Abstract
Cyanobacteria are generally assumed to be effective competitors at low CO2 levels because of their efficient CO2-concentrating mechanism (CCM), and yet how bloom-forming cyanobacteria respond to rising CO2 concentrations is less clear. Here, we investigate changes in CCM gene expression at ambient CO2 (400 ppm) and elevated CO2 (1,100 ppm) in six strains of the harmful cyanobacterium Microcystis. All strains downregulated cmpA encoding the high-affinity bicarbonate uptake system BCT1, whereas both the low- and high-affinity CO2 uptake genes were expressed constitutively. Four strains downregulated the bicarbonate uptake genes bicA and/or sbtA, whereas two strains showed constitutive expression of the bicA-sbtA operon. In one of the latter strains, a transposon insert in bicA caused low bicA and sbtA transcript levels, which made this strain solely dependent on BCT1 for bicarbonate uptake. Activity measurements of the inorganic carbon (Ci) uptake systems confirmed the CCM gene expression results. Interestingly, genes encoding the RuBisCO enzyme, structural carboxysome components, and carbonic anhydrases were not regulated. Hence, Microcystis mainly regulates the initial uptake of inorganic carbon, which might be an effective strategy for a species experiencing strongly fluctuating Ci concentrations. Our results show that CCM gene regulation of Microcystis varies among strains. The observed genetic and phenotypic variation in CCM responses may offer an important template for natural selection, leading to major changes in the genetic composition of harmful cyanobacterial blooms at elevated CO2.
INTRODUCTION
CO2 concentrations in the atmosphere may double during this century (1). In marine ecosystems, enhanced dissolution of atmospheric CO2 causes ocean acidification (2, 3). In freshwaters, however, CO2 concentrations may vary widely. In many lakes, the dissolved CO2 concentration exceeds the concentration expected from equilibrium with the atmosphere due to the input of large amounts of organic carbon from terrestrial systems (4, 5). Conversely, the photosynthetic activity of dense phytoplankton blooms can deplete the CO2 concentration far below atmospheric levels, which increases pH and makes bicarbonate the most abundant inorganic carbon species (6–8). Cyanobacteria are often considered to be very successful competitors at low CO2 levels (9, 10), and global warming is predicted to favor an expansion of cyanobacterial blooms in eutrophic waters (11–13). However, the response of bloom-forming cyanobacteria to elevated CO2 levels is not yet well understood.
Cyanobacteria typically use a CO2-concentrating mechanism (CCM) with up to five different uptake systems for inorganic carbon (Ci): three for bicarbonate and two for CO2 uptake (14). The two sodium-dependent bicarbonate uptake systems, BicA and SbtA, are present in some but not all freshwater cyanobacteria (15–17). BicA combines a low affinity for bicarbonate with a high flux rate, whereas SbtA usually has a high affinity for bicarbonate but a low flux rate (15, 18). Hence, BicA operates more effectively at high bicarbonate concentrations and vice versa SbtA at low bicarbonate concentrations (15, 17). The third bicarbonate system, BCT1, is present in most freshwater cyanobacteria and resembles SbtA with a high affinity for bicarbonate and a low flux rate. However, in contrast to SbtA, BCT1 does not require sodium and is directly ATP dependent (19). The two CO2 uptake systems, NDH-I3 and NDH-I4, are also present in most freshwater cyanobacteria and convert the passively diffusing CO2 inside the cell into bicarbonate via NADPH-driven electron flow (20). NDH-I3 combines a high affinity for CO2 with a low flux rate, whereas NDH-I4 combines a low affinity with a high flux rate (20, 21). Inside the cyanobacterial cells, bicarbonate accumulates in the cytoplasm and then diffuses into the carboxysomes, where it is converted back to CO2 via carbonic anhydrases to enable efficient carbon fixation by RuBisCO (14).
Microcystis is a harmful cyanobacterium that forms dense blooms in lakes all over the world (22–24). Moreover, many strains are capable of producing microcystin, which is a powerful hepatotoxin for birds, mammals, and humans (25, 26). Recently, it was shown that Microcystis strains show considerable genetic diversity in their Ci uptake system genes (17). Some strains lack the sbtA gene (genotype I), other strains lack bicA (genotype II), and again others contain the genes for all five Ci uptake systems (genotype III). Some strains also acquired transposon inserts in the bicA-sbtA operon (17), but it is unknown whether this affects the expression of bicA and sbtA in these strains. Although the expression of the CCM genes has been extensively studied in the model cyanobacterium Synechocystis sp. strain PCC 6803 (27–30), CCM gene expression patterns of the environmentally relevant cyanobacterium Microcystis have attracted only recent interest (31–33).
In this study, we compare expression of the Ci uptake genes in response to changing CO2 conditions among Microcystis strains representative of the different Ci uptake genotypes. Studies with Synechocystis PCC 6803 showed that genes for the high-affinity uptake systems for CO2 (NDH-I3) and bicarbonate (SbtA and BCT1) are induced at low CO2, whereas the expression of low-affinity Ci uptake systems (BicA and NDH-I4) remains unaltered (29, 30). It seems likely that the same expression patterns apply to Microcystis, although the presence of different Ci uptake genotypes could lead to variation in gene expression among Microcystis strains. We therefore defined two hypotheses for our study: (i) high-affinity Ci uptake genes are downregulated at elevated CO2 (1,100 ppm), whereas (ii) low-affinity Ci uptake genes are expressed constitutively in Microcystis. To investigate these hypotheses, we compared the CCM gene expression of six Microcystis strains at ambient and elevated CO2 levels. Furthermore, we measured O2 evolution of the strains exposed to different Ci conditions to compare the activity of their Ci uptake systems. The results reveal an unexpected diversity of CO2 responses within the genus Microcystis.
MATERIALS AND METHODS
Microcystis strains.
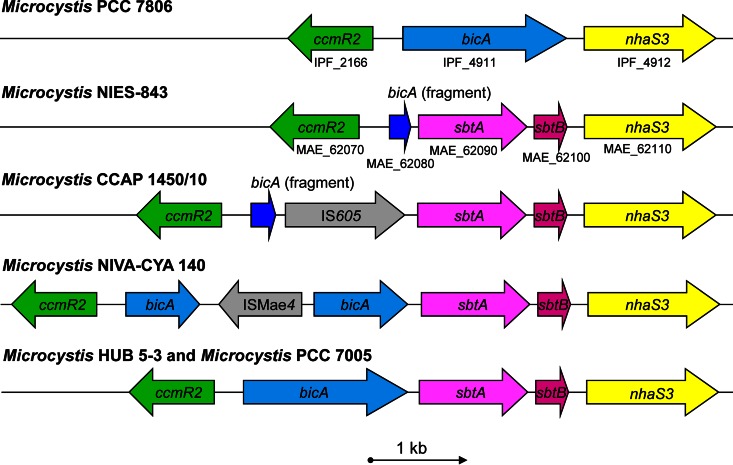
We studied six Microcystis strains with different Ci uptake systems (Fig. 1). The Ci uptake genotypes of these strains were described in a previous study (17). Strain PCC 7806 contains the bicA gene but lacks the sbtA gene and belongs to Ci uptake genotype I. Strains NIES-843 and CCAP 1450/11 contain the sbtA gene but lack a complete bicA gene and hence belong to genotype II. Strain NIVA-CYA 140 combines sbtA with a complete bicA gene that is no longer functional because of a transposon insert, and therefore this strain was also assigned to genotype II. Strains HUB 5-3 and PCC 7005 contain both the bicA and sbtA genes in the same operon and are therefore assigned to genotype III (Fig. 1). The four genes encoding the high-affinity bicarbonate transporter BCT1 (cmpABCD), as well as the genes encoding the high-affinity CO2 uptake system NDH-I3 (chpY, ndhD3, ndhF3, and other ndh genes) and the low-affinity CO2 uptake system NDH-I4 (chpX, ndhD4, ndhF4, and other ndh genes), were present in all six strains (17). Moreover, genome-wide microarray analysis showed that Microcystis PCC 7806 expresses all its CCM genes (32).
FIG 1.
The bicA-sbtA operon of the six Microcystis strains of the present study. Gene ccmR2 encodes a LysR transcriptional regulator that most likely regulates the expression of the bicA-sbtA operon. Some strains contain a complete bicA gene, whereas others only contain a small fragment. The gray arrows (IS605 and ISMae4) indicate transposon inserts. Gene sbtB encodes a protein that regulates the activity of SbtA (18). Gene nhaS3 encodes a proton/sodium antiporter. The IPF numbers are the locus tags of PCC 7806, and the MAE numbers are the locus tags of NIES-843. The protein sequence identity between the Microcystis strains is 99% for BicA and 97 to 99% for SbtA.
