Abstract
Owing to its capability of discriminating subtle mass-altering structural differences such as double bonds or elongated acyl chains, MALDI-based imaging MS (IMS) has emerged as a powerful technique for analysis of lipid distribution in tissue at moderate spatial resolution of about 50 μm. However, it is still unknown if MS1-signals and ion intensity images correlate with the corresponding apparent lipid concentrations. Analyzing renal sulfated glycosphingolipids, sulfatides, we validate for the first time IMS-signal identities using corresponding sulfatide-deficient kidneys. To evaluate the extent of signal quenching effects interfering with lipid quantification, we surgically dissected the three major renal regions (papillae, medulla, and cortex) and systematically compared MALDI IMS of renal sulfatides with quantitative analyses of corresponding lipid extracts by on-target MALDI TOF-MS and by ultra-performance LC-ESI-(triple-quadrupole)tandem MS. Our results demonstrate a generally strong correlation (R2 > 0.9) between the local relative sulfatide signal intensity in MALDI IMS and absolute sulfatide quantities determined by the other two methods. However, high concentrations of sulfatides in the papillae and medulla result in an up to 4-fold signal suppression. In conclusion, our study suggests that MALDI IMS is useful for semi-quantitative dissection of relative local changes of sulfatides and possibly other lipids in tissue.
Keywords: liquid chromatography, matrix-assisted laser desorption/ionization-time-of-flight, electrospray ionization-mass spectrometry, tandem mass spectrometry, galactosylceramide I3-sulfate, lactosylceramide II3-sulfate, cortex, medulla, papillae
In recent years, MALDI imaging MS (IMS) has become a common method in lipid analytics. It enables visualization of the differential distribution of lipids varying only in their acyl chain with good spatial resolution (≥7 μm/pixel), as well as with high mass accuracy (<3 ppm rms) and high mass resolution (R = 100.000 at m/z 200) (1). Although much work has been put into increasing the sensitivity of this method by the use of different matrix substances (2–7) or improved sample preparation (8–14) that achieves more homogeneous cocrystallization (15), reliability of the semi-quantitative read out remains a matter of debate: Differences in the tissue structure (16) as well as specific matrix analyte interaction (17) are suspected to cause ion suppression followed by unreliable signal response (18). Ionization behaviors of specific molecules are hard to predict because they are influenced by a number of parameters (16, 19). Initially, Burnum et al. (2) picked up this issue in their work on spatial and temporal alterations of phospholipids during mouse embryo implantation. They compared the MALDI IMS data with LC-ESI-MS2 data obtained in positive or negative mode after laser microdissection (2). Shortly afterwards, Hankin and Murphy (20) demonstrated the direct correlation between MALDI IMS ion intensities and the concentrations of phosphatidylcholines determined after laser microdissection and LC-ESI-MS2 quantification from different parts of rat brain. In this case, using dihydroxybenzoic acid as MALDI-matrix in positve mode, a good correlation between the two methods was observed for molecules with high abundance in the sample. Also, quantities of olanzapine, determined by LC-ESI-MS2 out of tissue homogenates, could be recovered by MALDI IMS in homogeneous liver tissue (21). However, it remains to be seen if this behavior can be transferred directly to other tissues with structural diversity, e.g., kidney, to other compounds, to the negative ionization mode, and to different matrices. One of these substances is 9-aminoacridine (9-AA), which is described to prefer sulfated glycosphingolipids in the negative ionization process (17) because of its basic character (22).
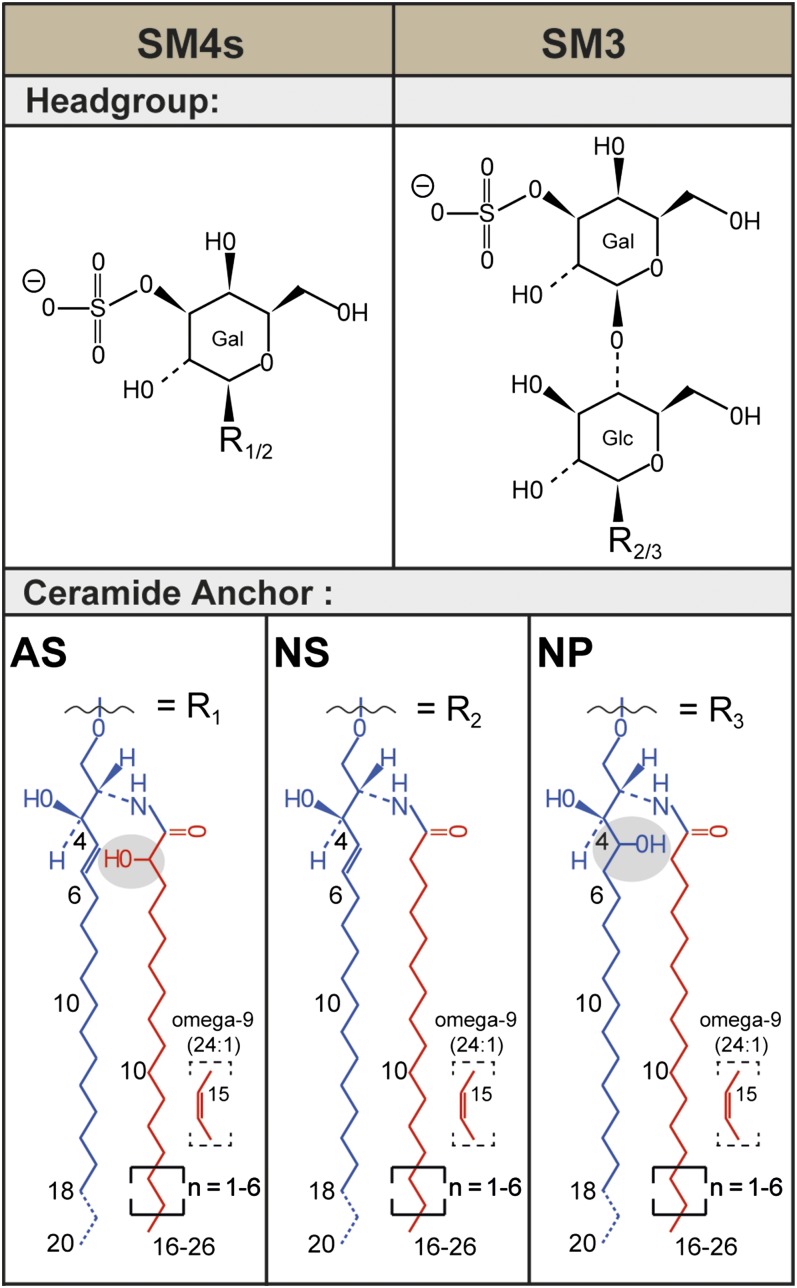
Recently, several publications especially focused on sulfatides using different imaging mass spectrometric methods (5, 23–28). Most of these studies addressed sulfatides of the central and peripheral neural system that predominantly contain simple galactosylceramide I3-sulfate (SM4s). Mammalian kidneys are rich in sulfatides. Like human kidneys, mouse or rat kidneys also contain the two simple sulfatides SM4s and lactosylceramide II3-sulfate (SM3) (Fig. 1). However, mouse and rat additionally express more complex sulfatides (29). By far, the most abundant sulfatide is in mouse SM4s, followed by SM3 and minor amounts of gangliotetraosylceramide II3-, IV3-bis sulfate (30). Depending on their sulfated glycan moiety, sulfatides are differentially distributed in the kidney (31–33). However, the large structural diversity of mammalian renal sulfatides mainly stems from differences in their ceramide anchor compositions, which cannot be distinguished by immunohistochemistry. Using MALDI IMS instead, a diverse sulfatide distribution to the papillae, medulla, and cortex was elucidated recently, especially with respect to their ceramide anchor composition (5, 34). Similar observations have been obtained in the brain, where distinct distributions of nonhydroxylated sulfatides [ceramide anchor with a nonhydroxy acyl chain and a sphingosine (NS)] and hydroxylated sulfatides [ceramide anchor with an alpha-hydroxy acyl chain and a sphingosine (AS)] were also observed using MALDI IMS (26).
