Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a common and important cause of colonization and infection in medical intensive care units (ICU). The aim of this study was to assess association factors between MRSA nasal colonization and subsequent infections in medical ICU patients by clinical investigation and molecular genotyping.
A prospective cohort observational analysis of consecutive patients admitted to medical ICUs between November 2008 and May 2010 at a tertiary teaching hospital were included. To detect MRSA colonization, the specimens from the nares were obtained within 3 days of admission to the ICU and again 1 week following admission to the ICU. Genetic relatedness for colonized and clinical isolates from each study patient with MRSA infection were analyzed and compared.
A total of 1266 patients were enrolled after excluding 195 patients with already present MRSA infections. Subsequent MRSA infection rates were higher in patients with nasal colonization than in those without (39.1% versus 14.7%, respectively). Multivariate Poisson regression analysis demonstrated that nasal MRSA colonization (relative risk [RR]: 2.50; 95% confidence interval [CI]: 1.90–3.27; P < 0.001) was independent predictors for subsequent MRSA infections. History of tracheostomy, however, was a protective predictor in all patients (RR: 0.38; 95% CI: 0.18–0.79; P = 0.010) and in patients with MRSA nasal colonization (RR: 0.22; 95% CI: 0.55–0.91; P = 0.037). Molecular genetics studies revealed that most MRSA isolates were healthcare-associated clones and that nasal and clinical isolates exhibited up to 75% shared identity.
Methicillin-resistant S. aureus nasal colonization was significantly associated with subsequent MRSA infection among medical ICU patients. Previous MRSA infection was associated with subsequent MRSA infections, and history of tracheostomy associated with reducing this risk. Most MRSA isolates were healthcare-associated strains that were significantly correlated between nasal and clinical isolates.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a common and important cause of infection in the intensive care unit (ICU) setting. Preceding MRSA colonization is a risk factor for subsequent MRSA infections.1 The colonizing bacterial strains may serve as endogenous reservoirs for overt clinical infections or may spread to other patients.2–7 Several studies have demonstrated a link between S. aureus carriage and subsequent infection in continuous peritoneal dialysis patients, nonsurgical patients, critical neonates, and medical ICU patients.8–12 The routine MRSA surveillance to prevent MRSA infections among ICU patients, however, is still a controversial policy.13 Controversy also exists about eliminating nasal MRSA carriage to prevent consequent MRSA infections. It is important to confirm the linkage between nasal carriage and clinical MRSA isolates to develop the best strategy to avoid systemic infection by decolonization methods. In 2000, 53% to 83% of S. aureus isolates attributed to nosocomial infections in 12 Taiwanese major hospitals were resistant to methicillin.14 In our adult ICUs, MRSA accounted for 77% of nasal S. aureus isolates with a high colonization rate (up to 32%) during a surveillance study in 2010.15
A prospective cohort observational study of medical ICU patients was undertaken to examine MRSA nasal colonization status and the development of MRSA infection. Our research goals were to determine the clinical association between nasal carriage of MRSA and subsequent MRSA infections and to identify additional risk factors associated with MRSA infection. The relationship between nasal and clinical isolates was also investigated using pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) analysis.
MATERIALS AND METHODS
Study Settings and Design
Chang-Gung Memorial Hospital, Lin-Kuo branch, is a university-affiliated 3700-bed tertiary teaching hospital in northern Taiwan that provides healthcare ranging from primary to tertiary care. A prospective cohort observational analysis of consecutive patients admitted to the 2 medical ICUs (44 beds) between November 2008 and May 2010 was performed. The Institutional Review Board of Chang-Gung Memorial Hospital reviewed and approved the study (IRB No.: 96–0104B) and the requirement for written informed consent was waived.
For patients with multiple medical ICU admissions, only the first admission was included in the analysis. Nasal surveillance specimens for MRSA were collected. To detect MRSA colonization, the specimens from the nares were obtained within 3 days of admission to the ICU and again 1 week following admission to the ICU. Methicillin-resistant S. aureus isolates recovered from clinical diagnostic samples (beyond survey culture specimens) submitted to the clinical microbiology laboratory were defined as clinical isolates. True MRSA infections were defined by the following criteria. Bloodstream infection required a positive blood culture. Pneumonia required a positive respiratory culture, a compatible chest radiograph with symptoms and signs of lower respiratory tract infection and a decision to treat. Urinary tract infection required a positive urine culture and either a decision to treat or the growth of >100,000 CFU/ml plus at least 50 leukocytes per high-power field. All other sites, including pleural effusion, ascites, skin, and soft tissue required a positive culture and a decision to treat.