Experimental setup.
We used the exponential phase of batch culture experiments exposed to ambient pCO2 (400 ppm) and elevated pCO2 (1100 ppm) in the gas flow to study gene expression and activity of the Ci uptake systems.
First, the six Microcystis strains were precultured in 1-liter Erlenmeyer flasks in modified BG11 medium (34; with 10 mmol liter−1 NaNO3 and no added Na2CO3/NaHCO3) for 1 week. The precultures were incubated at 25°C with 400 ppm of pCO2 and at 120 rpm in an orbital shaker incubator (Gallenkamp, Leicester, United Kingdom), with light provided by TL-D 30W/33-640 white fluorescent tubes (Philips, Eindhoven, The Netherlands) at 20 μmol photons m−2 s−1. Microscopy checks did not reveal any contaminations.
Subsequently, new 1-liter Erlenmeyer flasks with 400 ml of modified BG11 medium were inoculated with the exponentially growing precultures at an optical density at 750 nm (OD750) of ∼0.080. Four biological replicates were used for each strain. The Erlenmeyer flasks were topped with foam stoppers to allow gas exchange and placed in an Infors HT Multritron Pro incubator (Infors Benelux, Doetinchem, The Netherlands) at 25°C with 400 ppm of pCO2 and shaken at 120 rpm. Light was provided by white fluorescent tubes (Gro-lux F36W/Gro-T8; Havells-Sylvania Germany GmbH, Erlangen, Germany). It was recently shown that microcystins can bind to RuBisCO during oxidative stress (35), which may affect CO2 fixation. Therefore, we decided to use low light levels of 20 μmol photons m−2 s−1during the batch culture experiments to minimize possible effects of microcystins on carbon fixation. After 2 days at 400 ppm, the pCO2 in the incubator was increased to 1,100 ppm. The pCO2 concentration in the gas mixture was checked regularly with an environmental gas monitor for CO2 (EGM-4; PP Systems, Amesbury, MA). The flasks were sampled on a daily basis, and after 4 days the experiment was ended.
pH, dissolved inorganic carbon (DIC), and cell counts.
The pH was measured immediately after sampling with a Lab 860 pH meter in combination with a BlueLine 28 Gel pH electrode (Schott Instruments GmbH, Mainz, Germany). To determine the concentrations of DIC, culture samples were immediately pelleted (5 min at 4,000 × g and 20°C). Supernatant was filtered over 0.45-μm-pore-size 47-mm polyethersulfone membrane filters (Sartorius AG, Goettingen, Germany). The filtrate was transferred to sterile plastic urine analysis tubes (VF-109SURI; Terumo Europe N.V., Leuven, Belgium), which were filled completely (using a needle to leave all air out), and stored at 4°C until analysis. A TOC-VCPH TOC analyzer (Shimadzu, Kyoto, Japan) was used to determine the DIC (measured 3- to 5-fold per sample). DIC concentrations were converted to CO2(aq), bicarbonate, and carbonate concentrations using the measured pH of the samples (36). Cell numbers and biovolumes of samples from the different cultures were determined in triplicate using a Casy 1 TTC cell counter with a 60-μm capillary (Schärfe System GmbH, Reutlingen, Germany). Because the strains differed in cell size, we report the cyanobacterial abundances as biovolumes.
RNA extraction.
Just before and 20 h after increasing pCO2 from 400 to 1,100 ppm, 40-ml samples were taken for reverse transcription-quantitative PCR (RT-qPCR) analysis, immediately cooled on ice, and centrifuged for 5 min at 4,000 × g and 4°C in a precooled centrifuge. The pellets were immediately resuspended in 1 ml of TRIzol (Life Technologies, Grand Island, NY), frozen in liquid nitrogen, and stored at −80°C. Subsequent RNA extraction and purification was performed as described preciously (32). RNA concentrations were quantified by using a NanoDrop 1000 spectrophotometer (Thermo Scientific, San Jose, CA), and all RNA samples had A260/A280 and A260/A230 values above 1.8.
RT-qPCR analysis.
We investigated the expression of CCM genes using primers designed in this and previous studies (17, 32) (see also Table S1 in the supplemental material). The transcripts of the following CCM genes were targeted: cmpA (encoding a subunit of the high-affinity bicarbonate transporter BCT1), bicA and sbtA (the two sodium-dependent bicarbonate transporters), chpX (dehydration subunit of the low-affinity CO2 uptake system NDH-I4), chpY (dehydration subunit of the high-affinity CO2 uptake system NDH-I3), ccmR and ccmR2 (two CCM transcriptional regulators), rbcX (chaperone for RuBisCO), ccmM (structural component of the carboxysomes), ccaA1 and ccaA2 (two carboxysomal carbonic anhydrases), and ecaA (periplasmic carbonic anhydrase). In addition, we targeted transcripts of the 16S rRNA gene and the mcyB gene (microcystin synthetase).
Reverse transcription reactions were done as described previously (32), using Superscript III (Life Technologies, Grand Island, NY). Subsequently, the qPCR Maxima SYBR green master mix (2×; Thermo Fisher Scientific, Pittsburgh, PA) was applied with our primers to the obtained cDNA samples as described previously (32) to analyze PCR amplification in a ABI 7500 real-time PCR device (Applied Biosystems, Foster City, CA). The two-step cycling protocol was used, with a denaturation temperature of 95°C (15 s) and a combined annealing/extension temperature of 60°C (60 s) during 40 cycles. Melting-curve analysis was performed on all measured samples to rule out nonspecific PCR products. ROX solution (passive reference dye) was used to correct for any well-to-well variation.
We calculated relative changes in gene expression after 20 h of elevated pCO2 using the 16S rRNA gene as a reference gene. The LinRegPCR software tool version 2012.3 (37, 38) was used for baseline correction, calculation of quantification cycle (Cq) values and calculation of the amplification efficiency (E) of each individual run using linear regression (see Table S1 in the supplemental material). Amplification efficiencies of individual samples were between 1.8 and 2.0. The relative changes in gene expression were calculated with the comparative cycle threshold (CT) method (39).
One-tailed t tests were applied to identify significant changes in gene expression (n = 4 biological replicates), using the false discovery rate (FDR) to correct for multihypothesis testing (55). FDR-adjusted P values of <0.05 combined with log2 expression changes of <−0.8 or >0.8 were considered significant.
O2 evolution experiments.
We studied the activity of different Ci uptake systems of Microcystis strains acclimated to low or high CO2 levels using O2 measurements with an Oxy-4 mini O2 optode (PreSens GmbH, Regensburg, Germany). In mineral medium without nitrate, the initial O2 evolution rate reflects the Ci uptake rate of cyanobacteria (32, 40). Cells acclimated to low CO2 levels were obtained from batch cultures exposed to 400 ppm pCO2 for 4 days. As a result, the Ci availability of the mineral medium was low, CO2(aq) was depleted to 0.0018 ± 0.0004 μmol liter−1 the and the bicarbonate concentration was 76 ± 10 μmol liter−1 (see Fig. S1 in the supplemental material). Cells acclimated to high CO2 levels were obtained from batch cultures exposed to 400 ppm of pCO2 for 2 days and subsequently to 1,100 ppm of pCO2 for 2 days. The Ci availability in these cultures was high, the CO2(aq) value was 4.2 ± 1.3 μmol liter−1, and the bicarbonate concentration was 1,062 ± 43 μmol liter−1 (see Fig. S1 in the supplemental material).
Samples from these batch cultures were pelleted (4,000 × g for 5 min at 20°C), washed once, and then resuspended in Ci-deplete and N-deplete modified BG11 medium (no added NaCO3/NaHCO3 and NaNO3, but with added 0.1 mmol liter−1 NaCl and 10 mmol liter−1 CAPSO-KOH [pH 9.8]). The medium was aerated with N2 gas before usage. The response of the cells was studied at pH 9.8 to mimic dense blooms in which bicarbonate is the dominant Ci species. The OD750 of washed and resuspended samples was 0.300, and 3 ml of these samples was inserted into custom-made double-walled glass incubation chambers equipped with sensors connected to the O2 optode device. The glass chambers were connected to a RM6 water bath (Lauda, Postfach, Germany) to keep the temperature of the samples constant at 20.3°C. Magnetic stirring was used for mixing. The O2 optode sensors were calibrated with N2 gas (0% oxygen) and pressurized air (21% oxygen). A saturating amount of light was provided by KL1500 compact Schott lamps (Schott AG, Mainz, Germany). Saturating light levels lead to high O2 evolution rates, which facilitate detection of differences between the treatments. In pilot experiments, we found that the photosynthetic rates (expressed per chlorophyll a [chl a]) of strains PCC 7806 and PCC 7005 were saturated at ∼400 μmol photons m−2 s−1 and did not decrease up to 1,000 μmol photons m−2 s−1. Therefore, we used an incident light intensity (Iin) of 500 μmol photons m−2 s−1 for the O2 evolution experiments.
