Abstract
The compound 20-HETE is involved in numerous physiological functions, including blood pressure and platelet aggregation. Glucuronidation of 20-HETE by UDP-glucuronosyltransferases (UGTs) is thought to be a primary pathway of 20-HETE elimination in humans. The present study identified major UGT enzymes responsible for 20-HETE glucuronidation and investigated their genetic influence on the glucuronidation reaction using human livers (n = 44). Twelve recombinant UGTs were screened to identify major contributors to 20-HETE glucuronidation. Based on these results, UGT2B7, UGT1A9, and UGT1A3 exhibited as major contributors to 20-HETE glucuronidation. The Km values of 20-HETE glucuronidation by UGT1A3, UGT1A9, and UGT2B7 were 78.4, 22.2, and 14.8 μM, respectively, while Vmax values were 1.33, 1.78, and 1.62 nmol/min/mg protein, respectively. Protein expression levels and genetic variants of UGT1A3, UGT1A9, and UGT2B7 were analyzed in human livers using Western blotting and genotyping, respectively. Glucuronidation of 20-HETE was significantly correlated with the protein levels of UGT2B7 (r2 = 0.33, P < 0.001) and UGT1A9 (r2 = 0.31, P < 0.001), but not UGT1A3 (r2 = 0.02, P > 0.05). A correlation between genotype and 20-HETE glucuronidation revealed that UGT2B7 802C>T, UGT1A9 −118T9>T10, and UGT1A9 1399T>C significantly altered 20-HETE glucuronide formation (P < 0.05–0.001). Increased levels of 20-HETE comprise a risk factor for cardiovascular diseases, and the present data may increase our understanding of 20-HETE metabolism and cardiovascular complications.
Keywords: 20-hydroxyeicosatetraenoic acid, uridine diphosphate-glucuronosyltransferase, genetic polymorphisms, arachidonic acid
The compound 20-HETE is an arachidonic acid (ARA) metabolite generated through the cytochrome P450 (CYP) pathway (1). CYP4A11, CYP4F2, CYP4F3, CYP2U1, and CYP1A1 are known to metabolize ARA to 20-HETE (2, 3). In addition, 20-HETE plays a role in regulating cardiovascular hemostasis (4) as a vasoconstrictor and platelet activator (5, 6), as well as a hypertensive mediator (7, 8). Excess levels of 20-HETE are associated with an increased risk of cardiovascular diseases, such as hypertension (9). In addition, frequencies of functional variants in CYP4A11 and CYP4F2 genes, which are involved in the synthesis of 20-HETE, are higher among hypertensive patients (10, 11). In a study conducted on mice to identify biomarkers for rofecoxib-induced cardiotoxicity, 20-HETE increased by 120-fold after rofecoxib treatment compared with the control groups (6). This increase in 20-HETE was correlated with increased platelet aggregation and reduced bleeding time, suggesting that rofecoxib increases the levels of 20-HETE. Rofecoxib has been withdrawn from the market due to the increased risk of cardiovascular events. Furthermore, 20-HETE has been suggested to be a novel target molecule for the treatment of hypertension and cardiovascular complications (12). The compound 20-HETE is known to be excreted outside of the body in a glucuronidated form (9, 13). The excretion of 20-HETE glucuronide in urine differs widely among individuals by approximately 10-fold (14), and such variations in 20-HETE levels may affect blood hemostasis or other physiological functions. It was reported that the urinary excretion of 20-HETE glucoronide was increased in hypertensive patients (9, 11). However, there have been few studies in relation to the production of 20-HETE or levels of 20-HETE in the tissues of hypertensive patients (15). On the other hand, 20-HETE was found to decrease the blood pressure through natriuretic effect in the kidney (16), suggesting that organ-specific mechanisms of 20-HETE may be important in the regulation of cardiovascular hemostasis. Urinary excretion of 20-HETE glucoronide provides little information on the regulation of 20-HETE production and its levels in blood, which suggests that the metabolism of 20-HETE in the liver coupled with filtration by the kidney could be the most important determinant of the urinary excretion and circulating levels (17).
Glucuronidation is mediated through a superfamily of enzymes called UDP-glucuronosyltransferases (UGTs). These enzymes are classified into two major subfamilies in humans, namely, UGT1A and UGT2B (18). UGT enzymes are highly variable in their activities and are affected by several factors, such as gender, age, alcoholic status, and xenobiotic inhibitors or inducers (19). In addition, genetic polymorphisms in genes encoding UGTs have a strong impact on UGT expression and activity with clinical significance in the pharmacogenetics field (20). The glucuronidation of ARA and its metabolites were investigated and the majority of HETEs were found to be good substrates for UGT2B7, UGT1A1, UGT1A3, UGT1A4, UGT1A8, and UGT1A9 (21). However, detailed studies on 20-HETE glucuronidation among UGT enzymes are limited. Furthermore, genetic information on the formation of glucuronated metabolites of 20-HETE is lacking. Therefore, the objectives of the present study were to identify the major UGT enzymes responsible for 20-HETE glucuronidation and to investigate the correlation between genetic variants of the determined UGT enzymes and 20-HETE glucuronide formation using human liver microsomes (HLMs).
EXPERIMENTAL PROCEDURES
Materials
Alamethicin, uridine diphosphate glucuronic acid (UDPGA), saccharic acid-1,4-lactone, and diclofenac were obtained from Sigma-Aldrich (St. Louis, MO). The compound 20-HETE was acquired from Cayman (Ann Arbor, MI). The internal standards (estrone-glucuronide and 7-hydroxycoumarin-glucuronide) were purchased from Toronto Research Chemicals (Toronto, ON, Canada). Taq polymerase was from TaKaRa (Otsu, Shiga, Japan), and the AB BigDye™ Terminator v3.1 cycle sequencing kit was purchased from Applied Biosystems (Foster City, CA). Primary antibodies for UGT1A3 and UGT1A9 were bought from Abcam (Cambridge, MA). The primary antibody for β-actin and the chemiluminescence kit were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Recombinant human UGT isoforms, pooled HLMs, skimmed milk powder, and primary antibody for UGT2B7 were purchased from BD Biosciences (San Jose, CA). All other chemicals and organic solvents were of the highest grade available from commercial sources.
HLMs
HLMs from 44 Korean donors (48.7 ± 10.7 years old; mean ± SD) were obtained from the Biobank at Inje Pharmacogenomics Research Center (Inje University College of Medicine, Busan, Korea). The research protocol for the use of HLMs was approved by the institutional review board of Busan Paik Hospital (Busan, Korea) (22).
