Abstract
Mammalian kidneys are rich in sulfatides. Papillary sulfatides, especially, contribute to renal adaptation to chronic metabolic acidosis. Due to differences in their ceramide (Cer) anchors, the structural diversity of renal sulfatides is large. However, the underling biological function of this complexity is not understood. As a compound’s function and its tissue location are intimately connected, we analyzed individual renal sulfatide distributions of control and Cer synthase 2 (CerS)2-deficient mice by imaging MS (IMS) and by LC-MS2 (in controls for the cortex, medulla, and papillae separately). To explain locally different structures, we compared our lipid data with regional mRNA levels of corresponding anabolic enzymes. The combination of IMS and in source decay-LC-MS2 analyses revealed exclusive expression of C20-sphingosine-containing sulfatides within the renal papillae, whereas conventional C18-sphingosine-containing compounds were predominant in the medulla, and sulfatides with a C18-phytosphingosine were restricted to special cortical structures. CerS2 deletion resulted in bulk loss of sulfatides with C23/C24-acyl chains, but did not lead to decreased urinary pH, as previously observed in sulfatide-depleted kidneys. The reasons may be the almost unchanged C22-sulfatide levels and constant total renal sulfatide levels due to compensation with C16- to C20-acyl chain-containing compounds. Intriguingly, CerS2-deficient kidneys were completely depleted of phytosphingosine-containing cortical sulfatides without any compensation.
Keywords: ceramide synthase 2, imaging mass spectrometry, liquid chromatography, matrix-assisted laser desorption/ionization-time-of-flight, electrospray ionization-mass spectrometry, tandem mass spectrometry, galactosylceramide I3-sulfate, lactosylceramide II3-sulfate, cortex, medulla, papillae, sphingosine, serine palmitoyl-coenzyme A transferase small subunits, in source decay, knockout
Sulfatides, a class of amphiphilic anionic glycosphingolipids (GSLs), consist of a hydrophobic ceramide (Cer) membrane anchor and a polar sulfated carbohydrate head group. In the case of the prominent monosulfated forms like galactosylceramide I3-sulfate (SM4s) and lactosylceramide II3-sulfate (SM3), one sulfate ester is bound to the C3 atom of the terminal galactosyl residue. In contrast to the minor bis-sulfated form gangliotetraosylceramide-II3, IV3 bis-sulfate found in mouse kidney, SM4s as well as SM3 are present in both human and mouse kidneys (Fig. 1). Sulfatide SM4s is commonly known to stabilize the myelin sheaths of the central and peripheral nervous system. In glandular epithelia, sulfatide SM4s concentrations appear synchronized with Na+-K+-ATPase activities and are elevated upon NaCl loading (1–4). In the kidney, SM4s and SM3 are postulated to function as essential players in the transport of sodium chloride (5), but evidence is missing. Recently, analysis of mutant mice revealed an important function of sulfatides (SM4s and SM3) for the ammonia exchange process (6). A role in the process of osmotic balance, which had been postulated previously (1, 7), could be excluded in the recent study of mutant mice (6). With respect to their Cer anchor, sulfatides appear to be very diverse within the renal system.
Fig. 1.
