was quite small (less than 2-fold), and a larger increase in resistance (nearly 4-fold) was conferred by phage when cefoxitin selection was applied at 1/10 of the cefoxitin breakpoint concentration, perhaps indicating that a lack of adequate selection by the lower cefoxitin concentration simply failed to promote transduction; the 2-fold decrease might thus represent an anomaly. It would be interesting to test whether the transduction frequency also correlates with the incubation time, as precedent exists for transduction efficiency peaking after a defined time period: in early transduction experiments with soil, Zeph et al. (28) noted that optimal incubation times existed for P1 transduction of chloramphenicol resistance into E. coli recipients. It is possible that different antibiotics are selecting for transduction by different sets of phage, each of which yields peak transduction after specific periods of time. Shousha et al. (29) observed that of 243 bacteriophage randomly isolated from chicken meat, about a quarter was able to transduce resistance to one or more of the five antibiotics tested into E. coli. Resistance to kanamycin was transduced the most often, followed by that to chloramphenicol, while only a few phage transduced tetracycline or ampicillin resistance. It thus seems likely that some ARGs are subject to transduction less frequently than others. In our experiment, it is curious that ampicillin selection yielded no transduction, while cefoxitin selection did, since beta-lactamases or ampC promoter or attenuator mutations (33) might reasonably yield resistance to both antibiotics. One possibility is that selection with ampicillin did not yield an increase in transduction as robust as that seen for selection with cefoxitin. Thus, different types of selection might display different potentiating effects on the transduction frequency. It would be interesting to test whether selection with various antibiotics during transduction yields increased resistance to unrelated antibiotics in our experimental setup.
Overall, our results suggest that the application of subclinical concentrations of antibiotics can promote bacteriophage-mediated horizontal transfer of antibiotic resistance genes in agricultural soil microbiomes. Further work is required to determine if antibiotics entrained into soil through application of animal or human wastes might increase the risk of spreading antibiotic resistance to potential human pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by competitive funding through the AAFC Growing Forward 2 program. J.R. was supported by the NSERC Visiting Fellowship in Government Laboratories program.
We thank S. Gordon for valued technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02363-15.
REFERENCES
- 1.Martinez JL. 2009. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 157:2893–2902. doi: 10.1016/j.envpol.2009.05.051. [DOI] [PubMed] [Google Scholar]
- 2.Muniesa M, Colomer-Lluch M, Jofre J. 2013. Potential impact of environmental bacteriophages in spreading antibiotic resistance genes. Future Microbiol 8:739–751. doi: 10.2217/fmb.13.32. [DOI] [PubMed] [Google Scholar]
- 3.Marti E, Variatza E, Balcazar J. 2014. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol 22:36–41. doi: 10.1016/j.tim.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Frost L, Leplae R, Summers A, Toussaint A. 2005. Mobile genetic elements: the agents of open source evolution. Nat Rev Microbiol 3:722–732. doi: 10.1038/nrmicro1235. [DOI] [PubMed] [Google Scholar]
- 5.Lang AS, Zhaxybayeva O, Beatty JT. 2012. Gene transfer agents: phage-like elements of genetic exchange. Nat Rev Microbiol 10:472–482. doi: 10.1038/nrmicro2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sander M, Schmieger H. 2001. Method for host-independent detection of generalized transducing bacteriophages in natural habitats. Appl Environ Microbiol 67:1490–1493. doi: 10.1128/AEM.67.4.1490-1493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penadés J, Chen J, Quiles-Puchalt N, Carpena N, Novick R. 2015. Bacteriophage-mediated spread of bacterial virulence genes. Curr Opin Microbiol 23:171–178. doi: 10.1016/j.mib.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 8.Muniesa M, Imamovic L, Jofre J. 2011. Bacteriophages and genetic mobilization in sewage and faecally polluted environments. Microb Biotechnol 4:725–734. doi: 10.1111/j.1751-7915.2011.00264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsley LC, Consuegra EJ, Kakirde KS, Land AM, Harper WF, Liles MR. 2010. Identification of diverse antimicrobial resistance determinants carried on bacterial, plasmid, or viral metagenomes from an activated sludge microbial assemblage. Appl Environ Microbiol 76:3753–3757. doi: 10.1128/AEM.03080-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kenzaka T, Tani K, Nasu M. 2010. High-frequency phage-mediated gene transfer in freshwater environments determined at single-cell level. ISME J 4:648–659. doi: 10.1038/ismej.2009.145. [DOI] [PubMed] [Google Scholar]
- 11.Modi S, Lee H, Spina C, Collins J. 2013. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature 499:219–222. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calero-Cáceres W, Melgarejo A, Colomer-Lluch M, Stoll C, Lucena F, Jofre J, Muniesa M. 2014. Sludge as a potential important source of antibiotic resistance genes in both the bacterial and bacteriophage fractions. Environ Sci Technol 48:7602–7611. doi: 10.1021/es501851s. [DOI] [PubMed] [Google Scholar]
