Abstract
Arabinoxylan oligosaccharides (AXOS) are a promising class of prebiotics that have the potential to stimulate the growth of bifidobacteria and the production of butyrate in the human colon, known as the bifidogenic and butyrogenic effects, respectively. Although these dual effects of AXOS are considered beneficial for human health, their underlying mechanisms are still far from being understood. Therefore, this study investigated the metabolic interactions between Bifidobacterium longum subsp. longum NCC2705 (B. longum NCC2705), an acetate producer and arabinose substituent degrader of AXOS, and Eubacterium rectale ATCC 33656, an acetate-converting butyrate producer. Both strains belong to prevalent species of the human colon microbiota. The strains were grown on AXOS during mono- and coculture fermentations, and their growth, AXOS consumption, metabolite production, and expression of key genes were monitored. The results showed that the growth of both strains and gene expression in both strains were affected by cocultivation and that these effects could be linked to changes in carbohydrate consumption and concomitant metabolite production. The consumption of the arabinose substituents of AXOS by B. longum NCC2705 with the concomitant production of acetate allowed E. rectale ATCC 33656 to produce butyrate (by means of a butyryl coenzyme A [CoA]:acetate CoA-transferase), explaining the butyrogenic effect of AXOS. Eubacterium rectale ATCC 33656 released xylose from the AXOS substrate, which favored the B. longum NCC2705 production of acetate, explaining the bifidogenic effect of AXOS. Hence, those interactions represent mutual cross-feeding mechanisms that favor the coexistence of bifidobacterial strains and butyrate producers in the same ecological niche. In conclusion, this study provides new insights into the bifidogenic and butyrogenic effects of AXOS.
INTRODUCTION
Fermentation in the human colon is carried out by trillions of bacteria that contribute not only to health and well-being but also to disease (1–4). For instance, a decrease in the relative abundance of bifidobacteria in the human colon has been associated with antibiotic-associated diarrhea, irritable bowel syndrome, inflammatory bowel disease, allergies, and regressive autism (5, 6). Although it is currently not yet clear whether changes in microbial composition are a cause or a consequence of these disorders, there are indications that metabolites, in particular, the short-chain fatty acids (SCFAs), such as acetate, butyrate, and propionate, that are produced during fermentation in the colon play an important role in intestinal homeostasis (3, 7). Among the SCFAs produced in the human colon, butyrate has drawn the most attention, as it is an essential energy source for the colon epithelial cells and has a protective effect against inflammatory bowel disease and colon cancer (3, 8, 9). Most butyrate producers belong to the Firmicutes phylum and use a butyryl coenzyme A (CoA):acetate CoA-transferase in the final step of butyrate biosynthesis, which involves the net consumption of external acetate as a cosubstrate (10–12). This enzyme is present in the most abundant bacteria in the human colon, such as Faecalibacterium prausnitzii (up to 14% of the total fecal microbiota), Eubacterium rectale (up to 13%), and other butyrate producers, such as Roseburia spp., Anaerostipes caccae, and Eubacterium hallii (10–15).
One way to influence the activity and composition of the colon microbiota is by consuming nondigestible food compounds, termed prebiotics, such as inulin-type fructans (ITF), xylo-oligosaccharides (XOS), arabinoxylans (AX), and arabinoxylan oligosaccharides (AXOS) (12, 16, 17). AX and their hydrolysis products, AXOS, are a promising class of prebiotics, as they have the potential to stimulate bifidobacteria, as ITF do, a so-called bifidogenic effect (12, 18–24). Also, like ITF, they seem to stimulate the production of butyrate and an increase in the relative abundance of Eubacterium spp., Roseburia spp., and F. prausnitzii, a so-called butyrogenic effect (12, 18, 19, 21, 25–29).
Although the bifidogenic and butyrogenic effects of AX and AXOS are considered to be beneficial for human health, their overall underlying mechanisms and microbial backgrounds are still largely unknown. It is, however, probable that cross-feeding mechanisms like those that take place with ITF occur, in which case primary degraders such as bifidobacteria selectively and competitively degrade these fructose polymers to produce acetate and lactate, which are consumed by secondary degraders, such as roseburias, which produce butyrate (12, 30, 31). Recently, it has been suggested that the bifidogenic effect of AXOS is the result of specialization in the AXOS degradation mechanisms of bifidobacterial strains through complementary activities to avoid competition with each other and, hence, to be able to degrade these complex oligosaccharides by cooperation (12, 32). Several bifidobacterial strains have been grouped into five clusters according to the differences in their AXOS consumption patterns. Some strains prefer monosaccharides, while others consume the arabinose substituents of AXOS, whether or not this is followed by the consumption of the xylose backbones, either up to xylotetraose or longer xylose backbones and either intracellularly or extracellularly (32). However, it is currently not yet known if butyrate-producing colon bacteria can consume AX and AXOS and/or if AX- and AXOS-degrading colon bacteria participate in cross-feedings for butyrate production. For instance, in the case of ITF, colonic bacteria other than bifidobacteria may consume these fructose polymers either directly or through cross-feeding (10, 30).
This study aimed to provide an in-depth understanding of the metabolic interactions between Bifidobacterium longum subsp. longum NCC2705 (hereafter B. longum NCC2705), an acetate producer and arabinose substituent degrader of AXOS (32), and Eubacterium rectale ATCC 33656, an acetate-converting butyrate producer, during growth on AXOS. In particular, reverse transcription-quantitative PCR (RT-qPCR) was used to assess the differential expression of genes related to AXOS consumption, metabolite production, and potential associated stress and cellular responses between mono- and cocultures of both strains during various stages of their growth.
MATERIALS AND METHODS
Strains and media.
B. longum NCC2705, an isolate from infant feces kindly provided by F. Arigoni (Nestlé Research Center, Lausanne, Switzerland) and a representative of cluster II AXOS-degrading bifidobacterial strains, i.e., strains that consume the arabinose substituents of AXOS (32), was used throughout this study, as its whole-genome sequence has been annotated (33) and most of its annotated AX- and AXOS-degrading enzymes have been experimentally characterized (34). Moreover, this species is one of the dominant bifidobacterial species of the human gut microbiota (12). Eubacterium rectale ATCC 33656, an isolate from human feces purchased from the American Type Culture Collection (ATCC; Manassas, VA), was chosen as an abundant butyrate-producing strain with an annotated whole-genome sequence (35). Stock cultures were stored at −80°C in reinforced clostridial medium (RCM; Oxoid, Basingstoke, Hampshire, United Kingdom) supplemented with 25% (vol vol−1) glycerol (EMD Chemicals, Gibbstown, NJ) as a cryoprotectant. The authenticity of the strains was confirmed through 16S rRNA gene sequencing using the primers 63F and 1378R (36).
A medium for colon bacteria (MCB) (37) was used for the fermentation experiments. MCB was composed of the following compounds: Bacto peptone (BD Biosciences, Mississauga, ON, Canada), 6.5 g liter−1; Bacto soytone (Difco Laboratories, Detroit, MI), 5.0 g liter−1; yeast extract (BD Biosciences), 3.0 g liter−1; tryptone (BD Biosciences), 2.5 g liter−1; NaCl (Fisher Scientific, Fair Lawn, NJ), 4.5 g liter−1; K2HPO4 (J. T. Baker Chemical, Phillipsburg, NJ), 0.45 g liter−1; KH2PO4 (BDH, Toronto, ON, Canada), 0.45 g liter−1; MgSO4·7H2O (Fisher Scientific), 0.09 g liter−1; CaCl2 (Fisher Scientific), 0.07 g liter−1; cysteine-HCl (Acros Organics, Morris Plains, NJ), 0.4 g liter−1; NaHCO3 (BDH), 0.2 g liter−1; MnSO4·H2O (Fisher Scientific), 0.05 g liter−1; FeSO4·7H2O (Fisher Scientific), 0.005 g liter−1; ZnSO4·7H2O (Sigma-Aldrich, Oakville, ON, Canada), 0.005 g liter−1; hemin (Sigma-Aldrich), 0.005 g liter−1; menadione (Sigma-Aldrich), 0.005 g liter−1; Tween 80 (Sigma-Aldrich), 2.0 ml liter−1; and resazurin (0.1-g liter−1 solution; Acros Organics), 10.0 ml liter−1. In the case of monoculture fermentations with E. rectale ATCC 33656, MCB was supplemented with 4.0 g liter−1 of sodium acetate (Calbiochem, Darmstadt, Germany), as acetate is an indispensable cosubstrate for its growth (30, 31). This medium is further referred to as modified MCB (mMCB). MCB and mMCB supplemented with 1.0 g liter−1 of arabinose (Sigma-Aldrich) and 4.0 g liter−1 of xylose (Sigma-Aldrich) as the sole added energy sources, to mimic the typical arabinose/xylose ratio of 0.25 of AXOS (further referred to as MCB-AraXyl and mMCB-AraXyl, respectively), were used for inoculum buildup. All compounds were dissolved in demineralized water, and the medium was adjusted to pH 6.3 before sterilization. Wheat AXOS-5-0.27 (Fugeia, Leuven, Belgium) was used as the sole energy source and was added at a final concentration of 5.0 g liter−1 to assess its fermentation. AXOS-5-0.27 has an average degree of polymerization of 5, has an arabinose/xylose ratio of 0.27, and contains 1% (mass mass−1) ferulic acid. As it has previously been shown that AXOS-5-0.27 contains XOS as well (32, 38), the complete substrate is further referred to as (A)XOS, and the individual components are referred to as AXOS and XOS(A)XOS.
Mono- and coculture fermentation experiments.
Fermentations were carried out in 1-liter fermentors (système automatisé Biogénie quadruple; Biogénie, Quebec, QC, Canada) containing MCB or mMCB for the monoculture fermentation experiments with B. longum NCC2705 and E. rectale ATCC 33656, respectively, and containing MCB for the coculture fermentation experiments with both strains. After sterilization at 121°C with 1.1 × 105 Pa of overpressure for 20 min in an autoclave, the fermentors were put under anaerobic conditions by continuously sparging them with a mixture of 90% N2 and 10% CO2 (Praxair, Quebec, QC, Canada) in the case of monoculture fermentations with B. longum NCC2705 or with 100% N2 (Praxair) in the case of monoculture fermentations with E. rectale ATCC 33656 and coculture fermentations with both strains. This was followed by aseptic addition of the (A)XOS stock solutions that were filter sterilized (MediaKap-50 Plus high-performance hollow fiber membrane filters; pore size, 0.2 μm; Microgon, Rancho Dominguez, CA). The temperature, pH, and agitation of the fermentors were controlled online and kept constant at 37°C, pH 6.3 by automatic addition of a 5.0 M NaOH (EMD Chemicals) solution, and 100 rpm, respectively. The inocula (5.0% [vol vol−1]) were prepared through three subcultures. The stock culture cells of B. longum NCC2705 and E. rectale ATCC 33656 were first inoculated into 10 ml of RCM and incubated anaerobically in a glove box anaerobic chamber (Plas-Labs, Lansing, MI) containing an atmosphere of 80% N2, 10% H2, and 10% CO2 (Praxair) at 37°C for 12 h and 24 h, respectively. Next, 5.0% (vol vol−1) of each subculture was grown in 100 ml of MCB-AraXyl (B. longum NCC2705) or mMCB-AraXyl (E. rectale ATCC 33656), followed by anaerobic incubation at 37°C for 12 h. Subsequently, a third subculture of 5.0% (vol vol−1) was inoculated into 100 ml of the same medium and was incubated at 37°C for an additional 12 h.
