Abstract
Transcripts encoding a novel member of the lipoprotein receptor superfamily, termed LDL receptor-related protein (Lrp)13, were sequenced from striped bass (Morone saxatilis) and white perch (Morone americana) ovaries. Receptor proteins were purified from perch ovary membranes by protein-affinity chromatography employing an immobilized mixture of vitellogenins Aa and Ab. RT-PCR revealed lrp13 to be predominantly expressed in striped bass ovary, and in situ hybridization detected lrp13 transcripts in the ooplasm of early secondary growth oocytes. Quantitative RT-PCR confirmed peak lrp13 expression in the ovary during early secondary growth. Quantitative mass spectrometry revealed peak Lrp13 protein levels in striped bass ovary during late-vitellogenesis, and immunohistochemistry localized Lrp13 to the oolemma and zona radiata of vitellogenic oocytes. Previously unreported orthologs of lrp13 were identified in genome sequences of fishes, chicken (Gallus gallus), mouse (Mus musculus), and dog (Canis lupus familiaris). Zebrafish (Danio rerio) and Nile tilapia (Oreochromis niloticus) lrp13 loci are discrete and share genomic synteny. The Lrp13 appears to function as a vitellogenin receptor and may be an important mediator of yolk formation in fishes and other oviparous vertebrates. The presence of lrp13 orthologs in mammals suggests that this lipoprotein receptor is widely distributed among vertebrates, where it may generally play a role in lipoprotein metabolism.
Keywords: affinity purification, egg, endocytosis, oocyte, oogenesis, ovary, vitellogenesis, very low density lipoprotein receptor, yolk, low density lipoprotein receptor-related protein 13
The LDL receptor (LDLR) gene family is comprised of different genes encoding membrane receptors involved in endocytosis of a variety of ligands, most notably plasma lipoproteins. Characterized members of this family in vertebrates include LDLR (1), LDLR-related protein (LRP)1 (2), LRP2 (3), LRP3 (4), LRP4 (5), LRP5 (LRP7) (6), LRP6 (7), LRP8 (8), LRP10 (LRP9) (9), sortilin-related receptor LR11 (10), LRP11 (GenBank: NP_116221), LRP12 (11), and VLDL receptor (VLDLR) (12) or vitellogenin receptor (Vtgr) (13, 14). These receptors typically consist of unique configurations of epidermal growth factor precursor, class b YWxD (LDLb), and O-linked sugar domains, which define their identities, and class A ligand binding (LDLa) repeats, which determine their particular ligand specificities. Exceptional receptors also contain complement C1r/C1s, Uegf, Bmp1 (CUB) domains (i.e., LRP3, LRP10, LRP12) or motif at the N terminus with seven cysteines (MANEC) and polycystic kidney disease (PKD) domains (i.e., LRP11). With the exception of LRP4, LDLR family receptors are type I membrane proteins and all members contain transmembrane and cytoplasmic domains.
Acanthomorph fishes express three distinct lipoprotein yolk precursors, vitellogenins (Vtgs) (VtgAa, VtgAb, and VtgC) (15, 16). These Vtgs are produced by the liver, released into the circulatory system, and taken up specifically by growing oocytes via receptor-mediated endocytosis. We discovered four Vtgr proteins in white perch (Morone americana) ovary: a receptor greater than 212 kDa that binds only VtgAa (VtgAar), two receptors (116 kDa and 110.5 kDa) that preferentially bind VtgAb (VtgAbr), and a 150 kDa putative LDLR (pLDLR) that weakly and indiscriminately binds both VtgAa and VtgAb (17). The VtgC does not bind ovary Vtgr proteins in this species. We reported the molecular identity of a white perch Vtgr that is orthologous to mammalian VLDLR (18). This Vtgr is termed “Lr8−”, because it is a spliced variant gene transcript of vldlr that does not encode the O-linked sugar domain, as is characteristic of this form of Vldlr in fishes and chickens (19, 20). Herein, we refer to these Vldlr forms as Lr8+ and Lr8−, based on the presence or absence of the O-linked sugar domain, respectively. We have suggested that white perch Lr8− corresponds to one or both VtgAbr proteins based on the predicted molecular mass of the protein and on prior reports of fish and chicken Lr8− (17). However, the molecular identity of VtgAar remains unclear. Here, we describe the structure, expression, subcellular localization, and Vtg-binding properties of a novel lipoprotein receptor named ‘Lrp13’ that corresponds to VtgAar and show that the Lr8− corresponds to the VtgAbr.
MATERIALS AND METHODS
Sample collection
All experiments were conducted according to the Guide for the Care and Use of Laboratory Animals (21) and the procedures were approved by the North Carolina State University (NCSU) Institutional Animal Care and Use Committee. White perch (weight 425 ± 113 g; total length 265 ± 25 mm; all values are reported as mean ± standard deviation) were reared at NCSU (Raleigh, NC). Blood plasma was sampled from male white perch injected with estradiol-17β, as described previously (22), for purification of Vtgs. Ovaries were excised from vitellogenic white perch (n = 3; maximum oocyte diameter 536–552 μm) (23), for preparation of ovary membranes. Striped bass (Morone saxatilis) were reared at the NCSU Pamlico Aquaculture Field Laboratory (Aurora, NC). Ovary tissues from striped bass (n = 3) were collected by dissection or biopsy using a plastic cannula inserted through the urogenital pore at four time points: August (weight 1.90 ± 0.61 kg; total length 512 ± 18.0 mm), November (3.80 ± 0.14 kg; 625 ± 6.0 mm), February (4.40 ± 0.94 kg; 626 ± 46.6 mm), and April (4.54 ± 1.37 kg; 663 ± 74.5 mm). The most advanced oocytes during these time points represented one of four oocyte growth stages (24): early secondary growth (ESG, oocyte diameter 310 ± 21.8 μm), mid-vitellogenic growth (MVG, 503 ± 60.1 μm), late-vitellogenic growth (LVG, 831 ± 276 μm), and post-vitellogenic growth (PVG, 986 ± 33.0 μm), respectively. Ovary and liver samples were preserved in RNALater (Ambion, Austin, TX) for real-time quantitative RT-PCR. Ovary tissue was frozen in liquid nitrogen for quantitative tandem mass spectrometry. Ovary tissue from MVG and LVG striped bass was fixed for in situ hybridization in 0.2 M sodium phosphate buffer (pH 7.4) containing 4% paraformaldehyde, and for immunohistochemistry in Bouin’s solution (Sigma, Saint Louis, MO). Brain, heart, liver, ovary, foregut, muscle, and adipose from MVG striped bass were preserved in RNALater for semi-quantitative RT-PCR.
Molecular cloning
Primers (SB618 F1 and SB618 R1; see supplementary Table I for all primer sequences used in this study) were designed for striped bass contig 00618 (25). All DNA oligos were designed with Mac Vector (Accelrys Software, San Diego, CA) and obtained from Integrated DNA Technologies (Coralville, IA). Products were cloned from a Stratagene Uni-Zap XR cDNA library (La Jolla, CA) constructed from pooled white perch ovaries (18), and those from four colonies were bi-directionally sequenced according to our prior studies (16). Total RNA was extracted from pooled white perch ovaries using TRIzol reagent (Invitrogen, Carlsbad, CA) (25) and 3′ and 5′ rapid amplification of cDNA ends (RACE) were performed using FirstChoice RLM-RACE kit (Ambion). Primers for 5′RACE (WP618 R1 O and WP618 R1 N) and 3′RACE (WP618 F1 O, SB618 F0 O, WP618 F1 N, WP618 F2 N, and WP618 F3 N) with the suffix “O” or “N” were paired with outer and inner RACE primers, respectively. Sequences were assembled using MacVector and polypeptide domains were characterized using SMART (26). Eukaryotic Linear Motif (27) was used to identify putative functional motifs within the polypeptide domains.
