Abstract
High-level heat resistance of spores of Bacillus thermoamylovorans poses challenges to the food industry, as industrial sterilization processes may not inactivate such spores, resulting in food spoilage upon germination and outgrowth. In this study, the germination and heat resistance properties of spores of four food-spoiling isolates were determined. Flow cytometry counts of spores were much higher than their counts on rich medium (maximum, 5%). Microscopic analysis revealed inefficient nutrient-induced germination of spores of all four isolates despite the presence of most known germination-related genes, including two operons encoding nutrient germinant receptors (GRs), in their genomes. In contrast, exposure to nonnutrient germinant calcium-dipicolinic acid (Ca-DPA) resulted in efficient (50 to 98%) spore germination. All four strains harbored cwlJ and gerQ genes, which are known to be essential for Ca-DPA-induced germination in Bacillus subtilis. When determining spore survival upon heating, low viable counts can be due to spore inactivation and an inability to germinate. To dissect these two phenomena, the recoveries of spores upon heat treatment were determined on plates with and without preexposure to Ca-DPA. The high-level heat resistance of spores as observed in this study (D120°C, 1.9 ± 0.2 and 1.3 ± 0.1 min; z value, 12.2 ± 1.8°C) is in line with survival of sterilization processes in the food industry. The recovery of B. thermoamylovorans spores can be improved via nonnutrient germination, thereby avoiding gross underestimation of their levels in food ingredients.
INTRODUCTION
Bacillus endospores (spores) are widely present in nature and may contaminate food ingredients and food products. Due to the intrinsic stability of spores, which allows them to withstand environmental insults, sufficient inactivation of spores in commercially sterile food products is a major challenge for the food industry (1–4).
Bacillus thermoamylovorans produces spores with high-level heat resistance (3), and the spores are known to survive industrial food sterilization processes. The organism is facultatively anaerobic and has the ability to grow at temperatures between 40°C and 58°C (5, 6). In our experience, strains of B. thermoamylovorans are able to grow at 37°C but not at 30°C. The organism was first described as a nonsporogenous species (5, 7), but in an amended species description the formation of spores was reported (8). The occurrence of B. thermoamylovorans in a gelatin production plant and at dairy farms has been reported (3, 9). The genome sequence of one non-food-related B. thermoamylovorans strain from a biogas plant was published recently (6). Overall, the species has not been well characterized, and little is known about the spore properties that are important for control in foods, including spore resistance to various processing conditions and germination of spores that survive.
When spores exit dormancy via germination, food spoilage can occur upon outgrowth. These processes have been well studied in Bacillus subtilis (10–13). Germination can be induced by both nutrient and nonnutrient triggers, called germinants. Nutrients can initiate germination via interaction with germinant receptors (Ger receptors [GRs]), which are localized in the inner membrane of the spore and consist of three or four different subunits (A, B, C, and D) (14–17). The responsiveness of GRs to nutrient triggers can be enhanced by exposure of spores to sublethal temperatures during a so-called heat activation step (12, 18). In contrast, germination via the nonnutrient germinant dipicolinic acid chelated with Ca2+ ions (Ca-DPA) occurs by direct activation of the cortex lytic enzyme (CLE) CwlJ, thereby bypassing the requirement of GRs (19). Activated CwlJ then hydrolyzes the protective peptidoglycan cortex, resulting in rehydration of the spore core (19, 21). Ca-DPA-induced germination has been reported to be independent of a heat activation treatment (56). The germination behavior of spores is a heterogeneous process (22), which is reflected by varying germination kinetics and/or the emergence of so-called superdormant spores that do not respond to the applied germination trigger (23–26). For B. subtilis it has previously been described that spores superdormant to nutrients harbor lower numbers of germination receptor proteins (27), whereas spores that were superdormant to Ca-DPA showed decreased levels of CwlJ (25).
Improved understanding and control of bacterial food spoilage can be facilitated by combining experimental findings with in silico analysis of genome content (29). In this study, spore germination of four food isolates of B. thermoamylovorans (isolated from either acacia gum or milk) in response to nutrient and nonnutrient triggers was investigated, and the genome sequences of the strains were determined (30). The strain-specific spore germination data were linked with presence or absence of important germination-related genes. In addition, spore heat resistance kinetics were determined using standard plating techniques, with and without a Ca-DPA pretreatment, based on the insights into germination of this species obtained in this study. This approach led to a more accurate assessment of viable spore counts and heat resistance properties of spores of this species.
MATERIALS AND METHODS
Strains.
Four strains of B. thermoamylovorans isolated from different sources were used in this study for characterization of the spore properties. Strains B4064 and B4065 were isolated from acacia gum, whereas strains B4166 and B4167 were isolated from milk. For all strains, the genome sequences were determined (30).
Spore preparation.
Spores of B. thermoamylovorans were prepared as previously described for B. subtilis (31, 32) with slight modifications. The strains were precultured for 16 h at 45°C in brain heart infusion (BHI) broth supplemented with 1 mg/liter vitamin B12 (Merck) and subsequently spread on Schaeffer sporulation agar plates supplemented with 1 mg/liter vitamin B12 (3). These plates were incubated at 45°C for 7 days, and spores were harvested and washed successively in sterile water, as described before (31). Spore suspensions were stored at 4°C for 2 to 4 weeks prior to experiments. The purity of the spore suspensions (>95% phase-bright spores) was checked using phase-contrast microscopy (see below). For each strain, three independent spore crops were prepared.
Spore quantification.
Spore suspensions were enumerated in two ways, namely, by plate counting and flow cytometry. The spore counts were assessed by plate counting as follows. Spore suspensions were heat activated at 80, 90, and 100°C for 10 min, followed by pour plating in BHI agar (BHI-A) plates supplemented with 1 mg/liter vitamin B12 (in duplicate). Plates were incubated for 5 days at 45°C, after which CFU were enumerated. An increase in the heat activation temperature (up to 100°C for 10 min) did not affect the CFU counts; therefore, 80°C for 10 min was used routinely to assess the spore CFU counts. Based on the initial counts obtained, the spore suspensions were diluted to a working spore suspension of approximately 108 CFU/ml in phosphate-buffered saline (PBS) with a pH of 7.4.
Absolute spore counts were also determined by flow cytometry using a BD FACSAria II flow cytometer operated with BD FACSDiva software (version 6.0; BD Biosciences). Spore suspensions were diluted 100 times in sheath fluid (BD FACSFlow; BD Biosciences) to obtain event rates below 2,000 event s−1, and at least 20,000 events were measured for each spore crop (33). A predetermined amount of reference beads (Microsphere standard [6-μm diameter] Live/Dead BacLight bacterial viability and counting kit L34856) was added to each spore suspension, corresponding to 5 × 105 beads per ml. For each strain, three independent spore crops were measured in duplicate.
Spore germination.
Spore germination was studied and quantified using phase-contrast microscopy (see below). Prior to the experiments, spore crops were washed with ice-cold sterile Milli-Q water. If not stated otherwise, spores were heat activated at 80°C for 10 min or at 70°C for 30 min and subsequently cooled on ice and washed again with cold water. Heat-activated spores were diluted to a final optical density at 600 nm (OD600) of 1 in 200 μl of BHI or Luria-Bertani (LB) medium supplemented with vitamin B12 (1 mg/liter) and chloramphenicol (7.5 mg/liter) to prevent outgrowth of vegetative cells (34, 35). Alternatively, spores were diluted in mixtures of various nutrient-based germinants dissolved in 25 mM Tris-HCl, pH 7.4: (i) 100 mM l-alanine; (ii) l-asparagine, d-fructose, d-glucose, and KCl (all at 50 mM); (iii) l-alanine, l-arginine, l-asparagine, aspartic acid, l-cysteine-HCl, glutamic acid, l-glutamine, glycine, l-histidine, inosine, l-isoleucine, l-leucine, l-lysine, l-methionine, l-phenylalanine, l-proline, l-serine, l-threonine, l-tryptophan, and l-valine (all at 10 mM); or (iv) l-alanine, l-arginine, l-asparagine, aspartic acid, l-cysteine-HCl, glutamic acid, l-glutamine, glycine, l-histidine, inosine, l-isoleucine, l-leucine, l-lysine, l-methionine, l-phenylalanine, l-proline, l-serine, l-threonine, l-tryptophan, l-valine, d-fructose, d-glucose, and KCl (all at 10 mM) and chloramphenicol (7.5 mg/liter). For non-nutrient-induced germination experiments, non-heat-activated spores were diluted in equimolar mixtures of 20, 40, 60, or 80 mM CaCl2 and DPA (pH 7.4). A preliminary analysis indicated that 40 mM Ca-DPA was the most efficient concentration to trigger germination (data not shown), and this concentration was used in further experiments. As negative controls, both non-heat-activated and heat-activated spores were diluted in 25 mM Tris-HCl, pH 7.4. Spore dilutions were then incubated at 42°C while shaking (220 rpm). After 3, 6, and 24 h, the transition of phase-bright dormant spores to phase-dark germinated spores was monitored using phase-contrast microscopy. Microscopic imaging was performed using an IX71 microscope (Olympus) with a CoolSNAP HQ2 camera (Princeton Instruments), using a 100× phase-contrast objective and DeltaVision softWoRx 3.6.0 (Applied Precision) software. Images were taken using 32% APLLC white LED light and a 0.3-s exposure. The pixel size was 0.0643 μm, and binning was set to 1×1. Images obtained were analyzed using Fiji software (http://fiji.sc/Fiji) (36). For quantification of ratios of germinated and dormant spores, a minimum of 300 spores per condition were examined. All experiments were performed in duplicate using two independent spore crops.
