Abstract
Fluoroquinolone (FQ) agents are a potential resort to treat infection due to Enterobacteriaceae producing extended spectrum β-lactamase and susceptible to FQ. In a context of increase of non-susceptibility to carbapenems among Enterobacteriaceae, we characterized FQ resistance mechanisms in 75 Enterobacter cloacae isolates non-susceptible to ertapenem in North-Eastern France in 2012 and describe the population structure by pulsed field gel electrophoresis (PFGE) and multi-locus sequence typing (MLST). Among them, 14.7% (12/75) carried a carbapenemase-encoding gene. Except one isolate producing VIM-1, the carbapenemase-producing isolates carried the well-known IncL/M pOXA48a plasmid. Most of the isolates (59/75) harbored at least a FQ-R determinant. qnr genes were predominant (40%, 30/75). The MLST study revealed that E. cloacae isolates’ clonality was wide [24 different sequence types (STs)]. The more widespread STs were ST74, ST101, ST110, ST114, and ST133. Carbapenem MICs were higher for E. cloacae ST74 than for other E. cloacae isolates. Plasmid-mediated quinolone resistance determinants were more often observed in E. cloacae ST74 isolates. These findings showed that (i) pOXA-48a is spreading in North-Eastern France, (ii) qnr is preponderant in E. cloacae, (iii) E. cloacae comprised a large amount of lineages spreading in North-Eastern France, and (iv) FQ as an alternative to β-lactams to treat ertapenem non-susceptible Enterobacteriaceae are compromised.
Keywords: Fluoroquinolones, PMQR, QRDR, Enterobacter cloacae, Carbapenem, MLST
Introduction
Enterobacter cloacae complex is, together with Escherichia coli, very common human Enterobacteriaceae pathogens. They can be a reservoir for infection due to their intestine colonization in patients upon long-term hospitalization and antimicrobial treatment. Due to the β-lactams antibiotic intensive use, rate of Enterobacteriaceae resistant to lactams-lactams highly increased these last years in French hospitals (Carbonne et al., 2013). Carbapenems are one of the last lines to treat patients infected with multidrug resistant Enterobacteriaceae. Thus, decreased susceptibility to carbapenems represents a threat of therapeutic dead ends. Two main mechanisms lead to decrease the susceptibility to carbapenems in Enterobacteriaceae isolates: (i) production of carbapenemase or (ii) production of other β-lactamase in combination with decreased permeability (Nordmann et al., 2011).
In France, OXA-48 is the most prevalent carbapenem-hydrolyzing β-lactamase and is a public health concern (Poirel et al., 2012b; Robert et al., 2014).
Recently in France, fluoroquinolone (FQ) agents have been proposed as the first choice to treat FQ-susceptible extended-spectrum β-lactamase-producing enterobacterial (ESBL) isolates in pyelonephritis in order to spare carbapenems1. Then, FQ can be used to treat infections due to carbapenem non-susceptible isolates. However, plasmid-mediated quinolone resistances (PMQR) confer a low level of quinolone resistance and they were shown to significantly reduce the activity of ciprofloxacin in urinary (Allou et al., 2009; Guillard et al., 2013) and respiratory (Rodríguez-Martínez et al., 2008; Dominguez-Herrera et al., 2013) tract infection murine models. Multi-locus sequence typing (MLST) has been widely used to study population structure of E. coli and K. pneumoniae. Although MLST has also been described for E. cloacae, only few studies are available (Miyoshi-Akiyama et al., 2013; Izdebski et al., 2014).
The aim of this study was to characterize FQ resistance determinants carried by E. cloacae isolates showing a non-susceptibility to carbapenems and to assess the population structure of these isolates in order to better understand their spread in French hospitals.
Materials and Methods
Selection of Bacterial Strains
The survey was conducted in the five teaching hospitals (UH1: Besançon, UH2: Dijon, UH3: Nancy, UH4: Reims, and UH5: Strasbourg) and two general hospitals (GH1: Colmar and GH2: Troyes) in North-Eastern France from January 1st to June 30th, 2012. Medical data settings of the seven hospitals together were 10,311 beds and almost 540,000 admissions per year all through the period of investigation. After detection on chromID® ESBL agar plates (bioMérieux, Marcy l’Etoile, France), all non-duplicate E. cloacae isolates from clinical or screening samples non-susceptible to ertapenem (MIC ertapenem >0.5 μg/mL) were collected and sent to Reims Hospital laboratory.
Identification, Susceptibility Testing, and ESBL Detection
Identification of isolates was performed using MALDI-TOF (Bruker Daltonics, Bremen, Germany). Antibiotic susceptibilities were determined by the disk diffusion method according to European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines2 and ESBLs were detected by the double-disk synergy test as previously described (Jarlier et al., 1988; Duval et al., 2009). AmpC overexpression was detected by disk diffusion method performed on Mueller-Hinton agar supplemented with cloxacillin (250 μg/mL). Carbapenem (imipenem, ertapenem, doripenem, and meropenem) and FQ (norfloxacin, ofloxacin, nalidixic acid, and ciprofloxacin) MICs were determined using E test® strips (bioMérieux, Marcy l’Etoile, France) according to the manufacturer’s recommendations and interpreted according to EUCAST. Metallo-β-lactamase production was screened with the MβL E test (bioMérieux, Marcy l’Etoile, France). In addition, metallo-β-lactamase was detected with imipenem disk with and without EDTA (5 μL of 0.5 M EDTA per disk) as previously described (Liapis et al., 2014).
β-Lactamase Detection
Carbapenemase-encoding genes blaKPC, blaV IM, blaIMP, blaNDM, blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, and blaOXA-48-like were screened using multiplex PCRs and completed with simplex PCRs for blaIMI and blaGES as described elsewhere (Gharout-Sait et al., 2014). All the blaOXA-48-like detected were subsequently sequenced. Genes blaTEM, blaSHV, blaCTX-M, and blaOXA were detected by PCR using specific primers and sequenced as previously described for all the ESBL-producing strains (Guillard et al., 2014a). Plasmid-mediated AmpC-type genes blaACC, blaFOX, blaMOX, blaDHA, blaCMY, and blaMIR were screened using multiplex PCRs and sequenced as previously described (Gharout-Sait et al., 2014).
Quinolone Resistance Determining Region (QRDR) and PMQR Identification
The quinolone resistance determining region (QRDR) was amplified by PCR and sequenced in the gyrA, gyrB, parC, and parE genes, as described elsewhere (de Lastours et al., 2012). qnr, qepA, and oqxAB genes were detected by real-time PCR, aac(6’)-Ib-cr was detected by pyrosequencing as described elsewhere (Guillard et al., 2010, 2011, 2015).
Contribution of Efflux and Reduced Outer-membrane Permeability
For contribution of efflux and reduced outer-membrane permeability study, we randomly selected five strains that did not produce OXA-48 and three, which produced OXA-48.