At the start of the experiments, cells were allowed to take up all remaining Ci in the incubation chambers, which was monitored by a gradual decrease of the O2 evolution. Subsequently, the rate of O2 evolution (mg liter−1 min−1) was measured during 15-min intervals in a control treatment without additions and after adding 20, 300, or 10,000 μmol liter−1 of KHCO3 in the presence of different concentrations of NaCl and LiCl (Table 1). The different bicarbonate concentrations stimulate different Ci uptake systems. Sodium ions were added to stimulate the sodium-dependent bicarbonate transporters BicA and SbtA, whereas lithium ions were added to block bicarbonate uptake. In total, we applied six different treatments, which each activated or suppressed one or more different Ci uptake systems (Table 1). The units were converted to μmol O2 · mg−1 chl a · min−1 using data from chlorophyll a measurements that were acquired with HPLC as described previously (32). For each strain acclimated at low or high pCO2 levels, we tested whether O2 evolution rates were different between the treatments using one-way analysis of variance with post hoc comparison of the means based on Tukey's HSD test (α = 0.05; n = 4 per treatment).
TABLE 1.
Treatments in the O2 evolution experiments to study activity of the different Ci uptake systemsa
| Treatment |
Dissolved inorganic carbon |
Active Ci uptake system(s) | |||
|---|---|---|---|---|---|
| KHCO3 (μmol liter−1) | NaCl (mmol liter−1) | LiCl (mmol liter−1) | CO2(aq) (μmol liter−1) | HCO3− (μmol liter−1) | |
| 0 | 0.1 | 0 | 0 | 0 | None (control) |
| 20 | 0.1 | 0 | 0.006 | 15.80 | BCT1 |
| 20 | 25 | 0 | 0.006 | 15.80 | BCT1, SbtA |
| 300 | 25 | 0 | 0.090 | 236.7 | BCT1, SbtA, BicA |
| 300 | 25 | 25 | 0.090 | 236.7 | None |
| 10,000 | 25 | 25 | 3.000 | 7891 | NDH-I3, NDH-I4 |
Different concentrations of KHCO3, NaCl, and LiCl were added to induce or block the activity of specific Ci uptake systems. The resulting CO2(aq) and HCO3− concentrations expected at pH 9.8 and 20.3°C are shown. The last column indicates which Ci uptake systems are mostly active at the applied conditions.
RESULTS
Changes in DIC and pH at elevated CO2.
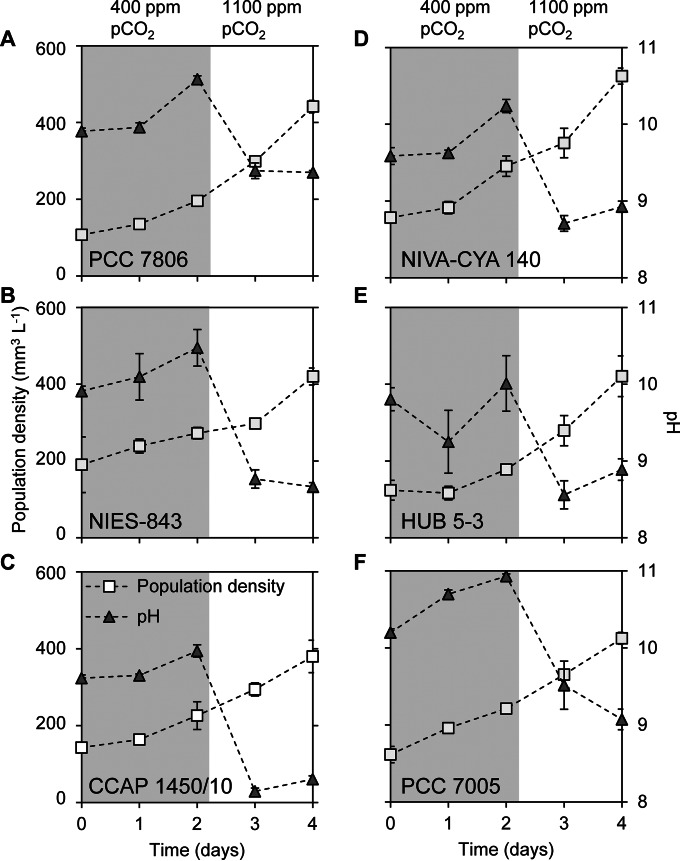
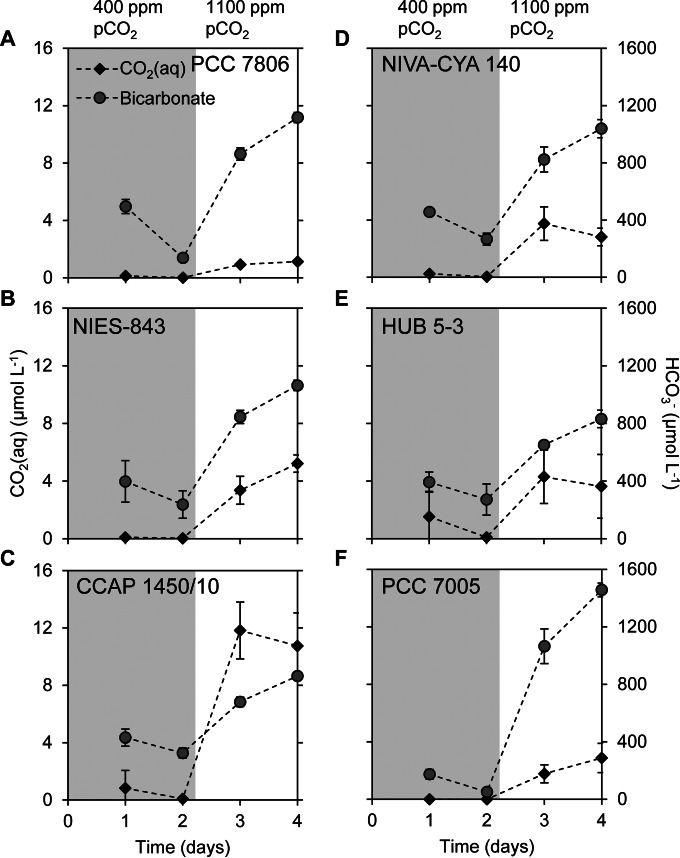
Six Microcystis strains (PCC 7806, NIES-843, CCAP 1450/10, NIVA-CYA 140, HUB 5-3, and PCC 7005) with different Ci uptake systems were grown under atmospheric pCO2 conditions of 400 ppm (Fig. 2). Assuming equilibrium with this atmospheric pressure, one would expect a CO2(aq) concentration of 13.5 μmol liter−1, and the pH of the mineral medium without cells would be ∼7. However, owing to the photosynthetic activity of the Microcystis population, the CO2(aq) concentration was depleted to <0.1 μmol liter−1, the bicarbonate concentration was <330 μmol liter−1, and the pH increased to 10 to 11 depending on the strain (Fig. 2 and 3). After 2 days, we raised the pCO2 in the gas flow to 1,100 ppm. As a consequence, the CO2(aq) concentration increased to 1 to 12 μmol liter−1 depending on the strain, the bicarbonate concentration increased to 800 to 1,500 μmol liter−1, and the pH dropped 1 to 2 U (Fig. 2 and 3).
FIG 2.
Growth and pH during the exponential phase in batch cultures of six Microcystis strains. (A) PCC 7806; (B) NIES-843; (C) CCAP 1450/10; (D) NIVA-CYA 140; (E) HUB 5-3; (F) PCC 7005. The batch cultures were exposed to 400 ppm of pCO2 in the gas flow for 2 days (shaded area), and thereafter the pCO2 concentration was increased to 1,100 ppm (unshaded area). The mineral medium contained 10 mmol liter−1 sodium ions. Error bars indicate the standard deviations (n = 4 biological replicates).