Spore heat inactivation.
Spore heat inactivation was determined using capillary tubes with two independent spore crops for each of the four strains, as previously described (31, 37). For all strains, the spore working suspensions (108 spores/ml in PBS) were heated at 110°C at 10 different time points up to 23 min. For strain B4064, the inactivation experiments were additionally performed at 115°C and 120°C to allow for detailed inactivation kinetics determination. Upon heat treatment, one part of the spore suspension was 10-fold serially diluted in peptone water, and appropriate dilutions were pour plated in duplicate in BHI agar plates supplemented with 1 mg/liter vitamin B12. The other part of the heated spore suspension was exposed to 40 mM Ca-DPA in sterile peptone water for 3 h at 45°C, followed by pour plating 10-fold serial dilutions (made in peptone water) in duplicate in BHI agar plates supplemented with 1 mg/liter vitamin B12. Per experiment, plating was performed in duplicate. After incubation for 5 days at 45°C, CFU were enumerated and recovery of spores determined.
The survivor count was plotted against the inactivation time, and based on the shape of the inactivation plot, a model was selected for fitting. Model fitting was performed with Microsoft Excel using the Solver Add-in.
For the experiments that included an incubation step with Ca-DPA prior to plating, the data were fitted with the log-linear model where the D value was determined from the equation log(Nt) = log(N0) − (t/D), where Nt is the surviving spore count at time t, N0 is the initial spore concentration, t is the time (time unit), and D is the decimal reduction time.
The inactivation plots of the experiments that did not included a Ca-DPA incubation step prior to plating showed the presence of a heat-sensitive population and a heat-resistant population; therefore, these were fitted with the biphasic Geeraerd model as described in the following equation (38)
Where Nt is the survivor count at time t, N0 is the initial spore count, (1 − f) and f are the heat-sensitive and heat-resistant fractions, respectively, ksen and kres are the inactivation rates (time unit−1) of the sensitive and the resistant populations, respectively, t is the time (time unit), and S is the duration of the shoulder (time unit). The D value was calculated by dividing the reciprocal of the inactivation rates by the natural logarithm of 10.
For the experiments with strain B4064 that included incubation with Ca-DPA prior to plating, additionally the z value, the increase in temperature required to achieve an additional log unit reduction, the reference D value (Dref) at the reference temperature (Tref) of 121.1°C, and the 95% prediction interval (PI) were calculated as previously described (39).
Genome mining.
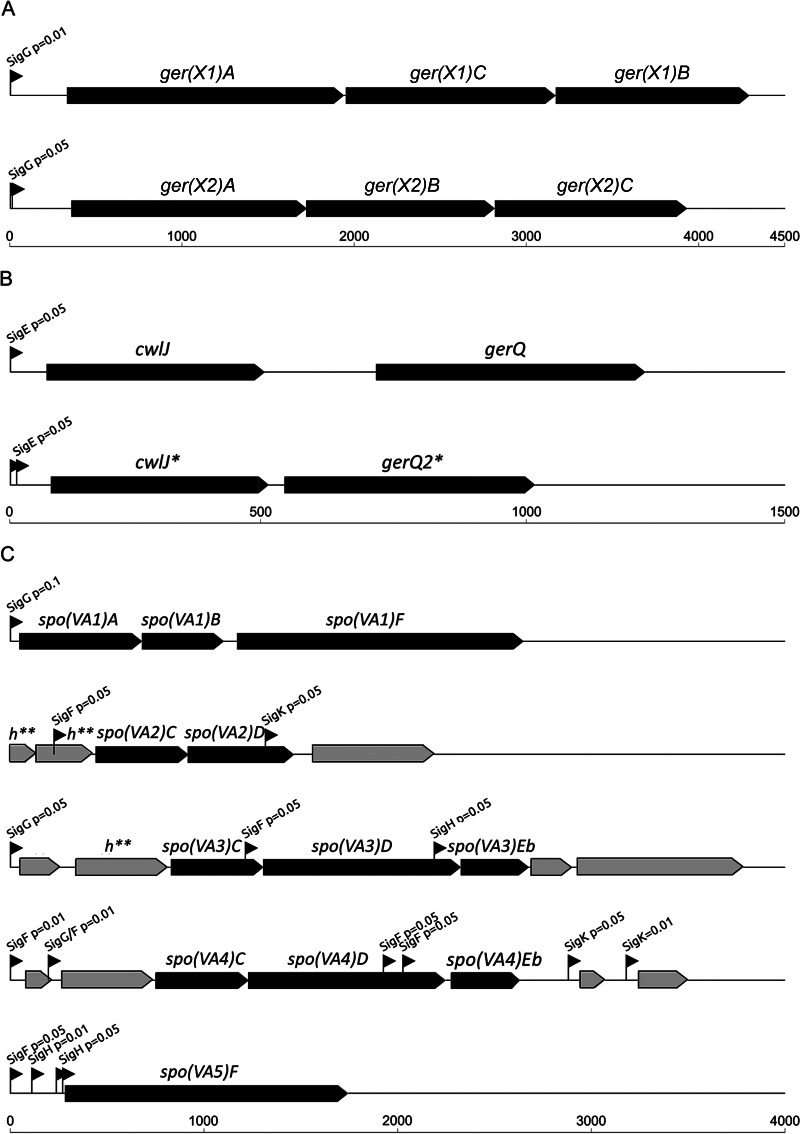
For all of the predicted protein sequences of the four B. thermoamylovorans strains and reference strain B. subtilis 168, an orthology prediction was performed using Ortho-MCL (40) (see Data Set S1 in the supplemental material). To find potential functional equivalents for a selection of germination-related genes (Table 1), corresponding protein sequence alignments were made using MUSCLE (41), followed by construction of a hidden Markov model (HMM) that was subsequently used to scan all genomes (42). For selected proteins found in this manner, maximum-likelihood trees were constructed using the maximum-likelihood phylogeny program PHYML (43). Phylogenetic trees were manually inspected for evolutionary relatedness of the proteins. Additionally, the genomic context was manually verified after visualization in the SEED Viewer (44) on the RAST annotation server (45). Prediction of binding sites for sporulation sigma factors (46) upstream of selected genes was performed with use of the database of transcriptional regulation in B. subtilis (DBTBS) (http://dbtbs.hgc.jp) (47). Schematic visualization of the predicted operon structures of genes ger(X1)ABC (where X1 is the first receptor), ger(X2)ABC, spoVAA to -AF, cwlJ, gerQ, cwlJ2, and gerQ2 (see Fig. 3) was made with the draw context tool on the Genome2D server (http://genome2d.molgenrug.nl) (48).
TABLE 1.