- 13.Colomer-Lluch M, Jofre J, Muniesa M. 2011. Antibiotic resistance genes in the bacteriophage DNA fraction of environmental samples. PLoS One 6:e17549. doi: 10.1371/journal.pone.0017549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marti E, Variatza E, Balcazar J. 2014. Bacteriophages as a reservoir of extended spectrum β-lactamase and fluoroquinolone resistance genes in the environment. Clin Microbiol Infect 20:O456–O459. doi: 10.1111/1469-0691.12446. [DOI] [PubMed] [Google Scholar]
- 15.Rahube T, Marti R, Scott A, Tien YC, Murray R, Sabourin L, Zhang Y, Duenk P, Lapen D, Topp E. 2014. Impact of fertilizing with raw or anaerobically digested sewage sludge on the abundance of antibiotic-resistant coliforms, antibiotic resistance genes, and pathogenic bacteria in soil and on vegetables at harvest. Appl Environ Microbiol 80:6898–6907. doi: 10.1128/AEM.02389-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marti R, Tien YC, Murray R, Scott A, Sabourin L, Topp E. 2014. Safely coupling livestock and crop production systems: how rapidly do antibiotic resistance genes dissipate in soil following a commercial application of swine or dairy manure? Appl Environ Microbiol 80:3258–3265. doi: 10.1128/AEM.00231-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marti R, Scott A, Tien Y-C, Murray R, Sabourin L, Zhang Y, Topp E. 2013. The impact of manure fertilization on the abundance of antibiotic resistant bacteria and frequency of detection of antibiotic resistance genes in soil, and on vegetables at harvest. Appl Environ Microbiol 79:5701–5709. doi: 10.1128/AEM.01682-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thurber R, Haynes M, Breitbart M, Wegley L, Rohwer F. 2009. Laboratory procedures to generate viral metagenomes. Nat Protoc 4:470–483. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 19.Muyzer G, Dewaal E, Uitierlinden A. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59:695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki M, Taylor L, DeLong E. 2000. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl Environ Microbiol 66:4605–4614. doi: 10.1128/AEM.66.11.4605-4614.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walsh F, Ingenfeld A, Zampicolli M, Hilber-Bodmer M, Frey J, Duffy B. 2011. Real-time PCR methods for quantitative monitoring of streptomycin and tetracycline resistance genes in agricultural ecosystems. J Microbiol Methods 86:150–155. doi: 10.1016/j.mimet.2011.04.011. [DOI] [PubMed] [Google Scholar]
- 22.Bert F, Branger C, Lambert-Zechovsky N. 2002. Identification of PSE and OXA beta-lactamase genes in Pseudomonas aeruginosa using PCR-restriction fragment length polymorphism. J Antimicrob Chemother 50:11–18. doi: 10.1093/jac/dkf069. [DOI] [PubMed] [Google Scholar]
- 23.United States Environmental Protection Agency. 2001. Method 1602: male-specific (F+) and somatic coliphage in water by single agar layer (SAL) procedure. United States Environmental Protection Agency, Washington, DC. [Google Scholar]
- 24.Ashelford K, Day M, Fry J. 2003. Elevated abundance of bacteriophage infecting bacteria in soil. Appl Environ Microbiol 69:285–289. doi: 10.1128/AEM.69.1.285-289.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Government of Canada. 2010. Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS) 2007. Public Health Agency of Canada, Guelph, ON, Canada. [Google Scholar]
- 26.Colomer-Lluch M, Jofre J, Muniesa M. 2014. Quinolone resistance genes (qnrA and qnrS) in bacteriophage particles from wastewater samples and the effect of inducing agents on packaged antibiotic resistance genes. J Antimicrob Chemother 69:1265–1274. doi: 10.1093/jac/dkt528. [DOI] [PubMed] [Google Scholar]
- 27.Chibani-Chennoufi S, Bruttin A, Dillmann M-L, Brüssow H. 2004. Phage-host interaction: an ecological perspective. J Bacteriol 186:3677–3686. doi: 10.1128/JB.186.12.3677-3686.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeph L, Onaga M, Stotzky G. 1988. Transduction of Escherichia coli by bacteriophage P1 in soil. Appl Environ Microbiol 54:1731–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shousha A, Awaiwanont N, Sofka D, Smulders FJ, Paulsen P, Szostak MP, Humphrey T, Hilbert F. 2015. Bacteriophages isolated from chicken meat and the horizontal transfer of antimicrobial resistance genes. Appl Environ Microbiol 81:4600–4606. doi: 10.1128/AEM.00872-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen H, Looft T, Bayles D, Humphrey S, Levine U, Alt D, Stanton T. 2011. Antibiotics in feed induce prophages in swine fecal microbiomes. mBio 2:e00260-11. doi: 10.1128/mBio.00260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perry J, Wright G. 2014. Forces shaping the antibiotic resistome. Bioessays 36:1179–1184. doi: 10.1002/bies.201400128. [DOI] [PubMed] [Google Scholar]
- 32.Smillie C, Smith M, Friedman J, Cordero O, David L, Alm E. 2011. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 480:241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 33.Mulvey M, Bryce E, Boyd D, Ofner-Agostini M, Land A, Simor A, Paton S. 2005. Molecular characterization of cefoxitin-resistant Escherichia coli from Canadian hospitals. Antimicrob Agents Chemother 49:358–365. doi: 10.1128/AAC.49.1.358-365.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.