Vertebrate lrp13 orthologs
The NCBI databases were queried for lrp13 orthologs and Lrp13 polypeptide sequences were collected from fishes, birds, and mammals along with sequences of other representative lipoprotein receptors. Sequences were aligned by ClustalW (28) to generate a dendrogram. Synteny of lrp13 and vldlr (lr8) were compared using zebrafish (Danio rerio) chromosome 5_9,505,781-9,678,348 and chromosome 10_15,283,756-15,333,718 (v. Zv9) and Nile tilapia (Oreochromis niloticus) scaffold GL831141.1_3,169,873-3,288,125 (reverse complemented) and scaffold GL831199.1_1,600,394-1,676,338 (v. Orenil1.0) available from Ensembl (29).
Protein-affinity chromatography
A mixture of VtgAa and VtgAb (VtgAa/b) was purified according to our studies (17, 30) and 20 mg of VtgAa/b was coupled to an equal volume (12 ml) of Affi-Gel 15 activated immunoaffinity support (Bio-Rad, Hercules, CA) following procedures adapted from Roehrkasten et al. (31). Affinity medium was poured into a 25 mm internal diameter column (12 ml) and the chromatography system was initialized with 10 column volumes of coupling buffer [20 mM HEPES, 150 mM NaCl, 1 mM CaCl2 (pH 7.4)] and equilibrated with 10 column volumes of binding buffer [20 mM Tris-HCl, 2 mM CaCl2, and 150 mM NaCl (pH 8.0), containing 1 mM phenylmethyl-sulfonyl fluoride and 4 IU/l aprotinin] at 4°C. Solubilized membrane proteins prepared from 25 g of vitellogenic white perch ovaries, according to our studies (17, 32), were recycled through the affinity media for 4.5 h at 4°C (1.0 ml/min flow rate). The column was washed with 15 vol of binding buffer at 4°C and buffer in the top reservoir was decanted and replaced with elution buffer [20 mM Tris-HCl, 10 mM suramin, 5 mM EDTA, 150 mM NaCl (pH 6.0)]. The column was equilibrated for 30 min at 4°C before eluting proteins with 5 vol of elution buffer. Fractions were collected and assayed for protein concentration using a BCA protein assay kit (Pierce, Rockford, IL) and a Bio-Rad 3550 microplate reader. Peak fractions were pooled and concentrated to 100 μg protein/ml with a Centricon YM-30 (Millipore, Billerica, MA). Affinity purified Vtgrs diluted 1:1 in Laemmli buffer were electrophoresed through 5% acrylamide precast Tris-HCl Ready Gels, as we described (17), and stained with Silver Stain Plus (Bio-Rad). Aliquots of solubilized ovary membrane proteins, affinity purified Vtgrs, and purified VtgAa/b were processed for LC-ESI-MS/MS using an LTQ linear ion trap mass spectrometer (Thermo, San Jose, CA) at the NCSU Genomic Sciences Laboratory (Raleigh, NC). Samples were analyzed in quadruplicate and ion fragmentation spectra were queried by MASCOT (Matrix Science, Boston, MA) against white perch VtgAa, VtgAb, VtgC (16), white perch Lr8− (18), and the translated striped bass ovary transcriptome (24, 25).
Preparation of α-Lrp13
A portion of cutthroat trout (Oncorhynchus clarki) CtLR13+1 (Lrp13) (33) was cloned into pET302/NT-His expression vector (Invitrogen) using In-Fusion Advantage PCR cloning kit (Clontech, Mountain View, CA) and primers CtLR13+1F and CtLR13+1R. Recombinant CtLR13+1 His-fusion proteins were expressed in Rosetta-gami B (DE3) pLysS (Novagen, Madison, WI) and recovered using BugBuster protein extraction reagent and Ni-charged His-Bind resin chromatography (Novagen). Superdex 200 gel purified recombinant CtLR13+1 was used to raise polyclonal α-CtLrp13 in rabbit as described by Hong et al. (34). Synthetic white perch Lrp13 peptides (CSLGYSGDSCQDHLLKT and TTLNESSQLRNLATQDC) and Lr8− peptides (CRPEANVSTSIQVDSTARGSA and CSVDLNGDNRKKVLQS) were used to raise polyclonal α-WpLrp13 and α-WpLr8− in chickens (GeneTel Laboratories, Madison, WI).
Ligand and Western blotting
Solubilized white perch ovary membrane proteins were separated by electrophoresis through 7.5 and 5% acrylamide precast Tris-HCl ready gels and subjected to ligand blotting with digoxigenin (DIG)-labeled VtgAa/b or Western blotting with α-WpLrp13 or α-WpLr8− at 1:10,000, as we previously described (17, 35).
Semi-quantitative RT-PCR
Primers were designed for striped bass lrp13 (SB618For1 and SB618Rev1) and ribosomal protein L9 (rpl9) [contig 10830 (25)] (RPl9For and RPl9Rev). The quality of total RNA extracted from brain, heart, liver, ovary, foregut, muscle, and adipose tissue was evaluated by NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE) OD260/OD280 and OD260/OD230 and agarose electrophoresis. Extracts were treated with DNA-Free (Applied Biosystems, Grand Island, NY) and cDNA was synthesized using SuperScript First-Strand synthesis system (Invitrogen). Amplifications were performed using PCR SuperMix (Invitrogen) and no template and no reverse transcription controls were incorporated into the assay.
Real-time quantitative RT-PCR
Primers for lr8− variant of vldlr (SBLR8For and SBLR8Rev) and lrp13 (SBLRX+1For and SBLRX+1Rev) were designed from striped bass contigs 04238 and 00618 (25). Priming sites of low complementarity between these striped bass contig sequences were chosen to ensure primer specificity. Total ovary and liver RNA was extracted and evaluated as described above and cDNA was synthesized with a high capacity cDNA synthesis kit (Applied Biosystems). Absolute real-time quantitative PCR assays were performed as previously described (36, 37) using Brilliant II SYBR Green QPCR Master Mix (Agilent Technologies, Santa Clara, CA). Samples were measured in triplicate using a 7300 real-time PCR system (Applied Biosystems) and gene expression was reported as copy number calculated from serially diluted plasmid DNA standard curves. Picha and colleagues (38, 39) show that normalization of target RNA to total RNA shows similar results to that for normalization to 18S RNA in hybrid striped bass, and we also show this trend when normalization of target RNA to total RNA is compared with normalization to ribosomal protein L9 in striped bass (37). Therefore, we used total RNA for normalization of lr8− and lrp13 gene expression. Melting curve analysis and agarose gel electrophoresis were performed to verify primer specificity and no template and no reverse transcription controls were employed in the assay.
In situ hybridization
DIG-labeled antisense and sense RNA probes were prepared by in vitro transcription of striped bass lrp13 using primers SB618ishF and SB618ishR (Roche, Indianapolis, IN). In situ hybridization of DIG-labeled probes was performed for 40 h at 65°C according to our previous report (40). Sections were photographed using a DXM1200F camera coupled to an 80iTUW-31-1 light microscope (Nikon, Tokyo, Japan).
Quantitative tandem mass spectrometry
Striped bass ovary extracts were subjected to reversed phase HPLC and tandem mass spectrometry (nanoLC-MS/MS) using an Eksigent (Dublin, CA) nanoLC-1D+ system with autosampler coupled to a hybrid Thermo Fisher LTQ Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA), as we previously reported (24). Spectral counts of the protein encoded by striped bass contig 00618 (25) (Lrp13) were normalized using ProteoIQ (NuSep, Bogart, GA) and these values were transformed: Log10(y + 1), y = normalized spectral count.