Assays of 20-HETE glucuronidation using HLMs and recombinant UGT enzymes
For in vitro 20-HETE glucuronidation assays, pooled HLMs or recombinant UGT was reconstituted in a reaction mixture containing 20 μM 20-HETE and 1.5 mg/ml alamethicin in 0.5 M Tris-HCl buffer (pH 7.4), which was preincubated on ice for 15 min, as described previously (23). Because there is limited information on kinetics for 20-HETE glucuronidation by a set of UGT enzymes, 20 μM 20-HETE was determined empirically for initial screening for UGT enzymes responsible for 20-HETE glucuronidation. The reaction was initiated by the addition of 5 mM UDPGA and incubated at 37°C for 4 h. The recombinant UGT isoforms used were UGT1A1, -1A3, -1A4, -1A6, -1A7, -1A8, -1A9, -1A10, -2B4, -2B7, -2B15, and -2B17. The reaction was terminated by adding 100 μl of acetonitrile containing 1.2 μM estrone-glucuronide as an internal standard. Concentrations of 20-HETE for kinetic analysis were 5, 10, 20, 50, 70, 100, 200, and 500 μM. Reconstitution conditions for the kinetics of 20-HETE glucuronidation by UGT1A3, -1A9, and -2B7 were identical to the assay described above. The kinetic parameters were estimated by plotting the reaction rates to the multiple concentrations of substrate and fitting these data to the Michaelis-Menten equation using SigmaPlot 8.0 (SPSS Inc., Chicago, IL). An API 3000 LC-MS/MS system (Applied Biosystems) coupled with an API 3000 LC-MS/MS system (Agilent Technologies, Santa Clara, CA) was used for the detection of glucuronide 20-HETE and the internal standard, as described previously with minor modifications (24). Briefly, compounds were separated on a reverse-phase Atlantis dC18 column (2.1 mm inner diameter × 150 mm, 3 μm particle size; Waters, Eschborn, Germany). The nebulizing gas flow was adjusted to 7 psi, curtain gas to a flow of 12 psi, and collision energy to −30 eV. The autosampler was set at 4°C and the injection volume was 5 μl. The mass transitions m/z for monitoring 20-HETE glucuronide and estrone-glucuronide were 495.3→319.3 and 445→269, respectively. Peak areas for all compounds were integrated using Analyst software (version 1.2; Applied Biosystems). Because commercial standards for 20-HETE glucuronide are not available, quantification of the glucuronide form of 20-HETE was accomplished using standard curves of 20-HETE. The glucuronidation of 20-HETE was compared with diclofenac glucuronidation among 44 HLMs of Koreans, as described previously (23, 25). Briefly, diclofenac (50 μM) was incubated with pooled HLMs in 0.5 M Tris-HCl buffered mixture (pH 7.4) containing 1.5 mg/ml alamethicin. Diclofenac glucuronide was detected using the API 3000 LC-MS/MS system in positive ionization mode. The m/z values for diclofenac-glucuronide and 7-hydroxycoumarin fragmentation were 424→298 and 339→163, respectively.
Genotyping
Genomic DNA was isolated from human liver tissues using the DNeasy kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Specific primers were designed to amplify the flanking region of UGT2B7 (802C>T and 211G>T), UGT1A9 (−118T9>T10 and 1399C>T), and UGT1A3 (31T>C, 133C>T, and 140T>C) (supplementary Table I). Excluding UGT2B7*71S, all PCR products were directly sequenced for the detection of the mutations. PCR was performed in a reaction volume of 30 μl that contained 100 ng of genomic DNA, 1× PCR buffer, 0.2 mM deoxyribonucleotide triphosphate, 0.2 μM of each primer, 1.5 mM MgCl2, and 1 unit of Taq polymerase. PCR was performed in the GeneAmp PCR 9700 (Applied Biosystems) with initial denaturation of 95°C for 5 min, followed by 30–35 cycles of denaturation at 95°C for 30 s, annealing at 51–61°C for 30 s, and extension at 72°C for 10–80 s. A final termination of the elongation step was performed at 72°C for 7 min. The PCR conditions and primers are summarized in supplementary Table I. Amplified PCR products were purified using a PCR purification kit and directly sequenced using an AB BigDye™ Terminator v3.1 cycle sequencing kit (ABI Prime 3130 Genetic Analyzer; Life Technologies, Carlsbad, CA). A Taqman SNP genotyping assay was used to detect UGT2B7*71S (rs12233719: assay ID C_45181106_10) using the ABI HT 7900 (Applied Biosystems) according to the manufacturer’s instructions.
Immunoblot analysis
HLMs were prepared as described previously (22). Liver microsomal proteins were boiled at 95°C for 5 min in loading buffer [0.1 M Tris-HCl (pH 6.8), 4% SDS, 1.5% bromophenol blue, 20% glycerol, and 5% mercaptoethanol]. The denatured microsomal proteins (15 μg per lane) were separated on a 13% SDS-polyacrylamide gel electrophoresis and then transferred to a nitrocellulose membrane in buffer containing 25 mM Tris-HCl, 192 mM glycine, and 20% (v/v) methanol. The membrane was blocked with 5% skimmed milk in Tris-buffered saline supplemented with 0.1% Tween 20 solution at room temperature for 1 h. Polyclonal anti-UGT1A3 IgG and anti-UGT1A9 IgG, monoclonal anti-UGT2B7 IgG, and monoclonal anti-GADPH IgG were used as primary antibodies overnight at 4°C. The immunoreactive protein was detected using the ECL method (GE Healthcare Bio-Sciences, Little Chalfont, Buckinghamshire, UK) and visualized by autoradiography on an LAS 3000 (Fuji Film Life Science, Stamford, CT). Band densities were quantified using a MultiGauge (Fuji Photo Film Corporation, Science Laboratory, Tokyo, Japan). The relative UGT protein levels in Western blotting were normalized to GAPDH, which was immunoblotted as a loading control. Gel to gel variations in band densities were normalized using the reference amount of microsomal protein (15 μg per lane) for each electrophoresis (22).
Statistical analysis
All enzyme activity values represent the mean ± SD of triplicate reactions. The correlation coefficients between the formation rates of glucuronide metabolite of 20-HETE and UGT protein expression among 44 HLMs were calculated using Pearson’s correlation coefficient. The effect of genetic polymorphisms in UGTs and demographic data on 20-HETE glucuronidation was assessed using a one-way ANOVA. All statistical analysis was performed using the SAS program (version 9.1.3; SAS Institute, Cary, NC). Statistically significant changes compared with the control group are indicated as *P < 0.05, **P < 0.01, and ***P < 0.001.