Quantitative real-time reverse transcriptase PCR (qRT-PCR) was performed to determine the expression of ompF and ompC porin genes and the acrB efflux pump gene, relative to the rpoB housekeeping gene. From bacteria grown to mid-exponential growth phase in LB, total RNA was extracted using RNeasy Kit (Qiagen, Courtaboeuf, France) as recommended by the manufacturer. Residual chromosomal DNA was removed by treating samples with the RNAase free DNase I (Qiagen) and TURBO DNA-free kit (Life Technologies). DNase-treated RNA samples were quantified using the easy-to-use QubitTM 3.0 Fluorometer (ThermoFisher Scientific). qRT-PCR experiments were performed using the KAPA SYBR® FAST One-Step qRT-PCR Kit Universal (Kapa Biosystems, USA) and the LightCycler 480 (Roche Molecular Diagnostics, Germany) according to manufacturers’ recommendations. Each experiment was performed in triplicate. RNA transcript levels were calculated using the 2-ΔΔCt method following the MIQE checklist (Bustin et al., 2009), and are expressed relative to levels in the E. cloacae CIP 60.85T control. Primers used are presented in Supplementary Table S1.
Bacterial outer-membrane proteins (OMPs) were detected by SDS/PAGE as previously described (Martínez-Martínez et al., 2002). Briefly, bacterial cells were broken by sonication, and membranes were collected by ultracentrifugation at 100,000 × g for 1 h. The inner membrane was solubilized with 1% sodium N-lauroylsarcosinate. Proteins in the outer membrane were separated by SDS/PAGE and gels were stained with Coomassie blue for visualization.
Plasmids Characterization
Plasmids carrying blaOXA-48 were studied as described elsewhere (Guillard et al., 2014b). Briefly, plasmid extraction was performed by the Kieser method. Transfer of the plasmids was then studied by conjugation assays using azide-resistant E. coli J53 as a recipient cell and counter-selection with ertapenem 0.5 μg/mL, rifampicin 250 μg/mL, and sodium azide 100 μg/mL. Incompatibility groups of the plasmids were eventually determined using the PCR-based replicon typing (PBRT) for the paired strains and transconjugants (Carattoli et al., 2005). The incL/M group was characterized using the primers described by Poirel et al. (2012a). Genetic environment of blaOXA-48 was studied by PCR mapping to seek for an environment such as Tn1999.2 using primers listed in Supplementary Table S1.
Genotyping
Pulsed-field gel electrophoresis (PFGE) was performed on E. cloacae isolates using the XbaI restriction enzyme as previously described and results were interpreted according to international criteria (Tenover et al., 1995).
Analysis of MLST Data
Multi-locus sequence typing was performed according to published protocols (Diancourt et al., 2005; Miyoshi-Akiyama et al., 2013). Clonal complexes (CC) were defined as a group of STs sharing at least five loci. In order to build a dendrogram with the 373 sequence types (STs) available at the time of the study (including the new ST described in this collection), we concatenated the sequences of seven MLST genes to form a 3,511-bp sequences alignment, defining 914 polymorphic positions. The best-fit nucleotide substitution model for this data was GTR + G + I, as determined with jModelTest 0.1.1. We used the Enterobacter aerogenes KCTC2190 as the outgroup strain. Maximum likelihood tree was constructed with RAxML 7.2.8 and visualized with Dendroscope 3.2.10 (Stamatakis, 2006; Huson and Scornavacca, 2012). In every case, 1000 bootstrap repetitions gave values above 900 for most branches.
Statistical Analysis
Qualitative variables were analyzed with the Chi2 test and the two-tailed Fisher exact test. Quantitative variables were compared using the Mann–Whitney test. The results were considered statistically significant when P < 0.05.
Results
Species Distribution of the Ertapenem Non-susceptible Enterobacteriaceae Isolates
The 75 E. cloacae isolates were distributed as follow in the different centres: 21 (28.0%) in UH4, 18 (24.0%) in UH3, 13 (17.3%) in GH2, 10 (13.3%) in UH5, 9 (12.0%) in UH1, 3 (4.0%) in UH2, and 1 (1.3%) in GH1. They were isolated from clinical samples (65.3%; n = 49: 22 (29.3%) urines, 5 (6.7%) blood, 22 (29.3 %) other infectious sites) or from 26 screening samples (34.7%; stools or rectal swabs). The global prevalence of carbapenem-non-susceptible isolates in E. cloacae species was 6.05% (0.91–8.36 according to the centre).
Antibiotic Susceptibility and Level of Resistance
All isolates but three were intermediate or resistant to piperacillin and all but one to ticarcillin–clavulanic acid. Ten point seven percent were susceptible to piperacillin–tazobactam, 4.0% to aztreonam, 6.7% to cefotaxime, 22.7% to cefpirome, 46.7 to cefepime, 33.3% to tobramycin, 38.7% to gentamicin, 96.0% to amikacin, 52.7% to tetracycline, 45.9% to chloramphenicol, 17.3% to nalidixic acid, 18.7% to ofloxacin, 26.7% to ciprofloxacin, 18.7% to norfloxacin, 57.3% to co-trimoxazole, 92.0% to fosfomycin, 93.3% to imipenem, 97.3% to meropenem, and 96.0% to doripenem. The frequency of isolates susceptible to FQ agents among clinical isolates did not differ from that of screening isolates (Table 1).
Table 1.
Fluoroquinolones MIC (μg/mL) among the clinical and the screening Enterobacter cloacae isolates.
| Nalidixic acid |
Norfloxacin |
Ciprofloxacin |
Ofloxacin |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | MIC50 | MIC90 | Range | |
| Screening isolates | 26 | >256 | >256 | 3->256 | >256 | >256 | 0,125–>256 | >32 | >32 | 0,016–>32 | > 32 | >32 | 0,125–>32 |
| Clinical isolates | 49 | >256 | >256 | 0,19->256 | 12 | >256 | 0,064–>256 | 2 | >32 | 0,012–>32 | 8 | >32 | 0,094–>32 |
| Urines | 22 | >256 | >256 | 2->256 | 9 | >256 | 0,064–>256 | 2 | >32 | 0,012–>32 | 6 | >32 | 0,094–>32 |
β-Lactam Resistance Determinants
Carbapenemases were not the major determinant leading to decrease susceptibility to carbapenems in E. cloacae isolates. Indeed, among them, 84% (63/75) did not produce a carbapenemase. Nonetheless, among the carbapenemase-producing isolates, OXA-48 (11/12 for E. cloacae) was the most prevalent carbapenemase, and interestingly the majority of OXA-48-producing isolates co-harbored CTX-M-15 and TEM-1 (11/11). One isolate produced VIM-1.