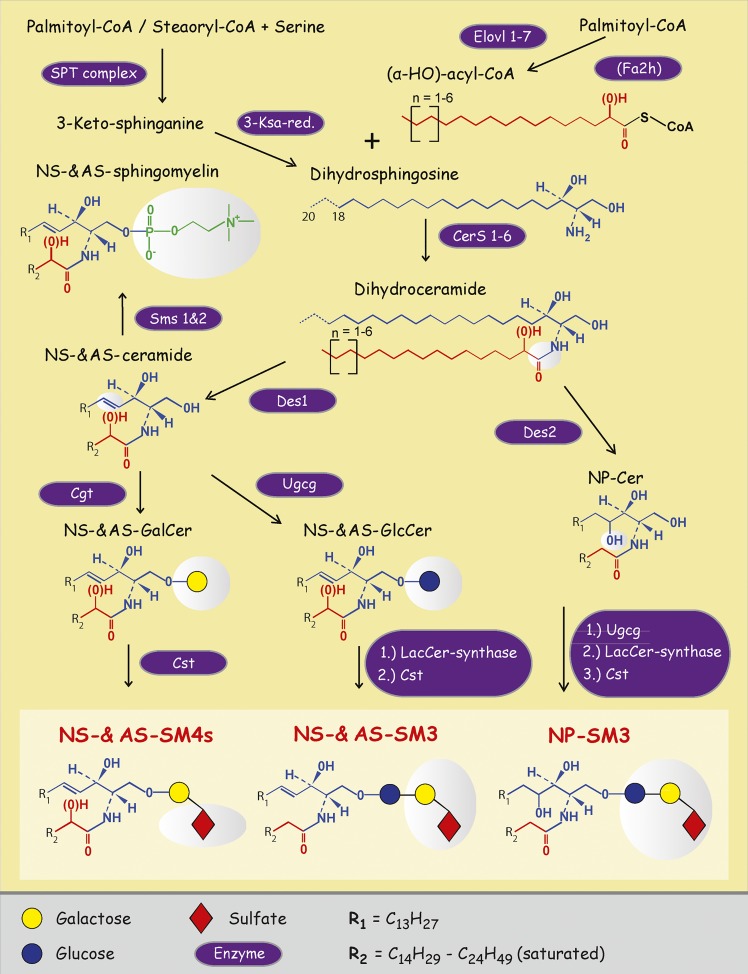
Anabolic pathway of renal sulfatides. SL synthesis starts out at the endoplasmic reticulum with the condensation of serine and a long chain acyl-CoA, in most cases palmitoyl-CoA. This is also the rate-limiting step, regulated by ORMDL proteins (ORM1-like proteins). After reduction of 3-keto-sphinganine to sphinganine, condensation with a variety of acyl-CoAs yields dihydroceramides, which are modified by dihydroceramid-Δ4-desaturase (Des)1 or Des2 to result in Cers or phyto-Cers, respectively, by that creating the huge diversity of Cer anchors found in renal sulfatides SM4s and SM3. Gray areas mark the structural changes in the molecules caused by the previous enzymatic step. 3-KSa-red., 3-ketosphinganine reductase; Sms 1 and 2, sphingomyelin synthase 1 and 2; LacCer-synthase, lactosylceramide synthase; Ugcg, UDP-glucose ceramide glucosyltransferase.
Modifications by α-hydroxylation of the acyl chain, as well as variations in its chain length from C16 up to C26, are commonly known. Next to the major sphingoid base C18-sphingosine (d18:1),3 C18-phytosphingosine (t18:0) also appears in SM3. As described in our previous work (6), hydroxylated forms [Cer anchor with an alpha-hydroxy acyl chain and a sphingosine (AS)] of SM4s are mainly located to the renal medulla while nonhydroxylated forms [Cer anchor with a nonhydroxy acyl chain and a sphingosine (NS)] show the highest concentration in the renal papillae. Until now, there was no systematic description of sulfatide location in the kidney with respect to the different fatty acid acyl chain and sphingoid base lengths. On one hand, the length of the acyl moiety depends on acyl-CoA availability as substrates for sphingolipid (SL) synthesis and, on the other hand, on the substrate specificity of the individual Cer synthases (CerSs). Seven members of mammalian elongases [elongation of very long chain fatty acids (Elovl)1–7] regulate differentially, in a cell type-specific manner, the elongation degree for saturated and polyunsaturated fatty acids, and by that determine the acyl-CoA pool available for SL synthesis (8). In mammals, six CerSs (CerS1–6) condense these acyl-CoAs with sphingoid bases to form (dihydro)ceramides. Whereas human and mouse CerS1, -5, and -6 seem to be restricted to a certain set of long chain acyl-CoAs (C14–C20), CerS2 and -4 accept very long acyl-CoAs (≥C22) up to 26 C-atoms in length (9–13), and CerS3, being rather promiscuous, further on can introduce ultra-long acyl-CoAs with up to 38 C-atoms in length (12). The length of the sphingoid base is guided by the serine palmitoyl-CoA transferase (SPT) complex (14–16) and impacts the final Cer anchor patterns observed. Hence, the acyl chain- and sphingoid base length-dependent localization of SLs to different parts of an organ should correlate with the regional specific expressions of elongases, CerS, and the SPT complex composition. Indeed, CerS2, for example, is expressed in myelin-forming cells where sulfatides with very long Cer anchors are found, but not in neurons which synthesize gangliosides with long chain Cer anchors (17). On the other hand, Cers3 is expressed in differentiating epidermis and spermatocytes, where unique ultra-long chain acyl moieties are found in respective SLs (10, 12, 18–20). However, the majority of ultra-long chain acyl moieties found in the epidermis are of saturated and monounsaturated nature (21), whereas in spermatocytes these ultra-long acyl chains are polyunsaturated with up to six double bonds (22, 23). One reason for this is differential expressions of Elovl enzymes, Elovl1 and -4 in the epidermis and Elovl5 and -2 in spermatocytes (20, 21, 24–27). Recently, it was demonstrated that the small subunits of the SPT guide the selectivity of the SPT complex for the acyl-CoA. In this context, a single amino acid residue, Met25 in ssSPTa and Val25 in ssSPTb, was identified, which confers specificity for palmitoyl- or stearoyl-CoA (15). Here we demonstrate the abundant and selective expression of C20-sphingosine-containing sulfatides/SLs, which correlates with mRNA expression of ssSPTb in the papillae.