RESULTS
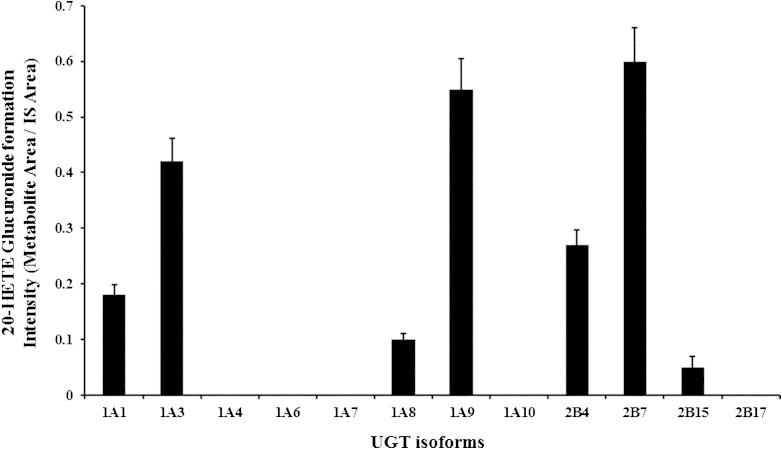
Assays for 20-HETE glucuronidation were performed in a reaction containing 20 μM 20-HETE, 15 μg of pooled HLMs, and 1.5 mg/ml of alamethicin in 0.5 M Tris-HCl buffer (pH 7.4), and 20-HETE glucuronide was detected using LC-MS/MS. To ascertain the experimental conditions, 20-HETE glucuronide formation was tested and compared with reactions lacking UDPGA. The 20-HETE glucuronide was detected at a retention time of 2.91 min in samples incubated with UDPGA. The product ion spectra of the glucuronidated 20-HETE peak showed m/z values of 495, 319, and 175, which were representative of 20-HETE glucuronide, 20-HETE aglycon, and glucuronic acid, respectively (24). Twelve recombinant UGT enzymes were screened to determine the major contributors of UGT enzymes for 20-HETE glucuronide formation. UGT2B7, UGT1A9, and UGT1A3 showed strong 20-HETE glucuronidation, followed by UGT2B4, UGT1A1, UGT1A8, and UGT1B15 in a decreasing order (Fig. 1). The kinetic parameters Km, Vmax, and CLint for 20-HETE glucuronidation with UGT1A3, -1A9, and -2B7 were determined as presented in Table 1. The Km values of UGT1A9 and -2B7 were 22.2 and 14.8 μM, respectively. However, UGT1A3 exhibited a higher Km value (78.4 μM) when compared with those of UGT1A9 and UGT2B7, indicating that UGT1A3 has a lower affinity toward 20-HETE glucuronidation than the UGT1A9 and -2B7. The Vmax values for 20-HETE glucuronidation by UGT1A3, -1A9, and -2B7 were similar. The intrinsic clearance values were evaluated under Michaelis-Menten equation, resulting in the highest value obtained from UGT2B7, followed by UGT1A9, and UGT1A3.
Fig. 1.
Screening for UGT enzymes responsible for the formation of 20-HETE glucuronide using recombinant human UGT isoforms. The glucuronide form of 20-HETE was biosynthesized by incubating 20 μM 20-HETE with the indicated recombinant UGT isoform proteins (15 μg) (BD Biosciences), 1.5 mg/ml alamethicin in 0.5 M Tris-HCl buffer (pH 7.4), and 5 mM UDPGA. The glucuronidated metabolite of 20-HETE was detected by scanning daughter ions of the deprotonated glucuronide at m/z 495.Our results represent one independent data set from two separate experiments. Values represent the mean ± SD of triplicates. UGT1A3, -1A9, and -2B7 metabolized 20-HETE more efficiently than other UGTs. Further details are described in the Experimental Procedures.
TABLE 1.
Kinetic analysis of 20-HETE analysis by UGT1A3, UGT1A9, and UGT2B7 isoforms
| Kinetic Parameters | |||
| UGT Isoform | Km (μM) | Vmax (nmol/min/mg protein) | CLint (Vmax/Km) (μl/min/mg protein) |
| UGT1A3 | 78.4 | 1.33 | 17.0 |
| UGT1A9 | 22.2 | 1.78 | 84.5 |
| UGT2B7 | 14.8 | 1.62 | 109.6 |
The best fit of data was determined by the examination of required parameters using SigmaPlot 8.0. The formation of 20-HETE glucuronide showed a fit to Michaelis-Menten kinetics. Details are described in the Experimental Procedures.
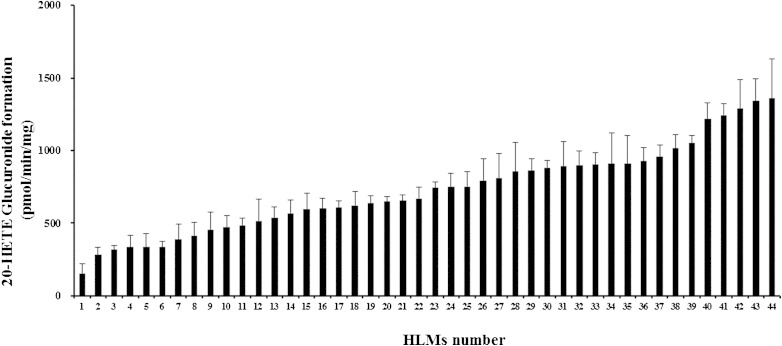
We then determined the levels of 20-HETE glucuronide in 44 HLMs (Fig. 2). A wide variation was observed in the levels of 20-HETE glucuronidation among HLMs. The mean ± SD of 20-HETE glucuronidation was 728 ± 100 pmol/min/mg. However, an over 10-fold variation in 20-HETE glucuronidation was observed in human liver samples. Because UGT activity is affected by sex, age, and alcoholic status (19), the impact of the demographic data of the HLM donors on 20-HETE glucuronidation was examined (supplementary Table II). Neither sex nor age affected the formation of 20-HETE glucuronide. Although the rate of 20-HETE glucuronidation was higher among alcoholics (832 ± 335 pmol/min/mg) than nonalcoholics (659 ± 273 pmol/min/mg), the difference in glucuronidation activity failed to reach statistical significance (P > 0.05).
Fig. 2.
The compound 20-HETE glucuronidation by HLMs (n = 44). HLMs (15 μg per reaction) were preincubated with 20 μM 20-HETE in a reaction mixture containing 1.5 mg/ml alamethicin and 0.5 M Tris-HCl buffer (pH 7.4) on ice for 15 min. The reaction was initiated by adding 5 mM UDPGA at 37°C for 4 h and terminated with 100 μl of acetonitrile. The formation of 20-HETE glucuronide was analyzed using HPLC coupled with MS. Further details are described in the Experimental Procedures.