Presence and absence of germination genes in four B. thermoamylovorans isolatesa
| Orthologous group | Locus tag in B. subtilis 168 | Gene name in B. subtilis | Locus tag in B. thermoamylovorans isolate: |
Gene name in B. thermoamylovorans | Predicted function (reference[s]) | |||
|---|---|---|---|---|---|---|---|---|
| B4064 | B4065 | B4166 | B4167 | |||||
| OG_1412 | BSU03700 | gerKA | B4064_2025 | B4065_1012 | B4166_2487 | B4167_2413 | ger(X1)A | Germinant receptor subunit A (14, 16) |
| OG_1411 | BSU03720 | gerKB | B4064_2027 | B4065_1014 | B4166_2485 | B4167_2411 | ger(X1)B | Germinant receptor subunit B (14, 16) |
| OG_1413 | BSU03710 | gerKC | B4064_2026 | B4065_1013 | B4166_2486 | B4167_2412 | ger(X1)C | Germinant receptor subunit C (14, 16) |
| OG_3211 | BSU17750, BSU33050, BSU35800 | yndD, gerAA, gerBA | NA | Germinant receptor subunit A (14, 16) | ||||
| OG_3212 | BSU17760, BSU33060, BSU35810 | yndE, gerAB, gerBB | NA | Germinant receptor subunit B (14, 16) | ||||
| OG_3213 | BSU17770, BSU33070, BSU35820 | yndF, gerAC, gerBC | NA | Germinant receptor subunit C (14, 16) | ||||
| OG_5565 | BSU07760 | yfkT | NA | Germinant receptor subunit A (14, 16) | ||||
| OG_5568 | BSU07780 | yfkR | NA | Germinant receptor subunit B (14, 16) | ||||
| OG_5567 | BSU07790 | yfkQ | NA | Germinant receptor subunit C (14, 16) | ||||
| OG_2571 | NA | B4064_3197 | B4065_2779 | B4166_2035 | B4167_3629 | ger(X2)A | Germinant receptor subunit A (14, 16) | |
| OG_2572 | NA | B4064_3196 | B4065_2778 | B4166_2036 | B4167_3630 | ger(X2)B | Germinant receptor subunit B (14, 16) | |
| OG_2573 | NA | B4064_3195 | B4065_2777 | B4166_2037 | B4167_3631 | ger(X2)C | Germinant receptor subunit C (14, 16) | |
| OG_1063 | BSU01550 | gerD | B4064_2622 | B4065_1779 | B4166_1566 | B4167_2160 | gerD | Nutrient germination, required for clustering of GRs in the spore inner membrane (63, 51) |
| OG_1510 | BSU34990 | gerF | B4064_1353 | B4065_2064 | B4166_2701 | B4167_3142 | gerF | Nutrient germination, prelipoprotein diacylglycerol transferase (53) |
| OG_1028 | BSU10720 | gerPA | B4064_3055 | B4065_2324 | B4166_1505 | B4167_2306 | gerPA | Nutrient germination, spore coat permeability to nutrients (52) |
| OG_1029 | BSU10710 | gerPB | B4064_3054 | B4065_2325 | B4166_1506 | B4167_2307 | gerPB | Nutrient germination, spore coat permeability to nutrients (52) |
| OG_1030 | BSU10700 | gerPC | B4064_3053 | B4065_2326 | B4166_1507 | B4167_2308 | gerPC | Nutrient germination, spore coat permeability to nutrients (52) |
| OG_1031 | BSU10690 | gerPD | B4064_3052 | B4065_2327 | B4166_1508 | B4167_2309 | gerPD | Nutrient germination, spore coat permeability to nutrients (52) |
| OG_1032 | BSU10680 | gerPE | B4064_3051 | B4065_2328 | B4166_1509 | B4167_2310 | gerPE | Nutrient germination, spore coat permeability to nutrients (52) |
| OG_1033 | BSU10670 | gerPF | B4064_3050 | B4065_2329 | B4166_1510 | B4167_2311 | gerPF | Nutrient germination, spore coat permeability to nutrients (52) |
| OG_459 | BSU23440 | spoVAA | B4064_0505 | B4065_0471 | B4166_0596 | B4167_0663 | spo(VA1)A | Ca-DPA release (11, 12) |
| OG_460 | BSU23430 | spoVAB | B4064_0506 | B4065_0472 | B4166_0597 | B4167_0662 | spo(VA1)B | Ca-DPA release (11, 12) |
| OG_1852 | BSU23420 | spoVAC | B4064_3111 | B4065_2010 | B4166_3483 | B4167_3718 | spo(VA4)C | Ca-DPA release (11, 12) |
| OG_2783 | NA | B4064_1741 | B4065_2822 | B4166_2586 | B4167_2402 | spo(VA2)C | Ca-DPA release (11, 12) | |
| OG_2872 | NA | B4064_1750 | B4065_2831 | B4166_2823 | B4167_2485 | spo(VA3)C | Ca-DPA release (11, 12) | |
| OG_1543 | BSU23410 | spoVAD | B4064_1751 | B4065_2832 | B4166_2822 | B4167_2484 | spo(VA3)D | Ca-DPA release (11, 12) |
| OG_2782 | NA | B4064_1742 | B4065_2823 | B4166_2585 | B4167_2401 | spo(VA2)D | Ca-DPA release (11, 12) | |
| OG_3063 | NA | B4064_3112 | B4065_2009 | B4166_3482 | B4167_3717 | spo(VA4)D | Ca-DPA release (11, 12) | |
| OG_3436 OG_3868 | BSU23402 | spoVAEb | B4064_1752 | B4065_2833 | B4166_2944 | B4167_2035 | spo(VA3)Eb* | Ca-DPA release (11, 12) |
| OG_3062 | NA | B4064_3113 | B4065_2008 | B4166_3481 | B4167_3716 | spo(VA4)Eb | Ca-DPA release (11, 12) | |
| OG_5841 | BSU23401 | spoVAEa | NA | Ca-DPA release (11, 12) | ||||
| OG_461 | BSU23390 | spoVAF | B4064_0507 | B4065_0473 | B4166_0598 | B4167_0661 | spo(VA1)F | Ca-DPA release (11, 12) |
| OG_2204 | NA | B4064_0396 | B4065_0417 | B4166_0988 | B4167_1024 | spo(VA5)F | Ca-DPA release (11, 12) | |
| OG_59 | BSU02600 | cwlJ | B4064_1307 | B4065_1231 | B4166_1228 | B4167_1271 | cwlJ | Cortex lytic enzyme (21) |
| B4064_2405 | B4065_2977 | cwlJ2 | Cortex lytic enzyme (21) | |||||
| OG_3282 | BSU37920 | gerQ | B4064_2404 | B4065_2978 | gerQ2 | Required for CwlJ localization in the spore coat (56) | ||
| OG_2275 | NA | B4064_1308 | B4065_1230 | B4166_1229 | B4167_1270 | gerQ | Required for CwlJ localization in the spore coat (56) | |
| OG_485 | BSU22930 | sleB | B4064_0540 | B4065_0503 | B4166_0631 | B4167_0629 | sleB | Cortex lytic enzyme (21) |
| OG_486 | BSU22920 | ypeB | B4064_0541 | B4065_0504 | B4166_0632 | B4167_0628 | ypeB | Required for SleB presence in the spore (21) |
| OG_615 | BSU28380 | gerM | B4064_1518 | B4065_3302 | B4166_0779 | B4167_0814 | gerM | Role in cortex hydrolysis (64) |
| OG_710 | BSU25540 | gpr | B4064_0330 | B4065_0354 | B4166_0924 | B4167_0959 | gpr | Germination protease, SASP degradation (65) |
The table shows locus tags of the genes present in each B. thermoamylovorans strain that belong to the orthologous groups (OGs) corresponding to different germination genes. For reference, a model laboratory strain, B. subtilis 168, also was included in the set, together with locus tags and names of its germination genes. Empty cells indicate absence of genes in the respective strains. In some cases, multiple genes, listed by different locus tags, belong to one OG in the individual strains. For multiple spoVA genes, which occur in five different operons in B. thermoamylovorans genomes, the numbers that indicate operon affiliation of individual genes have been arbitrarily added to the gene names. An asterisk indicates that these genes were combined in one OG based on the multiple-sequence alignment and phylogenetic tree. Abbreviations: NA, not applicable; SASP, small acid-soluble spore protein.
FIG 3.