Immunohistochemistry
The α-CtLrp13 was used in immunohistochemistry of striped bass ovary at 1:500 according to our previous report (35). Sections were enclosed using ProLong Gold with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Fluorescence microscopy was performed using a DMI6000 B inverted microscope equipped with an N PLAN 10.0 × 0.25 DRY objective and filter for TexasRed coupled to a DFC 360FX camera (Leica, Tokyo, Japan) and a TCS-SP5 spectral confocal microscope fitted to a DMI6000 B inverted microscope equipped with HCX PL APO CS 63 × 1.4 oil immersion objective (Leica).
Data analyses
Comparisons were performed by one-way ANOVA and post hoc Tukey-Kramer honestly significant difference (HSD) with α = 0.05 (SAS Institute, Cary, NC).
RESULTS
Molecular cloning
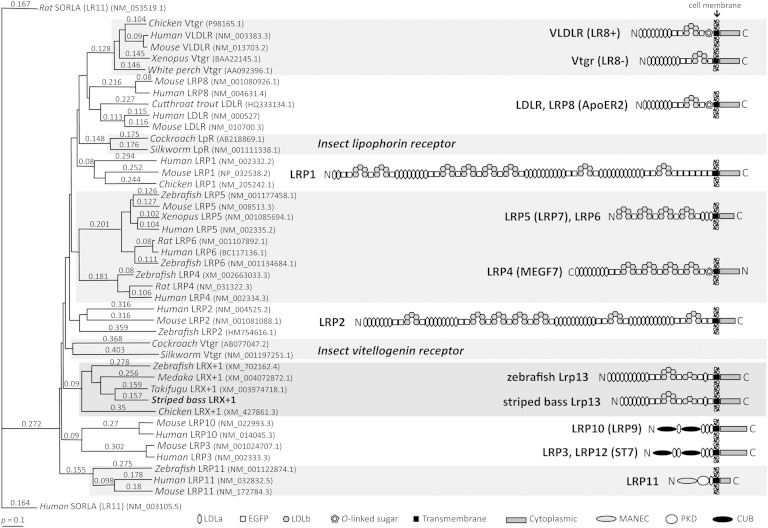
White perch Lrp13 (GenBank: KF387534) is orthologous to the protein encoded by striped bass contig 00618. The polypeptide sequences were 96% identical and the domain structures were conserved (supplementary Fig. I). The Lrp13 is a lipoprotein receptor, however it has a unique domain structure and is not an isoform of any previously described superfamily member (Fig. 1). Several serine and threonine phosphorylation sites and a proline-rich class IV WW domain interaction motif (AETTPSKQPSPV) are predicted within the Lrp13 cytoplasmic domain. The predicted molecular mass of Lrp13 is ∼130 kDa.
Fig. 1.
Left: ClustalW dendrogram showing relationships between low-density lipoprotein receptor family polypeptide sequences. GenBank accession numbers are provided. Numbers above each branch are pairwise- (p-)distances. Right: Models representing linear domain structures of LDLR family members as defined at the bottom.
Vertebrate lrp13 orthologs
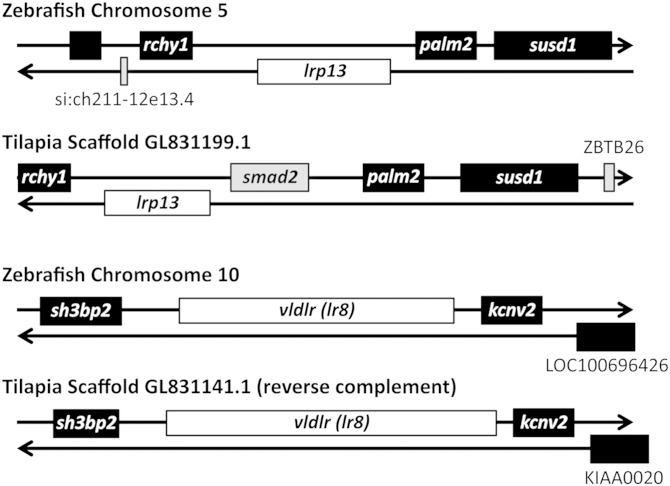
Putative lrp13 orthologs were identified in several vertebrates (GenBank HomoloGene: 132117). The lrp13 is predicted in genome sequences of zebrafish, medaka (Oryzias latipes), Takifugu rubripes, and chicken (Gallus gallus) and these polypeptide sequences are represented in Fig. 1. Additional lrp13 orthologs were identified in genome sequences of dog (Canis lupus familiaris) and mouse (Mus musculus) (GenBank: XP_532820 and XP_146277.8). Expressed sequence tags were identified in stickleback (Gasterosteus acculeatus), yellow perch (Perca flavescens), and Antarctic (Dissostichus mawsoni) fish (GenBank: CD505810, CD505014, CD500408, GO660341, FE225905, and FE226320). Syntenic arrangements of zebrafish and Nile tilapia lrp13 and vldr (lr8) loci are shown in Fig. 2.
Fig. 2.
Syntenic arrangements of zebrafish and Nile tilapia lrp13 and vldr (lr8) loci aligned to illustrate orthologies between the two species.
Protein-affinity chromatography
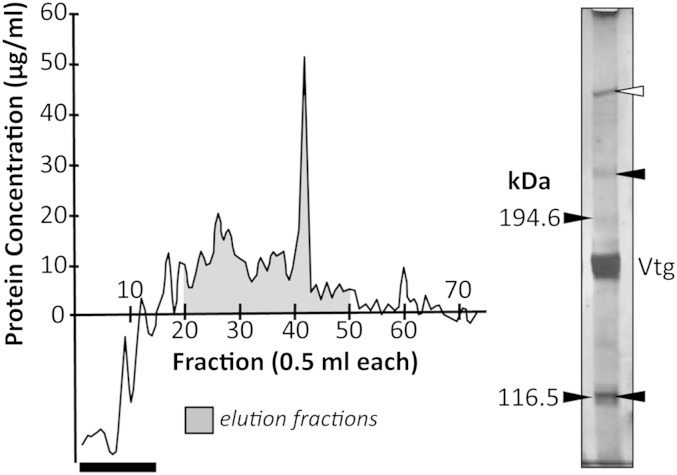
Thirty-one fractions (average 10.7 μg protein/ml) were collected as affinity purified Vtgrs (Fig. 3) and four LC-ESI-MS/MS peptide spectra were identified that matched to white perch Lr8−, accounting for ∼8.9% polypeptide sequence coverage (supplementary Table II). Positions of these peptides are shown in supplementary Fig. I. Additionally, 18 peptides were identified that match to white perch Lrp13, accounting for ∼18.5% polypeptide sequence coverage. These spectra were not detected in purified VtgAa/b nor were they detected among solubilized ovary membrane proteins prior to protein-affinity purification. The two most abundant proteins among both affinity purified Vtgrs and purified VtgAa/b were white perch VtgAa and VtgAb, which is not unexpected. Figure 3 shows a silver stained nonreducing gel of affinity purified Vtgrs with positions of putative VtgAar and VtgAbr indicated at ∼223 and 117 kDa, respectively. The molecular mass of VtgAar was estimated because it migrates to a position above the largest marker protein. A putative protein aggregate was observed at the 4% stacking gel margin. The VtgAa/b intensely appears at 165–177 kDa in the gel, near the molecular mass of VtgAa and the VtgAb monomer (16, 30).
Fig. 3.
Left: Chromatograph of protein-affinity purified white perch Vtgrs. The dark bar indicates column pass through and transition to elution buffer. Right: Silver stained nonreducing 5% acrylamide gel of affinity purified Vtgrs. Numbers to the left indicate the sizes of molecular mass markers (kDa). Positions of VtgAa/b (Vtg) and ovary Vtgrs (arrows) are indicated to the right. The white arrow indicates a putative protein aggregate.