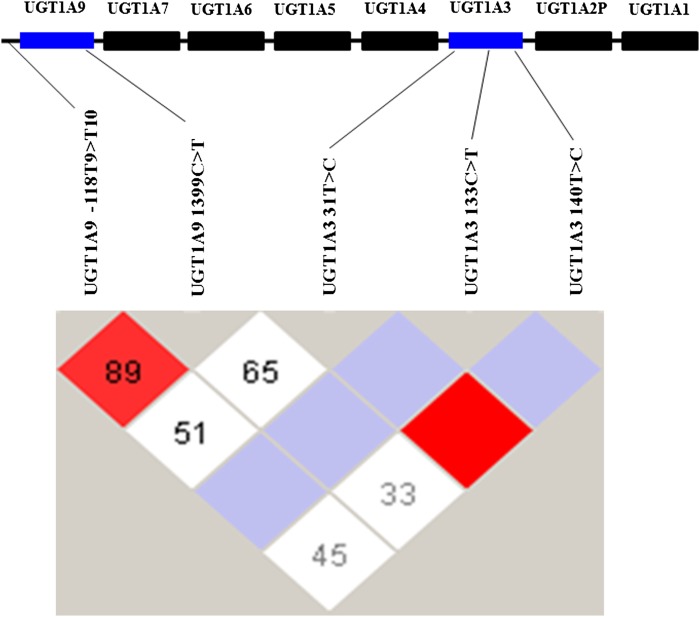
Because UGT2B7, -1A9, and -1A3 are critical UGT enzymes involved in the glucuronidation of 20-HETE (Fig. 1), the primary genetic polymorphisms in UGT2B7, -1A9, and -1A3 genes known to alter the metabolic activity of these UGTs among Asian ethnic groups were identified (26–28). The major functional polymorphisms were UGT2B7 (802C>T and 211G>T), UGT1A9 (−118T9>T10 and 1399C>T), and UGT1A3 (31T>C, 133C>T, and 140T>C).The frequencies of these polymorphisms among 44 HLMs are presented in Table 2. All analyzed polymorphisms were within Hardy-Weinberg equilibrium. A high degree of linkage disequilibrium (D’ = 0.879, r2 = 0.734) was observed between the two analyzed genetic variants, −118T9>T10 and 1399C>T on the UGT1A9 gene and between 31T>C and 140T>C on the UGT1A3 gene (Fig. 3). The results showed that UGT1A9 −118T9>T10, UGT1A9 1399C>T, and UGT2B7 802C>T genetic variations significantly influenced 20-HETE glucuronidation (P < 0.05–0.001) (Table 3). The rates of 20-HETE glucuronidation among UGT1A9 −118T9/T9, −118T9/T10, and −118T10/T10 were 375 ± 135, 696 ± 264, and 929 ± 270 pmol/min/mg, respectively. The rates of 20-HETE glucuronidation among UGT1A9 1399C/C, 1399C/T, and 1399T/T were 489 ± 338, 714 ± 305, and 811 ± 257 pmol/min/mg, respectively. The respective rates of 20-HETE glucuronidation among UGT2B7 802C/C, 802C/T, and 802T/T were 874 ± 242, 702 ± 315, and 619 ± 293 pmol/min/mg. The frequency of UGT1A3 (31T>C, 133C>T, and 140T>C) genotypes were low in the studied HLMs and no significant impact on the glucuronidation of 20-HETE was observed among the analyzed UGT1A3 genetic polymorphisms in this study.
TABLE 2.
The frequencies of UGT1A3, UGT1A9, and UGT2B7 genetic variants among HLMs (n = 44)
| Gene | Variantsa | Region | Reference | Enzyme Function | Total | wt/wt | wt/mt | mt/mt | Frequencyb (%) |
| UGT1A3 | 140T>C | Exon 1 | rs6431625 | Decrease | 44 | 38 | 6 | 0 | 7 |
| 31T>C | Exon 1 | rs3821242 | Decrease | 44 | 27 | 15 | 2 | 22 | |
| 133C>T | Exon 1 | rs45625338 | Increase | 44 | 38 | 5 | 1 | 8 | |
| UGT1A9 | −118T9>T10 | Promotor | rs3832043 | Increase | 44 | 5 | 26 | 13 | 59 |
| 1399C>T | Intron 1 | rs2741049 | Increase | 44 | 15 | 24 | 5 | 39 | |
| UGT2B7 | 211G>T | Exon 1 | rs12233719 | Decrease | 44 | 30 | 12 | 2 | 18 |
| 802C>T | Exon 2 | rs7439366 | Decrease | 44 | 13 | 18 | 13 | 50 |
The reference number used was GenBank accession NT_030640.1.
Position is indicated in relation to the start codon ATG of the UGT genes; the A in ATG is +1.
All of the frequencies are within Hardy-Weinberg equation (X2, P > 0.05).
Fig. 3.
Linkage disequilibrium map of UGT1A9 and -1A3 SNPs within the UGT1A gene locus genotyped in a Korean population (n = 44). The linkage disequilibrium was generated using Haploview 4.1 from the data related to five SNPs that are known to influence UGT1A9 and -1A3 activity. The linkage disequilibrium was analyzed using the statistics lD’l and r2 values. Red depicts a significant linkage while white indicates weak linkage between the pairs of SNPs. The numbers inside the squares indicate the D’ value multiplied by 100.
TABLE 3.
The effect of UGT1A3, UGT1A9, and UGT2B7 genetic variants on 20-HETE glucuronidation
| 20-HETE Glucuronidation (pmol/min/mg) | |||||
| Gene | Genetic Variation | Genotype | n | Mean ± SD | P |
| UGT1A3 | 140T>C | T/T | 40 | 784 ± 230 | Reference |
| T/C | 4 | 836 ± 315 | 0.678 | ||
| C/C | 0 | 0 | — | ||
| 31T>C | T/T | 27 | 730 ± 281 | Reference | |
| T/C | 15 | 732 ± 296 | 0.983 | ||
| C/C | 2 | 685 | 0.845 | ||
| 133C>T | C/C | 38 | 756 ± 230 | Reference | |
| C/T | 5 | 571 ± 230 | 0.098 | ||
| T/T | 1 | 472 | 0.231 | ||
| UGT1A9 | −118T9>T10 | T9/T9 | 5 | 375 ± 135 | Reference |
| T9/T10 | 26 | 696 ± 264 | 0.014* | ||
| T10/T10 | 13 | 929 ± 270 | 0.001** | ||
| 1399C>T | C/C | 5 | 489 ± 338 | Reference | |
| C/T | 21 | 714 ± 305 | 0.294 | ||
| T/T | 18 | 811 ± 257 | 0.031* | ||
| UGT2B7 | 211G>T | G/G | 30 | 728 ± 304 | Reference |
| G/T | 12 | 738 ± 326 | 0.925 | ||
| T/T | 2 | 678 ± 106 | 0.821 | ||
| 802C>T | C/C | 13 | 874 ± 242 | Reference | |
| C/T | 18 | 702 ± 315 | 0.111 | ||
| T/T | 13 | 619 ± 293 | 0.023* | ||
Significant difference in 20-HETE glucuronidation rates compared with the wild genotypes is represented by *P < 0.05 and **P < 0.01.