Fig. 1.
Structural diversity of renal sulfatides. Sulfatides are composed of a lipophilic ceramide anchor linked via a β-glycosidic bond to a polar glycan head group, which is sulfated. The most prominent murine and human renal sulfatides are sulfatide SM4s with a monosulfated galactosyl head group and sulfatide SM3 with a monosulfated lactosyl moiety. In mouse kidneys, a variety of ceramide anchors are found in sulfatides. In SM4s, combinations of the major sphingoid base C18-sphingosine (d18:1) with nonhydroxyl acyl chains (NS) and with α-hydroxyl acyl chains (AS) are found. [The ceramide anchor of sphingolipids often is detailed in parentheses following the abbreviation for ceramide (Cer), e.g., Cer(d18:1;16:0) or Cer(t18:0;h16:0). In parentheses, the description of the sphingoid base is followed by the description of the N-linked acyl chain after the semicolon. The sphingoid base is described first with a letter referring to the amount of hydroxy groups within the base (m, mono; d, di; t, tri; and te, tetra). Then the number of C-atoms follows, separated by a colon to the number of double bonds within the sphingoid base. After the semicolon, the fatty acid is described in analogy, but “h” indicates here the presence of one hydroxylation.] SM3 also appears with an NS-ceramide anchor, but, in addition, also anchors combining a C18-phytosphingosine (t18:0) with nonhydroxyl acyl chains (NP) are detected. Ceramide anchors with α-hydroxyl acyl chains are not observed for SM3.
With regard to the sulfatide-preferring properties of 9-AA, the complexity of renal sulfatide distribution, and the possibility of dissecting mouse kidney into the three different regions for classical analysis, the investigation of renal sulfatides with 9-AA in different parts of the kidney seemed to be an interesting model system for evaluating MALDI IMS quantitatively.
MATERIALS AND METHODS
Chemicals and reagents
Chemicals and solvents (acetonitrile, chloroform, methanol) used for lipid extraction and preparation of solutions and samples were generally obtained from Merck (Darmstadt, Germany) in the highest available purity. Sodium carbonate for lipid extraction was obtained from Fluka (part of Sigma-Aldrich, Taufkirchen, Germany). Either deionized water [Milli-Q, Millipore (part of Merck)], fully desalinated water (B. Braun AG, Melsungen, Germany), or water with HPLC-grade (Sigma-Aldrich) was used throughout this study. The 9-AA used as MALDI matrix was purchased from Merck. The semisynthetic glycosphingolipids, SM4s(d18:1;14:0), SM4s(d18:1;19:0), SM4s(d18:1;27:0), SM3(d18:1;14:0), SM3(d18:1;19:0), and SM3(d18:1;27:0), used as internal standard, have been described elsewhere (30).
Animals
All animal procedures were performed in accordance with the guidelines for the care and use of laboratory animals and were approved by Department 35 of the Regierungspräsidium Karlsruhe. Female wild-type (wt) C57BL/6N mice were obtained from the German Cancer Research Center. Kidneys of a specific knockout of cerebroside sulfotransferase (Cst) in renal tubular epithelial cells [Cstf/f Pax8 Cre (−/−)] mice have been described before (34).
Preparation of lipid extracts from kidneys of wt and mutant mice
Lipids were extracted according to a previously reported procedure (35), with slight modifications. Briefly, one frozen kidney (C57BL/6N mouse) was homogenized for 2 min on ice in 2 ml methanol and 300 μl water with an Ultra Turrax T25 basic (IKA Labortechnik, Staufen, Germany) at 24,000 rpm in a 15 ml polypropylene vial. The homogenate was transferred into a glass vial and the polypropylene vial was rinsed two times with 500 μl methanol. Afterwards, 3 ml of chloroform were given to the homogenate to get a solvent mixture of chloroform/methanol/water (10:10:1, v/v/v). The extract was centrifuged for 10 min at 3,000 rpm, and the supernatant was collected in a separate glass vial. Extraction was completed by repetition of this procedure twice, one time with 3 ml chloroform/methanol/water (10:10:1, v/v/v) and once with a 30:60:8 (v/v/v) mixture. Every extraction step included 2 min of sonication. The pooled extracts were dried under nitrogen stream (37°C). Finally, the extract was dissolved in 100 μl chloroform/methanol/water (10:10:1, v/v/v) per 100 mg kidney wet weight and stored at −20°C. For lipid extraction from separated renal regions (papillae, medulla, cortex), fresh kidneys (C57BL/6N) were rapidly prepared after euthanization. Between 7.4 mg and 18.4 mg of each region were carefully separated under a light microscope. Lipids were extracted from the freeze-dried homogenate as described above. The pooled extracts were dried under nitrogen stream (37°C). Finally, the extracts were dissolved in 100 μl chloroform/methanol/water (10:10:1, v/v/v) per 100 mg kidney wet weight and stored at −20°C.
For lipid extraction of kidneys from Cstf/f Pax8 Cre (−/−) mice, one frozen kidney was homogenized six times for 2 min on ice in 1 ml sodium carbonate buffer (200 mM, pH 9.3) with an Ultra Turrax T25 basic (IKA Labortechnik) at 24,000 rpm with pauses of 2 min in between. The homogenate was freeze-dried overnight (−49°C; 0.2 mbar) and the dry weight was determined. Glycosphingolipids were extracted from the freeze-dried homogenate twice with 500 μl chloroform/methanol/water (10:10:1, v/v/v) and once with a 30:60:8 (v/v/v) mixture. The pooled extracts were dried with a rotary evaporator at 50°C, dried under nitrogen stream (37°C), and stored at −20°C.
MALDI IMS
Kidneys from female (C57BL/6N) and Cstf/f Pax8 Cre (−/−) mice were frozen without prior perfusion and sliced into 10 μm sections using a Leica CM1510S cryostat (Leica Biosystems, Nussloch, Germany) at −20°C. To prevent interference of embedding medium (OCT; Sergipath, Richmond, IL) with mass spectrometric analysis, the frozen kidneys were fixed on the carrier by carefully dipping only one side of the organ in the embedding medium. Thereby, the side from which sections were obtained was free of embedding medium. Tissue sections were mounted onto indium tin oxide-coated conductive glass slides (Bruker Daltonics, Bremen, Germany), dried by vacuum desiccation without prior washing steps, and used immediately or stored at −80°C. The 9-AA [20 mM in acetonitrile/water (80:20, v/v)] was deposited on slides using an ImagePrep matrix sprayer (Bruker Daltonics). Mass spectrometric measurements were performed using an Autoflex III MALDI TOF/TOF instrument (Bruker Daltonics) with the adjustments described in the quantification section of the Materials and Methods. Mass spectra were obtained in negative ion-reflector mode in the m/z range from 700 to 1,200 Da. Images were acquired at a spatial resolution of 50 μm with 200 laser shots per position. Spectra were saved and the images constructed using flexImaging 3.0 software (Bruker Daltonics). Mass filters were chosen with a width of 0.2 Da. In all analyses, blood-derived lipids were not separately taken into account. However, serum sulfatide levels are less than 0.2% of kidney sulfatide levels and therefore negligible (36).