Ligand and Western blotting
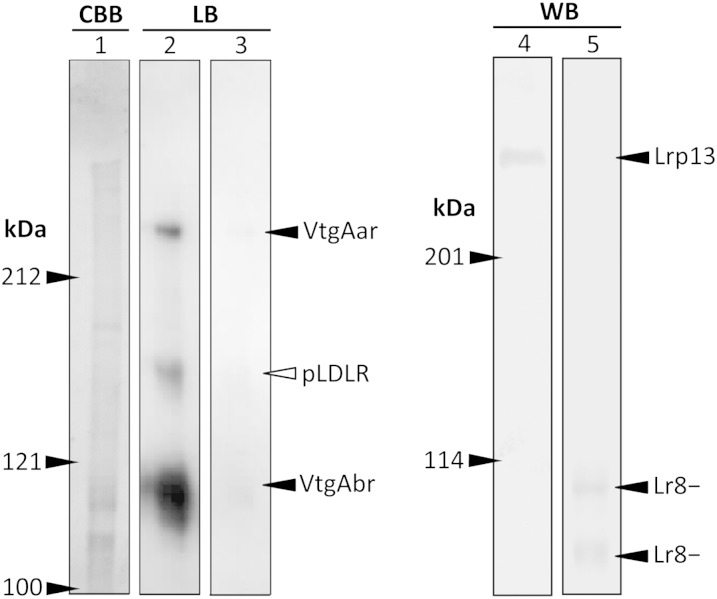
Ligand blotting revealed a ∼246 kDa VtgAar, a weakly binding 160 kDa pLDLR, and an intensely binding 104–119 kDa VtgAbr in white perch ovary (Fig. 4). Binding of DIG-VtgAa/b to these proteins was displaced by a 200-fold excess molar ratio of VtgAa/b. Western blotting revealed a ∼266 kDa Lrp13 and a pair of Lr8− proteins at 109 and 117 kDa. The molecular masses of VtgAar and Lrp13 were estimated as described above.
Fig. 4.
Left: Nonreducing 7.5% acrylamide gel stained with Coomassie brilliant blue (CBB) and ligand blot (LB) of white perch ovary membrane proteins. The LB in lane 2 was prepared using 0.25 μg/ml of DIG-labeled VtgAa/b and the LB in lane 3 was performed in the presence of a 200-fold excess molar ratio of unlabeled VtgAa/b. Right: Nonreducing 5% acrylamide gel Western blot (WB) of white perch ovary membrane proteins. The WB in lane 4 was prepared with α-WpLrp13 and the WB in lane 5 was performed with α-WpLr8−. Numbers to the left of gels or blots indicate the sizes of molecular mass markers (kDa). Positions of VtgAar, VtgAbr, pLDLR, Lr8−, and Lrp13 are indicated.
Semi-quantitative RT-PCR
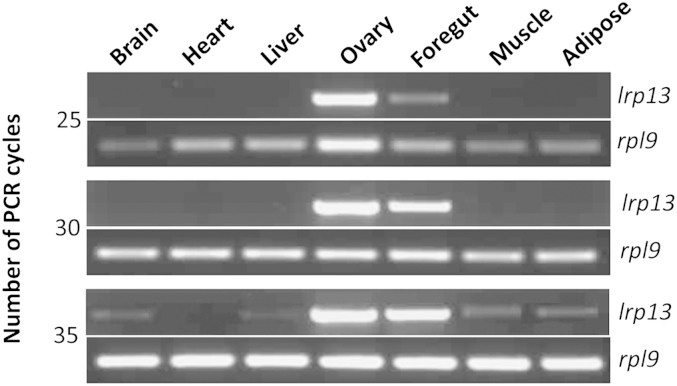
Expression of lrp13 is predominant in striped bass ovary, however, foregut is another site of expression, albeit lesser than that of ovary (Fig. 5). Faint lrp13 products were also detected by 35 cycles in striped bass brain, liver, muscle, and adipose tissue.
Fig. 5.
Semi-quantitative RT-PCR of lrp13 and rpl9 in striped bass tissues. The PCRs were terminated after 25, 30, and 35 cycles (indicated at the left).
Real-time quantitative RT-PCR
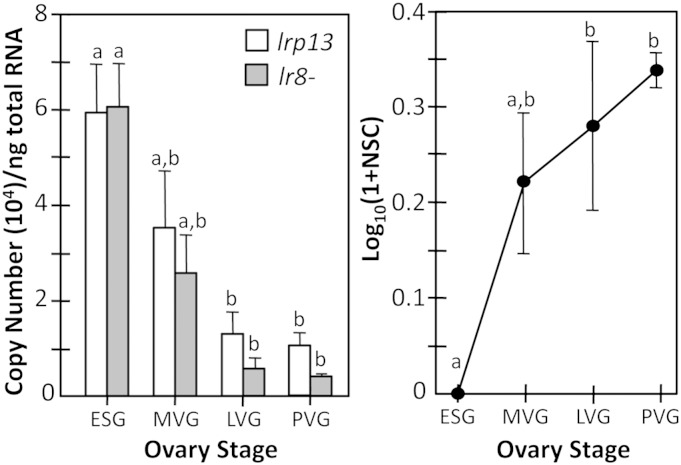
Peak lrp13 expression occurs in striped bass ovary during ESG and levels decline by LVG and PVG (P < 0.0059) (Fig. 6). A similar pattern is observed for lr8− (P < 0.0014). Transcripts encoding both receptors were detected in liver; however, these were considered to be trace levels, as ovary expression of each was on average 10,000-fold greater.
Fig. 6.
Real-time quantitative RT-PCR of lrp13 and lr8− (left) and quantitative mass spectrometry of Lrp13 (right) in ESG, MVG, LVG, and PVG striped bass ovary. Vertical bars and data points represent mean values and brackets indicate standard errors of the mean. Mean values bearing the same letter superscript are not significantly different (P < 0.05 ANOVA and Tukey-Kramer HSD).
Quantitative tandem mass spectrometry
NanoLC-MS/MS data for all proteins are reported in Reading et al. (24). The Lrp13 in the ovary during LVG and PVG was elevated compared with ESG (P < 0.0402) (Fig. 6).
In situ hybridization
Expression of lrp13 is predominant in striped bass ESG oocytes and is not detected in vitellogenic oocytes (Fig. 7). Transcripts stain throughout the ooplasm of ESG oocytes, but are not present in the follicular epithelium. No signal was observed when the sense in situ probe was used and this is shown in supplementary Fig. II.
Fig. 7.
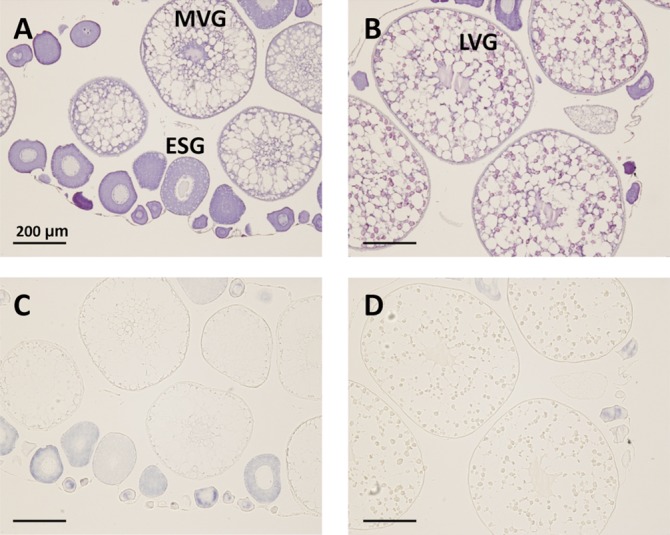
In situ hybridization of lrp13 in striped bass ovary. Hematoxylin-eosin stain (A, B) and adjacent sections (C, D) probed with antisense lrp13. ESG, MVG, and LVG oocytes are indicated in (A) and (B). Blue signal in (C) and (D) represents lrp13 localization.