All fermentations were performed in triplicate (these are further referred to as biological repetitions). Samples were taken after 0, 3, 6, 9, 12, 15, 24, 30, and 48 h of fermentation. Samples selected for metabolite analyses were immediately centrifuged at 5,000 × g for 15 min at 4°C and stored at −20°C until analysis.
Monitoring of bacterial growth. (i) OD measurements.
The optical density (OD) at 600 nm (OD600) was determined with a visible light spectrophotometer (Genesys 20; Thermo Scientific, Waltham, MA). Ultrapure water was used as a blank. Samples were diluted with water until the optical density was less than 0.3 unit. The resultant reading was multiplied by the dilution factor to give the correct population density, expressed in OD600 units. Each measurement was performed in triplicate. Averages and standard errors were calculated on the basis of the three biological repetitions.
(ii) Plate count determinations.
To determine the viable (cultivable) bacterial counts, expressed as the number of CFU per milliliter, samples were immediately diluted 10-fold in sterile physiological saline (0.85% [mass vol−1] NaCl). Platings were performed on RCM agar (1.5% [mass vol−1] agar) in triplicate. The plates were incubated at 37°C under anaerobic conditions (glove box anaerobic chamber) for 24 h. Averages and standard errors were calculated on the basis of the three biological repetitions. As E. rectale ATCC 33656 was very oxygen sensitive, plate counts were determined only for B. longum NCC2705.
(iii) qPCR and PMA-qPCR analyses.
To determine the concentrations of both the dead and viable (cultivable and noncultivable) cells of B. longum NCC2705 and E. rectale ATCC 33656, strain-specific quantitative PCR (qPCR) experiments targeting the elongation factor Tu (tuf) gene were performed. Both strains contained only one copy of this gene in their genomes (39), allowing the determination of their cell concentrations by quantification of the tuf copy number.
To distinguish viable and dead cells, qPCR in combination with propidium monoazide (PMA) (PMA-qPCR) was used (40). Therefore, samples were immediately diluted 1:10 with sterile peptone water (1 g liter−1 of Bacto peptone, 0.5 g liter−1 of cysteine-HCl, pH 6.8) and microcentrifuged at 15,000 × g for 15 min at 4°C. In the case of PMA-qPCR, the samples were first treated with 50 μM PMA (Biotium, Hayward, CA) and shaken in the dark for 5 min at room temperature. Next, the PMA-treated cell suspensions were placed in an LED-Active Blue system (Ingenia Biosystems, Barcelona, Spain) for 15 min, followed by microcentrifugation at 15,000 × g for 15 min at 4°C. Cell pellets were stored at −80°C until DNA extraction.
Genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Mississauga, ON, Canada) according to the manufacturer's protocol for Gram-positive bacteria, with minor modifications; i.e., 2.5 μl ml−1 of mutanolysin (20 U ml−1; Sigma-Aldrich) was added to the lysis buffer, and an additional incubation with DNase-free RNase (Roche, Laval, QC, Canada) was performed at 37°C for 60 min. The extracted genomic DNA was stored at −20°C until qPCR amplification.
Primers (see Table S1 in the supplemental material) were designed using Geneious Pro R6 software (Biomatters, San Francisco, CA). The presence of hairpins, self-dimers, and dimer pairs was checked using the same software program. The primers were synthesized by Integrated DNA Technologies (Coralville, IA). The gene and strain specificity of the primers was confirmed in silico by Primer-BLAST analysis (41) and in vitro by qPCR analysis of genomic DNA of both strains (see Table S1 in the supplemental material). qPCR was performed using a 7500 Fast real-time PCR system (Applied Biosystems, Carlsbad, CA) equipped with 96-well plates. Each PCR assay mixture of 20 μl contained 10 μl of Fast SYBR green master mix (Life Technologies, Burlington, ON, Canada), 1 μl of the extracted genomic DNA, 1 μl of each primer at its optimal concentration (see Table S1 in the supplemental material), and 8 μl of sterile nuclease-free water (Qiagen). The qPCR amplification program consisted of an initial denaturation step at 95°C for 20 s, followed by 40 two-step cycles at 95°C for 3 s and at 60°C for 30 s. In each run, negative controls (without genomic DNA) and positive controls (with genomic DNA extracted from B. longum NCC2705 or E. rectale ATCC 33656) were included for the two primer sets used. At the end of each run, a melting curve analysis was performed to check for the presence of nonspecific product formation and primer dimers. The cycle threshold (CT) values were determined using 7500 software (v2.0.6; Applied Biosystems), thereby averaging the automatically determined thresholds for each primer set over all runs performed and using this average during a reanalysis of all qPCR runs. All qPCR assays were performed in triplicate.
Finally, the tuf copy numbers per milliliter were calculated on the basis of the CT values obtained, using the corresponding standard curve. Averages and standard errors were calculated on the basis of the three biological repetitions. The standard curves for the tuf genes of B. longum NCC2705 and E. rectale ATCC 33656 were constructed according to a method previously described (42). Briefly, this method involves a PCR assay (see Table S2 in the supplemental material), using a TProfessional Basic gradient thermocycler (Biometra, Montreal, QC, Canada), with primers binding to sequences up- and downstream of the qPCR primer-binding sites of the tuf gene to obtain PCR amplicons containing the target sequence, followed by a subsequent cleanup (QIAquick PCR purification kit; Qiagen) of the amplification product to create qPCR standards. After performing a 10-fold serial dilution of the qPCR standards and a qPCR assay with the primers described above (see Table S1 in the supplemental material), a standard curve was made by plotting the CT values obtained from the serial dilutions of the qPCR standards as a linear function of the logarithm of the calculated copy numbers per milliliter (equation 1):
| (1) |
where the concentration of the qPCR standard is in grams per milliliter, 6.022 × 1023 is the number of molecules per mole, and the amplicon molecular mass is in grams per mole. The concentrations of the diluted qPCR standards were determined with a Qubit (v2.0) fluorometer and a Qubit double-stranded DNA high-sensitivity assay kit (Life Technologies). The molecular masses of the amplicons are listed in Table S2 in the supplemental material.
Monitoring of (A)XOS consumption and metabolite production. (i) Determination of carbohydrate concentrations.
To monitor the consumption of the (A)XOS substrate [consisting of arabinose, xylose, XOS(A)XOS (xylobiose, xylotriose, xylotetraose, xylopentaose, xylohexaose, and, possibly, also longer xylo-oligosaccharides), and AXOS], quantitative and qualitative analyses were performed.
The quantitative analysis of arabinose, xylose, and XOS(A)XOS (xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose) breakdown was performed by high-performance anion-exchange chromatography (HPAEC) with pulsed amperometric detection (PAD), as described before (32). Briefly, cell-free supernatants obtained through centrifugation at 5,000 × g for 15 min at 4°C were deproteinized using the Carrez A and Carrez B reagents and injected into a CarboPac PA100 analytical column (Thermo Scientific) of an ICS-3000 chromatograph (Thermo Scientific). A mixture of arabinose (Sigma-Aldrich), xylose (Sigma-Aldrich), xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose (all from Megazyme International, Bray, Ireland) was used as an external standard to perform quantifications. The concentrations of XOS(A)XOS molecules with chain lengths longer than the chain length of xylohexaose could not be determined, as there are no commercial standards available for these molecules. All samples were analyzed in triplicate. Averages and standard errors were calculated on the basis of the three biological repetitions.
As no commercial standards are available for AXOS molecules, they could be analyzed only qualitatively (32). Qualitative analysis of AXOS breakdown was performed by HPAEC-PAD, as described before (32, 38). Briefly, cell-free supernatants were deproteinized using the Carrez A and Carrez B reagents and injected into a CarboPac PA200 analytical column (Thermo Scientific) of an ICS-3000 chromatograph (Thermo Scientific). To be able to identify the AXOS molecules in the HPAEC-PAD chromatograms, enzymatic reference degradation chromatograms were used. These were generated through degradation of (A)XOS in solution by α-arabinofuranosidases with known specificities, as described previously (38).
(ii) Determination of metabolite concentrations.
Acetate, lactate, formate, ethanol, and butyrate concentrations were determined through high-performance liquid chromatography with a Waters chromatograph (Waters, Milford, MA) equipped with a differential refractive index detector (Hitachi, Foster City, CA), a 600E controller, a column oven (40°C), and a cooled 717 Plus autosampler. An ICSep ION-300 column (Transgenomic, Omaha, NE) was used with an 8.5 mM H2SO4 solution as the mobile phase at a flow rate of 0.4 ml min−1. Samples were diluted two times with ultrapure water, microcentrifuged at 20,000 × g for 15 min at 4°C, and filtered (pore size, 0.45 μm; Chromspec syringe filter; Chromatographic Specialties, Brockville, ON, Canada) before injection (25 μl) into the column. A mixture of acetate, lactate, formate, and butyrate (all from Sigma-Aldrich) plus ethanol (JT Baker Chemical) was used as an external standard to perform the quantifications. All samples were analyzed in triplicate. Averages and standard errors were calculated on the basis of the three biological repetitions.
Relative gene expression.
The expression of genes involved in metabolite production, (A)XOS degradation, and stress and cellular responses (see Tables S3 and S4 in the supplemental material) in B. longum NCC2705 and E. rectale ATCC 33656 relative to their expression in two controls (monoculture fermentations with B. longum NCC2705 and E. rectale ATCC 33656) was studied after 3, 6, 9, and 24 h of coculture fermentation.
(i) Primer and probe design.
TaqMan gene expression assays (containing both primers and probes; see Tables S3 and S4 in the supplemental material) for determination of the expression of 40 target genes involved in metabolite production, (A)XOS degradation, and stress and cellular responses as well as for 10 reference genes were manually designed with Geneious Pro R6 software (Biomatters). These assays were based on the whole-genome sequences of B. longum NCC2705 (National Center for Biotechnology Information [NCBI] accession number NC_004307, accessed in January 2014) and E. rectale ATCC 33656 (NCBI accession number NC_012781, accessed in January 2014). Gene functions (see Tables S3 and S4 in the supplemental material) were manually controlled on the basis of literature data (33–35) and information from the NCBI database (39) and the pathway databases BioCyc (43) and Kyoto Encyclopedia of Gene and Genomes (KEGG) (44) (all accessed in August 2013). In addition, a functional prediction of the glycoside hydrolases possibly involved in (A)XOS degradation by E. rectale ATCC 33656 was performed on the basis of the sequences in the Carbohydrate-Active enZYmes database (45), as has been done before for B. longum NCC2705 (34) (performed in January 2014; see Material and Methods SM1 and Table S4 in the supplemental material). The presence of hairpins, self-dimers, and dimer pairs in the TaqMan gene expression assays was assessed using Geneious Pro R6 (Biomatters). The primers and probes for the TaqMan gene expression assays were synthesized by Life Technologies. The gene and strain specificities of the constructed TaqMan gene expression assays were confirmed in silico by Primer-BLAST analysis (41) and in vitro by qPCR analysis of genomic DNA of both strains (see Tables S3 and S4 in the supplemental material).