Schematic visualization of the predicted operon structures of ger(X1)ABC and ger(X2)ABC, encoding putative germinant receptors (A), cwlJ, gerQ, cwlJ2, and gerQ2, involved in Ca-DPA germination (B), and five spoVA operons, spo(VA1), spo(VA2), spo(VA3), spo(VA4), and spo(VA5) (C). The sigma factor binding sites, together with the threshold P values used for their prediction, are indicated with black arrows. The asterisk indicates that cwlJ2 and gerQ2 are present only in strains B4064 and B4065 (A). The spoVA operons 2, 3, and 4 [spo(VA2), spo(VA3), and spo(VA4)] next to spoVA genes also contain genes encoding hypothetical proteins, indicated with light gray arrows, and predicted internal sigma factor binding sites (C). Operon spo(VA2), containing the spo(VA2)C and spo(VA2)D genes, is located on the edge of the contig in genomes of all B. thermoamylovorans food isolates. Thus, the nucleotide sequence and predicted promoters upstream of the two genes that encode hypothetical proteins (indicated as h**) are unknown. However, as the nucleotide sequence of the two h** genes encoding hypothetical proteins in the operon spo(VA2) is highly similar to the sequence of the h** gene in front of the spo(VA3)C gene in the operon spo(VA3), it is probable that the sequences upstream of the operons spo(VA2) and spo(VA3) are similar (C). Scales below each part of the figure indicate distances in nucleotide base pairs.
RESULTS
Quantification of spores.
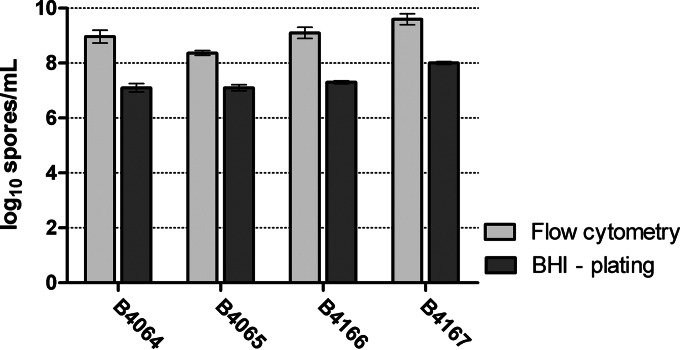
Spores, prepared on Schaeffer agar plates, were characterized with respect to spore germination and heat resistance. The number of spores in 1 ml of the working spore suspension was quantified using flow cytometry or CFU enumeration. The obtained numbers of spores per ml were strikingly different depending on the quantification technique used (Fig. 1). Using flow cytometry, the absolute number of spores in spore suspensions of strains B4064, B4065, B4166, and B4167 were 1.9, 1.3, 1.8, and 1.6 log units higher, respectively, than when enumerated using plating in BHI-A with vitamin B12. Since CFU plate counting enumeration depends on spore germination and outgrowth, this discrepancy indicates that only a small fraction (1.3% ± 1.0%, 5.3% ± 2.4%, 1.6% ± 0.9%, and 2.5% ± 2.1% for spores of strains B4064, B4065, B4166, and B4167, respectively) of the absolute number of spores undergoes germination and subsequent outgrowth on the BHI-A plates.
FIG 1.
Comparison of the spore count (log10 CFU/ml) obtained per strain by flow cytometry and plating on BHI after a heat treatment at 80°C for 10 min. Mean counts from three independent experiments were plotted, and error bars display the standard deviations.
Germination with nutrient germinants.
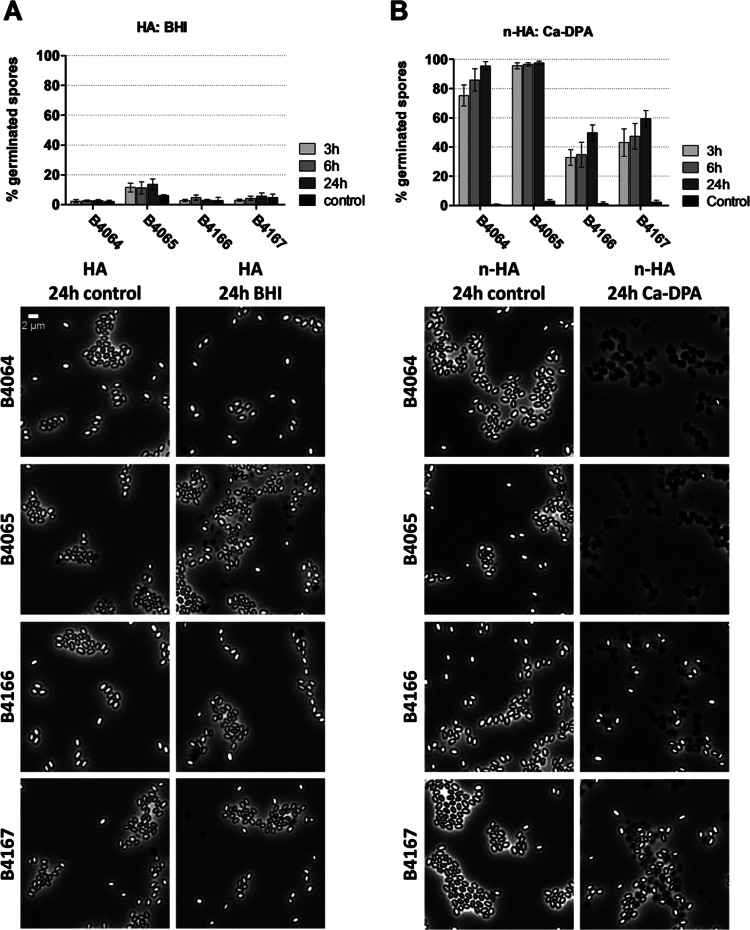
To establish whether the discrepancy between absolute spore counts and CFU counts was caused by inefficient germination, the germination efficiency of the heat-activated spores in the nutrient-rich BHI medium was assessed using phase-contrast microscopy. The analysis showed that the fraction of spores that germinated in BHI did not exceed 2.6% ± 0.8%, 13.6% ± 3.6%, 4.8% ± 0.5%, and 5.8% ± 2.1% for strains B4064, B4065, B4166, and B4167, respectively (Fig. 2). These numbers hardly exceeded the percentage of phase-dark spores in the negative controls (2.2% ± 0.7%, 6.2% ± 0.5%, 2.7% ± 2.2%, and 4.8% ± 2.3%, respectively) (Fig. 2). Moreover, the percentage of germinated spores did not increase significantly over time (Fig. 2). This implies that spores of B. thermoamylovorans germinate very poorly in BHI. In addition, spore germination was assessed in LB and simple nutrient mixtures i, ii, iii, and iv (see details in Materials and Methods), which resulted in similar observations (data not shown). Likewise, altering or omitting the heat activation treatment did not increase germination efficiency in rich medium (data not shown). Altogether, these results indicated that the tested nutrient germinants were not triggering germination of B. thermoamylovorans spores efficiently.
FIG 2.
Quantification of spore germination efficiency using phase-contrast microscopy. Spores were either heat activated (HA) or not (n-HA) and exposed to BHI plus vitamin B12 (A), Ca-DPA (B), or Tris buffer (control). Germination was calculated as the percentage of phase-dark spores on phase-contrast microscopic images made after 3, 6, and 24 h of incubation with germinant (images are shown for 24 h only). Mean percentages from two independent experiments were plotted, including error bars based on standard deviations. Scale bar, 2 μm.
Germination with Ca-DPA.
Besides germination of spores in response to nutrients, which requires the presence of GRs in the spores (11, 12), germination can also be induced by nonnutrient germinants (for instance, Ca-DPA) or by very high hydrostatic pressure (400 to 800 MPa) via mechanisms that are independent of GRs (11, 12, 49, 50). A weak germination response of B. thermoamylovorans spores was observed for nutrient triggers (Fig. 2). In addition, the germination of spores in response to addition of the nonnutrient germinant Ca-DPA was assessed; this type of germination does not require GRs (19). Exposure of spores to 40 mM Ca-DPA for 3 h resulted in very efficient spore germination for strains B4064 and B4065 (75.3% ± 7.2% and 95.7% ± 2.1% of germinated spores, respectively) and moderately efficient germination for strains B4166 and B4167 (32.8.7% ± 5.4% and 43.0% ± 9.4%, respectively) (Fig. 2). After 24 h of incubation, spore germination increased to 95.6% ± 3.0% and 97.6% ± 1.2% for strains B4064 and B4065, respectively, whereas it reached 49.7% ± 5.5% and 58.3% ± 5.6% for strains B4166 and B4167, respectively (Fig. 2). Altogether, these results indicate that Ca-DPA is an efficient germination trigger for B. thermoamylovorans spores, but the germination responses varied between the different isolates.