To identify potential risk factors for MRSA infection, the following data were collected from each patient: baseline demographics, characteristics, underlying and current diseases, clinical covariates, dates of previous and current hospitalizations, date of ICU admission, previous MRSA infection, already present MRSA infection, and subsequent MRSA infection. Previous infection was defined as MRSA infection diagnosed at least 2 weeks before the current hospitalization. Already present infection was defined as MRSA infection diagnosed during this hospitalization but before admission to medical ICU or within 24 hours after ICU admission.
Microbiology
Nasal and clinical isolates from each study patient with MRSA infection were genotyped and compared. Survey specimens for MRSA culture were obtained with a cotton swab, placed in transport medium (Venturi Transystem, Copan Innovation Ltd, Limmerick, Ireland), and processed in the microbiology laboratory within 4 hours. Coagulase tests were carried out by using rabbit plasma to make sure the identification of S. aureus after incubation and subcultivation. Based on the recommendation of Clinical and Laboratory Standards Institute (CLSI), Cefoxitin test was used to differentiate the MRSA from methicillin-susceptible S. aureus (MSSA).16
Molecular genotyping analysis was performed on a total of 65 clinical isolates and 43 nasal isolates from 36 randomly selected patients with MRSA infections. PFGE was used to fingerprint all MRSA isolates according to procedures described previously.12,17–19 Genotypes were allocated in alphabetical order, as in our prior studies; any new genotype, if identified, was allocated consecutively.12,17,18 Pulsed-field gel electrophoresis patterns with less than 4 band differences from an existing genotype were defined as subtypes of that genotype. Two isolates were regarded to be indistinguishable, highly related, or distinct if they had the similar subtype (no band difference), the same genotype (less than 4-band differences), or a different type (≥4-band differences), respectively. According to the measures described previously, Staphylococcal chromosome cassette mec (SCCmec) type and presence of Pantone-Valentine leukocidin (PVL) genes were established by polymerase chain reaction (PCR) assays.12,18,20,21 Multilocus sequence typing was performed for selective strains of representative PFGE patterns as described elsewhere.22
Statistical Analysis
Statistical analyses were performed using SPSS for Windows software version 21.0 (IBM, Armonk, N Y). Continuous data obtained were expressed as the mean ± standard deviation (SD) and frequencies were calculated for categorical data. Relative risk (RR) and 95% confidence interval (CI) were calculated using Poisson regression. Potential predictors of colonization with P < 0.05 in univariate analysis were further analyzed with a multivariate Poisson regression model in SPSS. Two-sided P values<0.05 were considered significant for multivariate analysis.
RESULTS
A total of 1461 patients admitted to the medical ICUs were evaluated. Excluding 195 patients who had already present MRSA infections, 1266 patients were enrolled in this study. Of these 1266 study patients, the initial MRSA nasal colonization rate was 16.4% (207/1266), and the rate of subsequent MRSA infections was 39.1% (81/207) (Figure 1). The subsequent MRSA infection rate of non-MRSA colonization patients was significantly lower than MRSA colonization patients (14.7% versus 39.1%, OR: 3.72; 95% CI: 2.68–5.16; P < 0.001). Overall, the MRSA infection rate during hospitalization was 18.7% (237/1266). For the patients with MRSA nasal colonization, true infection occurred 12.3 days (median, 7 days; interquartile range, 3–15 days) after colonization, and 16.9 days (median, 12 days; interquartile range, 7–20 days) after ICU admission, respectively. There was a significant difference in the meantime from ICU admission to the true MRSA infection between patients with MRSA colonization (16.9 days) and those without colonization (24.3 days) (P = 0.002). Of these 1266 study patients, 939 patients (74.2%) were experiencing respiratory failure with mechanical ventilation. There were 119 patients (9.4%) who underwent tracheostomy, which were all done before ICU admission, and all need ventilator support.
FIGURE 1.
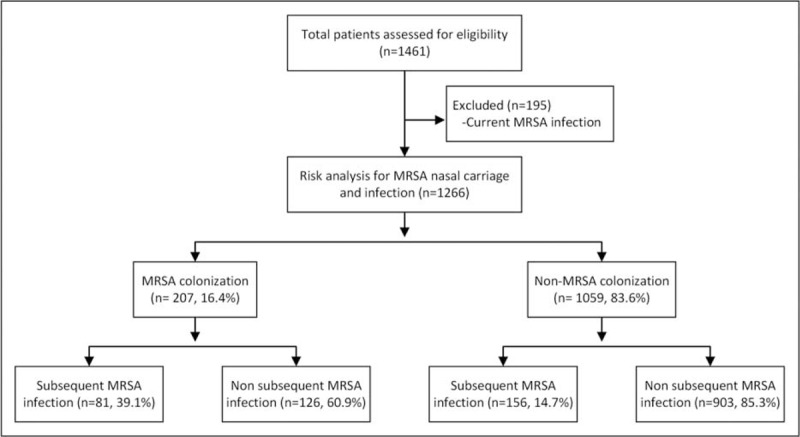
Schematic of analysis for medical intensive care unit patients by either MRSA infection or methicillin-resistant S. aureus nasal colonization. ICU, intensive care unit; MRSA, methicillin-resistant Staphylococcus aureus.