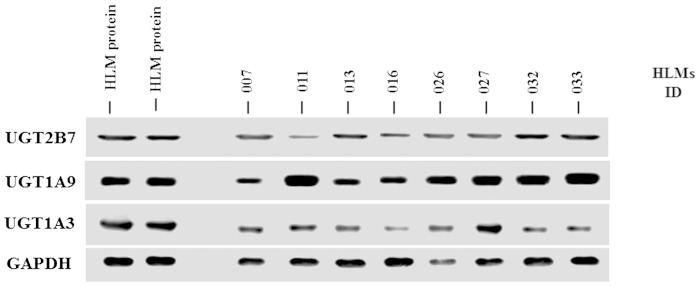
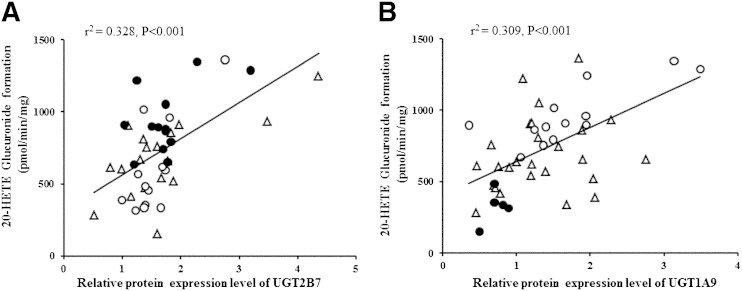
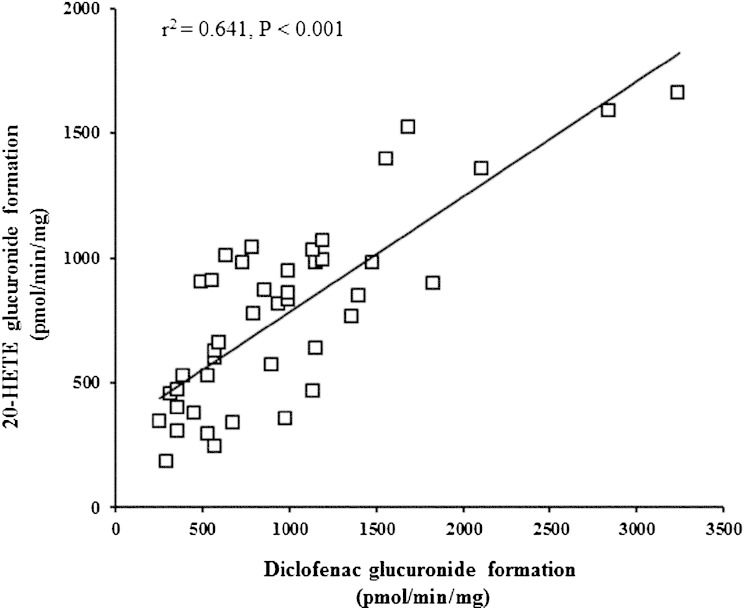
Table 4 shows that 20-HETE glucuronidation rates were the lowest among HLMs with UGT1A9 −118T9/1399C mutations and UGT2B7 802T genetic variation (P < 0.001, ANOVA). The rate of 20-HETE glucuronidation in HLMs combined with the UGT1A9 −118T9/1399C haplotype and UGT2B7 802T genetic variation was 346 ± 171 pmol/min/mg, while the rate of 20-HETE glucuronidation in HLMs combined with the UGT1A9 −118T10/1399C haplotype and UGT2B7 802C genetic variation was 1070 ± 247 pmol/min/mg. The relative protein expression of UGT2B7, -1A9, and -1A3 (the primary UGTs in 20-HETE glucuronidation) in HLMs was examined using Western blot analysis. The results showed high interindividual variation in protein expression of UGTs among HLM samples (Fig. 4). Therefore, the present study investigated the correlation between UGT1A3, -1A9, and -2B7 protein expression and 20-HETE glucuronidation. UGT1A9 and -2B7 protein expression levels were strongly correlated with 20-HETE glucuronide formation (P < 0.001), as shown in Fig. 5A, B. Liver microsomes with the UGT2B7 802T variant did not show lower UGT2B7 expression than UGT2B7 802C wild-type genotypes (Fig. 5A). Moreover, HLMs with homozygous UGT1A9 −118T9 showed lower UGT1A9 protein expression than UGT1A9 −118T10. This lower protein expression was correlated with a decreased 20-HETE glucuronidation rate (Fig. 5B). The formation of 20-HETE glucuronidation was proportional to the glucuronidation of diclofenac (r2 = 0.64, P < 0.001), which is known to be a prototype substrate of UGT2B7 and UGT1A9 (Fig. 6). This correlation further supports that UGT2B7 and UGT1A9 are involved in the formation of 20-HETE glucuronidation. Although UGT1A3 protein expression was correlated with 20-HETE glucuronidation (r2 = 0.022), the correlation coefficient value was not statistically significant (P > 0.05).
TABLE 4.
Combined effects of UGT1A9 and UGT2B7 variants on 20-HETE glucuronidation
| 20-HETE Glucuronidation (pmol/min/mg) | ||||
| UGT1A9 Genotype −118T9>T10 1399 C>T | UGT2B7 Genotype 802C>T | n | Mean ± SD | Pa |
| T9/T9 C/C | C/C | 0 | 0 | |
| C/T | 3 | 346 ± 171 | ||
| T/T | 2 | 419 | ||
| T9/T10 C/T | C/C | 9 | 786 ± 267 | |
| C/T | 8 | 700 ± 110 | <0.0001 | |
| T/T | 5 | 536 ± 330 | ||
| T10/T10 T/T | C/C | 3 | 1070 ± 247 | |
| C/T | 6 | 960 ± 301 | ||
| T/T | 3 | 920 ± 199 | ||
P values were calculated based on one-way ANOVA.
Fig. 4.
Representative Western blot analysis for UGT1A3, UGT1A9, and UGT2B7 in HLMs isolated from Korean liver donors. The HLM proteins (15 μg/lane) from liver tissues (lanes 3–10) were electrophoresed on 13% SDS-polyacrylamide gels. The presented results are randomly selected immunoblots of HLMs with the indication of band specificity in each UGT enzyme. The reference microsomal proteins (lanes 1 and 2) were used for normalization of the gel to gel variations in the band densities due to different exposure times in each film. Resolved proteins were transferred to a nitrocellulose membrane and immunoblotted using primary antibodies against UGT1A3, UGT1A9, and UGT2B7, as described in the Experimental Procedures. The immunoreactivity of GAPDH was used as an internal control for the relative protein expression in HLMs.
Fig. 5.
Correlation between 20-HETE glucuronidation and UGT protein amount with genotypes among HLMs (n = 44). A: Correlation between 20-HETE glucuronidation and UGT2B7 genotypes with its protein level. Solid circles, UGT2B7 802C/C; open triangles, UGT2B7 802C/T; open circles, UGT2B7 802T/T. B: Correlation between 20-HETE glucuronidation and UGT1A9 genotypes with its protein level. Solid circles, UGT1A9 −118T9/T9; open triangles, UGT1A9 −118T9/T10; open circles, UGT1A9 −118T10/T10. The formation of 20-HETE glucuronide was analyzed by HPLC coupled with MS. The protein expression was analyzed by Western blot. Genotyping of UGT2B7 802C>T and UGT1A9 −118T9>T10 variants was determined by direct DNA sequencing. Further details are described in the Experimental Procedures.