Immunohistochemistry
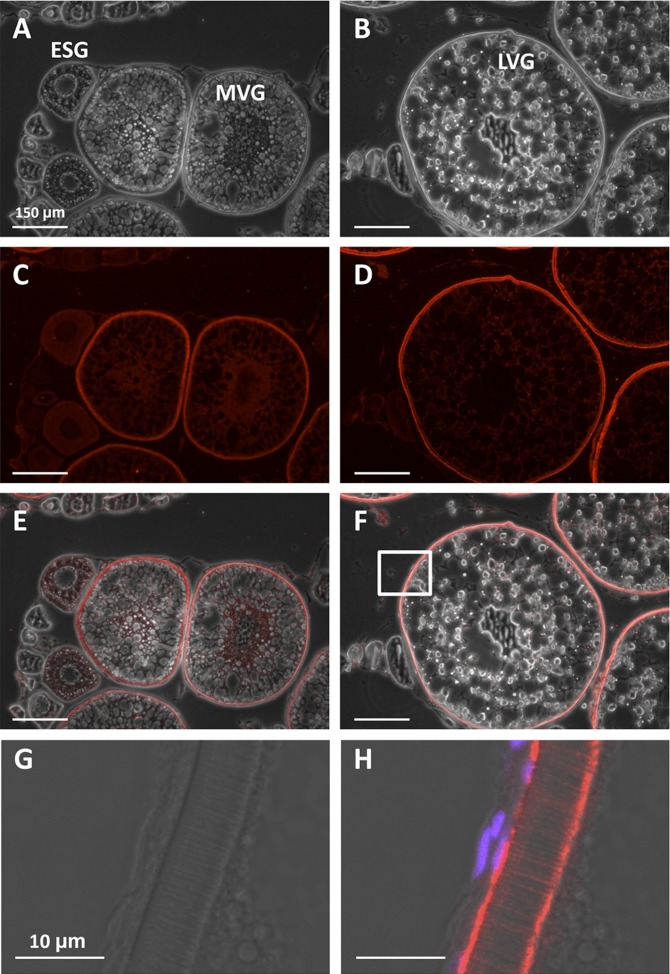
The Lrp13 was distributed weakly throughout the ooplasm of striped bass ESG oocytes and translocated to the periphery of MVG oocytes (Fig. 8). In LVG oocytes, Lrp13 was observed at the oolemma, throughout the zona radiata, and within the perivitelline space between the zona radiata externa and granulosa.
Fig. 8.
Immunohistochemistry of Lrp13 in striped bass ovary. Phase contrast (A, B), fluorescence (C, D), and image overlays (E, F) of phase contrast and fluorescence. The boxed area in (F) is shown at high magnification in phase contrast (G) and fluorescence (H). ESG, MVG, and LVG oocytes are indicated in (A) and (B). Red signal in (C–F) and (H) represents Lrp13 localization and magenta signal in (H) represents DAPI-stained nuclei.
DISCUSSION
We discovered a lipoprotein receptor that has not previously been reported in any vertebrate. Here, we propose inclusion of this gene in the lipoprotein receptor gene family with the formal name lrp13 according to the Zebrafish Information Network (ZFIN) nomenclature (41). Both Lrp13 and Lr8− bind Vtgs, as evidenced by protein-affinity chromatography. Neither receptor was detected in solubilized ovary membrane protein prior to purification nor in the VtgAa/b (ligand) preparation, thus they were selectively enriched by protein-affinity chromatography. Additionally, both receptors specifically eluted from immobilized VtgAa/b using buffer containing suramin, which is known to disrupt interactions between lipoprotein ligands and their receptors (42), including Vtgs and Vtgr (43). Therefore, Lrp13 binds VtgAa/b and is dissociated from it, similar to Lr8−.
The lrp13 is predominantly expressed in ovary (Fig. 5) and peak expression occurs during ESG, similar to lr8− (Fig. 6) (18). Weak gene expression was observed in striped bass somatic tissues, including foregut, liver, muscle, adipose tissue, and brain, as was previously reported for lr8− in white perch (18). Although lr8− is predominantly expressed in ovary, lr8− and lr8+ gene expression has been detected by RT-PCR in almost all somatic tissues of several other egg-laying vertebrates (13, 19, 20, 44, 45). Less sensitive methods of detection (i.e., Northern blotting) in these same studies failed to detect lr8− gene transcripts in somatic tissues, indicating that such ectopic expression is minor. Therefore, lr8 and lrp13 share similar expression patterns; however, spliced variants of lrp13 remain to be identified. In the case of the spliced variant forms of Lr8, there is no functional difference between the Lr8− and Lr8+ in chickens (19); thus, the physiological significance of the diverse tissue expression pattern of Vtgrs remains unclear and requires further investigation.
In situ hybridization provided an orthogonal confirmation of lrp13 expression in ESG oocytes (Fig. 7) and similar localization of lr8− expression has been shown in rainbow trout (Oncorhynchus mykiss) ovary (46). The inverse relationship between lrp13 gene transcript and Lrp13 protein levels during oocyte growth (Fig. 6) suggests that transcripts are stored and translated during vitellogenesis, as is characteristic of Lr8− in fishes (14) and chickens (13, 47). Therefore, lrp13 transcript levels peak prior to Lrp13 protein levels. Lrp13 translation occurs from ESG through MVG, indicated by localization of the protein throughout the ooplasm (Fig. 8). Slight centralized Lrp13 localization during MVG suggests active translation at the rough endoplasmic reticulum, which is a characteristic site of synthesis and posttranslational modification of transmembrane proteins. The Lrp13 translocates from the ooplasm during ESG to the oocyte periphery during MVG and oolemma and zona radiata during LVG, which is suggestive of an active role in receptor-mediated uptake of Vtgs. Collectively, these Vtg-binding properties and expression data suggest that Lrp13 functions as a Vtgr in fishes.
The VLDLRs and Lrp13 are not closely related by phylogenetic analysis (Fig. 1), indicating that these receptors are not paralogs. Zebrafish and tilapia lrp13 and vldlr (lr8) loci are discrete (Fig. 2); therefore, these gene transcripts are not spliced variants of a single locus. Although the loci share synteny between species, the lrp13 and vldlr loci are unrelated to one another. This suggests that these vtgrs have been distinctly retained in the fish lineages Ostariophysi and Acanthomorpha, which represent at least 115–200 million years of evolutionary divergence. This finding agrees with our present understanding of Vtg evolution in that the respective paralogous ligands (VtgAa and VtgAb) have existed in separate lineages that predate the divergence of ray-finned fishes and tetrapods (48, 49). Orthologs of lrp13 were identified in several fish species, suggesting that this gene is generally present among teleosts. The characteristic single LDLa domain adjacent to the transmembrane domain is conserved in all Lrp13 orthologs; however, the number of N-terminal LDLa varies from 7 to 10 between fish species (Fig. 1). Only white perch and striped bass full-length lrp13 transcripts have been verified; therefore, this variation may be an artifact of genome assembly or intron/exon prediction. Further sequence data will be required to verify this observation. While the cytoplasmic domain of Lr8− contains NPxY and YxxØ motifs (supplementary Fig. I) that are known to facilitate clathrin-mediated endocytosis and sorting of internalized vesicles (50), the cytoplasmic domain of Lrp13 contains neither of these tyrosine-based motifs. The cytoplasmic domain of Lrp13 does contain several phosphorylation sites within and near a putative proline-rich class IV WW interaction motif, a feature that has been shown in other membrane receptor proteins to mediate cellular processes including endocytosis, trafficking, signaling, and control of the cytoskeleton (51–54). Further investigation will be required to fully understand whether these features influence endocytosis and trafficking of the liganded Lrp13 receptor holocomplex; however, it appears that this process might differ from that of Lr8−.