MATERIALS AND METHODS
Chemicals and reagents
Chemicals and solvents (acetonitrile, chloroform, methanol) used for lipid extraction and preparation of solutions and samples were generally obtained from Merck (Darmstadt, Germany) in the highest available purity. Sodium carbonate for lipid extraction was obtained from Fluka (part of Sigma-Aldrich, Taufkirchen, Germany). Either deionized water [Milli-Q, Millipore (part of Merck)], fully desalinated water (B. Braun AG, Melsungen, Germany), or water with HPLC-grade (Sigma-Aldrich) was used throughout this study. The 9-aminoacridine (9-AA) used as MALDI matrix was purchased from Merck. The semisynthetic GSLs, SM4s (d18:1;14:0, d18:1;19:0, and d18:1;27:0) and SM3 (d18:1;14:0, d18:1;19:0, and d18:1;27:0), used as internal standard, have been described earlier (28).
Animals
All animal procedures were performed in accordance with the guidelines for the care and use of laboratory animals and were approved by Department 35 of the Regierungspräsidium Karlsruhe. Female wild-type (wt) mice (C57BL/6N) were obtained from the German Cancer Research Center. Cers2−/− mice have been described before (29), and were backcrossed prior to this study to a C57BL/6N background of 99.25%.
Preparation of lipid extracts from kidneys of wt and mutant mice
Lipids were extracted according to a previously reported procedure (30), with slight modifications. Briefly, one frozen kidney of either 11- or 6-week-old (fe)male Black-6 (C57BL/6N) or Cers2−/− mice were homogenized for 2 min on ice in 2 ml methanol and 300 μl water with an Ultra Turrax T25 basic (IKA Labortechnik, Staufen, Germany) at 24,000 rpm in a 15 ml polypropylene vial. For further extraction, each homogenate was transferred into a glass vial and the polypropylene vial was rinsed two times with 500 μl methanol. Afterwards, 3 ml of chloroform were added to obtain a solvent mixture of chloroform:methanol:water (10:10:1, v:v:v). The extract was centrifuged for 10 min at 3,000 rpm, and the supernatant was collected in a separate glass vial. Extraction was completed by repetition of this procedure twice, once with 3 ml chloroform:methanol:water (10:10:1, v:v:v) and once with a 30:60:8 (v:v:v) mixture. Every extraction step included 2 min of sonication. The pooled extracts were dried under nitrogen stream (37°C). Finally, the extract was dissolved in 100 μl chloroform:methanol:water (10:10:1, v:v:v) per 100 mg kidney wet weight and stored at −20°C. For lipid extraction from separated renal regions (papillae, medulla, cortex), fresh mouse kidneys (C57BL/6N) were rapidly prepared after euthanization. Between 7.4 and 18.4 mg of each region were carefully separated under a light microscope. Lipids were extracted from homogenate as described above. The pooled extracts were dried under nitrogen stream (37°C). Finally, the extracts were dissolved in 100 μl chloroform:methanol:water (10:10:1, v:v:v) per 100 mg kidney wet weight and stored at −20°C.