Focusing on isolates, which did not produce a carbapenemase, most of them (40/63) did not produce ESBL, and 29 out of 63 did not produce any acquired β-lactamases. These strains did not carry any plasmid-mediated AmpC-type genes except one harboring blaDHA-1. The eight strains, studied for efflux and porins expression, produced an overexpressed AmpC except one. This latter did not produce any acquired β-lactamases. Ertapenem MICs > 3 mg/L were related to a decreased expression of ompF and ompC, and for three strains with a lost a porin (Table 2).
Table 2.
Contribution of efflux and reduced outer membrane permeability to carbapenems for E. cloacae strains resistant to ertapenem.
| Strain | ST | β-lactamases | MIC (μg/mL) |
SDS PAGE | Relative expressiona |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ertapenem | Imipenem | Doripenem | Meropenem | ompF | ompC | acrB | ||||
| 12 01 033 | 133 | SHV-12, CTX-M-9, overexpressed AmpC | >32 | 16 | 4 | 6 | Loss of OmpC | 0.22 | 0.02 | 0.51 |
| 12 01 040 | 133 | SHV-12, CTX-M-9, overexpressed AmpC | >32 | 12 | 4 | 12 | Loss of OmpC | 0.23 | 0.03 | 0.7 |
| 12 05 006 | 175 | SHV-12, overexpressed AmpC | 32 | 2 | 0.75 | 1 | Loss of OmpF | 0.05 | 0.11 | 2.28 |
| 12 03 029 | 74 | CTX-M-15 | 2 | 2 | 0.5 | 2 | – | 0.44 | 0.22 | 2.76 |
| 12 06 020 | 102 | – | 1.5 | 2 | 1.5 | 1.5 | – | 0.52 | 0.36 | 2.12 |
| 12 05 004 | 74 | OXA-48, CTX-M-15 | 6 | 1.5 | 0.75 | 1.5 | – | 3.15 | 0.54 | 9 |
| 12 03 022 | 110 | OXA-48, CTX-M-15 | 3 | 1 | 0.75 | 0.5 | Modified porins | 3.23 | 0.009 | 6.57 |
| 12 03 028 | 74 | OXA-48, CTX-M-15 | 2 | 1 | 0.25 | 0.75 | - | 0.44 | 0.22 | 2.76 |
aRelative expression is calculated as –2ΔΔCt, where ΔΔCt = (Cttarget – Ctreference). The targets were each strain studied and the reference was the reference strain E. cloacae CIP 60.85T.
All the strains co-producing OXA-48 and CTX-M-15 were susceptible to carbapenems but ertapenem, with MICs close to the breakpoints. The three OXA-48 and CTX-M-15-producing strains, selected for efflux and outer membrane study, showed an acrB overexpression of at least a twofold change (Table 2). All these strains showed also a decreased ompC expression.
blaOXA-48 is Carried by an IncL/M Conjugative Plasmid
To study plasmids carrying blaOXA-48, we selected three strains to be studied among the E. cloacae OXA-48 producing-isolates according to antibiotic resistance genes characterization. Mating out assays and plasmid DNA analysis allowed identification of a 62-kb-long plasmid carrying blaOXA-48 but not blaTEM-1 and blaCTX-M-15. This conjugative plasmid belonged to the incL/M incompatibility group. Mapping of the genetic environment showed that blaOXA-48 was embedded in Tn1999.2 as previously described (Poirel et al., 2004). As expected, OXA-48-producing transconjugants showed a low level of resistance to carbapenems (data not shown).
FQ Resistance Determinants
Most of the E. cloacae (59/75, 78.7%) isolates harbored a FQ-R determinant. E. cloacae isolates carried in the same proportion either substitutions in QRDR only (37%, 28/75) or association of PMQR and substitutions in QRDR (36%, 27/75). Very few isolates produced PMQR solely (5%).
Focusing on PMQR harbored by the E. cloacae isolates, we showed that only qnr genes and aac(6’)-Ib-cr were detected. qnr genes were predominant (40%, 30/75) compared to aac(6’)-Ib-cr (23%, 17/75), (P < 0.02). Three qnr families were found (qnrA, qnrB, and qnrS) with a majority of qnrB (18/30). Looking more carefully at the alleles, we found 9 qnrA1, 16 qnrB1, 2 qnrB2 and 3 qnrS1. Of the 31 PMQR-harboring E. cloacae isolates, 13 (42%) carried qnr only and 17 (55%) qnr associated with aac(6’)-Ib-cr. One was AAC(6’)-Ib-cr producer only.
Substitutions in the QRDR were detected at high rate. Our findings showed that 73% (55/75) of isolates carried substitutions in QRDR. Majority of isolates (51%, 38/75) harbored substitutions in GyrA only. More precisely, E. cloacae isolates carried predominantly either one substitution in GyrA (48%, 36/75) or a combination of two substitutions in GyrA and one substitution in ParC (18.7%, 14/75). The GyrA substitutions Ser/Thr83Phe and Ser/Thr83Ile (49 and 36%, respectively) were the most frequent (Table 3). No substitutions were characterized in gyrB and parE.
Table 3.
Mutations observed in QRDR of E. cloacae isolates.
| Amino acid substitutions |
No. of isolates (Total of 75) | ||
|---|---|---|---|
| GyrA |
ParC |
||
| Ser/Thr83 | Asn87 | Ser80 | |
| – | – | – | 20 |
| Ile | – | – | 19 |
| Ile | – | Ile | 1 |
| Phe | – | – | 11 |
| Phe | – | Ile | 1 |
| Phe | Asn | – | 2 |
| Phe | Asn | Ile | 10 |
| Phe | Gly | Ile | 3 |
| Leu | – | Ile | 1 |
| Tyr | – | – | 6 |
| Tyr | His | Ile | 1 |
(–) Indicates no mutation found at the location.
As shown in Table 4, 82.6% of PMQR-producing E. cloacae had substitution in QRDR vs. 60.0% in PMQR-negative. But the number of substitutions in QRDR was higher in PMQR-negative isolates (e.g., 3 mutations: 32.5% vs. 0% for PMQR-negative and PMQR-producing isolates, respectively, P < 0.0001). For this analysis, we deleted duplicates in each PFGE if they carried same PMQR associated to same mutations pattern in QRDR.
Table 4.
Prevalence of QRDR mutations in PMQR-carrying and PMQR negative E. cloacae isolatesa.
| E. cloacae | PMQR+ | PMQR– | ||
|---|---|---|---|---|
| QRDR+ | 19 | 82.6% | 24 | 60.0% |
| QRDR– | 4 | 16.4% | 16 | 40.0% |
| 0 mutation | 4 | 17.4% | 16 | 40.0% |
| 1 mutation | 17 | 74.0% | 9 | 22.5% |
| 2 mutations | 2 | 8.6% | 2 | 5.0% |
| 3 mutations | 0 | 0.0% | 13 | 32.5% |
| Total | 23 | 40 | ||
aAfter exclusion of duplicates: one isolate was included in each PFGE harboring different PMQR and mutations pattern in QRDR compared to the other isolates included in the same PFGE profile.