(ii) RNA extraction.
Samples (1 ml) for RNA extraction were taken after 3, 6, 9, and 24 h of mono- and coculture fermentation and immediately treated with the RNAprotect Bacteria reagent (Qiagen) according to the manufacturer's instructions. Cell pellets were stored at −80°C until RNA extraction.
Total RNA was extracted using an RNeasy minikit (Qiagen) in the case of monoculture fermentations with B. longum NCC2705 and using an RNeasy midikit (Qiagen) in the case of monoculture fermentations with E. rectale ATCC 33656 and coculture fermentations with both strains and minor modifications of the manufacturer's instructions. Cell pellets were suspended in 100 μl (RNeasy minikit) or 200 μl (RNeasy midikit) of TE buffer (30 mM Tris base, 1 mM EDTA, pH 8.0) containing 15 mg ml−1 of lysozyme (Sigma-Aldrich), 100 µl ml−1 of proteinase K (20 mg ml−1; Sigma-Aldrich), and 100 µl ml−1 of mutanolysin (20 U ml−1; Sigma-Aldrich) and incubated at 37°C for 45 min. After addition of 700 μl (RNeasy minikit) or 1,500 μl (RNeasy midikit) of RLT buffer (Qiagen), cell suspensions were transferred into sterile microtubes containing 50 mg of zirconium beads (0.1 mm; BioSpec Products, Bartlesville, OK), shaken three times for 60 s each time in a Mini-Beadbeater-16 cell disrupter (BioSpec Products) with chilling on ice for 1 min between each step to avoid RNA degradation, and microcentrifuged at 20,000 × g for 15 s to remove cell debris. DNA was removed during RNA isolation by performing an on-column RNase-free DNase I (Qiagen) treatment according to the manufacturer's instructions. After elution of the RNA with 50 μl (RNeasy minikit) or 150 μl (RNeasy midikit) of RNase-free water, 2 μl (RNeasy minikit) or 6 μl (RNeasy midikit) of Protector RNase inhibitor (40 U μl−1; Roche) was added and a second RNase-free DNase I treatment (3 μl [RNeasy minikit] or 9 μl [RNeasy midikit] of DNase I [10 U μl−1; Roche]) was performed at 37°C for 30 min. DNase I was inactivated by adding 2 μl of EDTA (25 mM) to the reaction mixture and heating at 65°C for 15 min. The absence of DNA in the RNA samples was verified by qPCR using the tuf-specific primers designed for B. longum NCC2705 and E. rectale ATCC 33656 (see Table S1 in the supplemental material) on the basis of the conditions described above (see Table S1 in the supplemental material). RNA quality (expressed as the RNA integrity number [RIN]) and quantity were determined using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA). All extracted RNA was of good quality, and the RINs varied from 8.3 to 10.0 (average, 9.5 ± 0.4).
(iii) Monitoring of gene expression.
Total RNA (diluted to a standard concentration of 50 ng μl−1) was reverse transcribed into cDNA in a final volume of 100 μl with a high-capacity cDNA reverse transcription kit with RNase inhibitor (Applied Biosystems) according to the manufacturer's instructions, using a TProfessional Basic gradient thermocycler (Biometra). The reverse transcriptase reaction was applied using the following temperature profile: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min. cDNA samples were stored at −20°C for RT-qPCR analysis, and the remaining RNA was stored at −80°C.
A qPCR based on TaqMan fluorescence was performed using a ViiA7 real-time PCR system (Applied Biosystems) equipped with 384-well plates. Each PCR assay mixture of 10 μl contained 5 μl of TaqMan Fast Advanced master mix (Life Technologies), 4 μl of the 2.5× TaqMan gene expression assay mix (our own design in which the mixture contained both primers and a TaqMan probe), and 1 μl of cDNA. The qPCR amplification program consisted of an initial denaturation step at 95°C for 20 s, followed by 40 two-step cycles at 95°C for 1 s and at 60°C for 20 s. In each run, negative controls (without cDNA) and positive controls (with genomic DNA extracted from B. longum NCC2705 or E. rectale ATCC 33656) were included. The CT values were determined using ViiA7 software (v1.2.4; Applied Biosystems), the automatically determined thresholds for each TaqMan gene expression assay were averaged over all runs performed, and this average was used during a reanalysis of all qPCR runs. All qPCR assays were performed in triplicate.
(iv) Relative quantification of gene expression levels.
The relative expression ratio (RER), i.e., the level of expression of each target gene during coculture fermentations compared to its level of expression during monoculture fermentations in relation to the level of expression of multiple reference genes, was calculated by an efficiency-corrected relative quantification methodology implemented in REST 2009 software (Qiagen) (46) for each time point of the three biological repetitions. Briefly, this methodology consists of transformation of the CT value of each target gene (coculture fermentation) into ECT values (where E is the PCR amplification efficiency). This is followed by division of this value by the corresponding ECT value for the same target gene in the control sample (monoculture fermentation) (the ECT,control value) to obtain the EΔCT value. In a next step, the RER is determined by dividing the EΔCT value by a normalization factor, which is the geometric mean of the EΔCT values of the most stable reference genes. The E value for each TaqMan gene expression assay (target genes and reference genes) was determined by performing qPCR assays with a 5-log-unit dilution range of genomic DNA and plotting the logarithm of the genomic DNA concentrations as a linear function of the CT values obtained (47). From the slope of each standard curve, E was calculated as 10−1/slope. The E values of the TaqMan gene expression assays varied from 1.79 to 2.02 (see Tables S3 and S4 in the supplemental material). The CT cutoff value (the highest CT value) allowed was set at 32, as it was found that higher values did not fit the standard curve. For normalization, the selection of multiple reference genes with rather stable expression levels was aimed at for both mono- and coculture conditions. The software geNorm (48) was used to identify which reference genes out of five candidate genes for both B. longum NCC2705 and E. rectale ATCC 33656 (see Tables S3 and S4 in the supplemental material) were the most stable at each time point in the experimental setup applied. In the case of B. longum NCC2705, the genes coding for DNA-directed RNA polymerase subunit β (rpoB), FoF1 ATP synthase subunit β (atpD), and ATP-dependent DNA helicase PcrA (pcrA) were the most stable reference genes after 3 h of fermentation and rpoB, atpD, and the recombinase A gene (recA) were the most stable reference genes after 6, 9, and 24 h of fermentation. In the case of E. rectale ATCC 33656, the genes coding for FoF1 ATP synthase subunit β (EUBREC_0124), the DnaJ protein (EUBREC_1905), and DNA polymerase III subunits γ and τ (EUBREC_0328) were the most stable reference genes after 3 h of fermentation; the DNA-directed RNA polymerase subunit β gene (EUBREC_0379), the preprotein translocase subunit SecY gene (EUBREC_2512), and EUBREC_1905 were the most stable reference genes after 6 h of fermentation; EUBREC_2512, EUBREC_1905, and EUBREC_0328 were the most stable reference genes after 9 h of fermentation; and EUBREC_0379, EUBREC_2512, and EUBREC_0328 were the most stable reference genes after 24 h of fermentation. The statistical significance of the RERs was calculated using REST 2009 software (http://www.gene-quantification.de/rest-2009.html) on the basis of a pairwise fixed reallocation randomization test (46) with 2,000 permutations to determine whether the effects found were the result of the coculturing of B. longum NCC2705 and E. rectale ATCC 33656 or by chance only. P values of less than 0.050 were considered statistically significant. Gene expression corresponding to a negative RER is further referred to as underexpression (i.e., gene expression was lower during coculture fermentation than during monoculture fermentation), whereas gene expression corresponding to a positive RER is further referred to as overexpression (i.e., gene expression was higher during coculture fermentation than during monoculture fermentation).
RESULTS
Cocultivation of Bifidobacterium longum NCC2705 and Eubacterium rectale ATCC 33656. (i) Effect of cocultivation on bacterial growth.
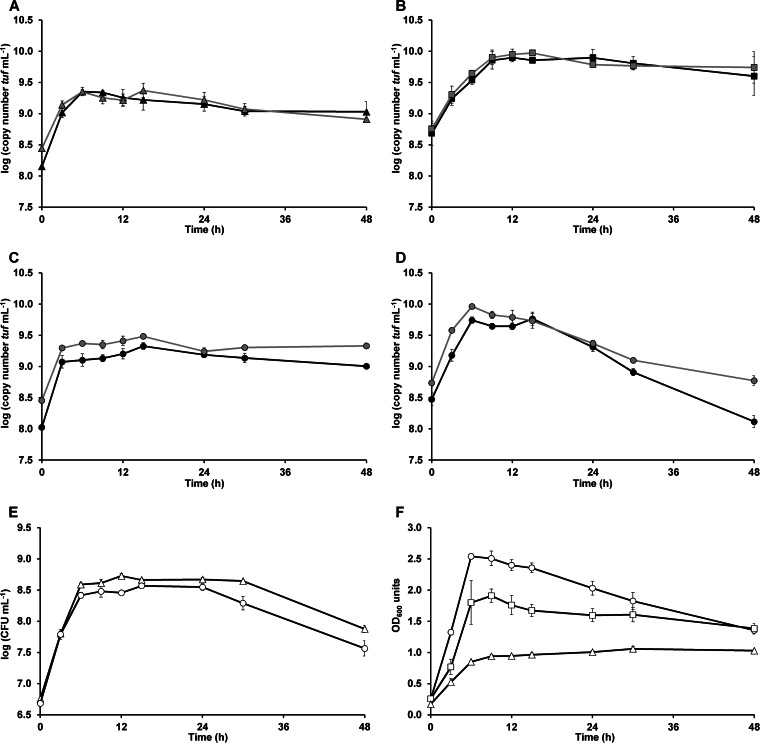
The growth curves of the strains B. longum NCC2705 and E. rectale ATCC 33656 obtained during the coculture fermentations showed patterns different from those obtained during the monoculture fermentations with either strain (Fig. 1). The qPCR and PMA-qPCR analyses unraveled that B. longum NCC2705 reached a first stationary growth phase after 3 h of coculture fermentation, after which it resumed its growth to reach a second stationary growth phase after 15 h of coculture fermentation (Fig. 1C). This growth profile was not seen during the monoculture fermentations with B. longum NCC2705 (Fig. 1A). During the second stationary growth phase of the coculture fermentations, the cell concentrations of B. longum NCC2705 were comparable to those during its stationary growth phase during the monoculture fermentations (Fig. 1A and C). The cell concentrations of B. longum NCC2705 obtained by PMA-qPCR were lower than those obtained by qPCR, indicating the presence of dead bacteria, but the differences in cell concentrations were larger during the coculture fermentations (Fig. 1C) than during the monoculture fermentations (Fig. 1A). In contrast to the stable cell concentrations of B. longum NCC2705 obtained by PMA-qPCR and qPCR, its plate counts decreased about 1.0 log unit (CFU milliliter−1) during the stationary growth phase after 30 h of monoculture fermentation and already after 24 h of coculture fermentation (Fig. 1E), indicating the presence of dead bacteria and viable but noncultivable bacteria.