MALDI imaging MS
Mouse kidneys from female (C57BL/6N) and Cers2−/− mice were frozen without prior perfusion and sliced into 10 μm sections using a Leica CM1510S cryostat (Leica Biosystems, Nussloch, Germany) at −20°C. To prevent interference of embedding medium with mass spectrometric analysis, the frozen kidneys were fixed on the carrier by carefully dipping only one side of the organ in the embedding medium. By doing that, the side from which sections were obtained was free of embedding medium. Tissue sections were mounted onto indium tin oxide-coated conductive glass slides (Bruker Daltonics), dried by vacuum desiccation without prior washing steps, and used immediately or stored at −80°C. The 9-AA [20 mM in acetonitrile:water (80:20, v:v)] was deposited on slides using an ImagePrep matrix sprayer (Bruker Daltonics). Mass spectrometric measurements were performed using an Autoflex III MALDI TOF/TOF instrument (Bruker Daltonics) equipped with a Smartbeam laser (200 Hz) and controlled by flexControl 3.4 software (Bruker Daltonics). The extraction voltage was 19 kV, and gated matrix suppression (<650 Da) was applied to prevent saturation of the detector with matrix ions. Mass spectra were obtained in negative ion-reflector mode in the m/z range from 700 to 1,200 Da using delayed extraction. Images were acquired at a spatial resolution of 50 μm with 200 laser shots per position. Spectra were saved and the images constructed using flexImaging 3.0 software (Bruker Daltonics). Mass filters were chosen with a width of 0.2 Da. In all analyses blood-derived lipids were not separately taken into account. However, serum sulfatide levels make up less than 0.2% of kidney sulfatide levels and may be negligible (31).
LC-MS2
Sulfatide analyses by ultra-performance (UP)LC-ESI-( triple-quadrupole)MS2 was carried out on an Aquity I system (Waters, Manchester, England) comprising an autosampler with cooled tray (14°C) coupled to a Xevo TQ-S mass spectrometer (Waters). The system was controlled by MassLynxs software v 4.1. Separation was performed on an Aquity UPLC bridged-ethylene-hybrid C18 1.7 μm column (2.1 × 50 mm) at 40°C. The flow rate was set to 500 μl/min. Elution solvent A consisted of methanol:water (95:5) and solvent B of isopropanol:water (99:1). The chromatographic conditions were as described in Table 1.
TABLE 1.
Gradient conditions for the separation of sulfatides on UPLC for negative electrospray ion mode detection
| Time (min) | Solvent A (%) | Solvent B (%) |
| Initial | 100 | 0 |
| 0.1 | 100 | 0 |
| 0.2 | 85 | 15 |
| 4.0 | 25 | 75 |
| 4.5 | 25 | 75 |
| 5.0 | 100 | 0 |
| 6.5 | 100 | 0 |
Injection volume was 10 μl. MS2 detection was performed with argon as collision gas at a flow rate of 0.15 ml/min. The source temperature was set to 150°C, while the desolvation temperature was set to 350°C. The spray was started by applying 2,500 V to the fixed capillary. Cone voltage was set to 50 V and the optimal collision energy for parent scan of 96.8 Da was 70 eV in negative ion mode. Quantification was performed by single reaction monitoring (SRM) (see supplementary Table I for the transition list). For quantification, nonendogenous species (d18:1,14:0; d18:1,19:0; d18:1,27:0) of SM4s and SM3 were used as internal standards (28). The response correction was performed as described in the first section (32), which is based on a previous mass spectrometric study of sulfatides (33). Retention times and corresponding correlation curves are plotted in supplementary Fig. I.
For the detection of neutral glycosphingolipids and sulfatide SM4s in the positive mode, the column was equilibrated with solvent A+, which consisted of methanol:water (95:5) with 1 mM ammonia formate and 0.05% formic acid. Compounds were eluted running a gradient with increasing percentage of solvent B+ [isopropanol:methanol (99:1) with 1 mM ammonia formate and 0.05% formic acid] as described in Table 2. A volume of 10 μl was injected per sample. MS2 detections were performed with argon as collision gas at a flow rate of 0.15 ml/min. The source temperature was set to 90°C, while the desolvation temperature was set to 250°C. The spray was started by applying 2.5 kV to the fixed capillary. The cone voltage was set to 50 V. The optimal collision energy for each transition is listed in Table 3.