Epidemiological Links of E. cloacae
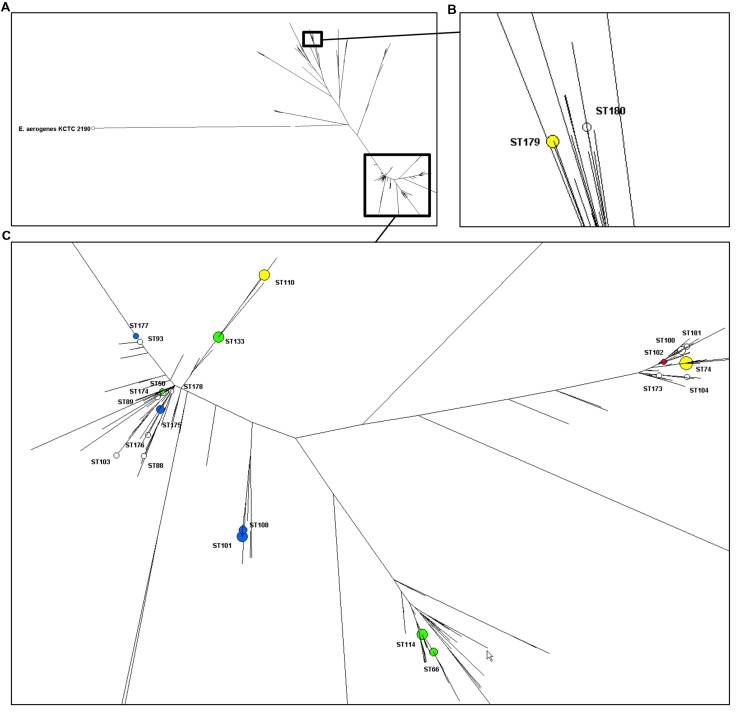
Typing results of E. cloacae isolates are shown in Table 5. Seventy-four isolates (one was untypeable with PFGE) were distributed into 40 pulsotypes and the 75 isolates clustered into 24 STs of which 12 were grouped into five CCs (CC42, CC74, CC114, CC133, and CC234). ST74 was the most frequent ST accounting for 16 out of 75 isolates (21%), followed by ST101 (nine isolates, 12%) ST110 and ST133 (eight isolates each, 11%) and ST114 (six isolates, 8%). Eight STs were split into two to five pulsotypes and the two most predominant STs, ST74 and ST101, were also the most diverse. The phylogenetic tree built from the sequence data of the seven MLST genes of the 373 STs of E. cloacae clearly showed that the great majority of our isolates clustered in one phylogenetic group (Figure 1).
Table 5.
E. cloacae STs identified in our study: clonal status, pulsotypes, centres, prevalence, carbapenem MIC and β-lactamases.
| CC (ST)a | PFGE profiles | Centres | Isolates (n) | Median MIC (range; μg/mL) |
β-lactamases (number of isolates)b,c | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Imipenem | Ertapenem | Doripenem | Meropenem | |||||||||
| 42 (101, 104, 108) | 2,8,10,14,17–20 | UH1, UH3, UH4, UH5, GH1, GH2 | 13 | 0.38 | (0.13–1) | 1 | (0.75–4) | 0.125 | (0.047–0.75) | 0.125 | (0.064–0.5) | SHV-12 (n = 1) |
| 74 (74) | 31–33,35,41 | UH2, UH3, UH4, UH5, GH2, | 16 | 0.62 | (0.13–2) | 2 | (0.75–12) | 0.25 | (0.064–1) | 0.44 | (0.064–2) | OXA-48 (n = 9), CTX-M-15 (n = 12) |
| 114 (66,114) | 3,5,40 | UH1, UH3, UH5 | 7 | 0.38 | (0.13–2) | 2 | (1–6) | 0.125 | (0.064–0.38) | 0.19 | (0.125–0.5) | CTX-M-9 (n = 1) CTX-M-15 (n = 3), TEM-24 (n = 1), SHV-12 (n = 1) |
| 133 (93, 103,133) | 15,21,23,38,39 | UH3, UH4, UH5, | 10 | 0.44 | (0.19–16) | 2 | (0.75–32) | 0.125 | (0.16–4) | 0.16 | (0.016–12) | CTX-M-9 (n = 7), SHV-12 (n = 3) |
| 234 (50,176,178) | 24,34,37 | UH2, UH3 | 3 | 0.5 | (0.38–1) | 2 | (0.75–12) | 0.25 | (0.064–0.5) | 0.38 | (0.064–0.5) | CTX-M-9 (n = 1), SHV-12 (n = 1) |
| ST110 | 28,29,30 | UH1, UH4, GH2 | 8 | 0.38 | (0.25–1) | 1.25 | (0.75–3) | 0.125 | (0.064–0.75) | 0.16 | (0.047–0.5) | OXA-48 (n = 1), CTX-M-15 (n = 1), SHV-12 (n = 2) |
| ST179 | ND,26,27 | UH3, GH2 | 4 | 0.5 | (0.5–0.75) | 1 | (0.75–4) | 0.094 | (0.064–0.38) | 0.094 | (0.064–0.75) | OXA-48 (n = 1), CTX-M-15 (n = 1) |
| ST100 | 6,7 | UH4 | 2 | 0.25 | (0.25–0.25) | 1.25 | (1–1.5) | 0.063 | (0.032–0.094) | 0.063 | (0.032–0.094) | – |
| ST175 | 16 | UH1, UH2 | 2 | 1.125 | (0.25–2) | 16.25 | (0.75–32) | 0.47 | (0.19–0.75) | 0.56 | (0.125–1) | SHV-12 (n = 1), DHA-1 (n = 1) |
| ST88,89, 102,173, 174,177, 180,181b | 1,4,9,11–13, 25,36 | UH1, UH4, UH5 | 10 | 0.25 | (0.13–1) | 0.88 | (0.75–32) | 0.1 | (0.047–0.75) | 0.079 | (0.064–2) | CTX-M-15 (n = 1), SHV-12 (n = 2), VIM-1 (n = 1) |
aFor E. cloacae, clonal complex determined according to Izdebski et al. (2014).
bCarbapenemase, extended spectrum β-lactamase, plasmid-mediated cephalosporinase. TEM-1 and TEM-2 have not been mentioned.
cOne isolate in each ST.
ND, Not determined; NT, Non-typeable.
FIGURE 1.
Distribution of the STs of 75 clinical isolates of Enterobacter cloacae (non-susceptible to carbapenems and isolated in North-Eastern France) on a dendrogram built with the data all known STs (n = 373). (A) Overview of the dendrogram with E. aerogenes as outgroup. (B) Close-up of the branch with ST179 and ST180. (C) Close-up the branch harboring the rest of the isolates described in this study. Enzymes in the different ST are represented with blue (SHV/TEM), yellow (OXA-48), green (CTX-M), and red (VIM) spots. Spots’ size depends on the amount of strains.