Fig. 6.
Correlation between 20-HETE glucuronidation and diclofenac glucuronidation in HLMs. The formation of 20-HETE glucuronide was analyzed by HPLC coupled with MS. Diclofenac (50 μM) was incubated with HLMs (n = 44) in 0.5 M Tris-HCl buffered mixture (pH 7.4) containing 1.5 mg/ml alamethicin for 1 h. Diclofenac glucuronide was detected by using HPLC coupled with MS in a positive ionization mode. The m/zs for diclofenac-glucuronide and 7-hydroxycoumarin (internal standard) fragmentation were 424→298 and 339→163, respectively. Further details are described in the Experimental Procedures.
DISCUSSION
The compound 20-HETE plays an important role in cardiovascular pathology and drug induced cardiotoxicity (5, 6). The glucuronidation reaction of 20-HETE is believed to inhibit 20-HETE and facilitate its elimination (24). However, factors contributing to variations in 20-HETE elimination remain poorly understood. To the best of our knowledge, this is the first study to investigate the effect of genetic polymorphisms in UGTs on 20-HETE glucuronidation activity. Our results demonstrated that UGT2B7 802C>T, UGT1A9 −118T9>T10, and UGT1A9 1399C>T genetic variants were associated with a significant change in 20-HETE glucuronide formation in HLMs. Our data suggest that these genetic variants may affect levels of 20-HETE in blood circulation, in addition to the genetic polymorphisms in 20-HETE-synthesizing CYPs, which showed a clinically significant association with hypertension development (10, 11). Therefore, UGT2B7 802C>T, UGT1A9 −118T9>T10, and UGT1A9 1399C>T variants may play a role in modulating platelet activation through differences in 20-HETE glucuronidation. Further studies are required to determine their contribution to platelet activation in a larger clinical study.
20-HETE is reportedly glucuronidated by UGT2B7, -1A1, and -1A3, and at a minor rate by UGT1A4 using UGT isoforms cloned and expressed in baculovirus-infected insect cells (24). In the present study, we screened the 20-HETE glucuronidation among 12 UGT isoforms using recombinant UGTs purchased from BD Gentest (Woburn, MA), which were also expressed in the baculovirus-infected insect cell system. In addition to UGT2B7, -1A1, and -1A3, our results demonstrated that UGT1A8, -1A9, -2B4, and -2B15 isoforms glucuronidated 20-HETE (Fig. 1). In particular, UGT1A9 strongly glucuronidated 20-HETE in addition to UGT1A1, -1A3, and -2B7. UGT1A9 has been shown to glucuronidate 5-, 12-, and 15-HETE, which are isomers of 20-HETE (21). In the present study, UGT1A3, -1A9, and -2B7 were determined as major contributors to the 20-HETE glucuronidation. The kinetic analyses were performed using these three enzymes. It was predicted that UGT2B7 and UGT1A9 exhibited 6- and 5-fold higher CLint values than the UGT1A3, respectively. This observation indicates that UGT2B7 and UGT1A9 might be highly specific for the glucuronidation of 20-HETE. Further studies would be needed to explore the consequences of these in vitro findings of the 20-HETE glucuronidation in vivo system.
Variation in 20-HETE glucuronidation among HLMs was observed in this report (Fig. 2). This observation agrees with a previous study that showed interindividual variation in the levels of 20-HETE elimination in human urine (14). Because 20-HETE is eliminated in human urine in the glucuronidated form, factors that affect UGT activity may affect 20-HETE elimination. Sex, age, alcoholic status, and genetic polymorphisms are known to affect UGT expression and activity (19). In the present study, the demographic data (sex, age, and alcoholic status) was not significantly associated with 20-HETE glucuronidation among HLMs (P > 0.05) (supplementary Table II). However, genetic polymorphisms (UGT2B7 802C>T, UGT1A9 −118T9>T10, and UGT1A9 1399C>T) influenced 20-HETE glucuronidation. These results suggest that genetic variations in UGT1A9 and -2B7 play major roles in interindividual variation in 20-HETE glucuronidation and elimination in humans.
Genotyping for the detection of UGT2B7 (802C>T and 211G>T), UGT1A9 (–118T9>T10 and 1399C>T), and UGT1A3 (31T>C, 133C>T, and 140T>C) polymorphisms were performed and analyzed to determine their contribution to 20-HETE glucuronidation and their effects on UGT protein expression. As shown in Table 2, all determined frequencies of UGT polymorphisms were within Hardy-Weinberg equilibrium. The frequencies of these variants were similar to those of other Asian populations (26–28). Moreover, UGT1A9 −118T9>T10 and 1399C>T genetic variations were found in high linkage disequilibrium in the present study (Fig. 3) and among Asians, but not Caucasians (29, 30).
Western blot analysis of UGT1A3, -1A9, and -2B7 showed that their protein expression varied significantly among the studied HLMs (Fig. 4). UGT1A9 and -2B7 protein expression levels were correlated strongly with 20-HETE glucuronidation among 44 HLMs from Koreans (P < 0.001) (Fig. 5A, B). These results suggest that factors affecting protein expression of UGT1A9 and -2B7, such as genetic variations, environmental inducers/inhibitors, and diseases, may affect 20-HETE glucuronidation. For example, the 20-HETE glucuronidation rate has been reported to be higher among patients with liver cirrhosis (31), and UGT expression was higher in liver cirrhosis than normal tissues (32), suggesting that UGTs play a role in 20-HETE glucuronidation in a protein-dependent manner.
In the present study, genetic polymorphisms UGT1A9 −118T9>T10 and 1399C>T significantly affected 20-HETE glucuronidation among HLMs (P < 0.001). The UGT1A9 −118T9 showed less 20-HETE glucuronidation activity than UGT1A9 −118T10. In addition, HLMs combined with the UGT1A9 −118T9/1399C haplotype and UGT2B7 802T genetic variants showed the lowest rate for 20-HETE glucuronidation (Table 4). UGT1A9 expression was higher in HLMs with the UGT1A9 −118T10/1399T than with −118T9/1399C, which correlated with increased 20-HETE glucuronidation rates. UGT1A9 −118T9>T10 is a promoter genetic variation, which showed increased UGT1A9 activity (33). The 1399C>T variant is an intronic polymorphism in the UGT1A9 gene, which was significantly correlated with UGT1A9 protein levels and activities (30). The molecular mechanism by which UGT1A9 −118T9>T10 and 1399C>T variants influence 20-HETE glucuronidation may be the result of increased transcriptional regulation and hence the increased availability of UGT1A9 enzymes for 20-HETE glucuronidation.