TABLE 2.
Gradient conditions for the separation of neutral glycosphingolipids and sulfatides on UPLC for positive electrospray ion mode detection
| Time (min) | Solvent A+ (%) | Solvent B+ (%) |
| Initial | 100 | 0 |
| 0.1 | 100 | 0 |
| 0.2 | 92 | 8 |
| 5.0 | 10 | 90 |
| 6.0 | 10 | 90 |
| 6.25 | 100 | 0 |
| 6.5 | 100 | 0 |
TABLE 3.
ESI-( triple-quadrupole)-MS2 transitions and collision energies
| Lipid class | Transition | Collision Energy (eV) |
| SM4s sulfatides | [M – SO3 + H]+ → [Sph – 2H2O + H]+ | 44 |
| [M – H2SO4 + H]+ → [Sph – 2H2O + H]+ | ||
| [Sph – 2H2O + H]+: m/z 264 or m/z 292 | ||
| HexCers | [M + H]+ → [Sph – 2H2O + H]+ | 44 |
| [M – H2O + H]+ → [Sph – 2H2O + H]+ | ||
| [Sph – 2H2O + H]+: m/z 264 or m/z 292 | ||
| Cers | [M + H]+ → [Sph – 2H2O + H]+ | 24 |
| [M – H2O + H]+ → [Sph – 2H2O + H]+ | ||
| [Sph – 2H2O + H]+: m/z 264 or m/z 292 | ||
| Sphingomyelins | [M + H]+ → [H2O3PO(CH2)2N(CH3)3]+ | 35 |
Quantification was performed by SRM (see supplementary Table I for the transition list). For quantification, the following nonendogenous species were used as described before (34): sphingomyelin (d18:1;12, d18:1;17:0, and d18:1;31:0), Cer (d18:1:14:0, d18:1;19:0, d18:1;25:0, and d18:1;31:0), and hexosylceramide (HexCer) (d18:1;14, d18:1;19:0, d18:1;25:0, and d18:1;31:0)
RNA isolation and quantitative real-time PCR
Kidneys from 6-week-old wt (C57BL/6N) mice were rapidly dissected into papillae, cortex, and medulla. Total RNA was extracted using Trizol reagent (Life Technologies) followed by DNase digest with TURBO DNA-free kit (Ambion), both according to the manufacturers’ instructions. Subsequently, RNA integrity was assessed using the Agilent Bioanalyzer1000 (Agilent) prior to reverse transcription with SuperscriptII (Life Technologies). The resulting cDNA was diluted 1:10 in diethylpyrocarbonate-treated water and analyzed by SYBR-green-based quantitative real-time PCR. Expression values were normalized to Gapdh and relative mRNA expression was calculated according to Livak and Schmittgen (35). Primers and annealing temperatures are specified in Table 4.
TABLE 4.