Evaluation of regions of interest in MALDI IMS
Representative regions of interest (ROIs) from each renal region, papillae, medulla, or cortex, containing 180–200 measurement points, were assigned in flexImaging 3.0 software (Bruker Daltonics). Average spectra of each ROI were calculated using ClinProTools version 3.0 (Bruker Daltonics).
Quantification by MALDI on-target
The 9-AA [20 mM in methanol/water (90:10, v/v)] was prepared freshly before use. Kidney lipid extracts were mixed with the internal standards of nonendogenous SM4s and SM3 species containing the ceramide anchors (d18:1;14:0), (d18:1;19:0), and (d18:1;27:0), dried under an air stream (37°C), and dissolved in methanol/water (90:10, v/v). Samples were mixed 1:1 (v/v) with matrix solution. Two microliters per spot were deposited on a steel target and dried under an air stream. All mass spectrometric measurements were performed using an Autoflex III MALDI TOF/TOF instrument (Bruker Daltonics) equipped with a Smartbeam laser (200 Hz) and controlled by flexControl 3.4 software (Bruker Daltonics).The extraction voltage was 19 kV, and gated matrix suppression (<650 Da) was applied to prevent saturation of the detector with matrix ions. Spectra were acquired in negative reflector mode using delayed extraction. On one sample spot, 20,000 laser shots were accumulated by random walk with one hundred shots per position. Baseline subtraction was performed, and peak intensities, signal-to-noise ratios, and spectral mass resolutions were determined with flexAnalysis 3.4 software (Bruker Daltonics). The mass spectrometer was calibrated internally using the theoretical masses of the nonendogenous standards SM4s(d18:1;14:0), SM4s(d18:1;19:0), SM4s(d18:1;27:0), SM3(d18:1;14:0), SM3(d18:1;19:0), and SM3(d18:1;27:0). These internal standards were also used for quantification.
Quantification by ultra-performance LC-ESI-MS2
Sulfatides were analyzed on a ultra-performance (UP)LC Aquity I system comprising an autosampler with cooled tray (15°C) (Waters, Manchester, England) coupled to a Xevo TQ-S triple-quadrupole mass spectrometer equipped with an ESI source (Waters). The system was controlled by MassLynxs software v4.1. Separation was performed on an Aquity UPLC bridged-ethylene-hybrid C18 1.7 μm column (2.1 × 50 mm) at 40°C. The flow rate was set to 500 μl/min and the column was equilibrated with methanol:water (95:5, v/v). A sample volume of 10 μl was injected and eluted with a gradient between solvent A, consisting of methanol:water (95:5), and solvent B, consisting of isopropanol:water (99:1). The chromatographic conditions were as described in Table 1.
TABLE 1.
Gradient conditions for the separation of sulfatides on UPLC
| Time (min) | Solvent A (%) | Solvent B (%) |
| Initial | 100 | 0 |
| 0.1 | 100 | 0 |
| 0.2 | 85 | 15 |
| 4.0 | 25 | 75 |
| 4.5 | 25 | 75 |
| 5.0 | 100 | 0 |
| 6.5 | 100 | 0 |
Tandem MS experiments were performed with argon as collision gas at a flow rate of 0.15 ml/min. The source temperature was set to 150°C, while the desolvation temperature was set to 350°C. The spray was started in negative ion mode by applying 2.5 kV to the fixed capillary; the cone voltage was set to 50 V. Deprotonated sulfatide ions were detected by single reaction monitoring with their transition to the negatively charged product ion m/z 96.8 (HSO4−) using a collision energy of 70 eV for the collision-induced dissociation (for the single reaction monitoring list see supplementary Table I.). The internal NS-sulfatide standards SM4s(d18:1;14:0), SM4s(d18:1;19:0), SM4s(d18:1;27:0), SM3(d18:1;14:0), SM3(d18:1;19:0), and SM3(d18:1;27:0), added prior to measurement, were used to determine calibration curves for retention times of sulfatide subclasses (supplementary Fig. I) and for quantification. As described earlier (37), additional product ions occur in AS-sulfatide (supplementary Fig. II), thereby reducing the probability for the transition to HSO4− in AS- as compared with NS-sulfatides. Therefore, transitions of AS-sulfatides to HSO4− were corrected with a factor of 1.114 prior to quantification using the NS-sulfatide standards.
Statistics
All statistic calculations (mean, SD, unpaired Student’s t-tests, one-way ANOVA post hocTukey tests, and correlation factors) were calculated with GraphPad Prism version 5.04 software. Isotope distributions were calculated with the isotope distribution calculator from the webpage of Scientific Instrument Service, Ringoes, New Jersey.
RESULTS
MALDI IMS of renal sulfatides, semi-quantitative depiction and signal validation
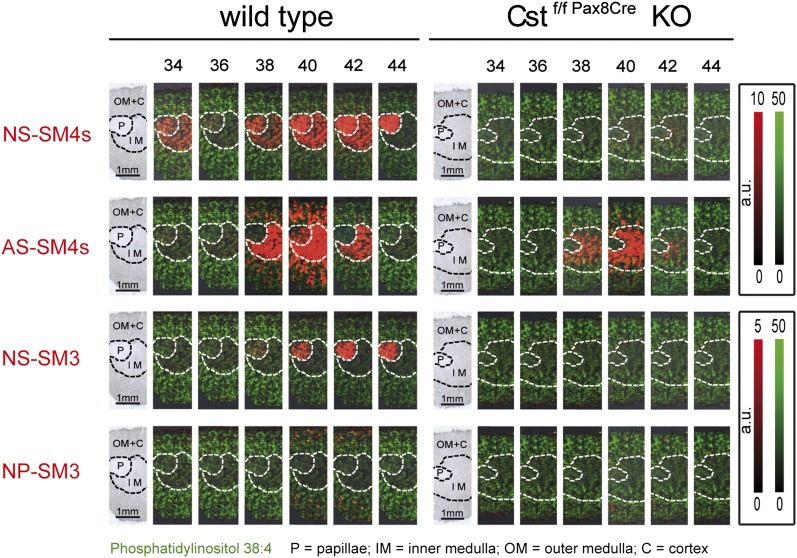
Our analysis started up with the question of normalization. In order to analyze not only the local distribution of a single sulfatide, but also to compare the relative concentrations of related sulfatides with each other by MALDI IMS, we had to normalize our data. As described before, in heterogeneous tissue like the kidney, normalization to the total ion count or vector normalization may generate misleading results (38). To avoid this problem, we normalized the color code of intensity for all sulfatide signals to the same absolute signal counts. To determine this maximum value for the color code of intensity, we picked the sulfatide with the highest signal from the average spectra of the three ROIs. For this compound, we identified the individual spectra/locations with highest intensity on the tissue section and set its value as maximum color code intensity for all sulfatides. With these settings, the resulting pictures suggested a prominent expression of sulfatides with C22- and C24-acyl chains3 (sulfatides with 42 and 44 C-atoms in their ceramide moiety), especially as compared with those with a C18-acyl chain (sulfatides with 36 C-atoms in their ceramide moiety). Furthermore, the pictures imply a similar high sulfatide concentration in the papillae and medulla, although the ceramide anchors contain a different type of acyl chain in these two regions (Fig. 2, wt). Finally, these pictures reflect the relative low abundance of SM3 over SM4 sulfatides in agreement with the literature (30, 39).