FIG 1.
Monitoring of the growth of Bifidobacterium longum NCC2705 (triangles) and Eubacterium rectale ATCC 33656 (squares) as a function of time in the presence of (A)XOS during monoculture fermentations in MCB and mMCB, respectively, and of both strains (circles) in the presence of (A)XOS during coculture fermentations in MCB. (A to D) qPCR (gray lines) and PMA-qPCR (black lines) [expressed as log (tuf copy number milliliter−1)] of B. longum NCC2705 (A and C) and E. rectale ATCC 33656 (B and D); (E) plate counts [expressed as log (CFU milliliter−1)] of B. longum NCC2705; (F) growth (expressed as OD600 units) of B. longum NCC2705 and E. rectale ATCC 33656. The averages of three biological repetitions with corresponding standard errors are shown.
As was seen for B. longum NCC2705, the PMA-qPCR and qPCR results showed larger differences in cell concentrations of E. rectale ATCC 33656 during the coculture fermentations (Fig. 1D) than during the monoculture fermentations (Fig. 1B), indicating the presence of a higher number of dead bacteria during coculture fermentation. This was especially the case during the late stationary growth phase after 24 h of coculture fermentation, whereby the results obtained by PMA-qPCR were 0.7 log unit (tuf copy number ml−1) lower than those obtained by qPCR (Fig. 1D). Moreover, during the coculture fermentations, the PMA-qPCR cell concentrations of E. rectale ATCC 33656 decreased 1.7 log unit (tuf copy number ml−1) between 15 h and 48 h of fermentation (Fig. 1D), whereas the cell concentrations decreased only slightly during the monoculture fermentations (Fig. 1B). A noteworthy finding was that in the case of the coculture fermentations, the total (dead and viable) cell concentrations of E. rectale ATCC 33656 also decreased during the stationary growth phase (Fig. 1D). Similarly, the decrease in the OD600 was more profound during the coculture fermentations than during the monoculture fermentations (Fig. 1F).
(ii) Effect of cocultivation on (A)XOS consumption and metabolite production.
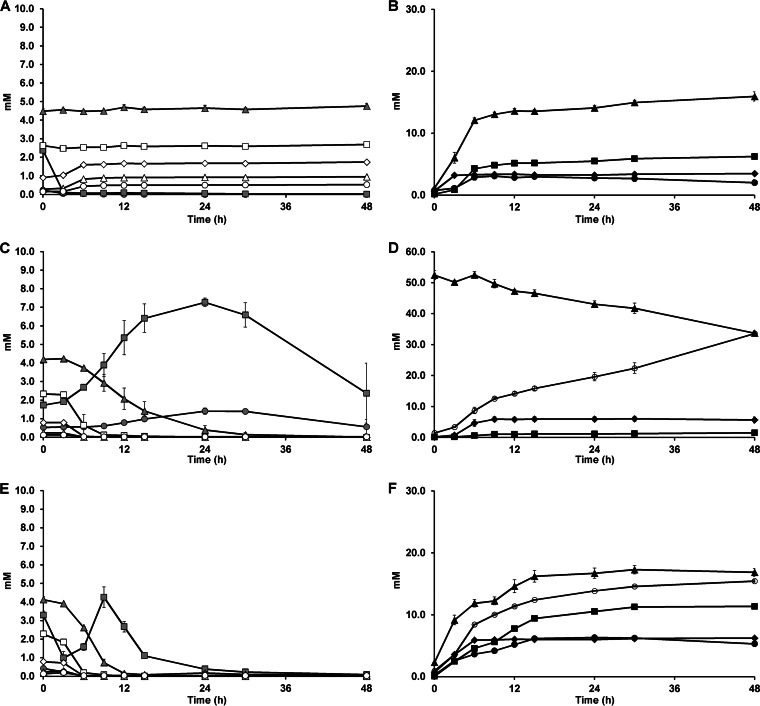
Differences in (A)XOS consumption and metabolite production kinetics were found between the mono- and coculture fermentations (Fig. 2 and 3). During the monoculture fermentations, B. longum NCC2705 did not consume XOS(A)XOS but consumed free arabinose and xylose and the arabinose substituents (both mono- and disubstituted) of AXOS. This was accompanied by an increase in the xylobiose, xylotriose, xylotetraose, xylopentaose, and xylohexaose concentrations (Fig. 2A) and possibly also the concentrations of longer XOS(A)XOS (Fig. 3A) in the extracellular medium. Bifidobacterium longum NCC2705 produced acetate, ethanol, formate, and lactate (Fig. 2B). Acetate, ethanol, and formate reached stable concentrations after 6 h of monoculture fermentation, when B. longum NCC2705 reached its stationary growth phase and the monosaccharides and arabinose substituents were exhausted. Lactate was no longer produced after 3 h of monoculture fermentation in favor of acetate, formate, and ethanol production.
FIG 2.
Quantitative analysis of arabinose, xylose, and XOS(A)XOS breakdown and metabolite production as a function of time with Bifidobacterium longum NCC2705 (A and B) and Eubacterium rectale ATCC 33656 (C and D) in the presence of (A)XOS during monoculture fermentations in MCB and mMCB, respectively, and with both strains (E and F) in the presence of (A)XOS during coculture fermentations in MCB. (Left) Monosaccharide and XOS(A)XOS consumption is represented as residual concentrations (in millimolar) of arabinose ( ), xylose (
), xylose ( ), xylobiose (
), xylobiose ( ), xylotriose (□), xylotetraose (♢), xylopentaose (Δ), and xylohexaose (○); (right) production (in millimolar) of the metabolites (and consumption of acetate) acetate (▲), butyrate (○), ethanol (●), formate (■), and lactate (◆). The averages of three biological repetitions with the corresponding standard errors are shown.
), xylotriose (□), xylotetraose (♢), xylopentaose (Δ), and xylohexaose (○); (right) production (in millimolar) of the metabolites (and consumption of acetate) acetate (▲), butyrate (○), ethanol (●), formate (■), and lactate (◆). The averages of three biological repetitions with the corresponding standard errors are shown.
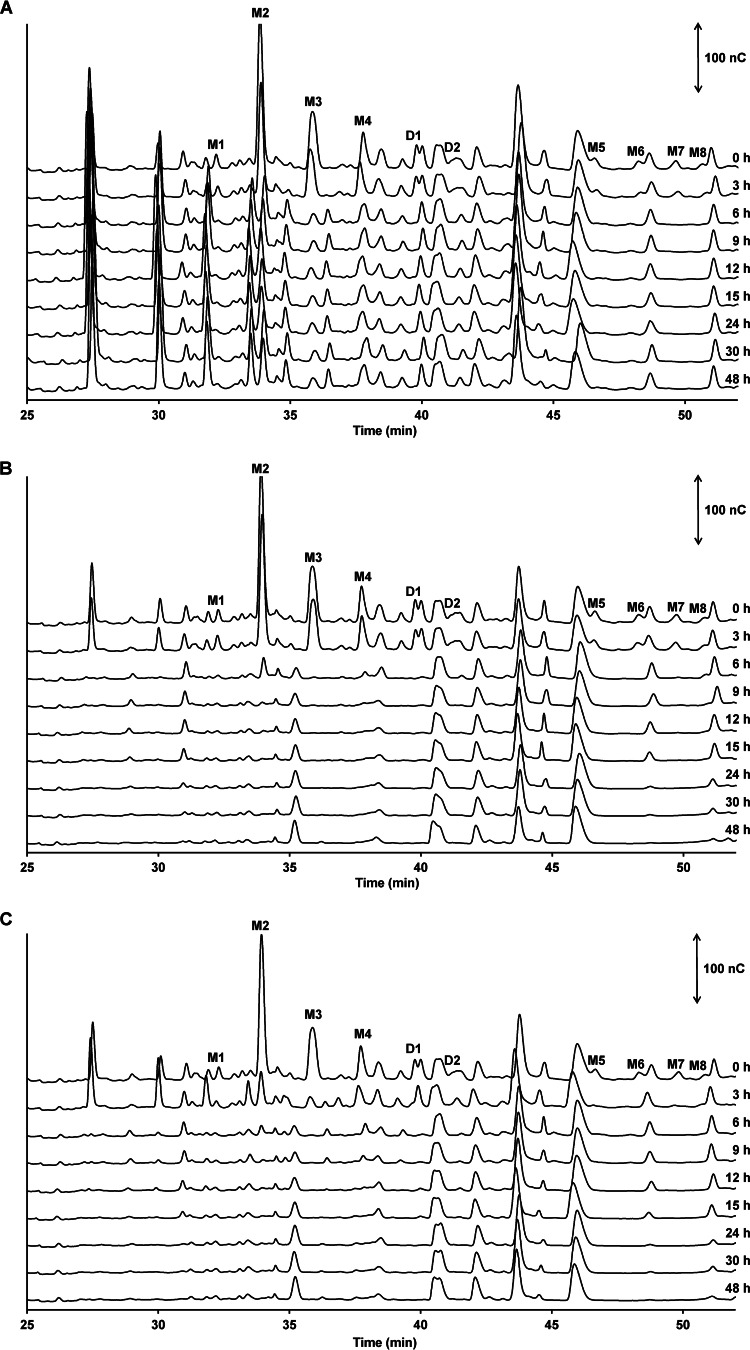
FIG 3.
Qualitative analysis of AXOS breakdown as a function of time by Bifidobacterium longum NCC2705 (A) and Eubacterium rectale ATCC 33656 (B) in the presence of (A)XOS during monoculture fermentations in MCB and mMCB, respectively, and by both strains (C) in the presence of (A)XOS during coculture fermentations in MCB. HPAEC-PAD chromatograms of AXOS indicate signal intensity (in nanocoulombs [nC]) at each retention time (minutes). Double-headed arrows represent the scale of the PAD response. Peaks D1 and D2 represent disubstituted AXOS molecules containing at least one disubstituted xylose residue; peaks M1 to M8 represent monosubstituted AXOS molecules containing at least one monosubstituted xylose residue. Other peaks possibly represent XOS(A)XOS molecules with chain lengths longer than the chain length of xylohexaose, for which no commercial standards are available. The data are representative of those from three biological repetitions of each of the fermentations.
Eubacterium rectale ATCC 33656 consumed AXOS (both mono- and disubstituted) and XOS(A)XOS, with arabinose and high concentrations of xylose being released into the extracellular medium during the monoculture fermentations (Fig. 2C and 3B). However, xylobiose was hydrolyzed less preferentially than the longer XOS(A)XOS. After 24 h of monoculture fermentation, when almost all (A)XOS molecules were degraded and no additional arabinose and xylose were produced, these monosaccharides were consumed only from that point on. During the consumption of the (A)XOS constituents, the acetate that was added to the medium was consumed and butyrate was concomitantly produced (33.6 ± 0.3 mM after 48 h of monoculture fermentation). Also, lactate and formate were produced, although at lower concentrations and only until 9 h of monoculture fermentation, after which the concentrations remained stable (Fig. 2D).