Genome mining for germination genes.
To explain the observed germination phenotypes, i.e., inefficient germination in response to nutrient triggers and different responses to Ca-DPA between strains, the presence of germination genes was evaluated in the genomes of the four B. thermoamylovorans isolates through genome mining (Table 1). In B. subtilis, nutrient-induced germination requires specific GRs that bind nutrient germinants (14, 16) and is facilitated specifically by several proteins, such as GerD (51), and GerPABCDEF (52). The analysis of the genomes of the four B. thermoamylovorans strains revealed the presence of GR genes which are deemed important for sensing nutrient germinants (Table 1), but despite their presence, only weak germination of spores was observed in the presence of rich media and various nutrients. The GR genes included two complete tricistronic operons, referred to further as ger(X1)ABC and ger(X2)ABC, both encoding putative GRs (Table 1 and Fig. 3). Both ger operons are predicted to be preceded by single [in the case of ger(X1)ABC] or double [in the case of ger(X2)ABC] binding sites for the sporulation sigma factor SigG (Fig. 3). In addition, the following genes encoding proteins that are expected to facilitate responses to nutrients (51–53) were found in the genomes of all four strains: gerD, gerF, gerPA, gerPB, gerPC, gerPD, gerPE, and gerPF (Table 1).
Genes other than the ones directly involved in sensing nutrients, but which play a role in subsequent germination events, were also found in the B. thermoamylovorans genomes (Table 1). These included the cwlJ, sleB, gerQ, and ypeB genes, which encode proteins that are important for lysis of the protective cortex layer, and nearly all of the spoVA genes (spoVAA, spoVAB, spoVAC, spoVAD, spoVAEb, and spoVAF), some of which encode proteins that are responsible for release of DPA from the spore core (11, 12). The germination gene spoVAEa was absent in the four sequenced B. thermoamylovorans strains, but SpoVAEa is considered to play only a minor role in germination (54). Interestingly, some spoVA genes, namely, spoVAC, spoVAD, and spoVAEb, occurred in multiple copies in the genomes of the sequenced strains of B. thermoamylovorans (Table 1). Thus, besides single spoVAA and spoVAB genes, all strains possessed three spoVAC and spoVAD genes as well as two spoVAEb and two spoVAF genes. The spoVA genes of B. thermoamylovorans were found in five different operons: (i) the spo(VA1) operon, comprising spoVAA-spoVAB-spoVAF; (ii) spo(VA2), consisting of the spoVAC-spoVAD genes; the (iii) spo(VA3) and (iv) spo(VA4) operons, both containing spoVAC-spoVAD-spoVAEb genes; and (v) spo(VA5), which comprises a single spoVAF gene (Table 1 and Fig. 3).
In B. subtilis, Ca-DPA initiates germination by direct activation of the cortex lytic enzyme CwlJ (19, 55), which requires GerQ for proper localization in the spore coat (56). Strains B4166 and B4167 contain a single cwlJ gene and gerQ gene, whereas strains B4064 and B4065 both carry two copies of cwlJ (further referred to as cwlJ and cwlJ2) and two copies of gerQ (referred to as gerQ and gerQ2) (Table 1). Both cwlJ and gerQ, as well as cwlJ2 and gerQ2, are adjacent to each other on the chromosome, possibly forming an operon preceded by the predicted SigE and SigK binding sites (Fig. 3). Pairwise amino acid alignments revealed 81% sequence identity between the CwlJ and CwlJ2 proteins. Moreover, both CwlJ and CwlJ2 contain the probable key catalytic glutamate 21 residue (E21) (see Fig. S1 in the supplemental material) (57). The same holds true for GerQ and GerQ2, which also exhibit high sequence identity (61%).
Spore heat inactivation and modeling.
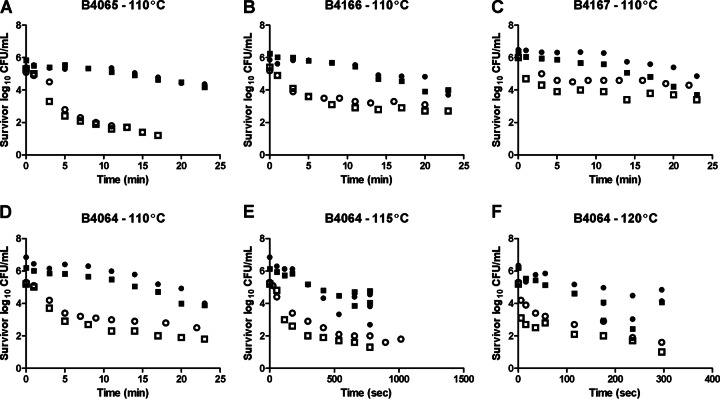
The heat inactivation of spores at 110°C was assessed for all strains, without a Ca-DPA treatment and with a Ca-DPA treatment to trigger nonnutrient germination before plating. The inactivation curve of spores that were not treated with Ca-DPA prior to plating showed biphasic behaviors with tailing, and data were fitted with a biphasic model from which the shoulder parameter was omitted (Fig. 4) (38). For these data sets, the D110°C values ranged from 0.7 (standard error, ±0.1) min to 3.3 ± 0.5 min for the heat-sensitive fraction of the spores and from 9.3 ± 1.9 min to 33.7 ± 2.0 min for the heat-resistant fraction (see Table S1 in the supplemental material). In contrast, tailing was not observed for any of the strains when spores were treated with Ca-DPA prior to dilution and plating. These inactivation curves were fitted with a log-linear model. For these data sets, the D110°C values ranged from 9.7 ± 0.5 min to 26.1 ± 3.2 min (see Table S1 in the supplemental material). For strain B4064, the inactivation kinetics were determined at 110°C and in addition also at 115°C and 120°C. This allowed for the determination of the z value (12.2 ± 1.8°C) and calculation of a Dref (Tref = 121.1°C) of 1.4 min (upper 95% prediction interval, 2.9 min) (see Table S1 in the supplemental material).
FIG 4.
(A to D) Spore heat inactivation plots of B. thermoamylovorans strains B4065 (A), B4166 (B), B4167 (C), and B4064 (D) at 110°C. (E and F) For strain B4064, this spore inactivation was additionally determined at 115°C (E) and 120°C (F). For all strains, two independent spore crops were exposed to a heat treatment, followed by exposure to Ca-DPA (40 mM for 3 h) or not before enumeration of survivors. The open circles and open squares correspond to spore crops 1 and 2, respectively, without Ca-DPA treatment. Closed circles and closed squares correspond to spore crops 1 and 2, respectively, with Ca-DPA treatment.
The viable spore counts that were obtained on plates following exposure to Ca-DPA always exceeded the viable spore counts that were obtained upon direct plating (without Ca-DPA exposure). This was the case for the initial spore count and the counts after heat treatment (Fig. 4). The maximum difference in viable spore numbers after heat exposure with and without exposure to Ca-DPA was 3.4 log units for strain B4065. Notably, the difference in viable spore counts between the two different methods was more prominent in strains B4064 and B4065 than in strains B4166 and B4167.
DISCUSSION
Spores of B. thermoamylovorans can pose problems in commercial sterile foods due to their high-level heat resistance and unpredictable germination. To improve our understanding of this problematic species and identify possible leads for spoilage control, we combined a phenotypic characterization of the germination behavior of four different food-related isolates with in silico analysis of their genome sequences (30). Based on our new insights into the germination properties of spores of this species, we subsequently determined heat resistance properties of spores of individual strains.