Fig. 2.
MALDI IMS of renal sulfatides in wt (left) and sulfatide-deprived (right) mice. Kidneys were dissected vertically to the longitudinal axis so that sections contained papillae, medulla, and cortex. The first picture of each series represents light microscopy of the analyzed renal section. Areas attributed to papillae (P), inner medulla (IM), and outer medulla together with cortex (OM+C) are marked with dotted lines. Individual sulfatide species are plotted with the false color red and are sorted by head group and type of ceramide anchor, as well as by length of the acyl chain. MS signal intensities relate to color intensity, which was normalized for all sulfatides of the identical head group (either SM4s or SM3) to the same color intensity per absolute signal counts; see color code on the right. Likewise, the signal intensities for PI (38:4), represented with the false color green, were normalized to the same intensities in wt and mutant sections. Note the loss or strong reduction of signals attributed to sulfatides in mutant (right) as compared with wt kidney.
To validate the specificity of these MS1 MALDI IMS signals, we used mice lacking cell-specific cerebroside sulfotransferase (Cst) in the renal epithelium (34), an enzyme required for sulfatide formation (Fig. 2, right) (29). In these mice, most of the signals were gone, verifying their sulfatide origin. Only residual signals implying AS-SM4s with hydroxylated arachidic acid, behenic acid, or lignoceric acid remained in the inner medulla. Hence, these signals either corresponded to other compounds with the same nominal mass (m/z value) or were due to residual renal sulfatides in the cell not targeted by the corresponding cell-specific mouse model. To differentiate, corresponding renal lipid extracts were analyzed by UPLC-MS2. This method is more specific, as in addition to the m/z of the molecular ions, chromatographic retention times and the m/z of the specific fragment ions of sulfatides at the defined collision energy are also taken into account. Indeed, LC-MS2 analysis confirmed the presence of residual sulfatide amounts (roughly 10%) of exactly these three sulfatide species in the mutant kidneys (supplementary Fig. III), and by that verified the sulfatide specificity of the residual MALDI IMS signals. In addition, we were able to image a kidney from a mouse with systemic Cst deficiency. In this case, all sulfatide-related signals were absent (supplementary Fig. IV).
Correlation of relative amounts of individual sulfatides as determined by tissue MALDI IMS and MALDI on-target or LC-MS2 from extracts
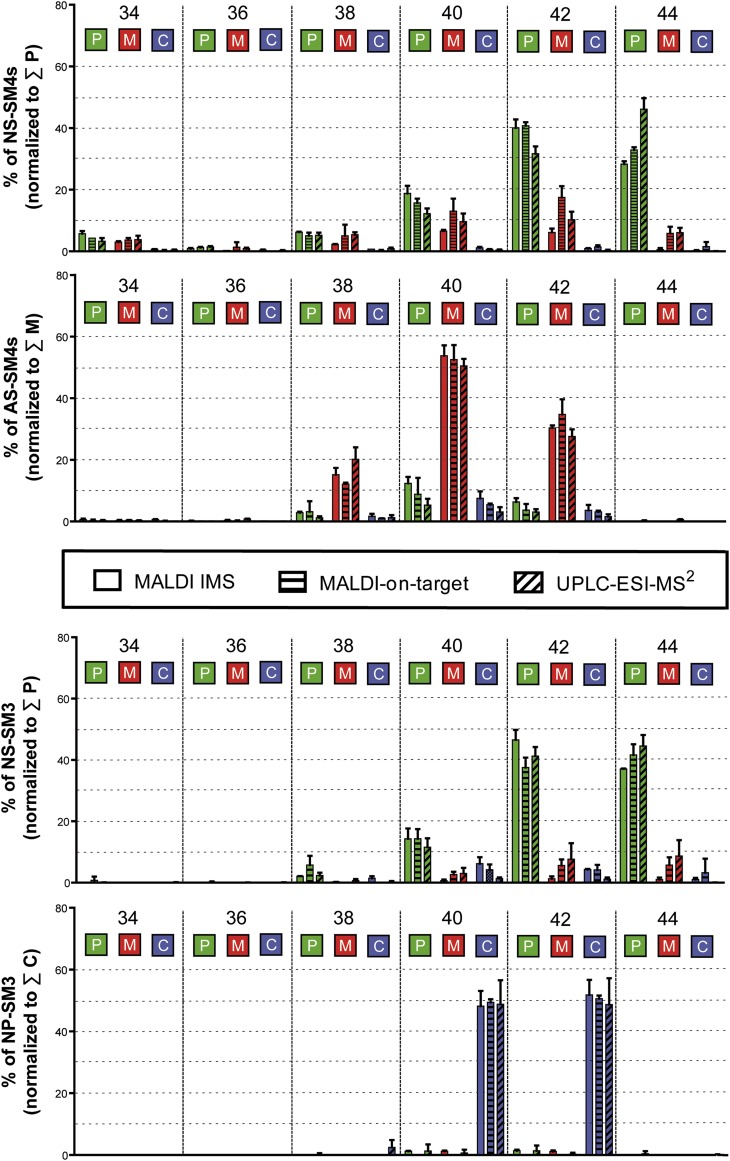
To obtain relative concentrations of sulfatides to each other by tissue MALDI IMS, the overall spectra from the MALDI IMS dataset were grouped into three ROIs, i.e., cortex, medulla, and papillae, and the average intensity per area for each compound and region was determined. To compare these results with the other two MS methods, MALDI on-target MS and UPLC-ESI-MS2, renal cortex, medulla, and papillae were dissected carefully with a scalpel followed by lipid extraction and quantification of the sulfatide compositions using internal sulfatide standards.
To compare the relative amounts obtained by MALDI IMS with the absolute amounts obtained by the other two MS methods, data of each method were normalized to the sum of values obtained within the kidney region, where the corresponding sulfatide subclass was most abundant; i.e., NS-SM4 and NS-SM3 were normalized to the sum of their signals in the papillae, AS-SM4s to the sum of their signals in the medulla, and ceramide anchor with a nonhydroxy acyl chain and a phytosphingosine (NP)-SM3 to the sum of their signals in the cortex (Fig. 3). As a result, all three methods reflect the abundance of very long chain (C40–C44) over long chain (C34–C38) sulfatides, as already suggested by the IMS pictures (Fig. 2). All three methods mirror the accumulation of nonhydroxylated very long chain sulfatides (NS-SM4 and NS-SM3) in the papillae, while hydroxylated sulfatides, AS-SM4, are highly expressed in medulla and nonhydroxylated, but phytosphingosine-containing sulfatides, NP-SM3s, are restricted to the cortical region (Fig. 3).
Fig. 3.
Comparison of relative sulfatide levels obtained by MALDI IMS, MALDI on-target MS, and UPLC-ESI-MS2 for papillae (P), medulla (M), and cortex (C) of mouse kidneys. Relative concentrations of individual sulfatides were normalized to the sum of all members of the corresponding sulfatide subgroup within papillae for NS-sulfatides, medulla for AS-sulfatides, or cortex for NP-sulfatides, which was set to 100% (n = 3).