A high molecular mass Vtgr has been observed in chicken ovary ligand blots (55). Although the identity of this protein was never reported, three derivative peptides, DDSGALLR, LLAQGLGxPTALALDLPTS, and LTAVDYMVK, match to the chicken Lrp13 ortholog. The lrp13 has predominant ovary expression (GenBank UniGene: Gga.13385); therefore, it may function as a Vtgr in chickens as well as in fishes.
The VtgAar and VtgAbr shown in Fig. 4 are similar in size to affinity purified Vtgrs (Fig. 3). The pLDLR was not evident in Fig. 3, suggesting that it may not have been affinity purified; however, VtgAa/b may also have obscured it in the gel. The Lrp13 is detected by Western blotting at a position corresponding to that of VtgAar (Fig. 4) and over 100 kDa higher than the predicted molecular mass of the polypeptide. The molecular mass of chicken Lrp13 predicted from the polypeptide is 259 kDa; however, the estimated size of the receptor in ligand blots is similarly exaggerated (∼380 kDa) (55). Anomalous migration of LDLR in ligand blots is also reported (56). The reason(s) why some lipoprotein receptors migrate to a position indicative of a higher molecular mass is unclear, but it may relate to the nonreducing electrophoresis required of ligand blotting. Therefore, for reasons unknown, Lrp13 may migrate to an anomalous position in ligand blots. The 104–119 kDa VtgAbr (Fig. 4) resolved as a distinct pair of proteins by Reading, Hiramatsu, and Sullivan (17), both of which are identified as Lr8− by Western blotting in the present report. The reason(s) for the apparent difference in molecular mass of these two proteins remains to be verified; however, they may represent spliced variants of the O-linked sugar domain in Lr8− and Lr8+. Further evidence is required to confirm this contention, as Lr8+ gene transcripts are not detected in white perch ovary (18); however, one of these proteins is generally considered to be Lr8− in chickens and rainbow trout (13, 14, 19). The protein aggregate (Fig. 3) was not observed in other gels or ligand blots (17) (Fig. 4) and, therefore, may represent VtgAa/b from the affinity column, as Vtgs are labile and fragments typically aggregate. We have previously attempted to definitively identify these Vtgr proteins by excising them from the gels and subjecting them to MALDI mass spectrometry or trypsin-digest nanospray liquid chromatography and tandem mass spectrometry, but these analyses did not return any peptide spectra, probably because the relevant membrane proteins were too low in abundance (17). However, our collective Western blotting results show that Lrp13 is the VtgAar and Lr8− is the VtgAbr.
Dog and mouse Lrp13 orthologs were identified. The function of this lipoprotein receptor is unexplored in mammals; however, the dog and mouse loci are expressed as gene transcripts (GenBank UniGene: Cfa.9209 and Mm.334491). The VLDLR acts as a Vtgr in oviparous vertebrates; however, it binds VLDL in placental mammals. It would be interesting to verify whether lrp13 has similarly retained protein-coding ability or has become a pseudogene, as have the vtgs in all mammals except for monotremes (49). Therefore, Lrp13 is a prime candidate for future research in mammals as well as in egg-laying vertebrates and, along with Vldlr, may provide an opportune model system for studying neofunctionalization of lipoprotein receptors and their ligands. The presence of Lrp13 orthologs in representative fishes, birds, and mammals suggests that it is widely distributed among vertebrates where it may generally play a role in lipoprotein metabolism. The Lrp13 appears to function as a Vtgr in fishes and is tentatively identified as the previously reported VtgAar of white perch. It may also be an important mediator of yolk deposition in other oviparous vertebrates including chickens.
For the largest group of fishes (Acanthomorpha), a system of dual ovarian Vtgrs (Lr8− and Lrp13) could explain, in part, the disparate accumulation and hydrolysis of egg yolk derived from the functionally distinct VtgAa and VtgAb, which are critical processes governing acquisition of proper egg buoyancy and provision of different types of nutrients to embryos and larvae (37, 57, 58). During maturation, yolk proteins derived from VtgAa are disproportionally hydrolyzed to free amino acids, which osmotically drive oocyte hydration and are nutrients selectively utilized by early embryos. As noted, the two forms of Vtg appeared sometime prior to the divergence of ray-finned fishes and tetrapods over 450 million years ago, with the selective VtgAa-derived yolk proteolysis and generation of an organic osmolyte pool of free amino acids being thought to have arisen approximately 55 million years ago as an adaptive response to spawning in seawater (48, 49). It has been suggested that the VtgAar and VtgAbr may deliver their ligands to separate compartments where their derived yolk proteins are subjected to the different degrees of hydrolysis during oocyte maturation (33). Coevolution and neofunctionalization of the VtgAar (Lrp13) and its ligand would be consistent with these scenarios, the lipoprotein receptor phylogeny shown in Fig. 1, and the observed differences in the intracellular domains of the two Vtgrs.
Finally, advancing our understanding of vertebrate lipoprotein receptor function is a crucial component of research on cardiovascular disease and other disorders related to lipidemias, and the Lrp13 is a new candidate that is yet to be explored.
Supplementary Material
Acknowledgments
The authors thank A. S. McGinty, M. S. Hopper, B. D. Ring, and J. Davis for husbandry of the research animals.
Footnotes
Abbreviations:
- DIG
- digoxigenin
- ESG
- early secondary growth
- LDLa
- class a ligand binding repeat
- LDLb
- class b YWxD repeat
- LDLR
- LDL receptor
- LRP
- LDL receptor-related protein
- Lr8−
- vitellogenin receptor that lacks an O-linked sugar domain
- Lr8+
- vitellogenin receptor that contains an O-linked sugar domain
- LVG
- late-vitellogenic growth
- MVG
- mid-vitellogenic growth
- nanoLC-MS/MS
- reversed phase HPLC and tandem mass spectrometry
- NCSU
- North Carolina State University
- pLDLR
- putative LDL receptor
- PVG
- post-vitellogenic growth
- RACE
- rapid amplification of cDNA ends
- VLDLR
- VLDL receptor
- Vtg
- vitellogenin
- VtgAar
- vitellogenin Aa receptor
- VtgAbr
- vitellogenin Ab receptor
- Vtgr
- vitellogenin receptor
This research was supported by grant awards to C.V.S and B.J.R. from North Carolina Sea Grant (R/AF-49, R/MG-0609, and R/12-SSS-3) and from the North Carolina Agricultural Foundation, Inc.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures and two tables.