This study has shown that poor recovery of spores of B. thermoamylovorans on standard rich cultivation media leads to a significant underestimation of the spore load that is actually present: enumeration of spores in spore suspensions using flow cytometry and plating on BHI-A showed that only a few percent (1.3% to 5.3%) of B. thermoamylovorans spores formed colonies (Fig. 1). Increasing the activation temperature did not improve spore counts (data not shown). This low number of colonies on BHI plates resulted mainly from inefficient spore germination in response to nutrients, as only 2.6% to 13.6% of spores germinated in the BHI broth (Fig. 2). Notably, the germination of B. thermoamylovorans spores was also very limited in the presence of LB broth and a variety of tested nutrient germinants, including l-alanine, a combination of l-asparagine, d-glucose, d-fructose, KCl, and a mixture of 19 individual amino acids and inosine with or without d-fructose, d-glucose, and KCl (data not shown). Based on these observations, the absence of genes encoding one or more germination proteins would provide a plausible explanation for a weak germination response of B. thermoamylovorans spores to nutrients. In B. subtilis it is known that the initial stages of nutrient germination require at least one functional germinant receptor plus the GerD protein (51), and the germination process is facilitated by the GerP proteins (52). At a later stage, some of the SpoVA proteins enable release of Ca-DPA from the spore core to the environment, and finally, at least one of the two lytic enzymes, CwlJ or SleB, is required for hydrolysis of the spore protective cortex layer (11, 12). In silico analysis of the genome sequences of B. thermoamylovorans showed that all known germination-specific genes, with the only exception of spoVAEa, were present in the genomes of the four strains, some of them in multiple copies (Table 1).
Two tricistronic operons encoding putative GRs were found on the chromosome, as well as the gerD gene (Table 1), indicative of a potential of B. thermoamylovorans spores to respond to nutrient germination triggers. Despite the fact that one of the GR operons in strains B4064 and B4065 encoded proteins that belong to the same orthologous group as the B. subtilis GerK receptor subunits, spores of B. thermoamylovorans displayed very little or no response to a nutrient mixture known to specifically trigger GerK (namely, l-asparagine, fructose, glucose, and potassium ions). In B. subtilis, activation of the GerK receptor in response to this mixture is also linked with the GerB receptor (58); our analysis indicated that subunits of the GerB receptor are absent in B. thermoamylovorans, which may explain the lack of germination in response to this mixture. Also, it is known from the literature that even small changes in a GR subunit sequence can alter or abolish activity of the GR complex in response to certain nutrients (20, 59, 60). For B. thermoamylovorans, no specific response could be detected in any of the nutrient combinations tested. The genomic sequences of the germinant receptor operons showed intact genes with predicted binding sites for the SigG transcription factor, which typically regulates expression of GR genes (46), upstream of the tricistronic operons. Assuming that these genes are expressed during sporulation and that the GR proteins are functional in the spore, the specificity of the two GRs present in B. thermoamylovorans remains to be determined.
All other key genes related to germination, including the spoVA operon (required for release of Ca-DPA from the core upon germination in B. subtilis [11, 12]), as well as those encoding the cortex lytic enzymes, sleB and cwlJ, were found in all four strains (Table 1). The only gene that was missing was spoVAEa, but the absence of this gene is in fact not uncommon for spore-forming species of the Bacillales and Clostridiales orders (54). In B. subtilis, deletion of spoVAEa has been associated with a slower nutrient-induced germination phenotype (54), but this fairly moderate decrease does not fully explain the dramatic loss in germination efficiency in B. thermoamylovorans (Fig. 2). In contrast, some other spoVA genes, namely, spoVAC, spoVAD, and spoVAEb, were found in multiple copies in the genomes (Table 1). The impact of this duplication is so far unclear, although it may alter the release of Ca-DPA from the spore core upon germination. On the whole, the poor nutrient germination response of B. thermoamylovorans cannot be linked directly to absence of key germination genes. Other explanations for the observed inefficient germination may be a weak penetration of nutrients through the coat layers, poor binding of nutrients to the GRs, lack of GR functionality, or lack of adequate signal transduction downstream of the germination receptors (11, 12).
Interestingly, despite very weak germination responses to nutrients, spores of all four B. thermoamylovorans strains germinated well in response to a nonnutrient germinant, namely, exogenous Ca-DPA. Ca-DPA is known to directly activate the cortex lytic enzyme CwlJ (19), which requires the GerQ protein for localization in the spore coat (56). CwlJ and GerQ have been shown to be essential for Ca-DPA-induced germination in B. subtilis and B. megaterium (19, 56, 61). Assuming that the germination process of spores of B. thermoamylovorans is similar to that in B. subtilis, our results suggest that B. thermoamylovorans nutrient germination is impaired not at the stage of peptidoglycan degradation and downstream events but at the stage preceding cortex hydrolysis. However, a clear difference in germination efficiency in response to Ca-DPA was observed between the strains, with germination of spores of B4064 and B4065 being highly efficient and that of spores of B4166 and B4167 being moderately efficient.
Analysis of the four genomes revealed the presence of two cwlJ and gerQ genes in strains B4064 and B4065 and single cwlJ and gerQ genes in strains B4166 and B4167. CwlJ and CwlJ2 on the one hand and GerQ and GerQ2 on the other hand displayed high amino acid sequence similarity (see Fig. S1 in the supplemental material), suggesting that both copies of each protein potentially play similar or identical roles in spore germination of strains B4064 and B4065. Spores of B4064 and B4065 showed higher germination efficiencies in response to Ca-DPA than spores of B4166 and B4167 (Fig. 2), but a direct link between the higher Ca-DPA germination efficiency in strains B4064 and B4065 than in strains B4166 and B4167 and the presence of two cwlJ and gerQ genes remains to be established.
Limited germination in response to nutrients has implications for counts obtained using standard plating techniques on rich media, as colony formation from single spores relies on efficient germination of spores and subsequent outgrowth. We demonstrated that enumeration on BHI plates strongly underestimates the number of viable spores. More efficient germination was observed following nonnutrient germination in response to Ca-DPA. To establish heat resistance, spores were subjected to heat treatments at 110°C and plated directly or after a Ca-DPA treatment. For all four strains, much higher recoveries were observed upon Ca-DPA exposure than after direct plating (up to 3.4-log-higher counts for spores of strain B4065), and tailing effects were absent. Heating was also performed at 115°C and 120°C for strain B4064, and at these temperatures, similar effects were observed (see Fig. 4).
Interestingly, the differences in viable spore counts with or without Ca-DPA exposure prior to plating were more prominent for strains B4064 and B4065 than for spores of strains B4166 and B4167. The latter two strains harbor only a single copy of the cwlJ and gerQ genes, and their spores showed less efficient germination with Ca-DPA than spores of strains B4064 and B4065, each harboring two cwlJ and gerQ genes (Table 1 and Fig. 2). Following heating at 110°C, the differences in recoveries with and without Ca-DPA treatments were less prominent for the strains harboring the single cwlJ and gerQ copies, which is likely due to the fact that spore germination was not complete for these spores, even following Ca-DPA exposure (Fig. 4). Even for spores that germinated best in response to Ca-DPA, germination was not 100% after incubation for 3 h with Ca-DPA (Fig. 2), indicating that counts on plates might still be underestimated.
B. thermoamylovorans was shown to produce highly heat-resistant spores compared to those of other spore-forming Bacillus spp. The decimal reduction times at 120°C (D120°C) were 1.9 ± 0.3 min and 1.3 ± 0.1 min for two independent spore crops of strain B4064 (see Table S1 in the supplemental material) obtained with an additional Ca-DPA treatment prior to plating. This is comparable to reported D values of B. subtilis strain A163, which is known to produce highly heat resistant spores (D120°C of 1.8 ± 0.1 min and 1.6 ± 0.1 min) (31). The spore heat resistance of the B. thermoamylovorans strains is only slightly lower than that of B. sporothermodurans, which is known to survive UHT processing and has reported D120°C values of 2.25 min (62). When spores of strain B4064 were directly plated on BHI, a heat resistant fraction (tailing) was observed, with D120°C values of 2.9 ± 0.3 min and 2.7 ± 0.5 min (see Table S1 in the supplemental material). The calculated D140°C for spores of strain B4064, based on the inactivation data obtained after plating preceded by a Ca-DPA treatment, was 2.3 s (upper 95% PI, 5.0 s), which is very high but still slightly below that of B. sporothermodurans spores, with reported D140°C values of 4.7 s and 5.0 s (2, 62). When comparing the heat resistance of spores of B. thermoamylovorans with that of spores of Geobacillus stearothermophilus, the Dref of 1.4 min calculated for strain B4064 is lower than the D121.1°C of 3.3 min that has been reported for G. stearothermophilus, based on literature data of 430 D values of this species (28).