During the coculture fermentations with B. longum NCC2705 and E. rectale ATCC 33656, acetate, butyrate (15.4 ± 0.2 mM after 48 h of fermentation), formate, ethanol, and lactate were produced (Fig. 2F). The E. rectale strain produced butyrate thanks to the acetate produced by the B. longum strain, but more acetate was produced by B. longum NCC2705 than was used by E. rectale ATCC 33656. Quantitative (Fig. 2E) and qualitative (Fig. 3C) analysis of (A)XOS degradation by B. longum NCC2705 and E. rectale ATCC 33656 during the coculture fermentations revealed breakdown profiles similar to those for E. rectale ATCC 33656 during the monoculture fermentations, although (A)XOS was depleted faster and lower concentrations of arabinose and xylose were elaborated into the extracellular medium. Maximum arabinose and xylose concentrations of 0.03 ± 0.02 mM and 4.3 ± 0.6 mM, respectively, were measured after 9 h of fermentation during the coculture fermentations (Fig. 2E), whereas maximum arabinose and xylose concentrations of 1.4 ± 0.2 mM and 7.3 ± 0.2 mM, respectively, were measured after 24 h of fermentation during the monoculture fermentations (Fig. 2C). In addition, a sudden decrease in the xylose concentrations was measured, and this was accompanied by an increase in acetate, ethanol, and formate (but not lactate) concentrations after 9 h of coculture fermentation (Fig. 2E and F).
Effect of cocultivation on gene expression. (i) Effect of cocultivation on gene expression in Bifidobacterium longum NCC2705.
For all 20 genes studied in B. longum NCC2705 (Fig. 4), significant (P < 0.050) differences in gene expression from that during the monoculture fermentations were found at one or multiple time points during the coculture fermentations with E. rectale ATCC 33656 (Table 1). The RERs ranged from −12.048 to +14.770.
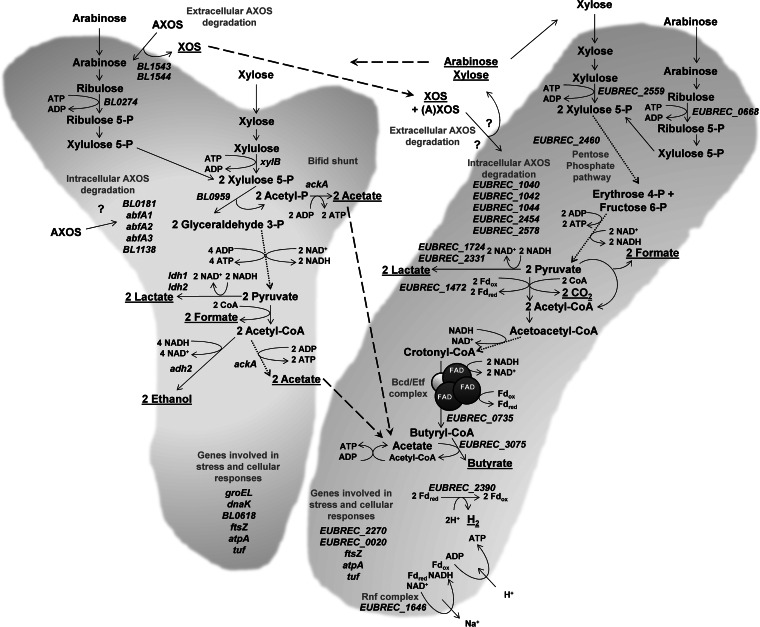
FIG 4.
Metabolic pathways and genes (reconstructed from the genome annotations based on information obtained from the NCBI, KEGG, and BioCyc databases) studied in Bifidobacterium longum NCC2705 and Eubacterium rectale ATCC 33656 by RT-qPCR. The mutual cross-feeding interactions between B. longum NCC2705 (represented by a bifid shunt on the left) and E. rectale ATCC 33656 (represented by a rod on the right) are represented as dashed lines. Dotted lines represent enzymatic conversions that involve different steps. Metabolites that are underlined are released extracellularly. Gene functions are described in Tables 1 and 2.
TABLE 1.
RERs of genes involved in the bifid shunt and metabolite production, (A)XOS degradation, and stress and cellular responses in Bifidobacterium longum NCC2705 at different time points during growth in MCB supplemented with (A)XOS in coculture with Eubacterium rectale ATCC 33656
| Metabolic pathway | Gene, proteina | 3 h |
6 h |
9 h |
24 h |
||||
|---|---|---|---|---|---|---|---|---|---|
| RERb | Pc | RER | P | RER | P | RER | P | ||
| Bifid shunt and metabolite production | BL0274, ribulokinase | 5.549 | 0.011 | −2.500 | 0.004 | −2.004 | <0.001 | 1.209 | 0.288 |
| xylB, xylulokinase | 1.897 | 0.018 | 5.098 | 0.003 | 14.770 | <0.001 | −1.346 | 0.055 | |
| BL0959, bifunctional xylulose-5-phosphate/fructose-6-phosphate phosphoketolase | −2.825 | 0.016 | −1.600 | 0.167 | −1.859 | 0.001 | 1.109 | 0.635 | |
| ldh1, lactate dehydrogenase | 1.945 | 0.017 | −1.325 | 0.055 | 3.185 | <0.001 | −1.011d | 0.951 | |
| ldh2, lactate dehydrogenase | 1.170 | 0.627 | −1.151 | 0.584 | 2.615 | <0.001 | −3.817 | <0.001 | |
| ackA, acetate kinase | 1.489 | 0.209 | −1.074 | 0.629 | 2.913 | <0.001 | 1.676 | 0.034 | |
| adh2, bifunctional acetaldehyde-CoA/alcohol dehydrogenase | 7.498 | <0.001 | −1.364 | 0.359 | 1.851 | <0.001 | −1.799 | 0.021 | |
| (A)XOS degradation | BL1543, extracellular α-arabinofuranosidase | 6.177 | 0.003 | −7.752 | <0.001 | 2.737 | 0.008 | −1.675d | 0.017 |
| BL1544, extracellular α-arabinofuranosidase | 3.794 | 0.006 | −12.048 | <0.001 | 2.909 | <0.001 | −1.538d | 0.043 | |
| BL0181, intracellular α-arabinofuranosidase | −1.385 | 0.239 | 1.093 | 0.733 | 1.352 | 0.036 | 1.083 | 0.554 | |
| abfA1, intracellular α-arabinofuranosidase | −1.965 | 0.006 | 1.768d | 0.001 | −2.882d | <0.001 | 1.246d | 0.111 | |
| abfA2, intracellular α-arabinofuranosidase | 8.708 | 0.006 | −1.984 | 0.081 | −1.475 | 0.103 | −1.307d | 0.334 | |
| abfA3, intracellular α-arabinofuranosidase | 2.284 | 0.004 | 1.809 | 0.062 | −1.410 | 0.103 | −2.070d | 0.006 | |
| BL1138, intracellular α-arabinofuranosidase | 1.358 | 0.349 | −1.144 | 0.606 | −2.618 | <0.001 | −4.505d | <0.001 | |
| Stress and cellular responses | groEL, chaperonin GroEL | −1.131 | 0.499 | 1.106 | 0.623 | −2.058 | <0.001 | −2.494 | 0.001 |
| dnaK, chaperone DnaK | 3.265 | <0.001 | 1.544 | <0.001 | 1.034 | 0.811 | 1.706 | 0.019 | |
| BL0618, Dps family protein | 1.316 | 0.574 | −1.070 | 0.717 | −2.058 | 0.015 | −2.088 | <0.001 | |
| ftsZ, cell division protein | 1.209 | 0.482 | 1.144 | 0.373 | 1.164 | 0.216 | −2.257 | <0.001 | |
| atpA, FoF1 ATP synthase, subunit α | −1.256 | 0.395 | 2.444 | <0.001 | 1.246 | 0.056 | −2.347 | 0.001 | |
| tuf, elongation factor Tu | −1.664 | 0.174 | −3.367 | <0.001 | −1.247 | 0.424 | 1.009 | 0.968 | |
Gene functions are described in Table S3 in the supplemental material.
Monoculture fermentation with B. longum NCC2705 in MCB with (A)XOS was used as a control. RERs depicted in bold showed a significant difference between the co- and monoculture fermentations (P < 0.050).
Statistical significance was determined using REST 2009 software on the basis of a pairwise fixed reallocation randomization test with 2,000 permutations.
RERs could not be interpreted, as the CT values obtained were above the CT cutoff value.
(a) Bifid shunt and metabolite production. Most genes involved in the bifid shunt and metabolite production were significantly differentially expressed after 3 h and 9 h of coculture fermentation (Table 1). The genes involved in the degradation of arabinose (ribulokinase, BL0274) and xylose (xylulokinase, xylB) to xylulose-5-phosphate for entry into the bifid shunt were overexpressed after 3 h of coculture fermentation, together with the genes involved in lactate (ldh1) and ethanol (adh2) production (see GenBank accession number NC_004307). The RERs of the xylulokinase gene (xylB) increased from 1.897 (P = 0.018) after 3 h of coculture fermentation to 5.098 (P = 0.003) and 14.770 (P < 0.001) after 6 h and 9 h of coculture fermentation, respectively, and decreased again, although not significantly (P = 0.055), after 24 h of coculture fermentation to −1.346. In contrast, despite the high level of increase in gene expression (RER = 5.549, P = 0.011) of the ribulokinase gene (BL0274) after 3 h of coculture fermentation, the RERs dropped to values below −2.000 after 6 h and 9 h of coculture fermentation. Moreover, all studied genes participating in acid and ethanol production were overexpressed after 9 h of coculture fermentation.
(b) (A)XOS degradation. After 3 h and 9 h of coculture fermentation, most genes involved in (A)XOS degradation were significantly differentially expressed (Table 1). Two extracellular α-arabinofuranosidase genes (BL1543 and BL1544) were overexpressed after 3 h of coculture fermentation. Also, after 3 h of coculture fermentation, two intracellular α-arabinofuranosidase genes (abfA2 and abfA3) were overexpressed, while the intracellular α-arabinofuranosidase gene (abfA1) was underexpressed. In contrast, after 6 h of coculture fermentation, BL1543 and BL1544 were underexpressed, and after 9 h of coculture fermentation, they were again overexpressed. After 9 h of coculture fermentation, the intracellular α-arabinofuranosidase genes, BL1138 and BL0181, were underexpressed and overexpressed, respectively, although for the latter gene the expression level during the monoculture fermentations (RER = 1.352, P = 0.036) was approximately the same as that during coculture fermentations.
(c) Stress and cellular responses. All stress and cellular response genes were significantly differentially expressed during the coculture fermentations at at least one time point, although only small changes in gene expression compared to that during the monoculture fermentations were found (RER range, −3.367 to+2.444; Table 1). The largest change in gene expression was found for the tuf gene (RER = −3.367, P < 0.001) after 6 h of coculture fermentation. The chaperonin gene (groEL) and the Dps family protein gene (BL0618) were underexpressed after 9 h and 24 h of coculture fermentation. The gene encoding the molecular chaperone DnaK was overexpressed after 3, 6, and 24 h of coculture fermentation. The cell division protein gene (ftsZ) and the FoF1 ATP synthase subunit α gene (atpA) were underexpressed after 24 h of coculture fermentation. Also, atpA was overexpressed after 6 h of coculture fermentation.