FIG 3.
Dissolved CO2 and bicarbonate concentration during the exponential phase in batch cultures of six Microcystis strains. (A) PCC 7806; (B) NIES-843; (C) CCAP 1450/10; (D) NIVA-CYA 140; (E) HUB 5-3; (F) PCC 7005. The batch cultures were exposed to 400 ppm of pCO2 in the gas flow for 2 days (shaded area), and thereafter the pCO2 concentration was increased to 1,100 ppm (unshaded area). The mineral medium contained 10 mmol liter−1 sodium ions. Error bars indicate the standard deviations (n = 4 biological replicates).
Changes in gene expression at elevated CO2.
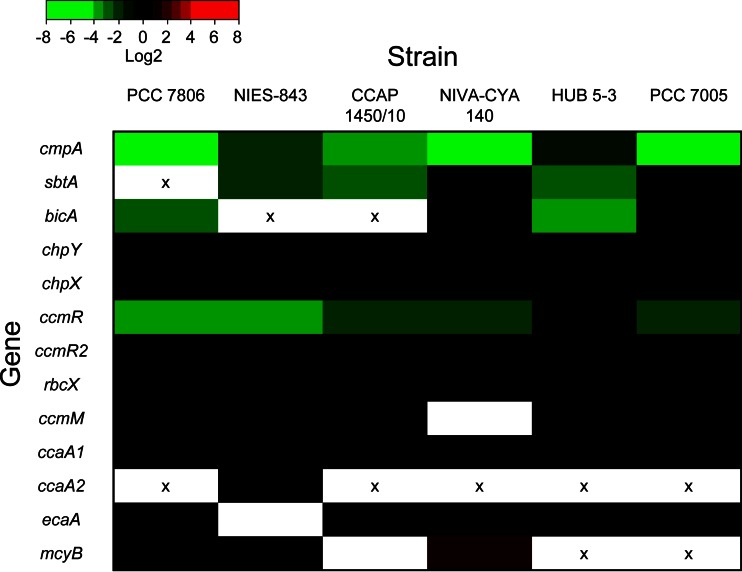
The expression of selected CCM genes was monitored before and 20 h after increasing the pCO2 in the gas flow (Fig. 4 and Table S2 in the supplemental material). At an elevated pCO2, all strains showed significant downregulation of cmpA, although four strains (PCC 7806, CCAP 1450/10, NIVA-CYA 140, and PCC 7005) showed a stronger downregulation than two others (NIES-843 and HUB 5-3). The sbtA gene encoding the high-affinity bicarbonate transporter SbtA was significantly downregulated in strains NIES-843, CCAP 1450/10, and HUB 5-3 but was constitutively expressed in strains NIVA-CYA 140 and PCC 7005. The bicA gene encoding the low-affinity bicarbonate transporter BicA was downregulated in strains PCC 7806 and HUB 5-3 but was also constitutively expressed in strains NIVA-CYA 140 and PCC 7005. None of the six strains showed significant changes in gene expression of the CO2 uptake genes chpX and chpY (Fig. 4).
FIG 4.
Heat map of changes in gene expression at elevated CO2 for each of the six Microcystis strains. Gene expression changes were obtained by RT-qPCR applied to samples taken before and 20 h after increasing the pCO2 level from 400 to 1,100 ppm. The color bar indicates log2 values. Significant downregulated genes are shown in green, significant upregulated genes are shown in red, and nonsignificant changes (P > 0.05) or log2 values between −0.8 and 0.8 are shown in black. Genes not measured are shown in white and genes absent in strains are marked with an “x”. Detailed RT-qPCR results are presented in Table S2 in the supplemental material.
Expression of the CCM transcriptional regulator ccmR was reduced significantly at elevated CO2 levels in all strains, except for strain HUB 5-3. In contrast, expression of the additional transcriptional regulator ccmR2 located upstream of the bicA-sbtA operon of Microcystis did not change significantly in any of the strains. The expression of several other CCM genes (rbcX, ccmM, ccaA1, ccaA2, and ecaA) was also not affected by elevated CO2 in any of the strains. The expression of mcyB was unaltered in PCC 7806 and NIES-843 and increased slightly but significantly in NIVA-CYA 140.
Ci uptake activity of low- and high-CO2 acclimated cells.
We studied O2 evolution of the Microcystis strains to compare the activity of their Ci uptake systems when the strains were acclimated to low- or high-CO2 conditions (Fig. 5). We applied six different treatments to activate different Ci uptake systems, as explained in Table 1. For the interpretation of these results, we note that the response of the O2 evolution rates to the treatments can be compared within each strain at a given pCO2 level (i.e., within the panels of Fig. 5) but cannot be compared quantitatively among the strains or among the two different pCO2 levels (i.e., among the panels of Fig. 5). The reason is that other factors also influence the O2 evolution rates, for example, the pigment concentrations and amounts of PSI and PSII may differ between strains and can also change with elevated CO2 (32). The results can be compared among strains in a relative sense, for example, two different strains can show a significant increase in O2 evolution after addition of 20 μmol liter−1 KHCO3, while a third strain does not. The O2 evolution rates of strain NIES-843 were highly variable among the biological replicates and declined strongly after 2 h. Repetition of the O2 evolution experiments with this strain did not improve the results, indicating that NIES-843 could not withstand the incubation conditions. Therefore, we only report the results for the other five Microcystis strains.
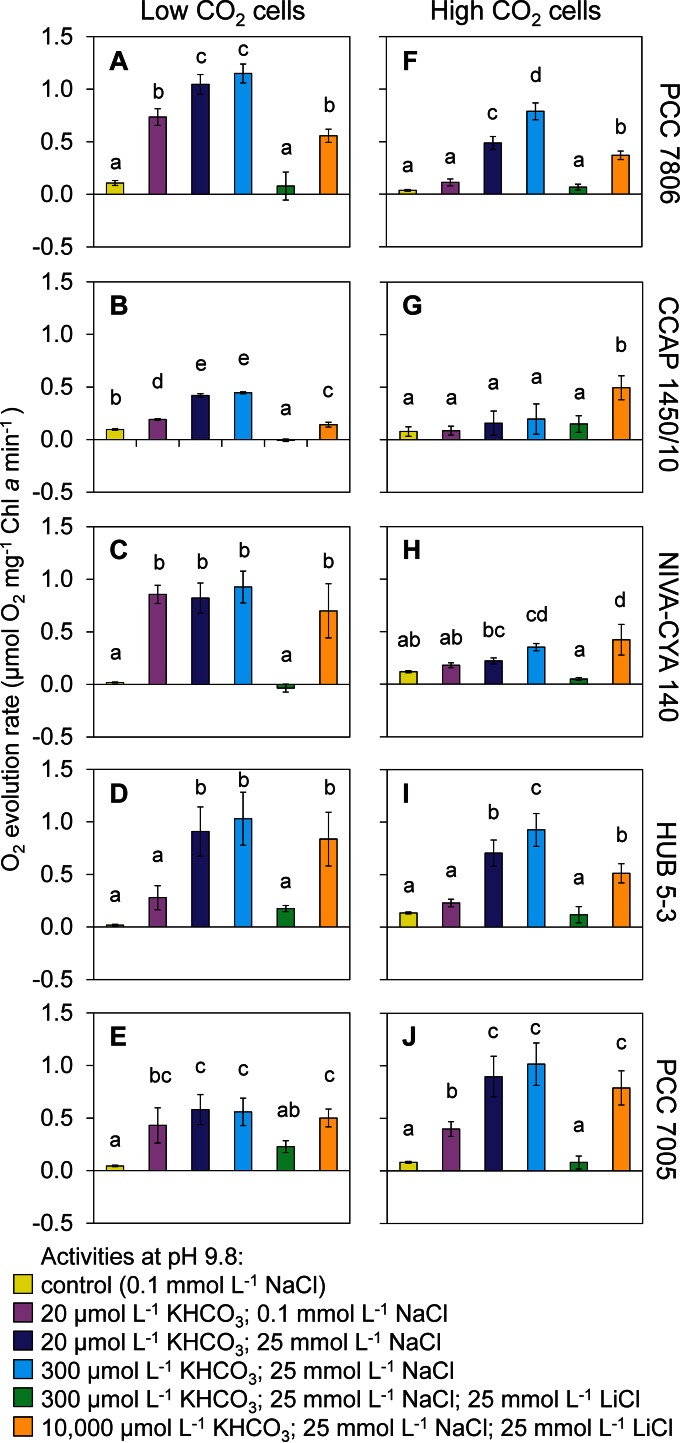
FIG 5.