Primers and annealing temperatures used for enhancing specifically each mRNA fragment
| Gene | Primer | Primer Sequence | Annealing Temperature (°C) |
| Cers1 | Forward | 5-TGA CTG GTC AGA TGC GTG A-3 | 60 |
| Reverse | 5-TCA GTG GCT TCT CGG CTT T-3 | ||
| Cers2 | Forward | 5-TCA TCA TCA CTC GGC TGG T-3 | 56 |
| Reverse | 5-AGC CAA AGA AGG CAG GGT A-3 | ||
| Cers3 | Forward | 5-ATC TCG AGC CCT TCT TCT CC-3 | 58 |
| Reverse | 5-CTG GAC GTT CTG CGT GAA T-3 | ||
| Cers4 | Forward | 5-TGC GCA TGC TCT ACA GTT TC-3 | 60 |
| Reverse | 5-CTC GAG CCA TCC CAT TCT T-3 | ||
| Cers5 | Forward | 5-TCC ATG CCA TCT GGT CCT A-3 | 56 |
| Reverse | 5-TGC TGC CAG AGA GGT TGT T-3 | ||
| Cers6 | Forward | 5-GGG TTG AAC TGC TTC TGG TC-3 | 56 |
| Reverse | 5-TTT CTT CCC TGG AGG CTC T-3 | ||
| Elovl1 | Forward | 5-GGT GGG GGA TAA AAA TTG CT-3 | 56 |
| Reverse | 5-CCA AGG GCA GAC AAT CCA TA-3 | ||
| Elovl2 | Forward | 5-GAC GCT GGT CAT CCT GTT CT-3 | 56 |
| Reverse | 5-GCT TTG GGG AAA CCA TTC TT-3 | ||
| Elovl3 | Forward | 5-TTT GCC ATC TAC ACG GAT GA-3 | 63 |
| Reverse | 5-CGT GTC TCC CAG TTC AAC AA-3 | ||
| Elovl4 | Forward | 5-TTT GGT GGA AGC GAT ACC TG-3 | 60 |
| Reverse | 5-ATG TCC GAG TGT AGA AGT TG-3 | ||
| Elovl5 | Forward | 5-CTC TCG GGT GGC TGT TCT T-3 | 56 |
| Reverse | 5-AGA GGC CCC TTT CTT GTT GT-3 | ||
| Elovl6 | Forward | 5-ACA ATG GAC CTG TCA GCA AA-3 | 56 |
| Reverse | 5-GTA CCA GTG CAG GAA GAT CAG T-3 | ||
| Elovl7 | Forward | 5-ATG GGA CCA GCC TAC CAG AA-3 | 63 |
| Reverse | 5-TTG CAG TCC TCC ATG AAG AA-3 | ||
| Fa2h | Forward | 5-TGA AGG CCC ACC ATG TCA AGC A-3 | 60 |
| Reverse | 5-TCC CAC AGT TTA GTG CTG ATG CCA-3 | ||
| Gcs | Forward | 5-TGG TCT TCT TCA TGT GCC ACT GCC-3 | 65 |
| Reverse | 5-GAT TCA CGG ATG AAC CAG GCC ACA-3 | ||
| Cgt | Forward | 5-GTA CAG GCA AAA GGC ATG GGG A-3 | 65 |
| Reverse | 5-AGC CCT CTG CCG ATA ACT GGG ATT-3 | ||
| Cst | Forward | 5-AAG CAG TTG GTG CTG AGG CCA T-3 | 60 |
| Reverse | 5-ACT GGA TTT CGG GCG TGA GCA T-3 | ||
| Sms1 | Forward | 5-ACG ATG GCC AAT CAG CAA GTG CT-3 | 60 |
| Reverse | 5-AAA TGG CCT GTA CCA CCA CAC CCT-3 | ||
| Sms2 | Forward | 5-ACA ACA CGG CTG TTT TGG TGG T-3 | 60 |
| Reverse | 5-AAC CAC CAA GCC CGA GAC AAG A-3 | ||
| Sptssa | Forward | 5-CCT GTG ATT GGT GAT TGG TG-3 | 60 |
| Reverse | 5-GCA TTT CAC AAC GCC TGT TA-3 | ||
| Sptssb | Forward | 5-CAA GCC TTG CAG CTT TTC TT-3 | 62 |
| Reverse | 5-TGC GTG CCA AAT ATC TTT CTT-3 | ||
| Sptlc1 | Forward | 5-CCA GAT GCC TCC GAA AAA TA-3 | 60 |
| Reverse | 5-GGT TCA CAT GAA CGC ACA TC-3 | ||
| Sptlc2 | Forward | 5-TTT TAG CCC TTG GCA TGT TC-3 | 61 |
| Reverse | 5-CAC TAT GCA GAC CAG GCT GA-3 | ||
| Sptlc3 | Forward | 5-TGG GTC AGG TCT TTT CAA GG-3 | 61 |
| Reverse | 5-CTG GAG CTT CGC ACA ATA CA-3 | ||
| Degs1 | Forward | 5-TCT TGA AGG GAC ACG AAA CC-3 | 60 |
| Reverse | 5-GTT GTA GTG CGG GAG GTC AT-3 | ||
| Degs2 | Forward | 5-AAC TTC GAG GGC TGG TTC TT-3 | 60 |
| Reverse | 5-ACC TTG GGG TTC ACA CAG AG-3 | ||
| Gapdh | Forward | 5-ACT CCC ACT CTT CCA CCT TC-3 | 60 |
| Reverse | 5-GGT CCA GGG TTT CTT ACT-3 |
Gcs, glucosylceramide synthase; Sms1 and -2, sphingomyelin synthase 1 and 2.