Table 1 shows the characteristics and risk factors of nasal MRSA colonization in these enrolled patients. The colonized MRSA patients were older than noncolonized patients (71.8 ± 15.7 versus. 68.9 ± 15.2, P = 0.023). In addition, the nasal MRSA patients had higher instances of hospitalization within the previous year (P = 0.029), old cerebrovascular accidents (p = 0.047), and MRSA infections previous to this admission (P < 0.001). Furthermore, multivariate Poisson regression analysis demonstrated that previous MRSA infections (RR: 2.19; 95% CI: 1.56–3.08; P < 0.001) were independent predictors for nasal MRSA colonization.
TABLE 1.
Risk Factors Associated With Nasal MRSA Colonization (N = 1266)
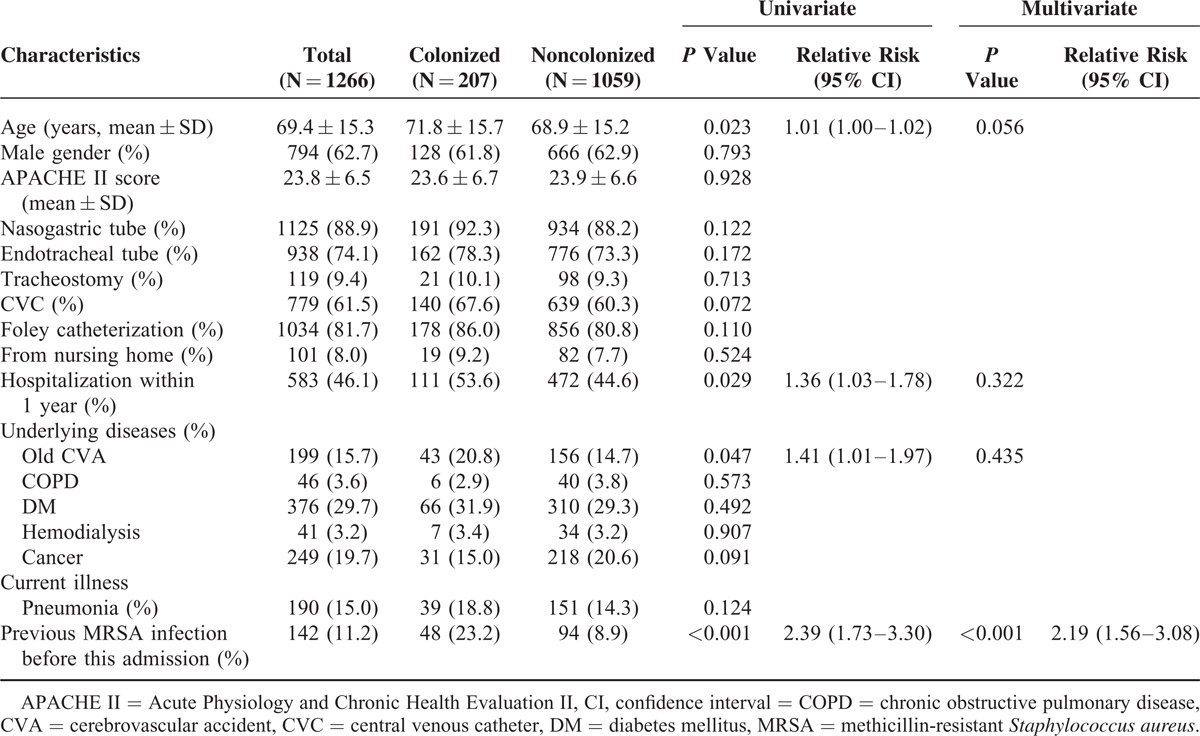
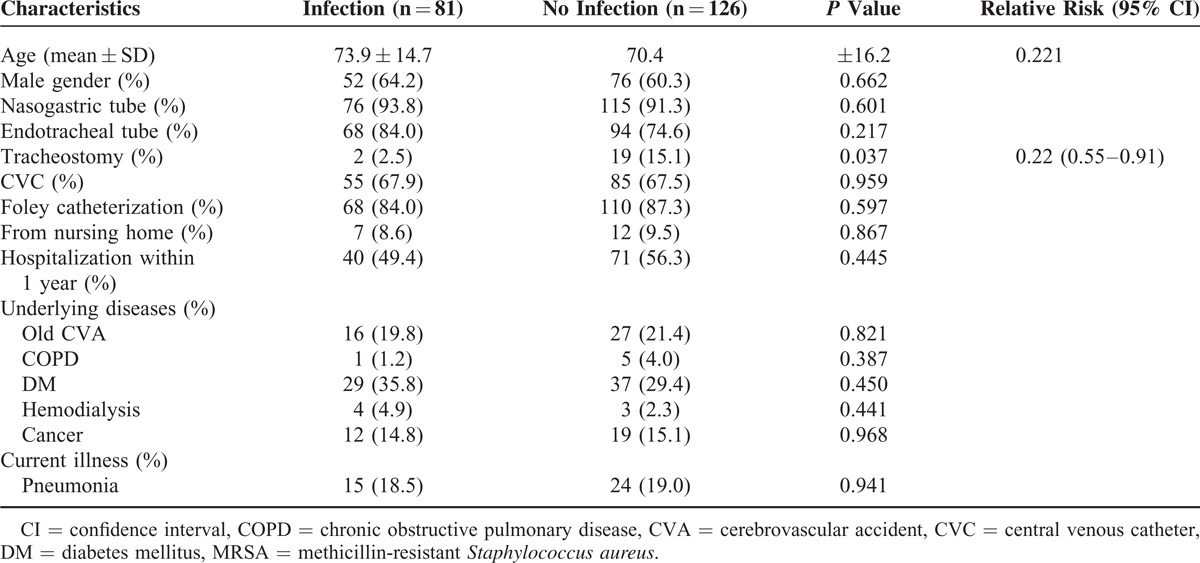