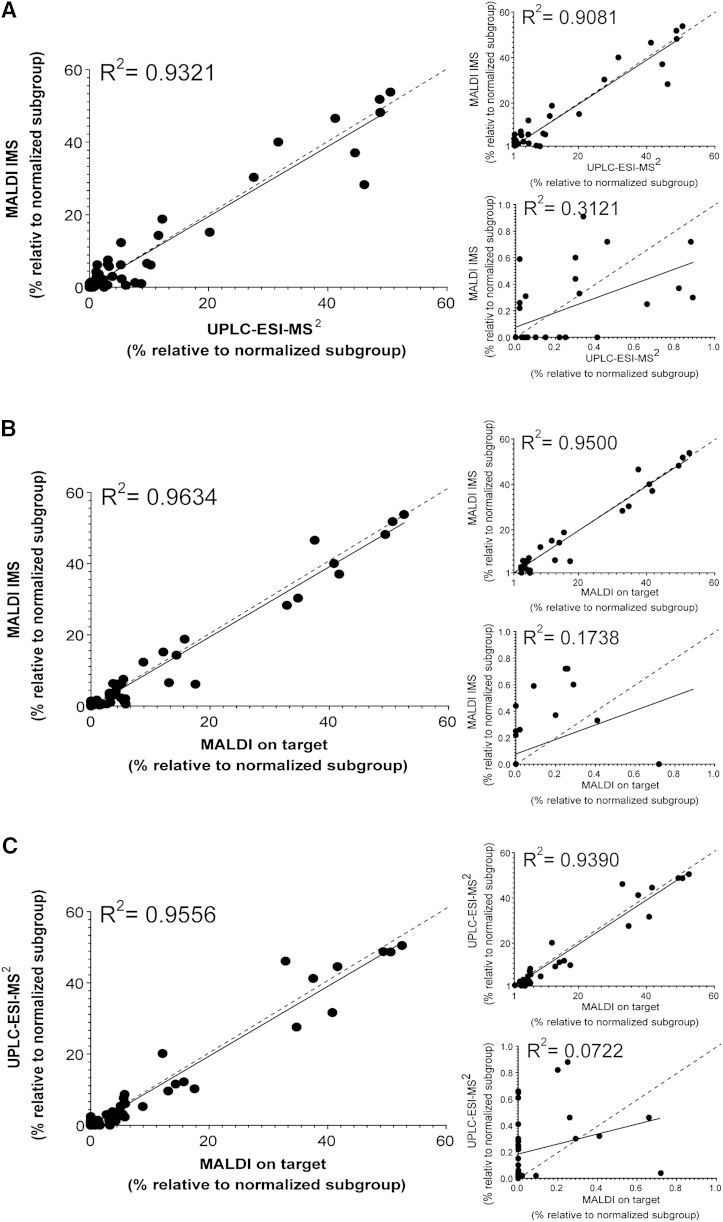
The overall relative data obtained by MALDI IMS correlate nicely with the corresponding UPLC-ESI-MS2 (R2 = 0.93, Fig. 4A) and MALDI on-target (R2 = 0.96, Fig. 4B) data. These correlations are similar to the one obtained between the data sets of MALDI on-target and UPLC-ESI-MS2 (R2 = 0.96, Fig. 4C). In general, the less abundant compounds of each group (<10%) varied more between methods than the more abundant group members (see small graphs for partial data sets in Fig. 4A–C).
Fig. 4.
Statistical correlation of the relative quantity of renal sulfatides (see Fig. 3) in between the mass spectrometric methods. Correlation of MALDI IMS and UPLC-ESI-MS2 (A), MALDI IMS and MALDI on-target (B), and UPLC-ESI-MS2 and MALDI on-target (C) data. A correlation of R2 > 0.9 was considered trustful. Linear regression (continuous line) and hypothetical linear regression of R2 = 1 (dotted line) are indicated for each dataset. The assignment of each point is shown in detail in the supplementary section (supplementary Figs. V–VII, supplementary Table V) (n = 3 for each, MALDI IMS, UPLC-ESI MS2, and MALDI on-target). Note, although the low intensity values alone do not correlate between the methods [lower small graph in (A–C)], they support the correlation of the more abundant values [upper small graph in (A–C)], which results in a better R2 value when all values are taken into account [main graph of (A–C)].
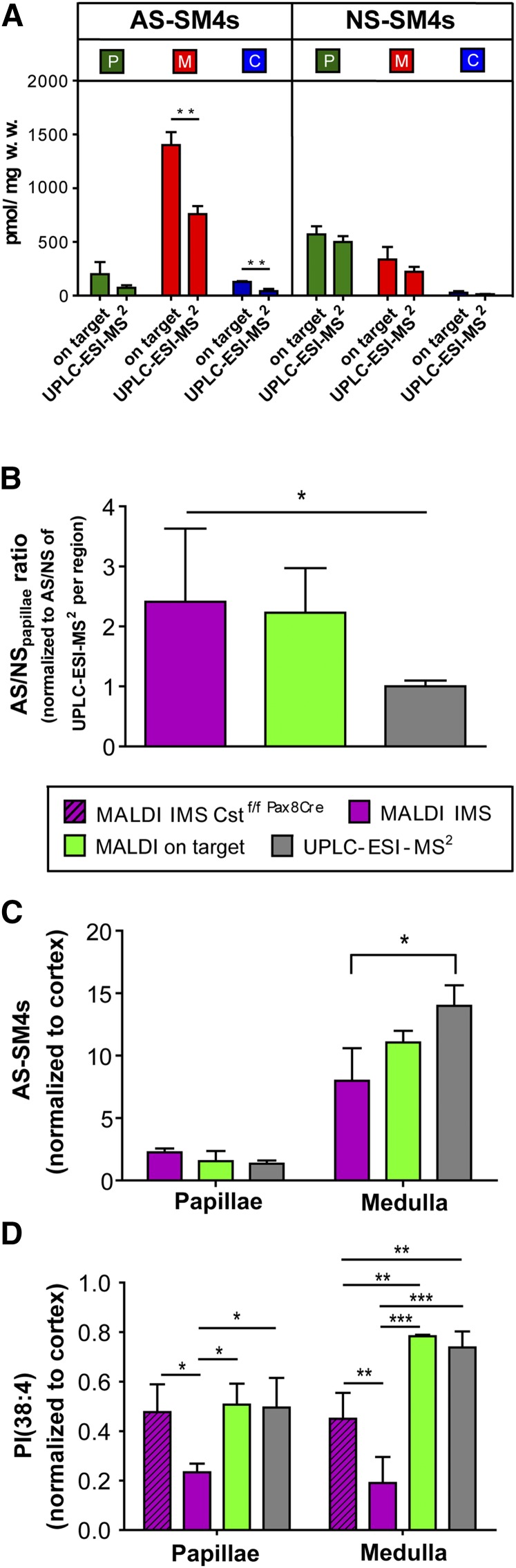
A quantitative comparison of AS-sulfatides with NS-sulfatides turned out to be more difficult, as significantly different results were obtained between MALDI and ESI methods. Using NS-SM4s internal standards for quantification, MALDI on-target MS delivered twice as high concentrations of AS-sulfatides as compared with analysis by UPLC-ESI-MS2, independent of the renal region investigated (Fig. 5A). Likewise, MALDI IMS analysis resulted in twice as high a ratio of AS- over NS-sulfatide signals, when compared with UPLC-ESI-MS2 data (Fig. 5B).
Fig. 5.