Enterobacter cloacae ST74 isolates were more often less susceptible than the other E. cloacae isolates to various antimicrobial agents (Table 6). They included nine of the 11 OXA-48 producing isolates observed.
Table 6.
Susceptibility rates of E. cloacae isolates to various antimicrobials according to the ST type.
| No. of susceptible isolates (%) |
|||||
|---|---|---|---|---|---|
| Antimicrobial agent | ST74 |
Other ST |
P-value | ||
| n | (%) | n | (%) | ||
| Ticarcillin + Clavulanic acid | 0 | (0.0) | 1 | (1.7) | NS |
| Piperacillin + Tazobactam | 2 | (12.5) | 6 | (10.2) | NS |
| Aztreonam | 0 | (0.0) | 3 | (5.1) | NS |
| Cefotaxime | 2 | (12.5) | 3 | (5.1) | NS |
| Ceftazidime | 2 | (12.5) | 2 | (3.4) | NS |
| Cefepime | 4 | (25.0) | 31 | (52.5) | 0.05 |
| Cefpirome | 3 | (18.8) | 14 | (23.7) | NS |
| Ertapenem | 0 | (0.0) | 0 | (0.0) | NS |
| Imipenem | 15 | (93.8) | 55 | (93.2) | NS |
| Meropenem | 16 | (100.0) | 57 | (96.6) | NS |
| Doripenem | 16 | (100.0) | 56 | (94.9) | NS |
| Tobramycin | 2 | (12.5) | 23 | (39.0) | 0.04 |
| Gentamicin | 3 | (18.8) | 26 | (44.1) | NS (0.06) |
| Amikacin | 14 | (87.5) | 58 | (98.3) | NS |
| Tetracycline | 10 | (62.5) | 29 | (50.0) | NS |
| Chloramphenicol | 3 | (18.8) | 31 | (53.4) | 0.05 |
| Nalidixic acid | 0 | (0.0) | 13 | (22.0) | NS |
| Ofloxacin | 1 | (6.3) | 13 | (22.0) | NS |
| Ciprofloxacin | 1 | (6.3) | 19 | (32.2) | 0.05 |
| Co-trimoxazole | 2 | (12.5) | 41 | (69.5) | <0.001 |
| Fosfomycin | 15 | (93.8) | 54 | (93.1) | NS |
Plasmid-mediated quinolone resistances determinants were more frequent in E. cloacae ST74 isolates than in non-ST74 (87% vs. 40% P = 0.001). 81% (13/16) of ST74 isolates harbored qnr vs. 29 % (17/58) of non-ST74 (P = 0.0002). Seventy-five percent (12/16) of ST74 isolates contained aac(6’)-Ib-cr vs. 9% (5/58) of non-ST74 (P = 4.10-7).
Ten of the 16 ST74 isolates were observed in GH2. They shared the same pulsotype 32, produced the identical β-lactamases and PMQR (CTX-M-15, TEM-1, QnrB1, and AAC(6’)-Ib-cr) and 8 of them produced OXA-48. It appears that the spread of blaOXA-48 in E. cloacae was mainly due to an epidemic strain.
Discussion
Although FQ use has been reported recently as a risk factor for infections due to carbapenem-resistant K. pneumoniae (Hussein et al., 2009), French guidelines for urinary tract infection treatment stated that FQ remains the first line treatment for pyelonephritis due to non-producing or producing-ESBL but FQ-susceptible enterobacterial isolates. These guidelines should help to decrease unnecessary carbapenem use to treat ESBL-producing enterobacterial isolates. So, we decided to study FQ resistance mechanisms in Enterobacteriaceae isolates non-susceptible to carbapenems in North-Eastern France: (i) we determined carbapenem susceptibility and β-lactamase detection; (ii) we characterized PMQR and mutations in QRDR for E. cloacae; and (iii) we defined the population structure of E. cloacae isolates.
β-Lactam Resistance Determinants
Carbapenemases were not the major determinant leading to decrease susceptibility to carbapenems in E. cloacae isolates. Nonetheless, OXA-48 was the most prevalent carbapenamase. All the strains co-harboring OXA-48 and CTX-M-15 were susceptible to carbapenems but ertapenem, with MICs close to the breakpoints. But interestingly, contribution of efflux and/or reduced outer membrane permeability was important in those strains. Reports about carbapenem non-susceptibility of E. cloacae were scarce and OXA-48-producing E. cloacae were rarely reported (Hammoudi et al., 2014; Majewski et al., 2014; Pantel et al., 2014). Then, the amount of OXA-48 E. cloacae isolates in our study showed increase of this carbapenemase in this species in France. In opposite to Hammoudi et al. (2014) report in Lebanon, carbapenemase production is not the major cause of carbapenem non-susceptibility in our isolates. The part played by ESBL or AmpC was previously reported and DHA-1 has been shown to reduce imipenem activity combined with loss of porins (Mammeri et al., 2010). This latter point is underscored by our results showing combination of overexpressed efflux and outer membrane impermeability for all the strains we sampled.
FQ Resistance Determinants
To our knowledge, few epidemiological studies have recently reported data about PMQR and/or substitutions in QRDR in E. cloacae, while huge data are available for E. coli. Majority of the E. cloacae isolates harbored a FQ-R determinant. E. cloacae isolates carried in the same proportion either substitutions in QRDR only (37%), or association of PMQR and substitutions in QRDR (36%). Very few isolates produced PMQR solely (5%). These findings are different from those reported by Ferjani et al. (2014) stating that E. cloacae strains, isolated in 2010 in Tunisia, carried predominantly qnr genes (50%) but less substitutions in QRDR (50%). Recently, it has been shown that E. cloacae non-susceptible to carbapenem isolated in China from 2009 to 2012 were highly PMQR carriers (68.9%), but no data about QRDR were available (Huang et al., 2012). Our results about efflux and outer membrane permeability confirmed that reduced outer membrane permeability and increased efflux are an important mechanism of FQ resistance in E. cloacae, as previously reported for this specie (Davin-Regli and Pagès, 2015; Li et al., 2015). Moreover as suggested by Davin-Regli and Pagès (2015) this particular property could be an asset for spreading in hospitals. We did not test all the strains for impermeability but regarding of the results we obtained from the sampled strains, overexpression of efflux pumps might be consider as a major resistance determinant in E. cloacae (Davin-Regli and Pagès, 2015). It has been proposed that OmpC disappears before OmpF in E. aerogenes in the presence of selective pressure from antibiotics and could be extend to other Enterobacteriaceae {de Champs et al., 1993ce, Lavigne et al., 2013wz}. It is noteworthy that our results are in agreement with this statement except for one strain, which lacked OmpF and had a decreased expression of OmpC. Similar results have been also reported for E. cloacae without any clear information about this discrepancy {Doumith et al., 2009ks}.
qnr is the Major PMQR Determinant in E. cloacae
Focusing on PMQR harbored by the E. cloacae isolates, we showed that only qnr genes and aac(6’)-Ib-cr were detected. qnr genes were predominant (40%, 30/75) and qnrB was the most predominant family.