UGT2B7 802C>T is a nonsynonymous variant that leads to the substitution of histidine to tyrosine in codon 268. In vitro experiments indicated that the UGT2B7 802C>T variant shows 2-fold higher activity compared with the wild-type allele (34). However, clinical studies demonstrated that UGT2B7 802C>T polymorphisms were associated with lower enzyme activity and decreased drug metabolism, such as epirubicin and diclofenac (35, 36). UGT2B7 802C>T was reported to be associated with promoter polymorphisms, which were related to the decreased activity of the UGT2B7 enzyme (37). In the present study, HLMs containing the UGT2B7 802C>T variant exhibited lower 20-HETE glucuronidation, as shown in Table 3, without demonstrating lower UGT2B7 expression compared with the wild-type (Fig. 5A). Although the number of HLMs in the present study was small (n = 44), the UGT2B7 802C>T variant appeared to be associated with decreased activity of 20-HETE glucuronidation without affecting expression levels. However, further studies are required to explore its influence on UGT2B7 protein expression, as well as its effect on 20-HETE glucuronidation.
Overall, we identified major UGT enzymes responsible for 20-HETE glucuronidation using recombinant UGT enzymes in vitro system. The effects of genetic polymorphisms in UGTs, UGT2B7 802C>T, UGT1A9 −118T9>T10, and UGT1A9 1399C>T variants were significant on the altered levels of 20-HETE glucuronide formation in human liver tissues. Further investigations are required to explore the consequences of these genotypes on the 20-HETE glucuronidation in vivo system. This study is the first to examine the effects of genetic polymorphisms in UGTs on 20-HETE glucuronidation using human liver tissues. Application of our genotype information to the clinical field with respect to 20-HETE levels may increase our understanding regarding the role of 20-HETE under pathophysiological conditions in humans, such as variable platelet activation and cardiovascular complications.
Supplementary Material
Footnotes
Abbreviations:
- ARA
- arachidonic acid
- CYP
- cytochrome P450
- HLM
- human liver microsome
- UDPGA
- uridine diphosphate glucuronic acid
- UGT
- UDP-glucuronosyltransferase
This work was supported by the National Research Foundation of Korea (NRF) Grant funded by the Korea government (MSIP) (R13-2007-023-00000-0) and by a grant of the Korean Health Technology Research and Development Project, Ministry of Health and Welfare, Republic of Korea (HI14C0067).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables.
REFERENCES
- 1.Zordoky B. N., El-Kadi A. O. 2010. Effect of cytochrome P450 polymorphism on arachidonic acid metabolism and their impact on cardiovascular diseases. Pharmacol. Ther. 125: 446–463. [DOI] [PubMed] [Google Scholar]
- 2.Konkel A., Schunck W. H. 2011. Role of cytochrome P450 enzymes in the bioactivation of polyunsaturated fatty acids. Biochim. Biophys. Acta. 1814: 210–222. [DOI] [PubMed] [Google Scholar]
- 3.Devos A., Lino Cardenas C. L., Glowacki F., Engels A., Lo-Guidice J. M., Chevalier D., Allorge D., Broly F., Cauffiez C. 2010. Genetic polymorphism of CYP2U1, a cytochrome P450 involved in fatty acids hydroxylation. Prostaglandins Leukot. Essent. Fatty Acids. 83: 105–110. [DOI] [PubMed] [Google Scholar]
- 4.Miyata N., Roman R. J. 2005. Role of 20-hydroxyeicosatetraenoic acid (20-HETE) in vascular system. J. Smooth Muscle Res. 41: 175–193. [DOI] [PubMed] [Google Scholar]
- 5.Wu C. C., Schwartzman M. L. 2011. The role of 20-HETE in androgen-mediated hypertension. Prostaglandins Other Lipid Mediat. 96: 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J. Y., Li N., Yang J., Qiu H., Ai D., Chiamvimonvat N., Zhu Y., Hammock B. D. 2010. Metabolic profiling of murine plasma reveals an unexpected biomarker in rofecoxib-mediated cardiovascular events. Proc. Natl. Acad. Sci. USA. 107: 17017–17022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baines A. D., Ho P. 2005. 20-HETE-mediated vasoconstriction by hemoglobin-O2 carrier in Sprague-Dawley but not Wistar rats. J. Appl. Physiol. 98: 772–779. [DOI] [PubMed] [Google Scholar]
- 8.Vasudevan H., Yuen V. G., McNeill J. H. 2012. Testosterone-dependent increase in blood pressure is mediated by elevated Cyp4A expression in fructose-fed rats. Mol. Cell. Biochem. 359: 409–418. [DOI] [PubMed] [Google Scholar]
- 9.Ward N. C., Rivera J., Hodgson J., Puddey I. B., Beilin L. J., Falck J. R., Croft K. D. 2004. Urinary 20-hydroxyeicosatetraenoic acid is associated with endothelial dysfunction in humans. Circulation. 110: 438–443. [DOI] [PubMed] [Google Scholar]
- 10.Mayer B., Lieb W., Gotz A., Konig I. R., Aherrahrou Z., Thiemig A., Holmer S., Hengstenberg C., Doering A., Loewel H., et al. 2005. Association of the T8590C polymorphism of CYP4A11 with hypertension in the MONICA Augsburg echocardiographic substudy. Hypertension. 46: 766–771. [DOI] [PubMed] [Google Scholar]
- 11.Liu H., Zhao Y., Nie D., Shi J., Fu L., Li Y., Yu D., Lu J. 2008. Association of a functional cytochrome P450 4F2 haplotype with urinary 20-HETE and hypertension. J. Am. Soc. Nephrol. 19: 714–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams J. M., Murphy S., Burke M., Roman R. J. 2010. 20-hydroxyeicosatetraeonic acid: a new target for the treatment of hypertension. J. Cardiovasc. Pharmacol. 56: 336–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watzer B., Reinalter S., Seyberth H. W., Schweer H. 2000. Determination of free and glucuronide conjugated 20-hydroxyarachidonic acid (20-HETE) in urine by gas chromatography/negative ion chemical ionization mass spectrometry. Prostaglandins Leukot. Essent. Fatty Acids. 62: 175–181. [DOI] [PubMed] [Google Scholar]
- 14.Ward N. C., Puddey I. B., Hodgson J. M., Beilin L. J., Croft K. D. 2005. Urinary 20-hydroxyeicosatetraenoic acid excretion is associated with oxidative stress in hypertensive subjects. Free Radic. Biol. Med. 38: 1032–1036. [DOI] [PubMed] [Google Scholar]