Activity of Ci uptake systems of five Microcystis strains, inferred from O2 evolution. The strains were acclimated to either low CO2 levels (A to E) or high CO2 levels (F to J). O2 evolution was measured after addition of different concentrations of KHCO3, NaCl, and LiCl to induce or block the activity of specific Ci uptake systems, as indicated in Table 1. Error bars indicate standard deviations (n = 4 biological replicates per treatment). Different letters above the bars indicate significant differences between the treatments, as tested by one-way analysis of variance with post hoc comparison of the means based on Tukey's HSD test (α = 0.05).
For cells acclimated to low-Ci conditions, the addition of 20 μmol liter−1 KHCO3 in the presence of only 0.1 mmol liter−1 NaCl induced significantly more O2 production than the control for all strains, except for strain HUB 5-3 which showed a nonsignificant response (Fig. 5A to E). This result indicates that BCT1 was active in all strains, although its activity was low in strain HUB 5-3. Application of 20 μmol liter−1 KHCO3 and 25 mmol liter−1 NaCl led to a significantly higher O2 production than at 20 μmol liter−1 KHCO3 and 0.1 mmol liter−1 NaCl for strains PCC 7806, CCAP 1450/10, and HUB 5-3. This result indicates that in response to the added sodium ions, these three strains activated their sodium-dependent bicarbonate transporter SbtA. However, strain PCC 7806 does not have SbtA, and its response might indicate activation of the other sodium-dependent bicarbonate transporter BicA. The O2 production at 300 μmol liter−1 KHCO3 and 25 mmol liter−1 NaCl was not significantly higher than at 20 μmol liter−1 KHCO3 and 25 mmol liter−1 NaCl for any of the strains. This result indicates that cells acclimated to low-Ci conditions relied largely on the high-affinity transporters BCT1 and SbtA for their bicarbonate uptake. Subsequent addition of 25 mmol liter−1 LiCl (in the presence of 300 μmol liter−1 KHCO3 and 25 mmol liter−1 NaCl) blocked bicarbonate uptake and, as a consequence, the O2 production of all strains was reduced to levels similar to that for the control treatment. Finally, the addition of 10,000 μmol liter−1 KHCO3 in the presence of 25 mmol liter−1 LiCl and 25 mmol liter−1 NaCl strongly increased the CO2(aq) concentration in the medium (see Table S1 in the supplemental material) and restored O2 production in all strains (Fig. 5A to E). Since lithium still blocked bicarbonate uptake, this result indicates that CO2 uptake was active in all strains.
For cells acclimated to high-Ci conditions, the addition of 20 μmol liter−1 KHCO3 in the presence of only 0.1 mmol liter−1 NaCl induced significantly more O2 production than the control only for strain PCC 7005 (Fig. 5F to J). This result indicates that the high-affinity bicarbonate transporter BCT1 was hardly active in any of the strains acclimated to high-Ci conditions, except for strain PCC 7005. Application of 20 μmol liter−1 KHCO3 and 25 mmol liter−1 NaCl led to a significantly higher O2 production than at 0.1 mmol liter−1 NaCl for strains PCC 7806, HUB 5-3, and PCC 7005, indicating that these strains activated their sodium-dependent bicarbonate transporter SbtA. Strain PCC 7806 does not have SbtA, and its response might indicate activation of the other sodium-dependent bicarbonate transporter BicA. The O2 production at 300 μmol liter−1 KHCO3 and 25 mmol liter−1 NaCl was significantly higher than at 20 μmol liter−1 KHCO3 and 25 mmol liter−1 NaCl for strains PCC 7806 and HUB 5-3, indicating that their low-affinity but high-flux bicarbonate transporter BicA was active when cells were acclimated to high-Ci conditions. Subsequent addition of 25 mmol liter−1 LiCl blocked bicarbonate uptake and, as a consequence, the O2 production of all strains was reduced to levels similar to that for the control treatment. Finally, addition of 10,000 μmol liter−1 KHCO3 in the presence of 25 mmol liter−1 LiCl and 25 mmol liter−1 NaCl increased O2 production in comparison to the control (Fig. 5F to J), indicating that CO2 uptake was active in all strains.
DISCUSSION
Evaluation of hypotheses.
Our results enable evaluation of the hypotheses that (i) high-affinity Ci uptake genes of Microcystis are downregulated at elevated CO2 (1,100 ppm), whereas (ii) low-affinity but high-flux Ci-uptake genes are constitutively expressed. Consistent with the first hypothesis, our results show that the cmpA gene encoding the bicarbonate-binding subunit of the high-affinity bicarbonate transporter BCT1 was downregulated in all strains. Downregulation of BCT1 at elevated CO2 could potentially be cost-effective for the cells, because bicarbonate uptake by BCT1 is expected to require 1 ATP molecule per molecule of bicarbonate (Table 2). However, the other high-affinity bicarbonate uptake gene, sbtA, was downregulated at elevated CO2 in only three of the five sbtA-containing strains. Furthermore, the gene chpY encoding the dehydration subunit of the high-affinity CO2 uptake system NDH-I3 was not downregulated at elevated CO2 in any of the strains. Hence, the first hypothesis applies to cmpA (BCT1) and partly to sbtA of the two high-affinity bicarbonate uptake systems but does not apply to chpY of the high-affinity CO2 uptake system.
TABLE 2.
Gene expression of the five different Ci uptake systems in Microcystis and three model cyanobacterial strainsa
| Ci uptake system (genes involved) | Energy cost (21, 51, 52) | Gene expressionb |
|||
|---|---|---|---|---|---|
| Microcystis strains (this study) | Synechocystis PCC 6803 (29, 30) | Synechococcus PCC 7002 (50) | Synechococcus PCC 7942 (45, 46) | ||
| BCT1 (cmpABCD) | 1 ATP per HCO3− | Inducible under low pCO2 | Inducible under low pCO2 | – | Inducible under low pCO2 |
| SbtA (sbtA) | 0.5 ATP per HCO3− | Constitutively expressed/inducible under low pCO2/– | Inducible under low pCO2 | Inducible under low pCO2 | Inducible under low pCO2 |
| BicA (bicA) | 0.25 ATP per HCO3− | Constitutively expressed/inducible under low pCO2/– | Constitutively expressed | Inducible under low pCO2 | – |
| NDH-I3 (chpY and others) | 1 NADPH per CO2-to-HCO3− conversion | Constitutively expressed | Inducible under low pCO2 | Inducible under low pCO2 | Inducible under low pCO2 |
| NDH-I4 (chpX and others) | 1 NADPH per CO2-to-HCO3− conversion | Constitutively expressed | Constitutively expressed | Constitutively expressed | Constitutively expressed |
The estimated energy costs of the different Ci uptake systems are indicated in terms of molecules of ATP or NADPH per molecule CO2 or HCO3−. The source or reference(s) are indicated in parentheses in the headings for each column of data. A dash (−) indicates that the Ci uptake system is absent.
The Microcystis strains, Synechocystis PCC 6803, and Synechococcus PCC 7942 were from freshwater (brackish water for Microcystis PCC 7806); Synechococcus PCC 7002 has a marine origin.
Consistent with the second hypothesis, the gene chpX encoding the dehydration subunit of the low-affinity CO2 uptake system NDH-I4 was constitutively expressed in all strains. However, the low-affinity bicarbonate uptake gene, bicA, was constitutively expressed only in strains NIVA-CYA 140 (where it is not functional because of a transposon insert; Fig. 1) and PCC 7005 but was downregulated at elevated CO2 in strains PCC 7806 and HUB 5-3. Hence, the second hypothesis is supported by chpX (NDH-I4), whereas the low-affinity bicarbonate uptake gene bicA shows a more variable response.
General observations.
Given that both hypotheses received only partial support, what general observations can still be obtained from the gene expression patterns of the Microcystis strains? First, it is noteworthy that several Ci uptake systems and one of their transcriptional regulators were regulated in response to elevated CO2, whereas other important CCM genes encoding the enzyme RuBisCO, structural components of the carboxysome, and carbonic anhydrases were not regulated at all (Fig. 4). Furthermore, a recent transcriptome study of Microcystis PCC 7806 found that expression of the ppc gene, encoding phosphoenolpyruvate carboxylase involved in an alternative Ci assimilation pathway, also remained constant under elevated-CO2 conditions (see Table S4 in the supplemental material in reference 32). These results indicate that the CCM genes of Microcystis respond to elevated CO2 mainly at the very first steps of the carbon fixation process by regulating the initial acquisition of inorganic carbon. Microcystis is a buoyant cyanobacterium that can develop dense blooms in eutrophic lakes, where it will be exposed to large fluctuations in CO2 availability at both daily and seasonal time scales (41). A highly specific response that mainly adjusts the initial Ci uptake systems, without large changes in expression of the carboxysome genes and genes of the downstream carbon assimilation pathways, could preserve energy and offer a robust strategy for a species that often experiences strongly fluctuating Ci conditions (32).