Table 2 shows that nasal MRSA colonization was an independent predictor for subsequent MRSA infections. Patients with nasal MRSA colonization had significantly higher rates of subsequent MRSA infections than those without colonization (RR: 2.50; 95% CI: 1.90–3.27; P < 0.001). History of tracheostomy (RR: 0.38; 95% CI: 0.18–0.79; p = 0.010), however, was an independent protective predictor for subsequent MRSA infection. Methicillin-resistant S. aureus nasal colonization was noted for 81 (34.2%) of 237 patients with MRSA infections. Methicillin-resistant S. aureus nasal colonization was also noted for 93 (65.5%) of 142 patients with previous MRSA infections. In patients with MRSA nasal colonization, subsequent MRSA infections decreased significantly only with the history of tracheostomy (RR: 0.22; 95% CI: 0.55–0.91; P = 0.037) (Table 3).
TABLE 2.
Risk Factors for Patients With Subsequent MRSA Infection
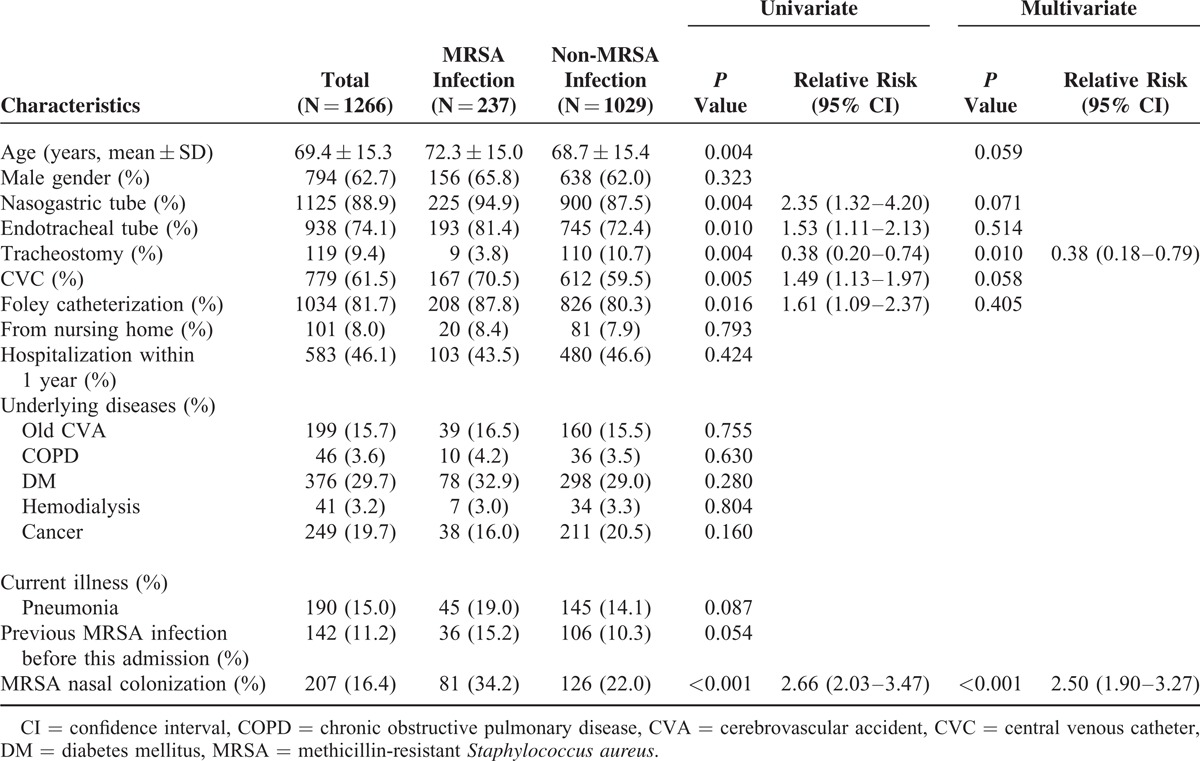
TABLE 3.