REFERENCES
- 1.Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., Russel D. W. 1984. The human LDL receptor: a cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 39: 27–38. [DOI] [PubMed] [Google Scholar]
- 2.Herz J., Hamann U., Rogne S., Myklebost O., Gausepohl H., Stanley K. K. 1988. Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 7: 4119–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saito A., Pietromonaco S., Loo A. K., Farquhar M. G. 1994. Complete cloning and sequencing of rat gp330/“megalin,” a distinctive member of the low density lipoprotein receptor gene family. Proc. Natl. Acad. Sci. USA. 91: 9725–9729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishii H., Kim D. H., Fujita T., Endo Y., Saeki S., Yamamoto T. T. 1998. cDNA cloning of a new low-density lipoprotein receptor-related protein and mapping of its gene (LRP3) to chromosome bands 19q12-q13.2. Genomics. 51: 132–135. [DOI] [PubMed] [Google Scholar]
- 5.Nakayama M., Nakajima D., Nagase T., Nomura N., Seki N., Ohara O. 1998. Identification of high-molecular-weight proteins with multiple EGF-like motifs by motif-trap screening. Genomics. 51: 27–34. [DOI] [PubMed] [Google Scholar]
- 6.Hey P. J., Twells R. C., Phillips M. S., Nakagawa Y., Brown S. D., Kawaguchi Y., Cox R., Xie G., Dugan V., Hammond H., et al. 1998. Cloning of a novel member of the low-density lipoprotein receptor family. Gene. 216: 103–111. [DOI] [PubMed] [Google Scholar]
- 7.Brown S. D., Twells R. C., Hey P. J., Cox R. D., Levy E. R., Soderman A. R., Metzker M. L., Thomas C. T., Todd J. A., Hess J. F. 1998. Isolation and characterization of LRP6, a novel member of the low density lipoprotein receptor gene family. Biochem. Biophys. Res. Commun. 248: 879–888. [DOI] [PubMed] [Google Scholar]
- 8.Kim D. H., Iijima H., Goto K., Sakai J., Ishii H., Kim H. J., Suzuki H., Kondo H., Saeki S., Yamamoto T. 1996. Human apolipoprotein E receptor 2. A novel lipoprotein receptor of the low density lipoprotein receptor family predominantly expressed in brain. J. Biol. Chem. 271: 8373–8380. [DOI] [PubMed] [Google Scholar]
- 9.Sugiyama T., Kumagai H., Morikawa Y., Wada Y., Sugiyama A., Yasuda K., Yokoi N., Tamura S., Kojima T., Nosaka T., et al. 2000. A novel low-density lipoprotein receptor-related protein mediating cellular uptake of apolipoprotein E-enriched beta-VLDL in vitro. Biochemistry. 39: 15817–15825. [DOI] [PubMed] [Google Scholar]
- 10.Jacobsen L., Madsen P., Moestrup S. K., Lund A. H., Tommerup N., Nykjaer A., Sottrup-Jensen L., Gliemannm J., Petersen C. M. 1996. Molecular characterization of a novel human hybrid-type receptor that binds the alpha2-macroglobulin receptor-associated protein. J. Biol. Chem. 271: 31379–31383. [DOI] [PubMed] [Google Scholar]
- 11.Qing J., Wei D., Maher V. M., McCormick J. J. 1999. Cloning and characterization of a novel gene encoding a putative transmembrane protein with altered expression in some human transformed and tumor-derived cell lines. Oncogene. 18: 335–342. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi S., Kawarabayasi Y., Nakai T., Sakai J., Yamamoto T. 1992. Rabbit very low density lipoprotein receptor: a low density lipoprotein receptor-like protein with distinct ligand specificity. Proc. Natl. Acad. Sci. USA. 89: 9252–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujo H., Hermann M., Kaderli M. O., Jacobsen L., Sugawara S., Nimpf J., Yamamoto T., Schneider W. J. 1994. Chicken oocyte growth is mediated by an eight ligand binding repeat member of the LDL receptor family. EMBO J. 13: 5165–5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davail B., Pakdel F., Bujo H., Perazzolo L. M., Waclawek M., Schneider W. J., Le Menn F. 1998. Evolution of oogenesis: the receptor for vitellogenin from the rainbow trout. J. Lipid Res. 39: 1929–1937. [PubMed] [Google Scholar]
- 15.Finn R. N., Kristoffersen B. A. 2007. Vertebrate vitellogenin gene duplication in relation to the “3R hypothesis”: correlation to the pelagic egg and the oceanic radiation of teleosts. PLoS ONE. 2: e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reading B. J., Hiramatsu N., Sawaguchi S., Matsubara T., Hara A., Lively M. O., Sullivan C. V. 2009. Conserved and variant molecular and functional features of multiple egg yolk precursor proteins (vitellogenins) in white perch (Morone americana) and other teleosts. Mar. Biotechnol. (NY). 11: 169–187. [DOI] [PubMed] [Google Scholar]
- 17.Reading B. J., Hiramatsu N., Sullivan C. V. 2011. Disparate binding of three types of vitellogenin to multiple forms of vitellogenin receptor in white perch. Biol. Reprod. 84: 392–399. [DOI] [PubMed] [Google Scholar]
- 18.Hiramatsu N., Chapman R. W., Lindzey J. K., Haynes M. R., Sullivan C. V. 2004. Molecular characterization and expression of vitellogenin receptor from white perch (Morone americana). Biol. Reprod. 70: 1720–1730. [DOI] [PubMed] [Google Scholar]
- 19.Bujo H., Lindstedt K. A., Hermann M., Dalmau L. M., Nimpf J., Schneider W. J. 1995. Chicken oocytes and somatic cells express different splice variants of a multifunctional receptor. J. Biol. Chem. 270: 23546–23551. [DOI] [PubMed] [Google Scholar]
- 20.Prat F., Coward K., Sumpter J. P., Tyler C. R. 1998. Molecular characterization and expression of two ovarian lipoprotein receptors in the rainbow trout, Oncorhynchus mykiss. Biol. Reprod. 58: 1146–1153. [DOI] [PubMed] [Google Scholar]
- 21.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. 2011. Guide for the Care and Use of Laboratory Animals. 8th edition. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 22.Heppell S. A., Jackson L. F., Weber G. M., Sullivan C. V. 1999. Enzyme-linked immunosorbent assay (ELISA) of vitellogenin in temperate basses (genus Morone): plasma and in vitro analyses. Trans. Am. Fish. Soc. 128: 532–541. [Google Scholar]
- 23.Jackson L. F., Sullivan C. V. 1995. Reproduction of white perch (Morone americana): the annual gametogenic cycle. Trans. Am. Fish. Soc. 124: 563–577. [Google Scholar]
- 24.Reading B. J., Williams V. N., Chapman R. W., Williams T. I., Sullivan C. V. 2013. Dynamics of the striped bass (Morone saxatilis) ovary proteome reveal a complex network of the translasome. J. Proteome Res. 12: 1691–1699. [DOI] [PubMed] [Google Scholar]
- 25.Reading B. J., Chapman R. W., Schaff J. E., Scholl E. H., Opperman C. H., Sullivan C. V. 2012. An ovary transcriptome for all maturational stages of the striped bass (Morone saxatilis), a highly advanced perciform fish. BMC Res. Notes. 5: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I., Doerks T., Bork P. 2012. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40: D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gould C. M., Diella F., Via A., Puntervoll P., Gemünd C., Chabanis-Davidson S., Michael S., Sayadi A., Bryne J. C., Chica C., et al. 2010. ELM: the status of the eukaryotic linear motif resource. Nucleic Acids Res. 38: D167–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson J. D., Higgins D. G., Gibson T. J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flicek P., Amode M. R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S., Fitzgerald S., et al. 2012. Ensembl 2012. Nucleic Acids Res. 40: D84–D90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hiramatsu N., Matsubara T., Hara A., Donato D. M., Hiramatsu K., Denslow N. D., Sullivan C. V. 2002. Identification, purification and classification of three forms of vitellogenin from white perch (Morone americana). Fish Physiol. Biochem. 26: 355–370. [Google Scholar]