Second, all of the Ci uptake genes investigated in the present study were either downregulated or remained unchanged at elevated CO2; none of them were upregulated. Hence, all Ci uptake systems that a strain was capable to produce were available for the cells at low CO2 levels, including the low-affinity Ci uptake systems. Third, the genes chpX and chpY of both CO2 uptake systems were expressed constitutively, which might again be an adaptation to fluctuating CO2 conditions. Constitutive expression of the high-affinity CO2 uptake system NDH-I3 might also be an adaptation to intercept low intracellular concentrations of CO2 leaking from the carboxysomes.
Methodological aspects.
Previously, the cellular response of strain PCC 7806 to elevated CO2 was investigated in highly controlled chemostats using whole-genome microarrays (32). In the present study, we simplified the experimental setup to batch cultures and limited our analysis to a smaller set of genes using RT-qPCR, which enabled investigation of a larger number of strains. We included strain PCC 7806 in our present study to check the consistency of the results.
Strain PCC 7806 downregulated expression of cmpA and bicA at elevated CO2 in both the previous and the present study (Fig. 4). Furthermore, both studies showed constitutive expression of the CO2 uptake, carboxysomal and RuBisCO genes, and downregulation of the transcriptional regulator gene ccmR, although ccmR2 was only downregulated in the previous study (32). Moreover, the O2 evolution data of PCC 7806 show that BCT1 and BicA were both active under low-pCO2 conditions (Fig. 5A), whereas BicA but not BCT1 was active under high-pCO2 conditions (Fig. 5F), in agreement with the findings of the previous study (32). Hence, the results of both studies are in good agreement, which gives confidence in the applied methods.
In our O2 evolution experiments, the cells were exposed to different salt treatments (Table 1). This could potentially bias the results because Microcystis strains differ in their salt tolerance and potassium ion sensitivity (42). In particular, strain PCC 7005 is very sensitive to elevated potassium ion concentrations and lacks several salt tolerance genes, whereas strain PCC 7806 is much more tolerant to potassium ions and can withstand 170 mmol liter−1 NaCl (42, 43). We therefore tried to minimize salt stress by exposing the cells to the different treatments for only 15 min. The results show that even the salt- and potassium-sensitive strain PCC 7005 maintained high O2 evolution rates in the treatment with the highest salinity (10 mmol liter−1 KHCO3, 25 mmol liter−1 LiCl and 25 mmol liter−1 NaCl; Fig. 5), and hence the applied salinities apparently did not hinder the activity of the cells during this short time interval.
Expression and activity of the sodium-dependent Ci uptake genes.
Expression of the sodium-dependent bicarbonate uptake genes bicA and sbtA varied widely among the strains. Even similar genotypes, such as strains HUB 5-3 and PCC 7005 that both belong to genotype III (bicA-sbtA), showed contrasting expression patterns for their bicarbonate uptake genes (Fig. 4). These two strains have bicA and sbtA located in one operon (17), and hence cotranscription explains why the expression patterns of these two genes are coupled (Fig. 4). The O2 evolution data indicate that HUB 5-3 mainly relies on its sodium-dependent bicarbonate uptake systems under low-Ci conditions (Fig. 5D), whereas PCC 7005 mainly relies on ATP-dependent bicarbonate uptake by BCT1 (Fig. 5E). Hence, a reduction of the cellular investments in bicarbonate uptake at elevated CO2 would be most effective by downregulation of the bicA-sbtA operon for HUB 5-3 and by downregulation of BCT1 for PCC 7005, a finding in line with the observed changes in gene expression (Fig. 4).
Strain NIVA-CYA 140 contains sbtA but has a transposon insert in the middle of a complete bicA gene (Fig. 1) and was therefore assigned to Ci uptake genotype II (no bicA) (17). At low pCO2 the transcription level of sbtA (relative to 16S rRNA) was ∼45× lower in this strain than in the other strains, indicating that the transposon insert interfered with transcription of the bicA-sbtA operon. Indeed, the strain depended strongly on BCT1 under low-pCO2 conditions, as evidenced from the lack of stimulation by added sodium ions in the O2 evolution experiments (Fig. 5C), whereas the strain depended mainly on CO2 uptake under high-pCO2 conditions (Fig. 5H). Hence, most likely this strain does not use the sodium-dependent bicarbonate uptake systems BicA and SbtA and therefore has a phenotype that deviates from all other Microcystis strains.
Strain CCAP 1450/10 also has a transposon insert, located between a small bicA fragment and a complete sbtA gene (Fig. 1). However, the gene expression results (Fig. 4) and the O2 evolution data (Fig. 5B) show that the transposon insert did not hinder expression of sbtA in this strain.
Comparison of CCM gene regulation of Microcystis with other cyanobacteria.
Previously, the CCM genes of the model cyanobacteria Synechocystis PCC 6803, Synechococcus PCC 7002, and Synechococcus 7942 were studied in detail (15, 19, 27–30, 44–48). Comparison of the CCM genes of our Microcystis strains with these model cyanobacteria reveals several similarities and differences (Tables 2 and 3).
TABLE 3.
Presence and function of the CCM transcriptional regulators in Microcystis and three model cyanobacterial strainsa
| Transcriptional regulator | Location in genome | Function |
|||
|---|---|---|---|---|---|
| Microcystis (17; this study) | Synechocystis PCC 6803 (29, 53) | Synechococcus PCC 7002 (50) | Synechococcus PCC 7942 (46, 54) | ||
| CcmR | Upstream of high-affinity CO2 uptake operon | Repressor/activator of cmpABCD operon (BCT1) | Repressor of sbtA and high-affinity CO2 uptake operon (not bicA) | Repressor of sbtA and bicA (possibly high-affinity CO2 uptake operon) | – |
| CcmR2 | Upstream of bicA-sbtA operon | Repressor/activator of bicA-sbtA operon | – | – | – |
| CmpR | Upstream of cmpABCD operon (BCT1) or separate location | – | Activator of cmpABCD operon | – | Activator of cmpABCD operon (possibly a repressor of sbtA and high-affinity CO2 uptake operon) |
The source or reference(s) are indicated in parentheses in the headings for each column of data. A dash (−) indicates that the transcriptional regulator is absent.
In all cyanobacteria investigated thus far, genes encoding the ATP-dependent high-affinity bicarbonate transporter BCT1 are induced under low-Ci conditions, whereas genes encoding the low-affinity CO2 uptake system NDH-I4 are constitutively expressed (Table 2). Hence, our two hypotheses do apply to the genes of these two uptake systems. The constitutive expression of genes encoding the high-affinity CO2 uptake system NDH-I3 in all Microcystis strains and the high-affinity bicarbonate transporter SbtA in some Microcystis strains deviates from the induction of these genes in the other three cyanobacteria. The presence and expression of bicA appears to be quite variable, not only in Microcystis but also in other cyanobacteria.
The CCM transcriptional regulators also differ among the cyanobacteria (Table 3). CcmR can regulate transcription of several Ci uptake genes. In Synechocystis PCC 6803, CcmR appeared to be a repressor of sbtA and the high-affinity CO2 uptake operon but not of bicA (29, 47). In contrast, in Synechococcus PCC 7002, CcmR appeared to be a repressor of bicA and sbtA and possibly the high-affinity CO2 uptake operon (48, 50). In Microcystis, CcmR probably regulates expression of the cmpABCD operon (encoding BCT1), since downregulation of ccmR at elevated pCO2 coincided with downregulation of the cmpA gene (Fig. 4). Synechocystis PCC 6803 and Synechococcus PCC 7942 use another transcriptional regulator, CmpR, for the cmpABCD operon (27, 47), which is absent from Microcystis. CcmR2 is the most likely transcriptional regulator for the bicA and sbtA genes in Microcystis, given the location of ccmR2 upstream of the bicA-sbtA operon (17).
Ecological implications.