In a study conducted on ESBL-producing Enterobacteriaceae in Tunisia, it has been shown that qnr genes were mainly detected in E. cloacae (50%) and aac(6’)-Ib-cr genes in E. coli (47.5%) isolates (Ferjani et al., 2014). Huang et al. studied the prevalence of PMQR among carbapenem non-susceptible E. cloacae isolated between 2009 and 2012 in China (Huang et al., 2012). From the 986 isolates, 35 (3.55%) were non-susceptible to carbapenem. In their study, qnr genes were detected in 75% of the isolates whereas only 15% carried aac(6’)-Ib-cr.
E. cloacae Isolates have a Wide Range of Substitutions Patterns
Substitutions in the QRDR were detected at high rate in the E. cloacae isolates (73%) studied here. Majority of isolates (51%) harbored substitutions in GyrA only, with Ser/Thr83Phe and Ser/Thr83Ile (49 and 36%, respectively) as the most frequent. These results differed from those previously reported for E. coli, stating that the GyrA substitution Ser83Leu is the most prevalent substitution in QRDR (Fendukly et al., 2003). As no substitutions were characterized in gyrB and parE, our findings underscored that gyrB and parE substitutions in E. cloacae are very rare. Indeed, these results were mostly described in E. coli isolates (Uchida et al., 2010), but our findings suggest a similar trend for E. cloacae.
GyrA substitutions were observed predominantly as single substitution in QRDR in E. cloacae, but ParC mutations were always combined with GyrA substitution. These results are in agreement with others studies and with the hypothesis that, in Enterobacteriaceae, DNA gyrase is the primary target enzyme where substitutions occur to resist against FQ. Topoisomerase IV, described as less sensitive to FQ than DNA gyrase, is the secondary target (Hooper, 2001). First-step quinolone resistance mutations occur in gyrA and mutations in parC occurred as a secondary event leading to highly resistant Enterobacteriaceae isolates.
The aims of our study were not to determine contribution of QRDR substitution to resistance, but it is well known that substitutions in hot spot of QRDR lead to high level of resistance as observed in this study (Hooper, 2001).
PMQR-negative E. cloacae Isolates Harbor Multiple Substitutions in QRDR
Impact of PMQR on substitution in QRDR under FQ pressure is still debating. Of course, in literature this issue was mainly studied for qnr. In one hand, it has been largely reported that Qnr proteins increase rate of substitution in QRDR by increasing the mutant selection concentration. But, in the other hand, it has been also observed in E. cloacae that fewer QRDR substitutions occurred in Qnr-producing isolates (Lascols et al., 2007). That was consistent with molecular analysis showing that Qnr protects QRDR, in gyrase and Topoisomerase IV, from substitution under FQ inhibition (Cesaro et al., 2008). So, we looked eventually at the prevalence of QRDR substitutions in PMQR-containing and PMQR-negative E. cloacae isolates trying to gain insight how QRDR substitutions are affected by presence of PMQR. Our findings seem to support the hypothesis that presence of PMQR may confer a low-level resistance promoting at least one substitution in QRDR. Although, harboring no PMQR seems to lead to multiple substitutions in QRDR. We could hypothesize that a strain carrying no PMQR may acquire one chromosomal substitution in GyrA under FQ pressure. Then, if the strain carries at least Qnr, this latter may avoid multiple substitutions by protecting QRDR domains.
Epidemiological Links of E. cloacae
Typing results of E. cloacae isolates showed ST74 as the most frequent ST with the lowest susceptibility against β-lactams and carrying PMQR frequently.
To our knowledge, very few E. cloacae clonality studies are available. Recently, a multinational study performed on 195 rectal carriage isolates, revealed several epidemic CC, such as CC74 and CC114 (Izdebski et al., 2014). The STs observed in our study were reported by Izdebski et al. (2014) except for ST110. Our study confirms the findings of Izdebski et al. (2014) showing that some E. cloacae lineages have an increased epidemic potential and may participate to the spread of the resistance to antibiotics, including carbapenems (with or without carbapenemase genes).
The large clonal diversity of E. cloacae would be linked to antibiotics pressure. Further studies might point out specific factors (e.g., colonization, ability to infect more easily) explaining these findings.
Conclusion
We combined here the molecular typing of carbapenem-non-susceptible E. cloacae isolates with the characterization of FQ resistance determinants. It will give new insights to the first multinational E. cloacae clonality study previously reported (Izdebski et al., 2014).
The MLST study revealed that E. cloacae comprised a large amount of lineages spreading in North-Eastern France. Nonetheless, E. cloacae ST74 was the most widespread ST, more often resistant to various antibiotics and carried more often PMQR. Carbapenemase production was not the major determinant leading to decrease susceptibility to carbapenem. Nonetheless carbapenemase-producing isolates carried the well-known IncL/M pOXA48a. In term of FQ-R, E. cloacae isolates harbored a wide range of pattern, from no substitution to multiple substitutions in QRDR. qnr was the major PMQR determinant found in E. cloacae.
These data underline, once again, the plausible role of the antibiotic pressure, in emergence and persistence of carbapenem and quinolones resistance genes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all healthcare professionals, hospital and microbiological laboratories involved in the study. CarbaFrEst Group participants: C. Eloy, General Hospital Troyes (France); D. De Briel, General Hospital Colmar (France); B. Jaulhac, University Hospital Strasbourg (France); A. Lozniewski, University Hospital Nancy (France); C. Neuwirth, University Hospital Dijon (France); P. Plesiat, University Hospital Besançon (France); M. Verdier, University Hospital Reims (France). We are also indebted with Talya Wagner Tammaro who read carefully and edited our manuscript.