- 31.Roehrkasten A., Ferenz H., Buschmann-Gebhardt B., Hafer J. 1989. Isolation of the vitellogenin-binding protein from locust ovaries. Arch. Insect Biochem. Physiol. 10: 141–149. [Google Scholar]
- 32.Hiramatsu N., Hara A., Hiramatsu K., Fukada H., Weber G. M., Denslow N. D., Sullivan C. V. 2002. Vitellogenin-derived yolk proteins of white perch, Morone americana: purification, characterization and vitellogenin-receptor binding. Biol. Reprod. 67: 655–667. [DOI] [PubMed] [Google Scholar]
- 33.Hiramatsu N., Luo W., Reading B. J., Sullivan C. V., Mizuta H., Ryu Y-W., Nishimiya O., Todo T., Hara A. 2013. Multiple ovarian lipoprotein receptors in teleosts. Fish Physiol. Biochem. 39: 29–32. [DOI] [PubMed] [Google Scholar]
- 34.Hong L., Fujita T., Wada T., Amano H., Hiramatsu N., Zhang X., Todo T., Hara A. 2009. Choriogenin and vitellogenin in red lip mullet (Chelon haematocheilus): purification, characterization, and evaluation as potential biomarkers for detecting estrogenic activity. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 149: 9–17. [DOI] [PubMed] [Google Scholar]
- 35.Mizuta H., Luo W., Ito Y., Mushiroba Y., Todo T., Hara A., Reading B. J., Sullivan C. V., Hiramatsu N. 2013. Ovarian expression and localization of vitellogenin receptor with eight ligand binding repeats in the cutthroat trout (Oncorhynchus clarki). Comp. Biochem. Physiol. B. 166: 81–90. [DOI] [PubMed] [Google Scholar]
- 36.Tipsmark C. K., Baltzegar D. A., Ozden O., Grubb B. J., Borski R. J. 2008. Salinity regulates claudin mRNA and protein expression in the teleost gill. Am. J. Physiol. Regul. Integr. Comp. Physiol. 294: R1004–R1014. [DOI] [PubMed] [Google Scholar]
- 37.Williams V. N., Reading B. J., Amano H., Hiramatsu N., Schilling J., Salger S. A., Islam Williams T., Gross K., Sullivan C. V. 2014. Proportional accumulation of yolk proteins derived from multiple vitellogenins is precisely regulated during vitellogenesis in striped bass (Morone saxatilis). J. Exp. Zool. A Ecol. Genet. Physiol. 321: 301–315. [DOI] [PubMed] [Google Scholar]
- 38.Picha M. E., Silverstein J. T., Borski R. J. 2006. Discordant regulation of hepatic IGF-I mRNA and circulating IGF-I during compensatory growth in a teleost, the hybrid striped bass (Morone chrysops x Morone saxatilis). Gen. Comp. Endocrinol. 147: 196–205. [DOI] [PubMed] [Google Scholar]
- 39.Picha M. E., Turano M. J., Tipsmark C. K., Borski R. J. 2008. Regulation of endocrine and paracrine sources of Igfs and Gh receptor during compensatory growth in hybrid striped bass (Morone chrysops X Morone saxatilis). J. Endocrinol. 199: 81–94. [DOI] [PubMed] [Google Scholar]
- 40.Luo W., Ito Y., Mizuta H., Massaki K., Hiramatsu N., Todo T., Reading B. J., Sullivan C. V., Hara A. 2013. Molecular cloning and partial characterization of an ovarian receptor with seven ligand binding repeats, an orthologue of low-density lipoprotein receptor, in the cutthroat trout (Oncorhynchus clarki). Comp. Biochem. Physiol. A Mol. Integr. Physiol. 166: 263–271. [DOI] [PubMed] [Google Scholar]
- 41.Bradford Y., Conlin T., Dunn N., Fashena D., Frazer K., Howe D. G., Knight J., Mani P., Martin R., Moxon S. A. T., et al. 2011. ZFIN: enhancements and updates to the zebrafish model organism database. Nucleic Acids Res. 39: D822–D829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown S. A., Via D. P., Gotto A. M., Jr, Bradley W. A., Gianturco S. H. 1986. Apolipoprotein E-mediated binding of hypertriglyceridemic very low density lipoproteins to isolated low density lipoprotein receptors detected by ligand blotting. Biochem. Biophys. Res. Commun. 139: 333–340. [DOI] [PubMed] [Google Scholar]
- 43.Tao Y., Berlinsky D. L., Sullivan C. V. 1996. Characterization of a vitellogenin receptor in white perch (Morone americana). Biol. Reprod. 55: 646–656. [DOI] [PubMed] [Google Scholar]
- 44.Li A., Sadasivam M., Ding J. L. 2003. Receptor-ligand interaction between vitellogenin receptor (VtgR) and vitellogenin (Vtg), implications on low density lipoprotein receptor and apolipoprotein B/E. J. Biol. Chem. 278: 2799–2806. [DOI] [PubMed] [Google Scholar]
- 45.Okabayashi K., Shoji H., Nakamura T., Hashimoto O., Asashima M., Sugino H. 1996. cDNA cloning and expression of the Xenopus laevis vitellogenin receptor. Biochem. Biophys. Res. Commun. 224: 406–413. [DOI] [PubMed] [Google Scholar]
- 46.Perazzolo L. M., Coward K., Davail B., Normand E., Tyler C. R., Pakdel F., Schneider W. J., Le Menn F. 1999. Expression and localization of messenger ribonucleic acid for the vitellogenin receptor in ovarian follicles throughout oogenesis in the rainbow trout, Oncorhynchus mykiss. Biol. Reprod. 60: 1057–1068. [DOI] [PubMed] [Google Scholar]
- 47.Shen X., Steyrer E., Retzek H., Sanders E. J., Schneider W. J. 1993. Chicken oocyte growth: receptor-mediated yolk deposition. Cell Tissue Res. 272: 459–471. [DOI] [PubMed] [Google Scholar]
- 48.Finn R. N., Kolarevic J., Kongshaug H., Nilsen F. 2009. Evolution and differential expression of a vertebrate vitellogenin gene cluster. BMC Evol. Biol. 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Babin P. J. 2008. Conservation of a vitellogenin gene cluster in oviparous vertebrates and identification of its traces in the platypus genome. Gene. 413: 76–82. [DOI] [PubMed] [Google Scholar]
- 50.Bonifacino J. S., Traub L. M. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72: 395–447. [DOI] [PubMed] [Google Scholar]
- 51.Ingham R. J., Colwill K., Howard C., Dettwiler S., Lim C. S., Yu J., Hersi K., Raaijmakers J., Gish G., Mbamalu G., et al. 2005. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 25: 7092–7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ilsley J. L., Sudol M., Winder S. J. 2002. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell. Signal. 14: 183–189. [DOI] [PubMed] [Google Scholar]
- 53.Sudol M., Sliwa K., Russo T. 2001. Functions of WW domains in the nucleus. FEBS Lett. 490: 190–195. [DOI] [PubMed] [Google Scholar]
- 54.Yaffe M. B., Elia A. E. 2001. Phosphoserine/threonine-binding domains. Curr. Opin. Cell Biol. 13: 131–138. [DOI] [PubMed] [Google Scholar]
- 55.Stifani S., Barber D. L., Aebersold R., Steyrer E., Shen X., Nimpf J., Schneider W. J. 1991. The laying hen expresses two different low density lipoprotein receptor-related proteins. J. Biol. Chem. 266: 19079–19087. [PubMed] [Google Scholar]
- 56.van Driel I. R., Davis C. G., Goldstein J. L., Brown M. S. 1987. Self-association of the low density lipoprotein receptor mediated by the cytoplasmic domain. J. Biol. Chem. 262: 16127–16134. [PubMed] [Google Scholar]
- 57.Matsubara T., Ohkubo N., Andoh T., Sullivan C. V., Hara A. 1999. Two forms of vitellogenin, yielding two distinct lipovitellins, play different roles during oocyte maturation and early development of barfin flounder, Verasper moseri, a marine teleost that spawns pelagic eggs. Dev. Biol. 213: 18–32. [DOI] [PubMed] [Google Scholar]
- 58.Williams V. N., Reading B. J., Hiramatsu N., Amano H., Glassbrook N., Hara A., Sullivan C. V. 2014. Multiple vitellogenins and product yolk proteins in striped bass, Morone saxatilis: molecular characterization and processing during oocyte growth and maturation. Fish Physiol. Biochem. 40: 395–415. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.