Variances in detection sensitivity arise from structural differences and regional total ion intensities. A, B: Structural differences influence detection sensitivity. A: Absolute quantities of NS- and AS-SM4s sulfatides determined in the different renal locations by MALDI on-target and UPLC-ESI-MS2 with the help of an internal NS-sulfatide standard. AS-SM4s values obtained with MALDI on-target are significantly higher than those determined with the UPLC-ESI-MS2 method. B: Relative ratio of AS- over NS-sulfatides as determined for all three methods. Intensities obtained for AS-sulfatides in each region were divided by those obtained for NS-sulfatides within the papillae. Then all AS/NS ratios were normalized to those obtained by UPLC-ESI-MS2 for each region, respectively. Note, both MALDI methods result in AS/NS levels that are twice as high as the ESI method. C, D: Regional differences in sensitivity. Values obtained for AS-sulfatides or for PI(38:4) in papillae (P) and medulla (M) were normalized to corresponding values obtained in the cortical (C) region, which contained the lowest total ion concentrations due to low sulfatide levels. The graphs reflect the relative regional intensities of AS-sulfatides (C) and PI, PI(38:4) (D), as obtained by the different MS techniques. With MALDI IMS, significantly lower values for PI were obtained in the sulfatide rich papillae and medulla as well as significant lower values for AS-sulfatides in the medulla as compared to UPLC-ESI-MS2. Note, this effect reversed when PI was analyzed in sulfatide-deprived kidney, having only residual AS-sulfatides in the medulla. (MALDI IMS Cstf/f Pax8Cre, n = 3; MALDI IMS, n = 3; UPLC-ESI MS2, n = 3; MALDI on-target, n = 3; *P < 0.05; **P < 0.01; ***P < 0.001).
The total average negative ion concentration turned out to be highest in the medulla, followed by the papillae, and was the lowest in the cortical region. Hence, it may be assumed that quenching effects due to competition of compounds for charge and detection are strongest in the medulla and are comparatively negligible in the cortex. Therefore, we normalized ion intensities determined in the papillae and the medulla to the average intensities measured in the cortex for each compound class and method. Indeed, as compared with UPLC-ESI-MS2, significantly fewer AS-sulfatide intensities were detected in the medulla by MALDI IMS (Fig. 5C). Likewise, 2- to 4-fold less phosphatidylinositol (PI) was measured in the papillae and medulla by IMS, as compared with UPLC-ESI-MS2 (Fig. 5D). As most of the ion counts were due to sulfatide signals, we compared these data with IMS data obtained from the kidneys of mice lacking sulfatides in the renal tubular epithelia. In line with our hypothesis, relative intensities for PI increased in mutant papillae to values similar to those obtained by the ESI method for wt kidneys. In mutant medullas, IMS values more than doubled, but reached only about two-thirds of the level determined by the ESI method for wt kidneys. However, it may be noted that mutant kidneys still contained residual AS-sulfatide levels in the medulla (Fig. 2, supplementary Fig. III). In order to exclude an influence of different osmolarities and ion compositions between renal regions and between wt and mutant mice, we repeated the IMS measurement with and without a washing procedure (40) prior to matrix deposition. However, the decreased PI signals in the medulla and papillae of wt as compared with mutant kidneys, remained after washing the sections with aqueous solutions, suggesting that ion suppression depended on sulfatides (supplementary Fig. VIII).
DISCUSSION
As described previously, negative ion mode analysis of mouse kidneys by MALDI IMS using 9-AA as matrix reflected, predominantly, signals of sulfated lipids. Besides these compounds, only a few signals corresponding to PI were observed. When recording IMS spectra in MS1 mode three questions came up: 1) Are the recorded signals specific for the compound attributed to them? 2) Does the ratio of signal intensities for two similar compounds reflect their ratio of concentrations within the same tissue spot? 3) Do signal intensities obtained at different locations directly reflect local relative compound concentrations?
The problem of signal specificity may be addressed in three ways: 1) use of a mass spectrometer with high accurate mass and high mass resolution, unless stereoisomers have to be differentiated; 2) use of tandem (or even MS3) MS screening for compound-specific fragments (here, drawbacks are strongly reduced sensitivity and eventually loss of information for other compounds); and 3) parallel analysis of a negative control. Because the mass resolution of the instrument used here was not sufficient for compound identification and because the MS2 mode was limited by a large precursor ion selector window and did not allow simultaneous screening of more than one compound by IMS, we decided to prove signal specificity with a negative control. For this, we took kidneys of mutant mice deficient in renal tubular epithelium sulfatides and compared them with wt sections on a single indium tin oxide-coated glass slide within one run. When such tissue is available, it is a perfect negative control independent of the m/z-resolution and potential tandem MS function of the IMS instrument used. The results demonstrate almost complete loss of sulfatide signals on mutant tissue and, hence, signal specificity. The residual medullar signals could have originated from other negatively charged compounds with the same nominal mass. However, residual amounts of sulfatides were also detected by UPLC-ESI-MS2 in the mutant kidneys (see supplementary Fig. III). Hence, it was rather likely that the residual IMS signals also reflected medullary hydroxyl-sulfatides originating from cells that simply did not respond to the PAX 8-driven Cre-recombinase. Pax8 drives expression in proximal tubular epithelial cells and expression of sulfatides in interstitial cells of the medulla would not be affected. Indeed, complete loss of sulfatides was analyzed in a mouse kidney with a systemic Cst deficiency (supplementary Fig. IV). These data are supported by an immunohistochemical study, which shows expression of sulfatides by interstitial medullary cells in rat kidneys (31).
Having validated signal identities, we turned to the quantitative aspects of IMS signal intensities. Quantitative data are quintessential for correct conclusions on pathway analysis, regulation, and pathological changes. In order to compare MALDI IMS data with those of accepted quantitative MS methods, IMS data had to be normalized. Besides the option of not normalizing data at all, the flexImaging software offers four normalization possibilities for IMS data: normalization of individual signals to 1) the total ion current within each spectrum; 2) the median ion intensity of all peaks; 3) the root mean square of all peaks; and 4) the total ion current of a freely chosen ion mass (or mass window) (38). As mentioned in the introduction, normalization methods 1 to 3 are prone to false positive detection in heterogeneous tissue. Because there was also no ion mass (matrix cluster, housekeeping molecule) within the chosen mass range, which could be trusted to be equally distributed across the tissue, we could also not apply normalization method 4. So we did not normalize our data with a software algorithm, but set the color code of intensity for all sulfatide signals to the same absolute maximal signal counts. However, no straightforward software tool was able to directly identify the spectrum with the highest intensity of a defined ion mass. Nevertheless, for closely related sulfatides, relative regional values were obtained, which were comparable (especially for more abundant ions) to data determined by the quantitative MS methods. Hence, quantitative comparison of closely related compounds can be performed directly from IMS data. However, subtle structural changes, like an additional α-hydroxyl group in sulfatides, may already lead to a distortion of the real relative concentrations, when directly comparing the intensities of such compounds with nonhydroxylated compounds in a defined location of the section.