Risk Factors for MRSA Infection in Patients With Nasal MRSA Colonization (N = 207)
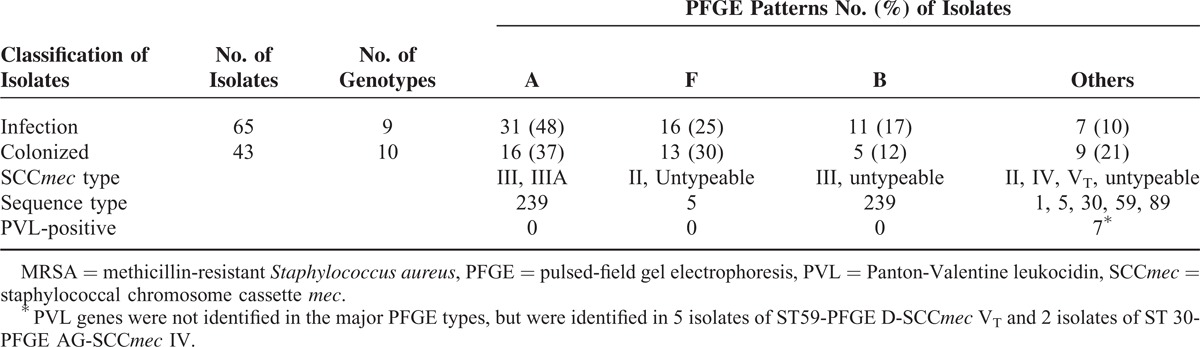
Molecular genotyping analysis was performed on a total of 65 clinical isolates and 43 nasal isolates from 36 patients with true MRSA infections (Table 4). Of these 36 patients, 14 patients (38.9%) had 2 to 7 clinical isolate for analysis. The source of the 65 clinical isolates included bloodstream (46 isolates), sputum (14 isolates), pleural effusion (3 isolates), and ascites (2 isolates). A total of 11 PFGE patterns were identified and 2 major patterns were type A and type F. Within 65 clinical isolates, the ratios of type A and type F are 48% and 25%, respectively. Of the 43 nasal isolates, types A and F accounted for 37.2% and 30.2% of the patterns, respectively. Seventeen isolates with various PFGE patterns were selected for MLST analysis and 2 sequence types (ST 239 and ST 5) were identified. Most isolates with PFGE type A and B sequenced as ST 239, whereas those with PFGE type F sequenced as ST 5. Overall, most MRSA isolates belonged to 1 of 2 major clones characterized as ST239/PFGE A/SCCmec III or IIIA/PVL negative (43.5%), and ST5/PFGE F/SCCmec II/PVL negative (26.9%). Both clones have been recognized as healthcare-associated clones in Taiwan.
TABLE 4.
Molecular Genotyping of 65 Clinical and 43 Nasal Colonized MRSA Isolates From 36 Patients With MRSA Infections
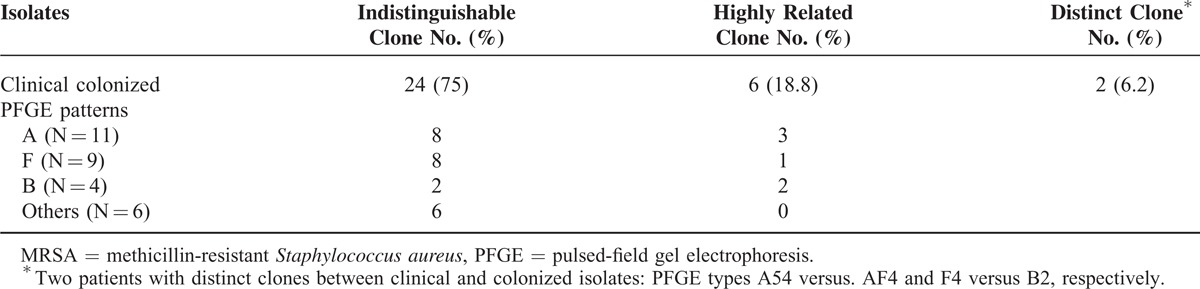
The genetic correlation between clinical and nasal colonized isolates was analyzed in 32 patients with MRSA infections after excluding 4 patients who had multiple nasal MRSA colonization or clinical isolates with different genotypes (Table 5). The infection sources in these 32 patients were: bacteremia (19), pneumonia (8), thoracic empyema (2), peritonitis (1), combined thoracic empyema with bacteremia (1), and combined pneumonia with peritonitis (1). The nasal and clinical isolates were indistinguishable in 24 patients (75.0%), highly related in 6 patients (18.8%), and distinct in 2 patients (6.2%) based on PFGE and MLST analyses.