In conclusion, our results reveal an unexpected diversity in CO2 responses of cyanobacteria. It was already known that Microcystis strains differ in their Ci uptake genes, which promotes variation in their CO2 response (17). Our results show that, on top of this genotypic diversity, there is also considerable phenotypic variation because strains with the same Ci uptake genes can show contrasting expression patterns and may differ widely in the activity of their Ci uptake systems. In other words, cyanobacterial strains differ in their adaptation to changing CO2 conditions not only because of variation in genetic composition but also because of further variation at the transcriptional and physiological level.
It is often argued that cyanobacteria generally have a very effective CCM and are therefore particularly strong competitors at low CO2 levels in comparison to eukaryotic phytoplankton (9). However, we know now that there is major variation in the CCM tactics among cyanobacteria and even among different strains within the same genus. Some Microcystis strains perform well at low CO2, whereas other strains are much better competitors under high-CO2 conditions (17, 49). This genetic and phenotypic variation in Ci uptake systems provides cyanobacterial communities with the potential for rapid adaptation and acclimation to changing CO2 conditions. These differential responses also indicate that the ongoing rise in atmospheric CO2 concentrations is likely to be more beneficial for some cyanobacterial strains than for others, which may lead to major changes in the genetic composition of harmful cyanobacterial blooms.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the reviewers for their helpful comments.
This research was supported by the Division of Earth and Life Sciences (ALW) of the Netherlands Organization for Scientific Research (NWO).
We declare that we have no conflict of interest related to the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02295-15.
REFERENCES
- 1.IPCC. 2014. Climate change 2014: synthesis report: contribution of working groups I, II, and III to the fifth assessment report of the Intergovernmental Panel on Climate Change. IPCC, Geneva, Switzerland. [Google Scholar]
- 2.Orr JC, Fabry VJ, Aumont O, Bopp L, Doney SC, Feely RA, Gnanadesikan A, Gruber N, Ishida A, Joos F, Key RM, Lindsay K, Maier-Reimer E, Matear R, Monfray P, Mouchet A, Najjar RG, Plattner GK, Rodgers KB, Sabine CL, Sarmiento JL, Schlitzer R, Slater RD, Totterdell IJ, Weirig MF, Yamanaka Y, Yool A. 2005. Anthropogenic ocean acidification over the twenty-first century and its impact on calcifying organisms. Nature 437:681–686. doi: 10.1038/nature04095. [DOI] [PubMed] [Google Scholar]
- 3.Doney SC, Fabry VJ, Feely RA, Kleypas JA. 2009. Ocean acidification: the other CO2 problem. Annu Rev Mar Sci 1:169–192. doi: 10.1146/annurev.marine.010908.163834. [DOI] [PubMed] [Google Scholar]
- 4.Cole JJ, Caraco NF, Kling GW, Kratz TK. 1994. Carbon dioxide supersaturation in the surface waters of lakes. Science 265:1568–1570. doi: 10.1126/science.265.5178.1568. [DOI] [PubMed] [Google Scholar]
- 5.Sobek S, Tranvik LJ, Cole JJ. 2005. Temperature independence of carbon dioxide supersaturation in global lakes. Global Biogeochem Cycles 19:GB2003. doi: 10.1029/2004GB002264. [DOI] [Google Scholar]
- 6.Talling JF. 1976. The depletion of carbon dioxide from lake water by phytoplankton. J Ecol 64:79–121. doi: 10.2307/2258685. [DOI] [Google Scholar]
- 7.Balmer MB, Downing JA. 2011. Carbon dioxide concentrations in eutrophic lakes: undersaturation implies atmospheric uptake. Inland Waters 1:125–132. doi: 10.5268/IW-1.2.366. [DOI] [Google Scholar]
- 8.Verspagen JMH, Van de Waal DB, Finke JF, Visser PM, Van Donk E, Huisman J. 2014. Rising CO2 levels will intensify phytoplankton blooms in eutrophic and hypertrophic lakes. PLoS One 9:e104325. doi: 10.1371/journal.pone.0104325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro J. 1997. The role of carbon dioxide in the initiation and maintenance of blue-green dominance in lakes. Freshwat Biol 37:307–323. doi: 10.1046/j.1365-2427.1997.00164.x. [DOI] [Google Scholar]
- 10.Low-Décarie E, Fussmann GF, Bell G. 2011. The effect of elevated CO2 on growth and competition in experimental phytoplankton communities. Glob Change Biol 17:2525–2535. doi: 10.1111/j.1365-2486.2011.02402.x. [DOI] [Google Scholar]
- 11.Paerl HW, Huisman J. 2008. Blooms like it hot. Science 320:57–58. doi: 10.1126/science.1155398. [DOI] [PubMed] [Google Scholar]
- 12.Davis TW, Berry DL, Boyer GL, Gobler CJ. 2009. The effects of temperature and nutrients on the growth and dynamics of toxic and non-toxic strains of Microcystis during cyanobacteria blooms. Harmful Algae 8:715–725. doi: 10.1016/j.hal.2009.02.004. [DOI] [Google Scholar]
- 13.O'Neil JM, Davis TW, Burford MA, Gobler CJ. 2012. The rise of harmful cyanobacteria blooms: the potential roles of eutrophication and climate change. Harmful Algae 14:313–334. doi: 10.1016/j.hal.2011.10.027. [DOI] [Google Scholar]
- 14.Price GD. 2011. Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth Res 109:47–57. doi: 10.1007/s11120-010-9608-y. [DOI] [PubMed] [Google Scholar]
- 15.Price GD, Woodger FJ, Badger MR, Howitt SM, Tucker L. 2004. Identification of a SulP-type bicarbonate transporter in marine cyanobacteria. Proc Natl Acad Sci U S A 101:18228–18233. doi: 10.1073/pnas.0405211101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rae BD, Forster B, Badger MR, Price GD. 2011. The CO2-concentrating mechanism of Synechococcus WH5701 is composed of native and horizontally acquired components. Photosynth Res 109:59–72. doi: 10.1007/s11120-011-9641-5. [DOI] [PubMed] [Google Scholar]
- 17.Sandrini G, Matthijs HCP, Verspagen JMH, Muyzer G, Huisman J. 2014. Genetic diversity of inorganic carbon uptake systems causes variation in CO2 response of the cyanobacterium Microcystis. ISME J 8:589–600. doi: 10.1038/ismej.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du J, Förster B, Rourke L, Howitt SM, Price GD. 2014. Characterisation of cyanobacterial bicarbonate transporters in Escherichia coli shows that SbtA homologs are functional in this heterologous expression system. PLoS One 9:e115905. doi: 10.1371/journal.pone.0115905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Omata T, Price GD, Badger MR, Okamura M, Gohta S, Ogawa T. 1999. Identification of an ATP-binding cassette transporter involved in bicarbonate uptake in the cyanobacterium Synechococcus sp. strain PCC 7942. Proc Natl Acad Sci U S A 96:13571–13576. doi: 10.1073/pnas.96.23.13571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maeda S, Badger MR, Price GD. 2002. Novel gene products associated with NdhD3/D4-containing NDH-1 complexes are involved in photosynthetic CO2 hydration in the cyanobacterium, Synechococcus sp. PCC7942. Mol Microbiol 43:425–435. doi: 10.1046/j.1365-2958.2002.02753.x. [DOI] [PubMed] [Google Scholar]
- 21.Price GD, Maeda S, Omata T, Badger MR. 2002. Modes of active inorganic carbon uptake in the cyanobacterium, Synechococcus sp. PCC7942 Funct Plant Biol 29:131–149. doi: 10.1071/PP01229. [DOI] [PubMed] [Google Scholar]
- 22.Verspagen JMH, Passarge J, Jöhnk KD, Visser PM, Peperzak L, Boers P, Laanbroek HJ, Huisman J. 2006. Water management strategies against toxic Microcystis blooms in the Dutch delta. Ecol Appl 16:313–327. doi: 10.1890/04-1953. [DOI] [PubMed] [Google Scholar]
- 23.Qin B, Zhu G, Gao G, Zhang Y, Li W, Paerl HW, Carmichael WW. 2010. A drinking water crisis in Lake Taihu, China: linkage to climatic variability and lake management. Environ Management 45:105–112. doi: 10.1007/s00267-009-9393-6. [DOI] [PubMed] [Google Scholar]