Based on the data obtained in this study, it can be concluded that the spores of B. thermoamylovorans are highly resistant and are potentially able to survive UHT treatments. When conventional plating techniques are used to determine the initial spore concentration and to estimate spore heat resistance, it is likely that predictions are not accurate, especially for noncharacterized species and strains. The lack of efficient nutrient germination of spores can lead to strong underestimations of counts, both of initial levels and of surviving spores after a heat treatment. When applied in a food processing setting, such large underestimations of the initial spore concentration can have detrimental effects on the safety boundaries of such processes.
In summary, we have demonstrated that spores of B. thermoamylovorans do not germinate efficiently upon nutrient-induced germination, despite the presence of the genes encoding two GRs. Spore germination was triggered upon exposure to Ca-DPA. Our results clearly show the importance of determining spore germination and outgrowth conditions prior to characterization of spore properties, including heat resistance, to avoid strong underestimation of viable spores that fail to germinate in response to regular nutrient germinants. The improved estimations of spore heat resistance obtained in this study will aid efforts in the food processing environment toward better control of spores of B. thermoamylovorans and ensuring sterility of food products.
Supplementary Material
ACKNOWLEDGMENTS
We declare that no competing interests exist.
This research was funded by TI Food and Nutrition, a public-private partnership on precompetitive research in food and nutrition.
The funding organization had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Esmée Janssen and Gerwin Kamstra for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01993-15.
REFERENCES
- 1.Brul S, van Beilen J, Caspers M, O'Brien A, de Koster C, Oomes S, Smelt J, Kort R, Ter Beek A. 2011. Challenges and advances in systems biology analysis of Bacillus spore physiology; molecular differences between an extreme heat resistant spore forming Bacillus subtilis food isolate and a laboratory strain. Food Microbiol 28:221–227. doi: 10.1016/j.fm.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 2.Scheldeman P, Herman L, Foster S, Heyndrickx M. 2006. Bacillus sporothermodurans and other highly heat-resistant spore formers in milk. J Appl Microbiol 101:542–555. doi: 10.1111/j.1365-2672.2006.02964.x. [DOI] [PubMed] [Google Scholar]
- 3.Scheldeman P, Pil A, Herman L, De Vos P, Heyndrickx M. 2005. Incidence and diversity of potentially highly heat-resistant spores isolated at dairy farms. Appl Environ Microbiol 71:1480–1494. doi: 10.1128/AEM.71.3.1480-1494.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Witthuhn M, Lucking G, Atamer Z, Ehling-Schulz M, Hinrichs J. 2011. Thermal resistance of aerobic spore formers isolated from food products. Int J Dairy Technol 64:486–493. doi: 10.1111/j.1471-0307.2011.00706.x. [DOI] [Google Scholar]
- 5.Combet-Blanc Y, Ollivier B, Streicher C, Patel BKC, Dwivedi PP, Pot B, Prensier G, Garcia J-L. 1995. Bacillus thermoamylovorans sp. nov., a moderately thermophilic and amylolytic bacterium. Int J Syst Bacteriol 45:9–16. doi: 10.1099/00207713-45-1-9. [DOI] [PubMed] [Google Scholar]
- 6.Koeck DE, Wibberg D, Maus I, Winkler A, Albersmeier A, Zverlov VV, Pühler A, Schwarz WH, Liebl W, Schlüter A. 2014. First draft genome sequence of the amylolytic Bacillus thermoamylovorans wild-type strain 1A1 isolated from a thermophilic biogas plant. J Biotechnol 192A:154–155. [DOI] [PubMed] [Google Scholar]
- 7.Combet-Blanc Y, Dieng MC, Kergoat PY. 1999. Effect of organic complex compounds on Bacillus thermoamylovorans growth and glucose fermentation. Appl Environ Microbiol 65:4582–4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coorevits A, Logan NA, Dinsdale AE, Halket G, Scheldeman P, Heyndrickx M, Schumann P, Van Landschoot A, De Vos P. 2011. Bacillus thermolactis sp. nov., isolated from dairy farms, and emended description of Bacillus thermoamylovorans. Int J Syst Evol Microbiol 61:1954–1961. doi: 10.1099/ijs.0.024240-0. [DOI] [PubMed] [Google Scholar]
- 9.De Clerck E, Vanhoutte T, Hebb T, Geerinck J, Devos J, De Vos P. 2004. Isolation, characterization, and identification of bacterial contaminants in semifinal gelatin extracts. Appl Environ Microbiol 70:3664–3672. doi: 10.1128/AEM.70.6.3664-3672.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moir A. 2006. How do spores germinate? J Appl Microbiol 101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 11.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Setlow P. 2014. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol 196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Setlow P. 2003. Spore germination. Curr Opin Microbiol 6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Paidhungat M, Setlow P. 2000. Role of Ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis. J Bacteriol 182:2513–2519. doi: 10.1128/JB.182.9.2513-2519.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramirez-Peralta A, Gupta S, Butzin XY, Setlow B, Korza G, Leyva-Vazquez M-A, Christie G, Setlow P. 2013. Identification of new proteins that modulate the germination of spores of Bacillus species. J Bacteriol 195:3009–3021. doi: 10.1128/JB.00257-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ross C, Abel-Santos E. 2010. The Ger receptor family from sporulating bacteria. Curr Issues Mol Biol 12:147–158. [PMC free article] [PubMed] [Google Scholar]
- 17.Stewart K-AV, Setlow P. 2013. Numbers of individual nutrient germinant receptors and other germination proteins in spores of Bacillus subtilis. J Bacteriol 195:3575–3582. doi: 10.1128/JB.00377-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keynan A, Murrell WG, Halvorson HO. 1962. Germination properties of spores with low dipicolinic acid content. J Bacteriol 83:395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paidhungat M, Ragkousi K, Setlow P. 2001. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca2+-dipicolinate. J Bacteriol 183:4886–4893. doi: 10.1128/JB.183.16.4886-4893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christie G, Lowe CR. 2008. Amino acid substitutions in transmembrane domains 9 and 10 of GerVB that affect the germination properties of Bacillus megaterium spores. J Bacteriol 190:8009–8017. doi: 10.1128/JB.01073-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chirakkal H, O'Rourke M, Atrih A, Foster SJ, Moir A. 2002. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology 148:2383–2392. doi: 10.1099/00221287-148-8-2383. [DOI] [PubMed] [Google Scholar]
- 22.Eijlander RT, Abee T, Kuipers OP. 2011. Bacterial spores in food: how phenotypic variability complicates prediction of spore properties and bacterial behavior. Curr Opin Biotechnol 22:180–186. doi: 10.1016/j.copbio.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 23.Ghosh S, Setlow P. 2009. Isolation and characterization of superdormant spores of Bacillus species. J Bacteriol 191:1787–1797. doi: 10.1128/JB.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh S, Setlow P. 2010. The preparation, germination properties and stability of superdormant spores of Bacillus cereus. J Appl Microbiol 108:582–590. doi: 10.1111/j.1365-2672.2009.04442.x. [DOI] [PubMed] [Google Scholar]
- 25.Perez-Valdespino A, Ghosh S, Cammett EP, Kong L, Li YQ, Setlow P. 2013. Isolation and characterization of Bacillus subtilis spores that are superdormant for germination with dodecylamine or Ca2+-dipicolinic acid. J Appl Microbiol 114:1109–1119. doi: 10.1111/jam.12125. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Palacios A, Lejeune JT. 2011. Moist-heat resistance, spore aging, and superdormancy in Clostridium difficile. Appl Environ Microbiol 77:3085–3091. doi: 10.1128/AEM.01589-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh S, Scotland M, Setlow P. 2012. Levels of germination proteins in dormant and superdormant spores of Bacillus subtilis. J Bacteriol 194:2221–2227. doi: 10.1128/JB.00151-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rigaux C, Denis JB, Albert I, Carlin F. 2013. A meta-analysis accounting for sources of variability to estimate heat resistance reference parameters of bacteria using hierarchical Bayesian modeling: estimation of D at 121.1 degrees C and pH 7, zT and zpH of Geobacillus stearothermophilus. Int J Food Microbiol 161:112–120. doi: 10.1016/j.ijfoodmicro.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Remenant B, Jaffrès E, Dousset X, Pilet M-F, Zagorec M. 2015. Bacterial spoilers of food: behavior, fitness and functional properties. Food Microbiol 45A:45–53. [DOI] [PubMed] [Google Scholar]