TABLE 5.
Genetic Correlation Between Clinical and Nasal Colonized MRSA Isolates From 32 Patients With MRSA Infections
DISCUSSION
In this study of medical ICUs patients, we found the subsequent MRSA infection rate of patients with nasal MRSA colonization to be significantly higher than that of patients without nasal MRSA colonization. History of hospitalization within 1 year, old cerebrovascular accidents, and previous MRSA infections were risk factors for nasal MRSA colonization. Nasal MRSA colonization was an independent predictor for subsequent MRSA infections. History of tracheostomy, however, was a protective predictor against subsequent MRSA infection. Molecular study revealed that most MRSA isolates were ST239/PFGE A/SCCmec III or IIIA/PVL (−) and ST5/PFGE F/SCCmec II/PVL (−), which have been classified as healthcare-associated clones.
Nosocomial S. aureus bacteremia is 3 times more frequent in S. aureus nasal carriers than in noncarriers (1.2% versus 0.4%).11 Methicillin-resistant S. aureus colonization was the leading risk factor for subsequent MRSA infection in patients with chronic skin ulcers during the same admission.7 In general, nasal MRSA colonization, either present at admission to the hospital or acquired during hospitalization, increases the risk of MRSA infection compared with patients without nasal colonization (25% versus 2%).2 Furthermore, the impact of colonization leading to infection may be prolonged up to 18 months after hospital discharge.23 In our study patients, we also found that patients with nasal MRSA colonization had higher rates of subsequent MRSA infections than patients without nasal colonization (39.1% versus 14.7%). Although 1 study showed that nasal MRSA colonization was a poor predictor of ICU-acquired MRSA infections requiring antibiotic treatment,24 a recent study observed that active surveillance cultures of MRSA yielded high specificity and negative predictive values for ventilator-associated pneumonia.25 In this study, we also found nasal MRSA colonization was 1 important risk factors of MRSA infection. One possible explanation for these different observations is that different ICU patient groups respond differently to the same strategy.
Furthermore, we found that tracheostomy, but not translaryngeal endotracheal tubes, was associated with lower risk for MRSA infection with or without previous nasal MRSA colonization. Previous studies have also found that early tracheostomy patients had significantly lower rates of mortality and pneumonia compared with patients with prolonged translaryngeal endotracheal tubes.26 Translaryngeal endotracheal tubes may provide a direct route from the upper airway—often colonized with pathologic organisms—to the lower airway. Translaryngeal endotracheal tubes keep the vocal cords open and that the bacteria from the back of the throat pass through the cords and then pass the cuff, get into the airway, reflux into the tube infect the biofilm in the endotracheal tubes tube, and finally translocate into the lungs. Tracheostomy is protective against nosocomial infection acquired from upper airway because of better hygiene and oral care. Therefore, tracheostomy, rather than a translaryngeal endotracheal tube to bypass the upper airway, may potentially decrease the risk for bacteria colonization and subsequent infection.