Statistics
All statistic calculations, the mean, SD, unpaired Student’s t-tests, and one-way ANOVA with post hoc Tukey tests, as well as the correlation factors, were calculated with GraphPad Prism version 5.04 software. Isotope distributions were calculated with the isotope distribution calculator from the webpage of Scientific Instrument Service, Ringoes, New Jersey.
RESULTS
Imaging MS reveals CerS2- and Cer anchor-dependent specific expression patterns for sulfatides in the cortex, medulla, and papillae
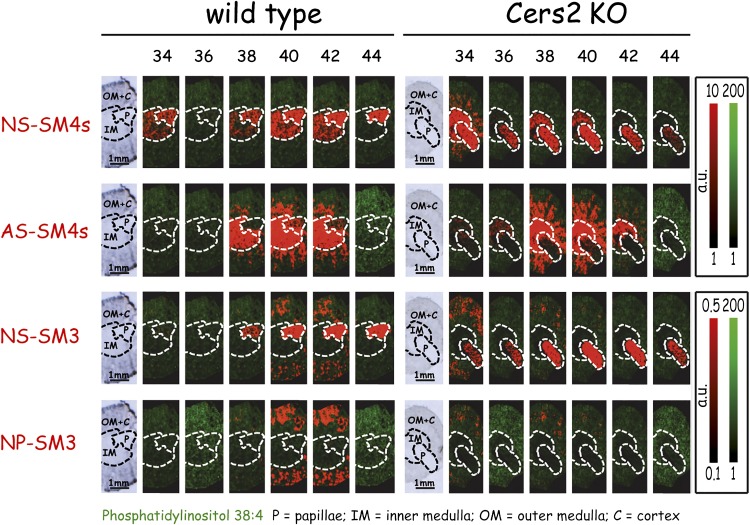
Analysis of mouse kidneys by imaging MS (IMS) demonstrated a Cer anchor-dependent expression of sulfatides within the major regions of the kidney. Sulfatides containing nonhydroxy fatty acids and phytosphingosine in their Cer anchor [Cer anchor with a nonhydroxy acyl chain and a phytosphingosine (NP)-SM3] were almost exclusively found in special cortical regions. The medulla, however, was about 4-fold (10- to 18-fold by LC-MS2) enriched in sulfatides carrying alpha-hydroxy acyl chains connected to sphingosines (AS-SM4s). These AS-SM4s appeared to emanate in stripes into the cortical regions. In correlation with the LC-MS2 data, signals for corresponding AS-SM3 were below detection limits in normal kidneys by IMS, which is in contrast to data obtained from kidneys of the storage disease ASA−/− mouse (36). Finally, sulfatides with nonhydroxy fatty acid moieties N-linked to sphingosines (NS-SM4s and NS-SM3) were most abundant in the papillae, but almost undetectable in the cortex (Fig. 2). The only major unsaturated sulfatide, NS-SM4s containing nervonic acid (C24:1), was distributed similarly to the saturated counterpart with lignoceric acid (C24:0; see supplementary Fig. III).
Fig. 2.
Representative MALDI IMS of renal sulfatides. To obtain comparable sections containing papillae, medulla, and cortex, mouse kidneys were dissected vertically. The first picture of each series represents light microscopy of the analyzed renal section. Areas attributed to papillae (P), inner medulla (IM), and outer medulla together with cortex (OM+C) are separated by dotted lines. Numbers above each column indicate the amount of C-atoms in the Cer anchor. Individual sulfatide species are plotted with the false color red. MS signal intensities relate to color intensity which was normalized for all sulfatides of the identical head group (either SM4s or SM3) to the same color intensity per absolute signal counts; see color code on the right. Likewise, the signal intensities for phosphatidylinositol (38:4), represented with the false color green, were normalized to the same intensities in wt and mutant sections. This experiment was repeated twice (see supplementary Fig. II).