Furthermore, we observed quenching effects with IMS. especially in regions of high total ion intensities. In the kidney, these are due to high sulfatide content in the medulla followed by the papillae. We exemplified the concomitant quenching effects for AS-sulfatides within the medulla and for the most prominent PI, PI(38:4), within the medulla and papillae. Compared with sulfatides (pKasulfatide ≈ −1.8), PIs (pKaPI = 2.5) are less acidic and, therefore, are more prone to quenching effects when competing for deprotonation. This may explain the stronger reduction of PI signals (26 and 47% of the UPLC-ESI-MS2 signal in the medulla and papillae, respectively) in regions of high total ion current as compared with AS-sulfatide signals (57% or no difference to the UPLC-ESI-MS2 signals in the medulla and papillae, respectively). The fact that PI signals almost reached the expected values in the papillae and medulla, when the kidneys strongly deprived of sulfatides were analyzed, proved sulfatides to quench the IMS signals for PI strongly in these regions. These quenching effects were not observed in MALDI on-target analysis, which may be due to the enrichment of lipids and the dilution of these compounds by on-target spotting. Even more importantly, the ratio of matrix to analyte drops from on-target to IMS analysis, and preferred matrix analyte interactions may lead to preferential cocrystalization of analytes (here AS-sulfatides), by that suppressing intensities of other compounds (here PI). These effects complicate the interpretation of IMS data: a direct conversion of signal intensities into regionally different concentrations may lead to severe misinterpretation. Differing with up to a factor of 4 for PI in the medulla from the expectations of the standard method (LC-ESI-MS), our data are in contrast to publications reporting variations of less than 30% from the expected values. For example, Takai et al. (41) had a recovery rate of 76–127% for a therapeutic peptide in liver and kidney. Furthermore, they published a significant correlation coefficient of R = 0.94 for IMS-based signal intensity of raclopride in whole-body sections compared with the concentration of the drug in the tissue samples of six different organs measured by LC-MS2 (42).
In summary, IMS is an invaluable tool to analyze the lipid distribution within tissue sections, primarily because no other tools are able to differentiate the lipid anchor moieties while addressing location. Our data demonstrate that the technique allows comparison of chemically similar compounds within defined regions. However, our results also point out that signal intensities may not directly correlate with analyte concentrations from region to region, especially when strong differences in total ion current occur. Therefore, we think it is still mandatory to confirm IMS results subsequently by other techniques, if possible.
Supplementary Material
Acknowledgments
The authors thank Benita von Tümpling-Radosta and Ulrike Rothermel, German Cancer Research Center (DKFZ) Heidelberg, for excellent technical assistance, as well as Björn Meyer, University of Applied Sciences Mannheim, for critical comments.
Footnotes
Abbreviations:
- 9-AA
- 9-aminoacridine
- AS
- ceramide anchor with an alpha-hydroxy acyl chain and a sphingosine
- Cst
- cerebroside sulfotransferase
- Cstf/f Pax8 Cre
- specific knockout of cerebroside sulfotransferase in renal tubular epithelial cells
- IMS
- imaging MS
- NP
- ceramide anchor with a nonhydroxy acyl chain and a phytosphingosine
- NS
- ceramide anchor with a nonhydroxy acyl chain and a sphingosine
- PI
- phosphatidylinositol
- ROI
- region of interest
- SM3
- lactosylceramide II3-sulfate
- SM4s
- galactosylceramide I3-sulfate
- UPLC
- ultra-performance LC
- wt
- wild-type
These acyl chain lengths are derived from the molecular ion weight when assuming the most prominent sphingoid base, C18-sphingosine, as part of the structure. Our recent ESI-MS2 data of the precursors ceramide and galactosylceramide support the abundant presence of C18-sphingosine in total renal extracts (43).
This work was supported by allocations to R.S. and H.-J.G. within a joint grant (Zentren für Angewandte Forschung an Hochschulen: Applied Biomedical Mass Spectrometry, “ZAFH ABIMAS”) from ZO IV by the Landesstiftung Baden-Württemberg and the Europäischer Fonds für regionale Entwicklung (EFRE) to C.H.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of text, eight figures, and five tables.
REFERENCES
- 1.Schober Y., Guenther S., Spengler B., Rompp A. 2012. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Chem. 84: 6293–6297. [DOI] [PubMed] [Google Scholar]
- 2.Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. 2009. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J. Lipid Res. 50: 2290–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fülöp A., Porada M. B., Marsching C., Blott H., Meyer B., Tambe S., Sandhoff R., Junker H. D., Hopf C. 2013. 4-Phenyl-α-cyanocinnamic acid amide: screening for a negative ion matrix for MALDI-MS imaging of multiple lipid classes. Anal. Chem. 85: 9156–9163. [DOI] [PubMed] [Google Scholar]
- 4.Le C. H., Han J., Borchers C. H. 2012. Dithranol as a MALDI matrix for tissue imaging of lipids by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 84: 8391–8398. [DOI] [PubMed] [Google Scholar]
- 5.Marsching C., Eckhardt M., Grone H. J., Sandhoff R., Hopf C. 2011. Imaging of complex sulfatides SM3 and SB1a in mouse kidney using MALDI-TOF/TOF mass spectrometry. Anal. Bioanal. Chem. 401: 53–64. [DOI] [PubMed] [Google Scholar]
- 6.Meriaux C., Franck J., Wisztorski M., Salzet M., Fournier I. 2010. Liquid ionic matrixes for MALDI mass spectrometry imaging of lipids. J. Proteomics. 73: 1204–1218. [DOI] [PubMed] [Google Scholar]
- 7.Thomas A., Charbonneau J. L., Fournaise E., Chaurand P. 2012. Sublimation of new matrix candidates for high spatial resolution imaging mass spectrometry of lipids: enhanced information in both positive and negative polarities after 1,5-diaminonapthalene deposition. Anal. Chem. 84: 2048–2054. [DOI] [PubMed] [Google Scholar]
- 8.Hankin J. A., Barkley R. M., Murphy R. C. 2007. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18: 1646–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aerni H. R., Cornett D. S., Caprioli R. M. 2006. Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 78: 827–834. [DOI] [PubMed] [Google Scholar]
- 10.Grove K. J., Frappier S. L., Caprioli R. M. 2011. Matrix pre-coated MALDI MS targets for small molecule imaging in tissues. J. Am. Soc. Mass Spectrom. 22: 192–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y., Liu Y., Allegood J., Wang E., Cachon-Gonzalez B., Cox T. M., Merrill A. H., Jr, Sullards M. C. 2010. Imaging MALDI mass spectrometry of sphingolipids using an oscillating capillary nebulizer matrix application system. Methods Mol. Biol. 656: 131–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouschen W., Schulz O., Eikel D., Spengler B. 2010. Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun. Mass Spectrom. 24: 355–364. [DOI] [PubMed] [Google Scholar]
- 13.Yang J., Caprioli R. M. 2011. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 83: 5728–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuerenberg M., Luebbert C., Deininger S. O., Ketterlinus R., Suckau D. 2007. MALDI tissue imaging: mass spectrometric localization of biomarkers in tissue slices. Nat. Methods. 4: iii–iv. [Google Scholar]