In this study, the rate of subsequent MRSA infection in patients with nasal MRSA colonization on admission was 39.1%. Clinical infection isolates were genotypically indistinguishable from nasal colonized isolates in 75.0% (24 in 32) of the cases based on PFGE patterns and MLST analysis. We demonstrated a significant association between nasal MRSA colonization and subsequent infections based on genotyping by PFGE in medical ICU patients. In other groups, such as mixed ICU and general ward patients, continuous peritoneal dialysis patients, surgical patients and neonates, the association rate ranged from 67% to 84.6%.8–12 One study also demonstrated that S. aureus nasal carriage and clinical isolates belong to similar genetic clusters using MLST, despite differences in sequence type assignments.27
There has been controversy about the benefits of reducing subsequent infection rates by decolonization. The Cochrane database of systematic reviews showed that application of intranasal mupirocin reduced infection rates only in dialysis and surgical patients.28 Bode et al showed that rapid screening to identify nasal carriers, followed by decolonization with both nasal mupirocin nasal ointment and chlorhexidine soap, could prevent hospital-acquired infection with S. aureus in surgical patients.29 Notably, a recent large-scale cluster-randomized trial among 74,256 patients concluded that universal decolonization was superior to both targeted decolonization and screening and isolation in both reducing rates of MRSA clinical isolates and bloodstream infection from any pathogen.30 Although this practice benefited from infection rate control and obviated the need for MRSA surveillance tests, further surveillance for mupirocin and chlorhexidine resistance remains an essential issue if the universal decolonization practice implemented widely.31,32
There are some limitations in this study. First, although this was a large study, these results reflected the association between nasal colonization and infection at a single medical center, and may not be generalized to other hospitals. Second, we did not obtain colonization specimens from anatomic sites other than nostrils, which may also contribute to subsequent MRSA infections. Third, our study was performed in 2 different ICUs. Differences in compliance to infection-control programs to prevent horizontal transmission of MRSA in these 2 ICUs may affect the occurrence of nasal MRSA colonization and subsequent infections. Finally, molecular genotyping analysis was performed randomly on a subset of the total strains, which could not represent all the strains.
In conclusion, nasal MRSA colonization was significantly associated with subsequent MRSA infection among medical ICU patients. Previous MRSA infection was associated with subsequent MRSA infections, and history of tracheostomy could reduce this risk. Molecular and genetic studies determined that most MRSA isolates were healthcare-associated strains with significant correlations between colonized nasal and clinical isolates. In future studies, this information may support a decolonization strategy for the medical ICU patients.
Acknowledgments
We also thank Dr. Horng-Chyuan Lin and Dr. Shu-Min Lin for their help with the study.
Footnotes
Abbreviations: CI = confidence interval, ICU = intensive care unit, MLST = multilocus sequence typing, MRSA = methicillin-resistant Staphylococcus aureus, MSSA = methicillin-susceptible Staphylococcus aureus, PCR = polymerase chain reaction, PFGE = pulsed-field gel electrophoresis, PVL = Pantone-Valentine leukocidin, RR = relative risk, SD = standard deviation.
Dr. Kao and Dr. Chen contributed equally to this article.
This study was supported by a grant from Chang Gung Memorial Hospital (CMRPG460113).
We thank Yu-Jr Lin for statistical consultation, which was supported by grants from Biostatistical Center for Clinical Research, Chang Gung Memorial Hospital (CLRPG3D0041.
The authors have no conflicts of interest to disclose.
REFERENCES
- 1.Ellis MW, Hospenthal DR, Dooley DP, et al. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin Infect Dis 2004; 39:971–979. [DOI] [PubMed] [Google Scholar]
- 2.Davis KA, Stewart JJ, Crouch HK, et al. Methicillin-resistant Staphylococcus aureus (MRSA) nares colonization at hospital admission and its effect on subsequent MRSA infection. Clin Infect Dis 2004; 39:776–782. [DOI] [PubMed] [Google Scholar]
- 3.Wenzel R, Perl T. The significance of nasal carriage of Staphylococcus aureus and the incidence of postoperative wound infection. J Hosp Infect 1995; 31:13–24. [DOI] [PubMed] [Google Scholar]
- 4.Kluytmans JA, Mouton JW, Ijzerman EP, et al. Nasal carriage of Staphylococcus aureus as a major risk factor for wound infections after cardiac surgery. J Infect Dis 1995; 171:216–219. [DOI] [PubMed] [Google Scholar]
- 5.Kluytmans J, Van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 1997; 10:505–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrouste-Orgeas M, Timsit JF, Kallel H, et al. Colonization with methicillin-resistant Staphylococcus aureus in ICU patients: morbidity, mortality, and glycopeptide use. Infect Control Hosp Epidemiol 2001; 22:687–692. [DOI] [PubMed] [Google Scholar]