- 15.Pat Kunert M., Drenjancevic I. 2011. 20-Hydroxyeicosatetraenoic acid, endothelial dysfunction and hypertension. Med. Glas. (Zenica). 8: 170–180. [PubMed] [Google Scholar]
- 16.Williams J. M., Sarkis A., Lopez B., Ryan R. P., Flasch A. K., Roman R. J. 2007. Elevations in renal interstitial hydrostatic pressure and 20-hydroxyeicosatetraenoic acid contribute to pressure natriuresis. Hypertension. 49: 687–694. [DOI] [PubMed] [Google Scholar]
- 17.Lohr J. W., Willsky G. R., Acara M. A. 1998. Renal drug metabolism. Pharmacol. Rev. 50: 107–141. [PubMed] [Google Scholar]
- 18.Tukey R. H., Strassburg C. P. 2000. Human UDP-glucuronosyltransferases: metabolism, expression, and disease. Annu. Rev. Pharmacol. Toxicol. 40: 581–616. [DOI] [PubMed] [Google Scholar]
- 19.Court M. H. 2010. Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system. Drug Metab. Rev. 42: 209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guillemette C. 2003. Pharmacogenomics of human UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 3: 136–158. [DOI] [PubMed] [Google Scholar]
- 21.Turgeon D., Chouinard S., Belanger P., Picard S., Labbe J. F., Borgeat P., Belanger A. 2003. Glucuronidation of arachidonic and linoleic acid metabolites by human UDP-glucuronosyltransferases. J. Lipid Res. 44: 1182–1191. [DOI] [PubMed] [Google Scholar]
- 22.Lee S. J., Lee S. S., Jung H. J., Kim H. S., Park S. J., Yeo C. W., Shin J. G. 2009. Discovery of novel functional variants and extensive evaluation of CYP2D6 genetic polymorphisms in Koreans. Drug Metab. Dispos. 37: 1464–1470. [DOI] [PubMed] [Google Scholar]
- 23.Seo K. A., Bae S. K., Choi Y. K., Choi C. S., Liu K. H., Shin J. G. 2010. Metabolism of 1’- and 4-hydroxymidazolam by glucuronide conjugation is largely mediated by UDP-glucuronosyltransferases 1A4, 2B4, and 2B7. Drug Metab. Dispos. 38: 2007–2013. [DOI] [PubMed] [Google Scholar]
- 24.Little J. M., Kurkela M., Sonka J., Jantti S., Ketola R., Bratton S., Finel M., Radominska-Pandya A. 2004. Glucuronidation of oxidized fatty acids and prostaglandins B1 and E2 by human hepatic and recombinant UDP-glucuronosyltransferases. J. Lipid Res. 45: 1694–1703. [DOI] [PubMed] [Google Scholar]
- 25.King C., Tang W., Ngui J., Tephly T., Braun M. 2001. Characterization of rat and human UDP-glucuronosyltransferases responsible for the in vitro glucuronidation of diclofenac. Toxicol. Sci. 61: 49–53. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y., Chen S., Li X., Wang X., Zeng S. 2006. Genetic variants of human UGT1A3: functional characterization and frequency distribution in a Chinese Han population. Drug Metab. Dispos. 34: 1462–1467. [DOI] [PubMed] [Google Scholar]
- 27.Hwang M. S., Lee S. J., Jeong H. E., Lee S., Yoo M. A., Shin J. G. 2010. Genetic variations in UDP-glucuronosyltransferase 2B7 gene (UGT2B7) in a Korean population. Drug Metab. Pharmacokinet. 25: 398–402. [DOI] [PubMed] [Google Scholar]
- 28.Yea S. S., Lee S. S., Kim W. Y., Liu K. H., Kim H., Shon J. H., Cha I. J., Shin J. G. 2008. Genetic variations and haplotypes of UDP-glucuronosyltransferase 1A locus in a Korean population. Ther. Drug Monit. 30: 23–34. [DOI] [PubMed] [Google Scholar]
- 29.Saito Y., Sai K., Maekawa K., Kaniwa N., Shirao K., Hamaguchi T., Yamamoto N., Kunitoh H., Ohe Y., Yamada Y., et al. 2009. Close association of UGT1A9 IVS1+399C>T with UGT1A1*28, *6, or *60 haplotype and its apparent influence on 7-ethyl-10-hydroxycamptothecin (SN-38) glucuronidation in Japanese. Drug Metab. Dispos. 37: 272–276. [DOI] [PubMed] [Google Scholar]
- 30.Girard H., Villeneuve L., Court M. H., Fortier L. C., Caron P., Hao Q., von Moltke L. L., Greenblatt D. J., Guillemette C. 2006. The novel UGT1A9 intronic I399 polymorphism appears as a predictor of 7-ethyl-10-hydroxycamptothecin glucuronidation levels in the liver. Drug Metab. Dispos. 34: 1220–1228. [DOI] [PubMed] [Google Scholar]
- 31.Sacerdoti D., Balazy M., Angeli P., Gatta A., McGiff J. C. 1997. Eicosanoid excretion in hepatic cirrhosis. Predominance of 20-HETE. J. Clin. Invest. 100: 1264–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debinski H. S., Mackenzie P. I., Lee C. S., Mashford M. L., Danks J. A., Tephly T. R., Green M., Desmond P. V. 1996. UDP glucuronosyltransferase in the cirrhotic rat liver. J. Gastroenterol. Hepatol. 11: 373–379. [DOI] [PubMed] [Google Scholar]
- 33.Yamanaka H., Nakajima M., Katoh M., Hara Y., Tachibana O., Yamashita J., McLeod H. L., Yokoi T. 2004. A novel polymorphism in the promoter region of human UGT1A9 gene (UGT1A9*22) and its effects on the transcriptional activity. Pharmacogenetics. 14: 329–332. [DOI] [PubMed] [Google Scholar]
- 34.Thibaudeau J., Lepine J., Tojcic J., Duguay Y., Pelletier G., Plante M., Brisson J., Tetu B., Jacob S., Perusse L., et al. 2006. Characterization of common UGT1A8, UGT1A9, and UGT2B7 variants with different capacities to inactivate mutagenic 4-hydroxylated metabolites of estradiol and estrone. Cancer Res. 66: 125–133. [DOI] [PubMed] [Google Scholar]
- 35.Parmar S., Stingl J. C., Huber-Wechselberger A., Kainz A., Renner W., Langsenlehner U., Krippl P., Brockmoller J., Haschke-Becher E. 2011. Impact of UGT2B7 His268Tyr polymorphism on the outcome of adjuvant epirubicin treatment in breast cancer. Breast Cancer Res. 13: R57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daly A. K., Aithal G. P., Leathart J. B., Swainsbury R. A., Dang T. S., Day C. P. 2007. Genetic susceptibility to diclofenac-induced hepatotoxicity: contribution of UGT2B7, CYP2C8, and ABCC2 genotypes. Gastroenterology. 132: 272–281. [DOI] [PubMed] [Google Scholar]
- 37.Duguay Y., Baar C., Skorpen F., Guillemette C. 2004. A novel functional polymorphism in the uridine diphosphate-glucuronosyltransferase 2B7 promoter with significant impact on promoter activity. Clin. Pharmacol. Ther. 75: 223–233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.