- 15.Luxembourg S. L., McDonnell L. A., Duursma M. C., Guo X., Heeren R. M. 2003. Effect of local matrix crystal variations in matrix-assisted ionization techniques for mass spectrometry. Anal. Chem. 75: 2333–2341. [DOI] [PubMed] [Google Scholar]
- 16.Pirman D. A., Kiss A., Heeren R. M., Yost R. A. 2013. Identifying tissue-specific signal variation in MALDI mass spectrometric imaging by use of an internal standard. Anal. Chem. 85: 1090–1096. [DOI] [PubMed] [Google Scholar]
- 17.Cheng H., Sun G., Yang K., Gross R. W., Han X. 2010. Selective desorption/ionization of sulfatides by MALDI-MS facilitated using 9-aminoacridine as matrix. J. Lipid Res. 51: 1599–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pirman D. A., Reich R. F., Kiss A., Heeren R. M., Yost R. A. 2013. Quantitative MALDI tandem mass spectrometric imaging of cocaine from brain tissue with a deuterated internal standard. Anal. Chem. 85: 1081–1089. [DOI] [PubMed] [Google Scholar]
- 19.Heeren R. M. A., Kukrer-Kaletas B., Taban I. M., MacAleese L., McDonnell L. A. 2008. Quality of surface: the influence of sample preparation on MS-based biomolecular tissue imaging with MALDI-MS and (ME-)SIMS. Appl. Surf. Sci. 255: 1289–1297. [Google Scholar]
- 20.Hankin J. A., Murphy R. C. 2010. Relationship between MALDI IMS intensity and measured quantity of selected phospholipids in rat brain sections. Anal. Chem. 82: 8476–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koeniger S. L., Talaty N., Luo Y., Ready D., Voorbach M., Seifert T., Cepa S., Fagerland J. A., Bouska J., Buck W., et al. 2011. A quantitation method for mass spectrometry imaging. Rapid Commun. Mass Spectrom. 25: 503–510. [DOI] [PubMed] [Google Scholar]
- 22.Shroff R., Svatos A. 2009. Proton sponge: a novel and versatile MALDI matrix for the analysis of metabolites using mass spectrometry. Anal. Chem. 81: 7954–7959. [DOI] [PubMed] [Google Scholar]
- 23.Pernber Z., Richter K., Mansson J. E., Nygren H. 2007. Sulfatide with different fatty acids has unique distributions in cerebellum as imaged by time-of-flight secondary ion mass spectrometry (TOF-SIMS). Biochim. Biophys. Acta. 1771: 202–209. [DOI] [PubMed] [Google Scholar]
- 24.Ageta H., Asai S., Sugiura Y., Goto-Inoue N., Zaima N., Setou M. 2009. Layer-specific sulfatide localization in rat hippocampus middle molecular layer is revealed by nanoparticle-assisted laser desorption/ionization imaging mass spectrometry. Med. Mol. Morphol. 42: 16–23. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y., Chen Y., Momin A., Shaner R., Wang E., Bowen N. J., Matyunina L. V., Walker L. D., McDonald J. F., Sullards M. C., et al. 2010. Elevation of sulfatides in ovarian cancer: an integrated transcriptomic and lipidomic analysis including tissue-imaging mass spectrometry. Mol. Cancer. 9: 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuki D., Sugiura Y., Zaima N., Akatsu H., Hashizume Y., Yamamoto T., Fujiwara M., Sugiyama K., Setou M. 2011. Hydroxylated and non-hydroxylated sulfatide are distinctly distributed in the human cerebral cortex. Neuroscience. 193: 44–53. [DOI] [PubMed] [Google Scholar]
- 27.Onishi S., Tatsumi Y., Wada K., Yang H. J., Sugiura Y., Setou M., Yoshikawa H. 2013. Sulfatide accumulation in the dystrophic terminals of gracile axonal dystrophy mice: lipid analysis using matrix-assisted laser desorption/ionization imaging mass spectrometry. Med. Mol. Morphol. 46: 160–165. [DOI] [PubMed] [Google Scholar]
- 28.Kadar H., Pham H., Touboul D., Brunelle A., Baud O. 2014. Impact of inhaled nitric oxide on the sulfatide profile of neonatal rat brain studied by TOF-SIMS imaging. Int. J. Mol. Sci. 15: 5233–5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishizuka I. 1997. Chemistry and functional distribution of sulfoglycolipids. Prog. Lipid Res. 36: 245–319. [DOI] [PubMed] [Google Scholar]
- 30.Sandhoff R., Hepbildikler S. T., Jennemann R., Geyer R., Gieselmann V., Proia R. L., Wiegandt H., Grone H. J. 2002. Kidney sulfatides in mouse models of inherited glycosphingolipid disorders: determination by nano-electrospray ionization tandem mass spectrometry. J. Biol. Chem. 277: 20386–20398. [DOI] [PubMed] [Google Scholar]
- 31.Trick D., Decker J., Groene H. J., Schulze M., Wiegandt H. 1999. Regional expression of sulfatides in rat kidney: immunohistochemical staining by use of monospecific polyclonal antibodies. Histochem. Cell Biol. 111: 143–151. [DOI] [PubMed] [Google Scholar]
- 32.Nagai K., Tadano-Aritomi K., Iida-Tanaka N., Yoshizawa H., Ishizuka I. 2005. Metabolism of sulfolipids in isolated renal tubules from rat. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 140: 487–495. [DOI] [PubMed] [Google Scholar]
- 33.Kikkawa Y., Mimura A., Inage Z. 1991. Regional distribution of sulfatide in human kidney, and anti-sulfatide antibodies in sera from patients with nephritis detected by TLC immunostaining [article in Japanese]. Nippon Jinzo Gakkai Shi. 33: 635–642. [PubMed] [Google Scholar]
- 34.Stettner P., Bourgeois S., Marsching C., Traykova-Brauch M., Porubsky S., Nordström V., Hopf C., Koesters R., Sandhoff R., Wiegandt H., et al. 2013. Sulfatides are required for renal adaptation to chronic metabolic acidosis. Proc. Natl. Acad. Sci. USA. 110: 9998–10003. [Erratum. 2013. Proc. Natl. Acad. Sci. USA. 110: 14813.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Folch J., Lees M., Sloane-Stanley G. H. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 36.Zhang X., Nakajima T., Kamijo Y., Li G., Hu R., Kannagi R., Kyogashima M., Aoyama T., Hara A. 2009. Acute kidney injury induced by protein-overload nephropathy down-regulates gene expression of hepatic cerebroside sulfotransferase in mice, resulting in reduction of liver and serum sulfatides. Biochem. Biophys. Res. Commun. 390: 1382–1388. [DOI] [PubMed] [Google Scholar]
- 37.Hsu F. F., Turk J. 2004. Studies on sulfatides by quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes that include an unusual internal galactose residue loss and the classical charge-remote fragmentation. J. Am. Soc. Mass Spectrom. 15: 536–546. [DOI] [PubMed] [Google Scholar]
- 38.Deininger S. O., Cornett D. S., Paape R., Becker M., Pineau C., Rauser S., Walch A., Wolski E. 2011. Normalization in MALDI-TOF imaging datasets of proteins: practical considerations. Anal. Bioanal. Chem. 401: 167–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tadano-Aritomi K., Hikita T., Fujimoto H., Suzuki K., Motegi K., Ishizuka I. 2000. Kidney lipids in galactosylceramide synthase-deficient mice. Absence of galactosylsulfatide and compensatory increase in more polar sulfoglycolipids. J. Lipid Res. 41: 1237–1243. [PubMed] [Google Scholar]
- 40.Angel P. M., Spraggins J. M., Baldwin H. S., Caprioli R. 2012. Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem. 84: 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takai N., Tanaka Y., Watanabe A., Saji H. 2013. Quantitative imaging of a therapeutic peptide in biological tissue sections by MALDI MS. Bioanalysis. 5: 603–612. [DOI] [PubMed] [Google Scholar]
- 42.Takai N., Tanaka Y., Inazawa K., Saji H. 2012. Quantitative analysis of pharmaceutical drug distribution in multiple organs by imaging mass spectrometry. Rapid Commun. Mass Spectrom. 26: 1549–1556. [DOI] [PubMed] [Google Scholar]
- 43.Imgrund S., Hartmann D., Farwanah H., Eckhardt M., Sandhoff R., Degen J., Gieselmann V., Sandhoff K., Willecke K. 2009. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 284: 33549–33560. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.