Funding. This work was supported by a grant from the Ministère de la Santé et des Sports (Projet Hospitalier de Recherche Clinique CARBAFREST, an annual grant from the Université de Reims Champagne-Ardenne (EA 4687) and an annual grant from the Université de Franche-Comté (UMR 6249).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2015.01186
References
- Allou N., Cambau E., Massias L., Chau F., Fantin B. (2009). Impact of Low-Level Resistance to fluoroquinolones due to qnrA1 and qnrS1 genes or a gyrA mutation on Ciprofloxacin bactericidal activity in a murine model of Escherichia coli urinary tract infection. Antimicrob. Agents Chemother. 53 4292–4297. 10.1128/AAC.01664-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert D., Naas T., Héritier C., Poirel L., Nordmann P. (2006). Functional characterization of IS1999, an IS4 family element involved in mobilization and expression of beta-lactam resistance genes. J. Bacteriol. 188 6506–6514. 10.1128/JB.00375-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S. A., Benes V., Garson J. A., Hellemans J., Huggett J., Kubista M., et al. (2009). The MIQE Guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55 611–622. 10.1373/clinchem.2008.112797 [DOI] [PubMed] [Google Scholar]
- Carattoli A., Bertini A., Villa L., Falbo V., Hopkins K. L., Threlfall E. J. (2005). Identification of plasmids by PCR-based replicon typing. J. Microbiol. Methods 63 219–228. 10.1016/j.mimet.2005.03.018 [DOI] [PubMed] [Google Scholar]
- Carbonne A., Arnaud I., Maugat S., Marty N., Dumartin C., Bertrand X., et al. (2013). National multidrug-resistant bacteria (MDRB) surveillance in France through the RAISIN network: a 9 year experience. J. Antimicrob. Chemother. 68 954–959. 10.1093/jac/dks464 [DOI] [PubMed] [Google Scholar]
- Cesaro A., Bettoni R. R. D., Lascols C., Merens A., Soussy C.-J., Cambau E. (2008). Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J. Antimicrob. Chemother. 61 1007–1015. 10.1093/jac/dkn077 [DOI] [PubMed] [Google Scholar]
- Davin-Regli A., Pagès J.-M. (2015). Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol. 6:392 10.3389/fmicb.2015.00392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Champs C., Henquell C., Guelon D., Sirot D., Gazuy N., Sirot J. (1993). Clinical and bacteriological study of nosocomial infections due to Enterobacter aerogenes resistant to imipenem. J. Clin. Microbiol. 31 123–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lastours V., Cambau E., Guillard T., Marcade G., Chau F., Fantin B. (2012). Diversity of individual dynamic patterns of emergence of resistance to quinolones in Escherichia coli from the fecal flora of healthy volunteers exposed to ciprofloxacin. J. Infect. Dis. 206 1399–1406. 10.1093/infdis/jis511 [DOI] [PubMed] [Google Scholar]
- Diancourt L., Passet V., Verhoef J., Grimont P. A. D., Brisse S. (2005). Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. J. Clin. Microbiol. 43 4178–4182. 10.1128/JCM.43.8.4178-4182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Herrera J., Velasco C., Docobo-Pérez F., Rodríguez-Martínez J. M., López-Rojas R., Briales A., et al. (2013). Impact of qnrA1, qnrB1 and qnrS1 on the efficacy of ciprofloxacin and levofloxacin in an experimental pneumonia model caused by Escherichia coli with or without the GyrA mutation Ser83Leu. J. Antimicrob. Chemother. 68 1609–1615. 10.1093/jac/dkt063 [DOI] [PubMed] [Google Scholar]
- Doumith M., Ellington M. J., Livermore D. M., Woodford N. (2009). Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J. Antimicrob. Chemother. 63 659–667. 10.1093/jac/dkp029 [DOI] [PubMed] [Google Scholar]
- Duval V., Maiga I., Maiga A., Guillard T., Brasme L., Forte D., et al. (2009). High Prevalence of CTX-M-Type β-Lactamases among Clinical Isolates of Enterobacteriaceae in Bamako, Mali. Antimicrob. Agents Chemother. 53 4957–4958. 10.1128/AAC.00675-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendukly F., Karlsson I., Hanson H. S., Kronvall G., Dornbusch K. (2003). Patterns of mutations in target genes in septicemia isolates of Escherichia coli and Klebsiella pneumoniae with resistance or reduced susceptibility to ciprofloxacin. APMIS 111 857–866. 10.1034/j.1600-0463.2003.1110904.x [DOI] [PubMed] [Google Scholar]
- Ferjani S., Saidani M., Amine F. S., Boutiba-Ben Boubaker I. (2014). Prevalence and characterization of plasmid-mediated quinolone resistance genes in extended-spectrum β-Lactamase-producing Enterobacteriaceae in a Tunisian Hospital. Microbial. Drug Resist. 21 158–166. 10.1089/mdr.2014.0053 [DOI] [PubMed] [Google Scholar]
- Gharout-Sait A., Alsharapy S.-A., Brasme L., Touati A., Kermas R., Bakour S., et al. (2014). Enterobacteriaceae isolates carrying the New Delhi metallo-β-lactamase gene in Yemen. J. Med. Microbiol. 63 1316–1323. 10.1099/jmm.0.073767-0 [DOI] [PubMed] [Google Scholar]
- Guillard T., Bertrand X., de Champs C., Cholley P., Bajolet O., Gbaguidi-Haore H. (2014a). aac(6’)-Ib-cr is the major plasmid-mediated quinolone resistance determinant inextended-spectrum. J. Global. Antimicrob. Resist. 2 111–113. 10.1016/j.jgar.2014.01.006 [DOI] [PubMed] [Google Scholar]
- Guillard T., Grillon A., de Champs C., Cartier C., Madoux J., Berçot B., et al. (2014b). Mobile insertion cassette elements found in Small non-transmissible plasmids in Proteeae may explain qnrD mobilization. PLoS ONE 9:e87801 10.1371/journal.pone.0087801.s002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard T., Cambau E., Chau F., Massias L., De Champs C., Fantin B. (2013). Ciprofloxacin treatment failure in a murine model of pyelonephritis due to an AAC(6’)-Ib-cr-producing Escherichia coli strain susceptible to ciprofloxacin in vitro. Antimicrob. Agents Chemother. 57 5830–5835. 10.1128/AAC.01489-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard T., Duval V., Moret H., Brasme L., Vernet-Garnier V., De Champs C. (2010). Rapid detection of aac(6’)-Ib-cr quinolone resistance gene by pyrosequencing. J. Clin. Microbiol. 48 286–289. 10.1128/JCM.01498-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard T., Lebreil A.-L., Hansen L. H., Kisserli A., Berger S., Lozniewski A., et al. (2015). Discrimination between native and Tn6010-associated oqxAB in Klebsiella spp., Raoultella spp. and other Enterobacteriaceae using a two-step strategy. Antimicrob. Agents Chemother. 59 5838–5840. 10.1128/AAC.00669-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard T., Moret H., Brasme L., Carlier A., Vernet-Garnier V., Cambau E., et al. (2011). Rapid detection of qnr and qepA plasmid-mediated quinolone resistance genes using real-time PCR. Diagn. Microbiol. Infect. Dis. 70 253–259. 10.1016/j.diagmicrobio.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Hammoudi D., Moubareck C. A., Aires J., Adaime A., Barakat A., Fayad N., et al. (2014). Countrywide spread of OXA-48 carbapenemase in Lebanon: surveillance and genetic characterization of carbapenem-non-susceptible Enterobacteriaceae in 10 hospitals over a one-year period. Int. J. Infect. Dis. 29 139–144. 10.1016/j.ijid.2014.07.017 [DOI] [PubMed] [Google Scholar]