(ii) Effect of cocultivation on gene expression of Eubacterium rectale ATCC 33656.
For all 20 genes studied in E. rectale ATCC 33656 (Fig. 4), significant increases and decreases in gene expression (P < 0.050) during the coculture fermentations with B. longum NCC2705 compared to those during the monoculture fermentations were found at at least one of the time points investigated (Table 2). The RERs varied from −11.494 to +16.138.
TABLE 2.
RERs of genes involved in the pentose phosphate pathway and metabolite production, (A)XOS degradation, and stress and cellular responses in Eubacterium rectale ATCC 33656 at different time points during growth in MCB supplemented with (A)XOS in coculture with Bifidobacterium longum NCC2705
| Metabolic pathway | Gene, proteina | 3 h |
6 h |
9 h |
24 h |
||||
|---|---|---|---|---|---|---|---|---|---|
| RERb | Pc | RER | P | RER | P | RER | P | ||
| Pentose phosphate pathway and metabolite production | EUBREC_0668, ribulokinase | 2.298 | 0.087 | 2.155 | 0.025 | 1.179 | 0.417 | −3.425 | <0.001 |
| EUBREC_2559, xylulokinase | 6.574 | <0.001 | 1.574 | 0.232 | 1.935 | 0.014 | −6.329 | <0.001 | |
| EUBREC_2460, transketolase domain protein | 16.138 | <0.001 | 2.426 | 0.043 | 1.444 | 0.255 | −7.692 | <0.001 | |
| EUBREC_1724, lactate dehydrogenase | 1.830 | <0.001 | 1.593 | 0.229 | −1.376 | 0.160 | −2.024 | 0.010 | |
| EUBREC_2331, lactate dehydrogenase | 4.653 | <0.001 | −1.808 | 0.002 | −1.126 | 0.497 | −4.831 | <0.001 | |
| EUBREC_ 1472, pyruvate:ferredoxin oxidoreductase | 3.594 | <0.001 | 1.165 | 0.535 | 1.300 | 0.115 | −1.852 | <0.001 | |
| EUBREC_0735, butyryl-CoA dehydrogenase belonging to the Bcd/Etf complex | 1.652 | 0.141 | 2.879 | <0.001 | −1.395 | 0.253 | −2.294 | 0.001 | |
| EUBREC_3075, butyryl-CoA:acetate CoA-transferase | 3.234 | 0.047 | 2.877 | 0.002 | 1.113 | 0.435 | −5.556 | 0.015 | |
| EUBREC_2390, ferredoxin hydrogenase | 2.631 | 0.002 | 1.092 | 0.361 | 1.579 | 0.089 | −3.344 | <0.001 | |
| EUBREC_1646, ferredoxin belonging to the Rnf complex | 4.030 | <0.001 | −1.502 | 0.017 | −2.008 | 0.016 | −3.436 | <0.001 | |
| (A)XOS degradation | EUBREC_1040, exo-oligoxylanase | 2.381 | 0.016 | −1.969 | 0.015 | 1.290 | 0.209 | 1.780 | 0.010 |
| EUBREC_1042, bifunctional β-xylosidase/α-arabinofuranosidase | 1.561 | 0.162 | 3.865 | <0.001 | 1.027 | 0.867 | −11.494 | <0.001 | |
| EUBREC_1044, β-xylosidase | 2.133 | 0.005 | −1.218 | 0.344 | −1.163 | 0.360 | −6.289 | <0.001 | |
| EUBREC_2454, α-arabinofuranosidase | 4.192 | <0.001 | −2.326 | 0.007 | −1.178 | 0.370 | 2.307 | 0.003 | |
| EUBREC_2578, α-arabinofuranosidase | 1.062 | 0.809 | −2.237 | 0.003 | −2.174 | 0.002 | −2.268 | 0.001 | |
| Stress and cellular responses | EUBREC_2270, chaperonin GroEL | −1.508 | 0.007 | −1.256 | 0.233 | 1.533 | 0.027 | −3.040 | <0.001 |
| EUBREC_0020, chaperone DnaK | −1.524 | 0.075 | 4.765 | <0.001 | 1.292 | 0.281 | −1.972 | 0.002 | |
| ftsZ, cell division protein | 2.075 | <0.001 | 1.533 | 0.050 | −1.133 | 0.249 | −2.222 | 0.027 | |
| atpA, FoF1 ATP synthase, α subunit | −1.567 | 0.110 | 3.950 | <0.001 | −2.096 | 0.014 | −1.536 | 0.004 | |
| EUBREC_0383, elongation factor Tu | 1.852 | 0.021 | 1.971 | <0.001 | −1.010 | 0.975 | −1.379 | 0.062 | |
Gene functions are described in Table S4 in the supplemental material.
Monoculture fermentations with E. rectale ATCC 33656 in mMCB with (A)XOS was used as a control. RERs depicted in bold showed a significant difference between the co- and monoculture fermentations (P < 0.050).
Statistical significance was determined using REST 2009 software on the basis of a pairwise fixed reallocation randomization test with 2,000 permutations.
(a) Pentose phosphate pathway and metabolite production. All genes involved in the pentose phosphate pathway and metabolite production were overexpressed after 3 h of coculture fermentation, except for the ribulokinase gene (EUBREC_0668) and the butyryl-CoA dehydrogenase gene (EUBREC_0735), for which no significant differences were found (Table 2). In contrast, those genes were significantly overexpressed after 6 h of coculture fermentation, together with the transketolase gene (EUBREC_2460) and the butyryl-CoA:acetate CoA-transferase gene (EUBREC_3075), while the lactate dehydrogenase gene (EUBREC_2331) and the ferredoxin gene (EUBREC_1646) from the membrane-bound ferredoxin oxidoreductase (Rnf) complex were underexpressed. After 9 h of coculture fermentation, only two genes, i.e., the xylulokinase gene (EUBREC_2559; RER = 1.935, P = 0.014) and the ferredoxin gene mentioned above (RER = −2.008, P = 0.016), were significantly differentially expressed. After 24 h of fermentation, all genes were underexpressed during the coculture fermentations compared to their levels of expression during the monoculture fermentations.
(b) (A)XOS degradation. Although all five genes involved in (A)XOS degradation were overexpressed after 3 h of coculture fermentation, only the genes coding for exo-oligoxylanase (EUBREC_1040), β-xylosidase (EUBREC_1044), and α-arabinofuranosidase (EUBREC_2454) were significantly differentially expressed compared to their levels of expression during the monoculture fermentations (Table 2). After 6 h of coculture fermentation, a significant increase in gene expression was found for the bifunctional β-xylosidase/α-arabinofuranosidase gene (EUBREC_1042), while the other genes were underexpressed. After 24 h of coculture fermentation, all five genes were significantly differentially expressed compared to their levels of expression during monoculture fermentations, and of these, two genes (EUBREC_1040 and EUBREC_2454) were overexpressed and the other three genes (EUBREC_1042, EUBREC_1044, and EUBREC_2578) were underexpressed.
(c) Stress and cellular responses. The genes involved in stress responses (the chaperone DnaK gene [EUBREC_0020] and the chaperonin GroEL gene [EUBREC_2270]) were either overexpressed or underexpressed at the different time points during the coculture fermentations (Table 2). The largest change in expression was found for the chaperone DnaK gene (RER = 4.765, P < 0.001) after 6 h of coculture fermentation. At that time, all genes involved in cellular functions (atpA, tuf, and ftsZ) were significantly overexpressed, while expression was gene dependent at the other time points.
DISCUSSION
The present study revealed the presence of complex cross-feeding interactions between B. longum NCC2705 (an acetate producer) and E. rectale ATCC 33656 (an acetate-converting butyrate producer) during coculture fermentation, an overview of which is represented in Fig. 4. Both B. longum NCC2705 and E. rectale ATCC 33656 used the (A)XOS substrate, although to different extents (Fig. 4). The B. longum strain displayed an extracellular arabinose substituent-oriented AXOS metabolism; i.e., it consumed the arabinose substituents of AXOS and released decorticated xylose backbones (XOS) into the extracellular medium (32) (Fig. 4). The E. rectale strain displayed a nonpreferential extracellular (A)XOS metabolism; i.e., it consumed the whole (A)XOS substrate, possibly at its cell surface, and released arabinose and xylose into the extracellular medium (Fig. 4). The cross-feeding interactions were mutually beneficial, as both strains benefited from each other's presence. The E. rectale strain produced butyrate by means of a butyryl-CoA:acetate CoA-transferase thanks to the acetate produced by the B. longum strain, resulting in a butyrogenic effect, while the B. longum strain took advantage of the monosaccharides released by the extracellular degradation of (A)XOS by the E. rectale strain, resulting in an increase in its cell concentration, the production of even more acetate, and, thus, a bifidogenic effect (Fig. 4).
Coculture fermentations with B. longum NCC2705 and E. rectale ATCC 33656 started with higher levels of expression of their genes involved in (A)XOS degradation and metabolite production compared to their levels of expression during monoculture fermentations. The B. longum strain overexpressed its two extracellular α-arabinofuranosidase genes (BL1543 and BL1544) and two of its five intracellular α-arabinofuranosidase genes (abfA2 and abfA3), and this was accompanied by the overexpression of the ribulokinase gene, which is involved in arabinose degradation to xylulose-5-phosphate, which is part of the bifid shunt. Overexpression of BL1543 and abfA2 (which for the latter, however, was not significant) occurred for the growth of B. longum NCC2705 on AX, too (49). However, the function of intracellular α-arabinofuranosidases in B. longum NCC2705 is unknown, as the degradation of AXOS, with a concomitant increase in the amount of AXOS-derived decorticated XOS in the medium, indicates an extracellular arabinose substituent-oriented AXOS metabolism (Fig. 4) (32). Moreover, until now, no (A)XOS transporters or intracellular XOS-degrading enzymes have been found in B. longum NCC2705 (32, 34). Consequently, it is likely that B. longum NCC2705 does not consume (A)XOS intracellularly. During coculture fermentation, the B. longum strain also overexpressed its alcohol dehydrogenase gene, which could be correlated with a higher level of production of ethanol. Indeed, in Bifidobacterium spp., pyruvate can be converted into formate, acetate, and ethanol (Fig. 4), at the expense of lactate, to respond to a higher energy demand when growing on complex carbohydrates, which results in a lower growth rate and a shift from lactate production to mixed acid production (12, 37, 50–52). The production of additional ethanol allowed the B. longum strain to regenerate more NAD+, which is important for continuation of the upper pathways. Similarly, the E. rectale strain significantly overexpressed its xylulokinase and transketolase genes and genes involved in metabolite production. Despite the overexpression of the genes involved in butyrate production, no more butyrate was produced during coculture fermentation than during monoculture fermentation. Also, the overexpression of three genes encoding intracellular (A)XOS-degrading enzymes was found. However, also in this case, their contribution to (A)XOS degradation is still unclear, as the extracellular release of arabinose and xylose by the E. rectale strain indicated the presence of extracellular (A)XOS-degrading enzymes (Fig. 4) (32). These findings suggest that other yet unidentified genes in the genome of E. rectale ATCC 33656 contribute to extracellular (A)XOS degradation or that (A)XOS are transported into the cell with a concomitant degradation and export of arabinose and xylose out of the cell. The latter mechanism seems to be less plausible, because this would be a waste of energy, as E. rectale would eventually use its own released arabinose and xylose molecules. Likewise, overexpression of the β-endoxylanase, β-xylosidase, exo-oligoxylanase, and α-arabinofuranosidase genes has been reported in Bacteroides thetaiotaomicron ATCC 29148 and B. longum NCC2705 when they are grown together in the cecum of germfree mice that are fed a chow diet (53). Also, cocultivation of B. longum NCIMB 8809 and Bifidobacterium breve NCIMB 8807 or Lactobacillus delbrueckii subsp. lactis 193 in de Man-Rogosa-Sharpe medium increased the production of enzymes related to carbohydrate metabolism (54, 55). Similarly, genes related to carbohydrate metabolism in Lactobacillus paracasei ATCC 334 and Lactococcus lactis subsp. cremoris SK11 are overexpressed when these organisms are grown together during cheese making (56). These data suggest that B. longum NCC2705 and E. rectale ATCC 33656 adjusted their carbohydrate metabolism to increase their ability to compete for (A)XOS during cocultivation.