- 24.Michalak AM, Anderson EJ, Beletsky D, Boland S, Bosch NS, Bridgeman TB, Chaffin JD, Cho K, Confesor R, Daloglu I, Depinto JV, Evans MA, Fahnenstiel GL, He L, Ho JC, Jenkins L, Johengen TH, Kuo KC, Laporte E, Liu X, McWilliams MR, Moore MR, Posselt DJ, Richards RP, Scavia D, Steiner AL, Verhamme E, Wright DM, Zagorski MA. 2013. Record-setting algal bloom in Lake Erie caused by agricultural and meteorological trends consistent with expected future conditions. Proc Natl Acad Sci U S A 110:6448–6452. doi: 10.1073/pnas.1216006110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jochimsen EM, Carmichael WW, An JS, Cardo DM, Cookson ST, Holmes CE, Antunes MB, de Melo Filho DA, Lyra TM, Barreto VS, Azevedo SM, Jarvis WR. 1998. Liver failure and death after exposure to microcystins at a hemodialysis center in Brazil. N Engl J Med 338:873–878. doi: 10.1056/NEJM199803263381304. [DOI] [PubMed] [Google Scholar]
- 26.Huisman JM, Matthijs HCP, Visser PM. 2005. Harmful cyanobacteria. Springer, Dordrecht, The Netherlands. doi: 10.1007/1-4020-3022-3. [DOI] [Google Scholar]
- 27.Omata T, Gohta S, Takahashi Y, Harano Y, Maeda S. 2001. Involvement of a CbbR homolog in low CO2-induced activation of the bicarbonate transporter operon in cyanobacteria. J Bacteriol 183:1891–1898. doi: 10.1128/JB.183.6.1891-1898.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGinn PJ, Price GD, Maleszka R, Badger MR. 2003. Inorganic carbon limitation and light control the expression of transcripts related to the CO2-concentrating mechanism in the cyanobacterium Synechocystis sp. strain PCC6803. Plant Physiol 132:218–229. doi: 10.1104/pp.019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang HL, Postier BL, Burnap RL. 2004. Alterations in global patterns of gene expression in Synechocystis sp. PCC 6803 in response to inorganic carbon limitation and the inactivation of ndhR, a LysR family regulator. J Biol Chem 279:5739–5751. doi: 10.1074/jbc.M311336200. [DOI] [PubMed] [Google Scholar]
- 30.Eisenhut M, von Wobeser EA, Jonas L, Schubert H, Ibelings BW, Bauwe H, Matthijs HCP, Hagemann M. 2007. Long-term response toward inorganic carbon limitation in wild type and glycolate turnover mutants of the cyanobacterium Synechocystis sp. strain PCC 6803. Plant Physiol 144:1946–1959. doi: 10.1104/pp.107.103341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penn K, Wang J, Fernando S, Thompson J. 2014. Secondary metabolite gene expression and interplay of bacterial functions in a tropical freshwater cyanobacterial bloom. ISME J 8:1866–1878. doi: 10.1038/ismej.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandrini G, Cunsolo S, Schuurmans JM, Matthijs HCP, Huisman J. 2015. Changes in gene expression, cell physiology and toxicity of the harmful cyanobacterium Microcystis aeruginosa at elevated CO2. Front Microbiol 6:401. doi: 10.3389/fmicb.2015.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steffen MM, Belisle BS, Watson SB, Boyer GL, Bourbonniere RA, Wilhelm SW. 2015. Metatranscriptomic evidence for co-occurring top-down and bottom-up controls on toxic cyanobacterial communities. Appl Environ Microbiol 81:3268–3276. doi: 10.1128/AEM.04101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rippka R, Deruelles J, Waterbury JB, Herdman M, Stanier RY. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111:1–61. doi: 10.1099/00221287-111-1-1. [DOI] [Google Scholar]
- 35.Zilliges Y, Kehr JC, Meissner S, Ishida K, Mikkat S, Hagemann M, Kaplan A, Börner T, Dittmann E. 2011. The cyanobacterial hepatotoxin microcystin binds to proteins and increases the fitness of Microcystis under oxidative stress conditions. PLoS One 6:e17615. doi: 10.1371/journal.pone.0017615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stumm W, Morgan JJ. 1996. Aquatic chemistry: chemical equilibria and rates in natural waters, 3rd ed Wiley-Interscience, New York, NY. [Google Scholar]
- 37.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 38.Ruijter JM, Ramakers C, Hoogaars WM, Karlen Y, Bakker O, Van den Hoff MJ, Moorman AF. 2009. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res 37:e45. doi: 10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Miller AG, Espie GS, Canvin DT. 1988. Active transport of inorganic carbon increases the rate of O2 photo-reduction by the cyanobacterium Synechococcus UTEX 625. Plant Physiol 88:6–9. doi: 10.1104/pp.88.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maberly SC. 1996. Diel, episodic and seasonal changes in pH and concentrations of inorganic carbon in a productive lake. Freshwat Biol 35:579–598. doi: 10.1111/j.1365-2427.1996.tb01770.x. [DOI] [Google Scholar]
- 42.Sandrini G, Huisman J, Matthijs HCP. 2015. Potassium sensitivity differs among strains of the harmful cyanobacterium Microcystis and correlates with the presence of salt tolerance genes. FEMS Microbiol Lett 362:fnv121. doi: 10.1093/femsle/fnv121. [DOI] [PubMed] [Google Scholar]
- 43.Tonk L, Bosch K, Visser PM, Huisman J. 2007. Salt tolerance of the harmful cyanobacterium Microcystis aeruginosa. Aquat Microb Ecol 46:117–123. doi: 10.3354/ame046117. [DOI] [Google Scholar]
- 44.Ohkawa H, Price GD, Badger MR, Ogawa T. 2000. Mutation of ndh genes leads to inhibition of CO2 uptake rather than HCO3− uptake in Synechocystis sp. strain PCC 6803. J Bacteriol 182:2591–2596. doi: 10.1128/JB.182.9.2591-2596.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shibata M, Katoh H, Sonoda M, Ohkawa H, Shimoyama M, Fukuzawa H, Kaplan A, Ogawa T. 2002. Genes essential to sodium-dependent bicarbonate transport in cyanobacteria. J Biol Chem 277:18658–18664. doi: 10.1074/jbc.M112468200. [DOI] [PubMed] [Google Scholar]
- 46.Woodger FJ, Badger MR, Price GD. 2003. Inorganic carbon limitation induces transcripts encoding components of the CO2-concentrating mechanism in Synechococcus sp. PCC7942 through a redox-independent pathway. Plant Physiol 133:2069–2080. doi: 10.1104/pp.103.029728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwarz D, Nodop A, Hüge J, Purfürst S, Forchhammer K, Michel KP, Bauwe H, Kopka J, Hagemann M. 2011. Metabolic and transcriptomic phenotyping of inorganic carbon acclimation in the cyanobacterium Synechococcus elongatus PCC 7942. Plant Physiol 155:1640–1655. doi: 10.1104/pp.110.170225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burnap RL, Hagemann M, Kaplan A. 2015. Regulation of CO2 concentrating mechanism in cyanobacteria. Life 5:348–371. doi: 10.3390/life5010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van de Waal DB, Verspagen JMH, Finke JF, Vournazou V, Immers AK, Kardinaal WEA, Tonk L, Becker S, Van Donk E, Visser PM, Huisman J. 2011. Reversal in competitive dominance of a toxic versus non-toxic cyanobacterium in response to rising CO2. ISME J 5:1438–1450. doi: 10.1038/ismej.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woodger FJ, Bryant DA, Price GD. 2007. Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp. strain PCC 7002: role of NdhR/CcmR. J Bacteriol 189:3335–3347. doi: 10.1128/JB.01745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Price GD, Badger MR, von Caemmerer S. 2011. The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol 155:20–26. doi: 10.1104/pp.10.164681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGrath JM, Long SP. 2014. Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol 164:2247–2261. doi: 10.1104/pp.113.232611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Figge RM, Cassier-Chauvat C, Chauvat F, Cerff R. 2001. Characterization and analysis of an NAD(P)H dehydrogenase transcriptional regulator critical for the survival of cyanobacteria facing inorganic carbon starvation and osmotic stress. Mol Microbiol 39:455–469. doi: 10.1046/j.1365-2958.2001.02239.x. [DOI] [PubMed] [Google Scholar]
- 54.Price GD, Badger MR, Woodger FJ, Long BM. 2008. Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59:1441–1461. doi: 10.1093/jxb/erm112. [DOI] [PubMed] [Google Scholar]
- 55.Benjamini Y, Yekutieli D. 2001. The control of false discovery rate in multiple testing under dependence. Ann Statist 29:1165–1188. doi: 10.1214/aos/1013699998. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.