- 7.Roghmann MC, Siddiqui A, Plaisance K, et al. MRSA colonization and the risk of MRSA bacteraemia in hospitalized patients with chronic ulcers. J Hosp Infect 2001; 47:98–103. [DOI] [PubMed] [Google Scholar]
- 8.von Eiff C, Becker K, Machka K, et al. Nasal carriage as a source of Staphylococcus aureus bacteremia. N Engl J Med 2001; 344:11–16. [DOI] [PubMed] [Google Scholar]
- 9.Nouwen J, Schouten J, Schneebergen P, et al. Staphylococcus aureus carriage patterns and the risk of infections associated with continuous peritoneal dialysis. J Clin Microbiol 2006; 44:2233–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perl TM, Cullen JJ, Wenzel RP, et al. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N Engl J Med 2002; 346:1871–1877. [DOI] [PubMed] [Google Scholar]
- 11.Wertheim HFL, Vos MC, Ott A, et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet 2004; 364:703–705. [DOI] [PubMed] [Google Scholar]
- 12.Huang YC, Chou YH, Su LH, et al. Methicillin-resistant Staphylococcus aureus colonization and its association with infection among infants hospitalized in neonatal intensive care units. Pediatrics 2006; 118:469–474. [DOI] [PubMed] [Google Scholar]
- 13.Huang SS, Rifas-Shiman SL, Warren DK, et al. Improving methicillin-resistant Staphylococcus aureus surveillance and reporting in intensive care units. J Infect Dis 2007; 195:330–338. [DOI] [PubMed] [Google Scholar]
- 14.Hsueh PR, Liu CY, Luh KT. Current status of antimicrobial resistance in Taiwan. Emerg Infect Dis 2002; 8:132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CB, Chang HC, Huang YC. Nasal meticillin-resistant Staphylococcus aureus carriage among intensive care unit hospitalised adult patients in a Taiwanese medical centre: one time-point prevalence, molecular characteristics and risk factors for carriage. J Hosp Infect 2010; 74:238–244. [DOI] [PubMed] [Google Scholar]
- 16.Wilker MA. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; sixteenth informational supplement. 16th edn.2006; Wayne, PA: Clinical and Laboratory Standards Institute, M100-S16. [Google Scholar]
- 17.Huang YC, Su LH, Wu TL, et al. Molecular epidemiology of clinical isolates of methicillin-resistant Staphylococcus aureus in Taiwan. J Clin Microbiol 2004; 42:307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang YC, Su LH, Wu TL, et al. Changing molecular epidemiology of methicillin-resistant Staphylococcus aureus bloodstream isolates from a teaching hospital in Northern Taiwan. J Clin Microbiol 2006; 44:2268–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol 1995; 33:2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lina G, Piemont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 1999; 29:1128–1132. [DOI] [PubMed] [Google Scholar]
- 21.Oliveira DC, de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2002; 46:2155–2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enright MC, Day NP, Davies CE, et al. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000; 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang SS, Platt R. Risk of methicillin-resistant Staphylococcus aureus infection after previous infection or colonization. Clin Infect Dis 2003; 36:281–285. [DOI] [PubMed] [Google Scholar]
- 24.Sarikonda KV, Micek ST, Doherty JA, et al. Methicillin-resistant Staphylococcus aureus nasal colonization is a poor predictor of intensive care unit-acquired methicillin-resistant Staphylococcus aureus infections requiring antibiotic treatment. Crit Care Med 2010; 38:1991–1995. [DOI] [PubMed] [Google Scholar]
- 25.Chan JD, Dellit TH, Choudhuri JA, et al. Active surveillance cultures of methicillin-resistant Staphylococcus aureus as a tool to predict methicillin-resistant S. aureus ventilator-associated pneumonia. Crit Care Med 2012; 40:1437–1442. [DOI] [PubMed] [Google Scholar]
- 26.Rumbak MJ, Newton M, Truncale T, et al. A prospective, randomized, study comparing early percutaneous dilational tracheotomy to prolonged translaryngeal intubation (delayed tracheotomy) in critically ill medical patients. Crit Care Med 2004; 32:1689–1694. [DOI] [PubMed] [Google Scholar]
- 27.Lamers RP, Stinnett JW, Muthukrishnan G, et al. Evolutionary analyses of Staphylococcus aureus identify genetic relationships between nasal carriage and clinical isolates. PLoS One 2011; 6:e16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Rijen M, Bonten M, Wenzel R, et al. Mupirocin ointment for preventing Staphylococcus aureus infections in nasal carriers. Cochrane Database Syst Rev 2008; 4:CD006216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bode LG, Kluytmans JA, Wertheim HF, et al. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med 2010; 362:9–17. [DOI] [PubMed] [Google Scholar]
- 30.Huang SS, Septimus E, Kleinman K, et al. Targeted versus universal decolonization to prevent ICU infection. N Engl J Med 2013; 368:2255–2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harbarth S, Dharan S, Liassine N, et al. Randomized, placebo-controlled, double-blind trial to evaluate the efficacy of mupirocin for eradicating carriage of methicillin resistant Staphylococcus aureus. Antimicrob Agents Chemother 1999; 43:1412–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robicsek A, Beaumont JL, Thomson RB, et al. Topical therapy for methicillin-resistant Staphylococcus aureus colonization: impact on infection risk. Infect Control Hosp Epidemiol 2009; 30:623–632. [DOI] [PubMed] [Google Scholar]