- 30.Krawczyk AO, Berendsen EM, Eijlander RT, de Jong A, Wells-Bennik MHJ, Kuipers OP. 2015. Draft genome sequences of four Bacillus thermoamylovorans strains isolated from milk and acacia gum, a food ingredient. Genome Announc 3:e00165–15. doi: 10.1128/genomeA.00165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berendsen EM, Zwietering MH, Kuipers OP, Wells-Bennik MHJ. 2015. Two distinct groups within the Bacillus subtilis group display significantly different spore heat resistance properties. Food Microbiol 45A:18–25. [DOI] [PubMed] [Google Scholar]
- 32.Schaeffer P, Millet J, Aubert JP. 1965. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A 54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gasol JM, Del Giorgio PA. 2000. Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar 64:197–224. [Google Scholar]
- 34.Hu H, Emerson J, Aronson AI. 2007. Factors involved in the germination and inactivation of Bacillus anthracis spores in murine primary macrophages. FEMS Microbiol Lett 272:245–250. doi: 10.1111/j.1574-6968.2007.00766.x. [DOI] [PubMed] [Google Scholar]
- 35.Smelt JPPM, Bos AP, Kort R, Brul S. 2008. Modelling the effect of sub(lethal) heat treatment of Bacillus subtilis spores on germination rate and outgrowth to exponentially growing vegetative cells. Int J Food Microbiol 128:34–40. doi: 10.1016/j.ijfoodmicro.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 36.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez J-Y, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu S, Labuza TP, Diez-Gonzalez F. 2006. Thermal inactivation of Bacillus anthracis spores in cow's milk. Appl Environ Microbiol 72:4479–4483. doi: 10.1128/AEM.00096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geeraerd AH, Valdramidis VP, Van Impe JF. 2006. GInaFiT, a freeware tool to assess non-log-linear microbial survivor curves. Int J Food Microbiol 110:297 (Erratum.) doi: 10.1016/j.ijfoodmicro.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 39.van Asselt ED, Zwietering MH. 2006. A systematic approach to determine global thermal inactivation parameters for various food pathogens. Int J Food Microbiol 107:73–82. doi: 10.1016/j.ijfoodmicro.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Stoeckert CJ Jr, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson LS, Eddy S, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 44.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eijlander RT, de Jong A, Krawczyk AO, Holsappel S, Kuipers OP. 2014. SporeWeb: an interactive journey through the complete sporulation cycle of Bacillus subtilis. Nucleic Acids Res 42:D685–D691. doi: 10.1093/nar/gkt1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Makita Y, Nakao M, Ogasawara N, Nakai K. 2004. DBTBS: database of transcriptional regulation in Bacillus subtilis and its contribution to comparative genomics. Nucleic Acids Res 32:D75–D77. doi: 10.1093/nar/gkh074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baerends R, Smits W, de Jong A, Hamoen L, Kok J, Kuipers O. 2004. Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol. 5:R37. doi: 10.1186/gb-2004-5-5-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Black EP, Wei J, Atluri S, Cortezzo DE, Koziol-Dube K, Hoover DG, Setlow P. 2007. Analysis of factors influencing the rate of germination of spores of Bacillus subtilis by very high pressure. J Appl Microbiol 102:65–76. doi: 10.1111/j.1365-2672.2006.03062.x. [DOI] [PubMed] [Google Scholar]
- 50.Doona CJ, Ghosh S, Feeherry FF, Ramirez-Peralta A, Huang Y, Chen H, Setlow P. 2014. High pressure germination of Bacillus subtilis spores with alterations in levels and types of germination proteins. J Appl Microbiol 117:711–720. doi: 10.1111/jam.12557. [DOI] [PubMed] [Google Scholar]
- 51.Pelczar PL, Igarashi T, Setlow B, Setlow P. 2007. Role of GerD in germination of Bacillus subtilis Spores. J Bacteriol 189:1090–1098. doi: 10.1128/JB.01606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Butzin XY, Troiano AJ, Coleman WH, Griffiths KK, Doona CJ, Feeherry FE, Wang G, Li Y-Q, Setlow P. 2012. Analysis of the effects of a gerP mutation on the germination of spores of Bacillus subtilis. J Bacteriol 194:5749–5758. doi: 10.1128/JB.01276-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Igarashi T, Setlow B, Paidhungat M, Setlow P. 2004. Effects of a gerF (lgt) mutation on the germination of spores of Bacillus subtilis. J Bacteriol 186:2984–2991. doi: 10.1128/JB.186.10.2984-2991.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Valdespino A, Li Y, Setlow B, Ghosh S, Pan D, Korza G, Feeherry FE, Doona CJ, Li Y-Q, Hao B, Setlow P. 2014. Function of the SpoVAEa and SpoVAF proteins of Bacillus subtilis spores. J Bacteriol 196:2077–2088. doi: 10.1128/JB.01546-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magge A, Granger AC, Wahome PG, Setlow B, Vepachedu VR, Loshon CA, Peng L, Chen D, Li Y-Q, Setlow P. 2008. Role of dipicolinic acid in the germination, stability, and viability of spores of Bacillus subtilis. J Bacteriol 190:4798–4807. doi: 10.1128/JB.00477-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ragkousi K, Eichenberger P, van Ooij C, Setlow P. 2003. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca2+-dipicolinate. J Bacteriol 185:2315–2329. doi: 10.1128/JB.185.7.2315-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Y, Butzin XY, Davis A, Setlow B, Korza G, Üstok FI, Christie G, Setlow P, Hao B. 2013. Activity and regulation of various forms of CwlJ, SleB, and YpeB proteins in degrading cortex peptidoglycan of spores of Bacillus species in vitro and during spore germination. J Bacteriol 195:2530–2540. doi: 10.1128/JB.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Atluri S, Ragkousi K, Cortezzo DE, Setlow P. 2006. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J Bacteriol 188:28–36. doi: 10.1128/JB.188.1.28-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cooper GR, Moir A. 2011. Amino acid residues in the GerAB protein important in the function and assembly of the alanine spore germination receptor of Bacillus subtilis 168. J Bacteriol 193:2261–2267. doi: 10.1128/JB.01397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Y, Catta P, Stewart K-AV, Dufner M, Setlow P, Hao B. 2011. Structure-based functional studies of the effects of amino acid substitutions in GerBC, the C subunit of the Bacillus subtilis GerB spore germinant receptor. J Bacteriol 193:4143–4152. doi: 10.1128/JB.05247-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Setlow B, Peng L, Loshon CA, Li YQ, Christie G, Setlow P. 2009. Characterization of the germination of Bacillus megaterium spores lacking enzymes that degrade the spore cortex. J Appl Microbiol 107:318–328. doi: 10.1111/j.1365-2672.2009.04210.x. [DOI] [PubMed] [Google Scholar]
- 62.Huemer IA, Klijn N, Vogelsang HWJ, Langeveld LPM. 1998. Thermal death kinetics of spores of Bacillus sporothermodurans isolated from UHT milk. Int Dairy J 8:851–855. doi: 10.1016/S0958-6946(98)00129-0. [DOI] [Google Scholar]
- 63.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. 2011. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol Microbiol 81:1061–1077. doi: 10.1111/j.1365-2958.2011.07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Slynn GM, Sammons RL, Smith DA, Moir A, Corfe BM. 1994. Molecular genetical and phenotypical analysis of the gerM spore germination gene of Bacillus subtilis 168. FEMS Microbiology Lett 121:315–320. doi: 10.1111/j.1574-6968.1994.tb07119.x. [DOI] [PubMed] [Google Scholar]
- 65.Sussman MD, Setlow P. 1991. Cloning, nucleotide sequence, and regulation of the Bacillus subtilis gpr gene, which codes for the protease that initiates degradation of small, acid-soluble proteins during spore germination. J Bacteriol 173:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.