Alternatively, the increase in gene expression that was found could also be the result of a synergistic and, thus, more efficient (A)XOS degradation pattern when both strains are present, as this degradation requires the cooperative action of several carbohydrate-active enzymes encompassing α-arabinofuranosidases, β-endoxylanases, β-xylosidases, and exo-oligoxylanases (57). During cocultivation, the B. longum strain took advantage of the set of (A)XOS-degrading enzymes encoded by the genome of the E. rectale strain (Fig. 4). Indeed, coculture fermentation with E. rectale ATCC 33656 gave B. longum NCC2705 access to the otherwise inaccessible decorticated XOS and XOS(A)XOS molecules of (A)XOS, and it thereby consumed the extracellular xylose released by the E. rectale strain through (A)XOS degradation (Fig. 4). This resulted in a regain in growth of the B. longum strain after it initially reached its stationary growth phase. In accordance with the presence of higher concentrations of free extracellular xylose during coculture fermentation, overexpression of the xylulokinase gene occurred in the B. longum strain throughout fermentation. The highest increase corresponded to the moment that a sudden decrease in the xylose concentration occurred, and this was accompanied by the production of additional acetate, ethanol, and formate (but not lactate) and overexpression of the corresponding genes in the B. longum strain. At this point, overexpression of the extracellular α-arabinofuranosidases (BL1543 and BL1544) occurred; however, this was in contradiction to the underexpression of the ribulokinase gene and the exhaustion of the arabinose substituents of the AXOS molecules. It has been shown that BL1543 can get overexpressed in B. longum NCC2705 during fermentation in the presence only of xylose (49). However, as B. longum NCC2705 consumed the arabinose and xylose liberated by E. rectale ATCC 33656 during cocultivation, there was a faster depletion of these monosaccharides than during fermentation with the E. rectale strain solely. This was accompanied by the underexpression of the ribulokinase, xylulokinase, and transketolase genes and genes involved in metabolite production in E. rectale ATCC 33656 toward the end of the fermentation. This in turn resulted in less butyrate production (and, thus, less ATP production) during coculture fermentation than during monoculture fermentation and led to a significant decrease in the viability of the E. rectale strain during its stationary growth phase. As the total cell concentrations, i.e., the concentrations of dead and viable cells, also decreased during coculture fermentation, it can be suggested that cell lysis of E. rectale ATCC 33656 took place. This could be beneficial for other still living cells by providing building blocks for their further growth and maintenance (58). Although the presence of B. longum NCC2705 led to less butyrate production and decreased viability, without B. longum NCC2705 and, thus, without its acetate production, E. rectale ATCC 33656 could not grow or produce butyrate.
The presence of the E. rectale strain did not represent a major stress for the B. longum strain, as rather small changes in the levels of expression of stress and cellular response genes in B. longum NCC2705 occurred during coculture fermentation. Indeed, underexpression of the BL0618 gene, thought to be involved in starvation stress and to be important for competition in the stationary growth phase during cocultivation, occurred (59, 60). The overexpression of the dnaK gene did not necessarily imply stress, as molecular chaperone-encoding genes are also expressed during common growth (i.e., under nonstressful conditions) to assist with the proper folding process of new proteins (61, 62). However, the B. longum strain reached the viable but noncultivable state in the stationary growth phase faster during coculture fermentation, which was reflected in the decreased expression of the gene encoding a cell division protein. The presence of more acids in the medium could explain why the B. longum strain was more stressed. The differential expression during coculture fermentation of all genes involved in stress and cellular functions in the E. rectale strain could not be linked directly to changes in the metabolism and viability of the E. rectale strain.
In conclusion, the present study showed that the bifidogenic and butyrogenic effects of AXOS are the result of AXOS degradation by bifidobacteria and acetate-converting butyrate-producing colon bacteria, respectively, as a result of mutual cross-feeding interactions. Hence, these cross-feeding interactions favor the coexistence of bifidobacterial strains and acetate-converting butyrate producers in the same ecological niche. The interactions between B. longum NCC2705 and E. rectale ATCC 33656 seem to be based on the reciprocal exchange of metabolites (acetate and xylose, respectively), as changes in gene expression could be linked to changes in carbohydrate consumption and concomitant metabolite production. However, the presence of other bacterial strains that consume AXOS in the human colon complicates the attempts to fully understand and predict the bifidogenic and butyrogenic effects of AXOS at the moment. Nevertheless, this work contributes to a better understanding of the complex bacterial interactions and stimulation of beneficial bacteria in the human colon ecosystem.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the financial support of the Research Council of the Vrije Universiteit Brussel (projects SRP, IRP, and IOF), the Hercules Foundation, and the Research Foundation-Flanders (FWO-Vlaanderen). Denis Roy is Canada Research Chair in Lactic Culture Biotechnology for Dairy and Probiotic Industries, and this research was undertaken, in part, thanks to funding from the Canada Research Chairs program. This work was also supported by a Natural Sciences and Engineering Council of Canada (NSERC) discovery grant awarded to Denis Roy. Audrey Rivière is the recipient of a Ph.D. fellowship from the Research Foundation-Flanders (FWO-Vlaanderen) and received an international mobility grant from FWO-Vlaanderen to stay 1 year at the Institut sur la Nutrition et les Aliments Fonctionnels (INAF).
We thank Marie Verheyde (INAF) for her contribution to the experimental work and Patricia Savard (INAF) for her scientific advice. We also thank Christophe Courtin (Katholieke Universiteit Leuven) for providing AXOS. Filip Timmermans (Thermo Scientific) is acknowledged for kindly providing the CarboPac PA200 column.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02089-15.
REFERENCES
- 1.Collins SM, Surette M, Bercik P. 2012. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 2.Everard A, Cani PD. 2013. Diabetes, obesity and gut microbiota. Best Pract Res Clin Gastroenterol 27:73–83. doi: 10.1016/j.bpg.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12:661–672. doi: 10.1038/nrmicro3344. [DOI] [PubMed] [Google Scholar]
- 4.Stecher B, Maier L, Hardt WD. 2013. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol 11:277–284. doi: 10.1038/nrmicro2989. [DOI] [PubMed] [Google Scholar]
- 5.Di Gioia D, Aloisio I, Mazzola G, Biavati B. 2014. Bifidobacteria: their impact on gut microbiota composition and their applications as probiotics in infants. Appl Microbiol Biotechnol 98:563–577. doi: 10.1007/s00253-013-5405-9. [DOI] [PubMed] [Google Scholar]
- 6.Grimm V, Westermann C, Riedel CU. 2014. Bifidobacteria-host interactions—an update on colonisation factors. Biomed Res Int 2014:960826. doi: 10.1155/2014/960826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK. 2014. Specialized metabolites from the microbiome in health and disease. Cell Metab 20:719–730. doi: 10.1016/j.cmet.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheppach W, Weiler F. 2004. The butyrate story: old wine in new bottles? Curr Opin Clin Nutr Metab Care 7:563–567. doi: 10.1097/00075197-200409000-00009. [DOI] [PubMed] [Google Scholar]
- 9.Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. 2008. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119. [DOI] [PubMed] [Google Scholar]
- 10.Falony G, Verschaeren A, De Bruycker F, De Preter V, Verbeke K, Leroy F, De Vuyst L. 2009. In vitro kinetics of prebiotic inulin-type fructan fermentation by butyrate-producing colon bacteria: implementation of online gas chromatography for quantitative analysis of carbon dioxide and hydrogen gas production. Appl Environ Microbiol 75:5884–5892. doi: 10.1128/AEM.00876-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis P, Flint HJ. 2009. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 12.De Vuyst L, Moens F, Selak M, Rivière A, Leroy F. 2014. Summer meeting 2013: growth and physiology of bifidobacteria. J Appl Microbiol 116:477–491. doi: 10.1111/jam.12415. [DOI] [PubMed] [Google Scholar]
- 13.Van den Abbeele P, Belzer C, Goossens M, Kleerebezem M, De Vos WM, Thas O, De Weirdt R, Kerckhof FM, Van de Wiele T. 2013. Butyrate-producing Clostridium cluster XIVa species specifically colonize mucins in an in vitro gut model. ISME J 7:949–961. doi: 10.1038/ismej.2012.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vital M, Howe AC, Tiedje JM. 2014. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio 5:e00889-14. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker AW, Duncan SH, Louis P, Flint HJ. 2014. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol 22:267–274. doi: 10.1016/j.tim.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 16.Gibson GR, Probert HM, Loo JV, Rastall RA, Roberfroid MB. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev 17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 17.Bindels LB, Delzenne NM, Cani PD, Walter J. 2015. Towards a more comprehensive concept for prebiotics. Nat Rev Gastroenterol Hepatol 12:303–310. doi: 10.1038/nrgastro.2015.47. [DOI] [PubMed] [Google Scholar]
- 18.Hughes SA, Shewry PR, Li L, Gibson GR, Sanz ML, Rastall RA. 2007. In vitro fermentation by human fecal microflora of wheat arabinoxylans. J Agric Food Chem 55:4589–4595. doi: 10.1021/jf070293g. [DOI] [PubMed] [Google Scholar]
- 19.Van Craeyveld V, Swennen K, Dornez E, Van de Wiele T, Marzorati M, Verstraete W, Delaedt Y, Onagbesan O, Decuypere E, Buyse J, De Ketelaere B, Broekaert WF, Delcour JA, Courtin CM. 2008. Structurally different wheat-derived arabinoxylooligosaccharides have different prebiotic and fermentation properties in rats. J Nutr 138:2348–2355. doi: 10.3945/jn.108.094367. [DOI] [PubMed] [Google Scholar]
- 20.Cloetens L, Broekaert WF, Delaedt Y, Ollevier F, Courtin CM, Delcour JA, Rutgeerts P, Verbeke K. 2010. Tolerance of arabinoxylan-oligosaccharides and their prebiotic activity in healthy subjects: a randomised, placebo-controlled cross-over study. Br J Nutr 103:703–713. doi: 10.1017/S0007114509992248. [DOI] [PubMed] [Google Scholar]