- Hooper D. C. (2001). Mechanisms of action of antimicrobials: focus on fluoroquinolones. Clin. Infect. Dis. 32 S9–S15. 10.1086/319370 [DOI] [PubMed] [Google Scholar]
- Huang S.-Y., Zhu X.-Q., Wang Y., Liu H.-B., Dai L., He J.-K., et al. (2012). Co-carriage of qnrS1, floR, and blaCTX-M-14 on a multidrug-resistant plasmid in Escherichia coli isolated from pigs. Foodborne Pathog Dis. 9 896–901. 10.1089/fpd.2012.1131 [DOI] [PubMed] [Google Scholar]
- Huson D. H., Scornavacca C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 61 1061–1067. 10.1093/sysbio/sys062 [DOI] [PubMed] [Google Scholar]
- Hussein K., Sprecher H., Mashiach T., Oren I., Kassis I., Finkelstein R. (2009). Carbapenem resistance among Klebsiella pneumoniae isolates: risk factors, molecular characteristics, and susceptibility patterns. Infect. Control Hosp. Epidemiol. 30 666–671. 10.1086/598244 [DOI] [PubMed] [Google Scholar]
- Izdebski R., Baraniak A., Herda M., Fiett J., Bonten M. J. M., Carmeli Y., et al. (2014). MLST reveals potentially high-risk international clones of Enterobacter cloacae. J. Antimicrob. Chemother. 70 48–56. 10.1093/jac/dku359 [DOI] [PubMed] [Google Scholar]
- Jarlier V., Nicolas M. H., Fournier G., Philippon A. (1988). Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10 867–878. 10.1093/clinids/10.4.867 [DOI] [PubMed] [Google Scholar]
- Lascols C., Robert J., Cattoir V., Bébéar C., Cavallo J.-D., Podglajen I., et al. (2007). Type II topoisomerase mutations in clinical isolates of Enterobacter cloacae and other enterobacterial species harbouring the qnrA gene. Int. J. Antimicrob. Agents 29 402–409. 10.1016/j.ijantimicag.2006.11.008 [DOI] [PubMed] [Google Scholar]
- Lavigne J.-P., Sotto A., Nicolas-Chanoine M.-H., Bouziges N., Pagès J.-M., Davin-Regli A. (2013). An adaptive response of Enterobacter aerogenes to imipenem: regulation of porin balance in clinical isolates. Int. J. Antimicrob. Agents 41 130–136. 10.1016/j.ijantimicag.2012.10.010 [DOI] [PubMed] [Google Scholar]
- Li X.-Z., Plésiat P., Nikaido H. (2015). The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin. Microbiol. Rev. 28 337–418. 10.1128/CMR.00117-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liapis E., Pantel A., Robert J., Nicolas-Chanoine M. H., Cavalie L., van der Mee-Marquet N., et al. (2014). Molecular epidemiology of OXA-48-producing Klebsiella pneumoniae in France. Clin. Microbiol. Infect. 20 O1121–O1123. 10.1111/1469-0691.12727 [DOI] [PubMed] [Google Scholar]
- Majewski P., Wieczorek P., Sacha P. T., Frank M., Juszczyk G., Ojdana D., et al. (2014). Emergence of OXA-48 carbapenemase-producing Enterobacter cloacae ST89 infection in Poland. Int. J. Infect. Dis. 25 107–109. 10.1016/j.ijid.2014.02.024 [DOI] [PubMed] [Google Scholar]
- Mammeri H., Guillon H., Eb F., Nordmann P. (2010). Phenotypic and Biochemical Comparison of the Carbapenem-Hydrolyzing Activities of Five Plasmid-Borne AmpC β-Lactamases. Antimicrob. Agents Chemother. 54 4556–4560. 10.1128/AAC.01762-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Martínez L., Pascual A., Conejo M. D. C., García I., Joyanes P., Doménech-Sánchez A., et al. (2002). Energy-dependent accumulation of norfloxacin and porin expression in clinical isolates of Klebsiella pneumoniae and relationship to extended-spectrum beta-lactamase production. Antimicrob. Agents Chemother. 46 3926–3932. 10.1128/AAC.46.12.3926-3932.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi-Akiyama T., Hayakawa K., Ohmagari N., Shimojima M., Kirikae T. (2013). Multilocus sequence typing (MLST) for characterization of Enterobacter cloacae. PLoS ONE 8:e66358 10.1371/journal.pone.0066358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann P., Naas T., Poirel L. (2011). Global spread of Carbapenemase-producing Enterobacteriaceae. Emerg. Infect. Dis. 17 1791–1798. 10.3201/eid1710.110655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel A., Boutet-Dubois A., Jean-Pierre H., Marchandin H., Sotto A., Lavigne J. P., et al. (2014). French regional surveillance program of carbapenemase-producing Gram-negative bacilli: results from a 2-year period. Eur. J. Clin. Microbiol. Infect. Dis. 33 2285–2292. 10.1007/s10096-014-2189-5 [DOI] [PubMed] [Google Scholar]
- Poirel L., Bonnin R. A., Nordmann P. (2012a). Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob. Agents Chemother. 56 559–562. 10.1128/AAC.05289-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Potron A., Nordmann P. (2012b). OXA-48-like carbapenemases: the phantom menace. J. Antimicrob Chemother. 67 1597–1606. 10.1093/jac/dks121 [DOI] [PubMed] [Google Scholar]
- Poirel L., Héritier C., Tolün V., Nordmann P. (2004). Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 48 15–22. 10.1128/AAC.48.1.15-22.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert J., Pantel A., Mérens A., Lavigne J.-P., Nicolas-Chanoine M.-H. ONERBA’s Carbapenem et al. (2014). Incidence rates of carbapenemase-producing Enterobacteriaceae clinical isolates in France: a prospective nationwide study in 2011-12. J. Antimicrob. Chemother. 69 2706–2712. 10.1093/jac/dku208 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Martínez J. M., Pichardo C., García I., Pachón-Ibañez M. E., Docobo-Pérez F., Pascual A., et al. (2008). Activity of ciprofloxacin and levofloxacin in experimental pneumonia caused by Klebsiella pneumoniae deficient in porins, expressing active efflux and producing QnrA1. Clin. Microbiol. Infect. 14 691–697. 10.1111/j.1469-0691.2008.02020.x [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Tenover F. C., Arbeit R. D., Goering R. V., Mickelsen P. A., Murray B. E., Persing D. H., et al. (1995). Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y., Mochimaru T., Morokuma Y., Kiyosuke M., Fujise M., Eto F., et al. (2010). Geographic distribution of fluoroquinolone-resistant Escherichia coli strains in Asia. Int. J. Antimicrob. Agents 35 387–391. 10.1016/j.ijantimicag.2009.12.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.