- 21.Damen B, Verspreet J, Pollet A, Broekaert WF, Delcour JA, Courtin CM. 2011. Prebiotic effects and intestinal fermentation of cereal arabinoxylans and arabinoxylan oligosaccharides in rats depend strongly on their structural properties and joint presence. Mol Nutr Food Res 55:1862–1874. doi: 10.1002/mnfr.201100377. [DOI] [PubMed] [Google Scholar]
- 22.Neyrinck AM, Possemiers S, Druart C, van de Wiele T, De Backer F, Cani PD, Larondelle Y, Delzenne NM. 2011. Prebiotic effects of wheat arabinoxylan related to the increase in bifidobacteria, roseburia and bacteroides/prevotella in diet-induced obese mice. PLoS One 6:e20944. doi: 10.1371/journal.pone.0020944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van den Abbeele P, Gerard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Kleerebezem M, Zoetendal EG, Smidt H, Verstraete W, Van de Wiele T, Possemiers S. 2011. Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ Microbiol 13:2667–2680. doi: 10.1111/j.1462-2920.2011.02533.x. [DOI] [PubMed] [Google Scholar]
- 24.Maki KC, Gibson GR, Dickmann RS, Kendall CWC, Chen CYO, Costabile A, Comelli EM, McKay DL, Almeida NG, Jenkins D, Zello GA, Blumberg JB. 2012. Digestive and physiologic effects of a wheat bran extract, arabino-xylan-oligosaccharide, in breakfast cereal. Nutrition 28:1115–1121. doi: 10.1016/j.nut.2012.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Gråsten S, Liukkonen KH, Chrevatidis A, El-Nezami H, Poutanen K, Mykkänen H. 2003. Effects of wheat pentosan and inulin on the metabolic activity of fecal microbiota and on bowel function in healthy humans. Nutr Res 23:1503–1514. doi: 10.1016/S0271-5317(03)00164-7. [DOI] [Google Scholar]
- 26.Sanchez JI, Marzorati M, Grootaert C, Baran M, Van Craeyveld V, Courtin CM, Broekaert WF, Delcour JA, Verstraete W, Van de Wiele T. 2009. Arabinoxylan-oligosaccharides (AXOS) affect the protein/carbohydrate fermentation balance and microbial population dynamics of the simulator of human intestinal microbial ecosystem. Microb Biotechnol 2:101–113. doi: 10.1111/j.1751-7915.2008.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pollet A, Van Craeyveld V, Van de Wiele T, Verstraete W, Delcour JA, Courtin CM. 2012. In vitro fermentation of arabinoxylan oligosaccharides and low molecular mass arabinoxylans with different structural properties from wheat (Triticum aestivum L.) bran and psyllium (Plantago ovata Forsk) seed husk. J Agric Food Chem 60:946–954. doi: 10.1021/jf203820j. [DOI] [PubMed] [Google Scholar]
- 28.Walton GE, Lu C, Trogh I, Arnaut F, Gibson GR. 2012. A randomised, double-blind, placebo controlled cross-over study to determine the gastrointestinal effects of consumption of arabinoxylan-oligosaccharides enriched bread in healthy volunteers. Nutr J 36:1–11. doi: 10.1186/1475-2891-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nielsen TS, Lærke HN, Theil PK, Sørensen JF, Saarinen M, Forssten S, Bach Knudsen KE. 2014. Diets high in resistant starch and arabinoxylan modulate digestion processes and SCFA pool size in the large intestine and faecal microbial composition in pigs. Br J Nutr 112:1837–1849. doi: 10.1017/S000711451400302X. [DOI] [PubMed] [Google Scholar]
- 30.Falony G, Vlachou A, Verbrugghe K, De Vuyst L. 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl Environ Microbiol 72:7835–7841. doi: 10.1128/AEM.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Vuyst L, Leroy F. 2011. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifidobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol 149:73–80. doi: 10.1016/j.ijfoodmicro.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 32.Rivière A, Moens F, Selak M, Maes D, Weckx S, De Vuyst L. 2014. The ability of bifidobacteria to degrade arabinoxylan oligosaccharide constituents and derived oligosaccharides is strain dependent. Appl Environ Microbiol 80:204–217. doi: 10.1128/AEM.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc Natl Acad Sci U S A 99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lagaert S. 2013. Study of the arabinoxylan and arabinoxylan oligosaccharide-degrading enzymes of Bifidobacterium adolescentis and Bifidobacterium longum. Ph.D. thesis. Katholieke Universiteit Leuven, Leuven, Belgium. [Google Scholar]
- 35.Mahowald MA, Rey FE, Seedorf H, Turnbaugh PJ, Fulton RS, Wollam A, Shah N, Wang C, Magrini V, Wilson RK, Cantarel BL, Coutinho PM, Henrissat B, Crock LW, Russell A, Verberkmoes NC, Hettich RL, Gordon JI. 2009. Characterizing a model human gut microbiota composed of members of its two dominant bacterial phyla. Proc Natl Acad Sci U S A 106:5859–5864. doi: 10.1073/pnas.0901529106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Dymock D, Wade WG. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van der Meulen R, Makras L, Verbrugghe K, Adriany T, De Vuyst L. 2006. In vitro kinetic analysis of oligofructose consumption by Bacteroides and Bifidobacterium spp. indicates different degradation mechanisms. Appl Environ Microbiol 72:1006–1012. doi: 10.1128/AEM.72.2.1006-1012.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rivière A, Eeltink S, Pierlot C, Balzarini T, Moens F, Selak M, De Vuyst L. 2013. A high-resolution ion-exchange chromatography method for monitoring of prebiotic arabinoxylan-oligosaccharide degradation in a complex fermentation medium. Anal Chem 85:4982–4990. doi: 10.1021/ac400187f. [DOI] [PubMed] [Google Scholar]
- 39.NCBI Resource Coordinators. 2014. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 42:D7–D17. doi: 10.1093/nar/gkt1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gagnon M, Savard P, Rivière A, LaPointe G, Roy D. 2015. Bioaccessible antioxidants in milk fermented by Bifidobacterium longum subsp. longum strains. Biomed Res Int 2015:169381. doi: 10.1155/2015/169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Junick J, Blaut M. 2012. Quantification of human fecal Bifidobacterium species by use of quantitative real-time PCR analysis targeting the groEL gene. Appl Environ Microbiol 78:2613–2622. doi: 10.1128/AEM.07749-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caspi R, Altman T, Billington R, Dreher K, Foerster H, Fulcher CA, Holland TA, Keseler IM, Kothari A, Kubo A, Krummenacker M, Latendresse M, Mueller LA, Ong Q, Paley S, Subhraveti P, Weaver DS, Weerasinghe D, Zhang P, Karp PD. 2014. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lombard V, Ramulu HG, Drula E, Coutinho PM, Henrissat B. 2014. The Carbohydrate-Active Enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfaffl MW. 2004. Quantification strategies in real-time PCR, p 87–120. In Bustin SA. (ed), The real-time PCR encyclopedia A-Z of quantitative PCR. International University Line, La Jolla, CA. [Google Scholar]
- 47.Applied Biosystems. 2004. Amplification efficiency of TaqMan® gene expression assays: application note 127AP05-02. Applied Biosystems, Foster City, CA. [Google Scholar]
- 48.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savard P, Roy D. 2009. Determination of differentially expressed genes involved in arabinoxylan degradation by Bifidobacterium longum NCC2705 using real-time RT-PCR. Probiotics Antimicrob Proteins 1:121–129. doi: 10.1007/s12602-009-9015-x. [DOI] [PubMed] [Google Scholar]
- 50.Van der Meulen R, Avonts L, De Vuyst L. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl Environ Microbiol 70:1923–1930. doi: 10.1128/AEM.70.4.1923-1930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Van der Meulen R, Adriany T, Verbrugghe K, De Vuyst L. 2006. Kinetic analysis of bifidobacterial metabolism reveals a minor role for succinic acid in the regeneration of NAD+ through its growth-associated production. Appl Environ Microbiol 72:5204–5210. doi: 10.1128/AEM.00146-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Falony G, Lazidou K, Verschaeren A, Weckx S, Maes D, De Vuyst L. 2009. In vitro kinetic analysis of fermentation of prebiotic inulin-type fructans by Bifidobacterium species reveals four different phenotypes. Appl Environ Microbiol 75:454–461. doi: 10.1128/AEM.01488-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonnenburg JL, Chen CT, Gordon JI. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol 4:e413. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ruiz L, Sánchez B, de Los Reyes-Gavilán CG, Gueimonde M, Margolles A. 2009. Coculture of Bifidobacterium longum and Bifidobacterium breve alters their protein expression profiles and enzymatic activities. Int J Food Microbiol 133:148–153. doi: 10.1016/j.ijfoodmicro.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 55.Sánchez B, Burns P, Ruiz L, Binetti A, Vinderola G, Margolles JRA, Ruas-Madiedo P, de los Reyes-Gavilán CG. 2013. Co-culture affects protein profile and heat tolerance of Lactobacillus delbrueckii subsp. lactis and Bifidobacterium longum. Food Res Int 54:1080–1083. doi: 10.1016/j.foodres.2013.01.026. [DOI] [Google Scholar]
- 56.Desfossés-Foucault E, LaPointe G, Roy D. 2014. Transcription profiling of interactions between Lactococcus lactis subsp. cremoris SK11 and Lactobacillus paracasei ATCC 334 during cheddar cheese simulation. Int J Food Microbiol 178:76–86. doi: 10.1016/j.ijfoodmicro.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Lagaert S, Pollet A, Courtin CM, Volckaert G. 2014. β-Xylosidases and α-l-arabinofuranosidases: accessory enzymes for arabinoxylan degradation. Biotechnol Adv 32:316–332. doi: 10.1016/j.biotechadv.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 58.Bayles KW. 2014. Bacterial programmed cell death: making sense of a paradox. Nat Rev Microbiol 12:63–69. doi: 10.1038/nrmicro3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nair S, Finkel SE. 2004. Dps protects cells against multiple stresses during stationary phase. J Bacteriol 186:4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klijn A, Mercenier A, Arigoni F. 2005. Lessons from the genomes of bifidobacteria. FEMS Microbiol Rev 29:491–509. doi: 10.1016/j.fmrre.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 61.Ruiz L, Ruas-Madiedo P, Gueimonde M, de Los Reyes-Gavilán CG, Margolles A, Sánchez B. 2011. How do bifidobacteria counteract environmental challenges? Mechanisms involved and physiological consequences. Genes Nutr 6:307–318. doi: 10.1007/s12263-010-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turroni F, Foroni E, Montanini B, Viappiani A, Strati F, Duranti S, Ferrarini A, Delledonne M, van Sinderen D, Ventura M. 2011. Global genome transcription profiling of Bifidobacterium bifidum PRL2010 under in vitro conditions and identification of reference genes for quantitative real-time PCR. Appl Environ Microbiol 77:8578–8587. doi: 10.1128/AEM.06352-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.