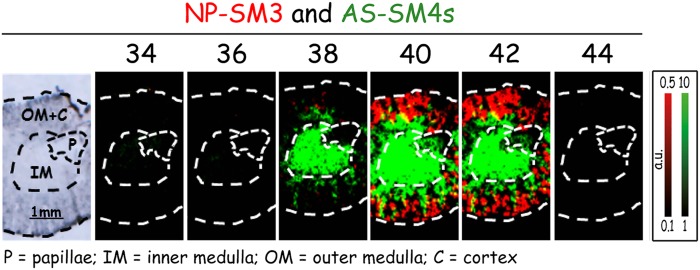
These results were validated by LC-MS2 analysis of surgically separated cortical, medullar, and papillar renal tissue (Fig. 3). IMS, however, is able to identify more subtle differences in distribution than LC-MS2. Thus, when plotting the sum of NP-sulfatide against the sum of AS-sulfatide signals within a single IMS picture, we could demonstrate that the cortical fraction of AS-sulfatides was restricted to renal structures distinct from those containing NP-sulfatides (Fig. 4). The data is consistent with the localization of AS-SM4s (d18:1;h24:0) and NP-SM3 (t18:0;24:0) that was published very recently (37).
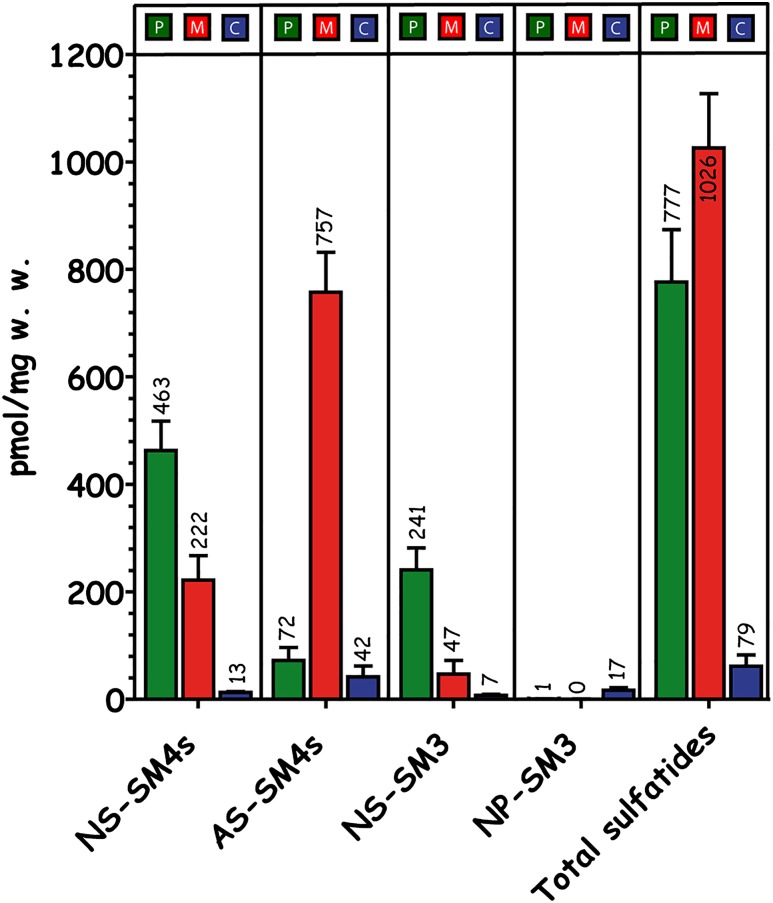
Fig. 3.
Quantitative distribution of renal sulfatides in the papillae (P), medulla (M), and cortex (C). Quantitative data were investigated by UPLC-ESI-MS2 (n = 3). The highest concentrations of sulfated GSLs were recorded in the medulla followed by the papillae (76% of the medulla) and more than 10-fold less sulfatide concentrations in the cortex (8% of the medulla). In the medulla, 74% of all sulfatides are AS-sulfatides, whereas in the papillae, 91% are NS-sulfatides. Finally the concentration of NP-sulfatides is 17-fold higher in the cortex than in any other region.
Fig. 4.
Distribution of the sum of AS-SM4s and of NP-SM3. Note that there is no overlap of these compounds, also not in the outer medulla and the cortex.
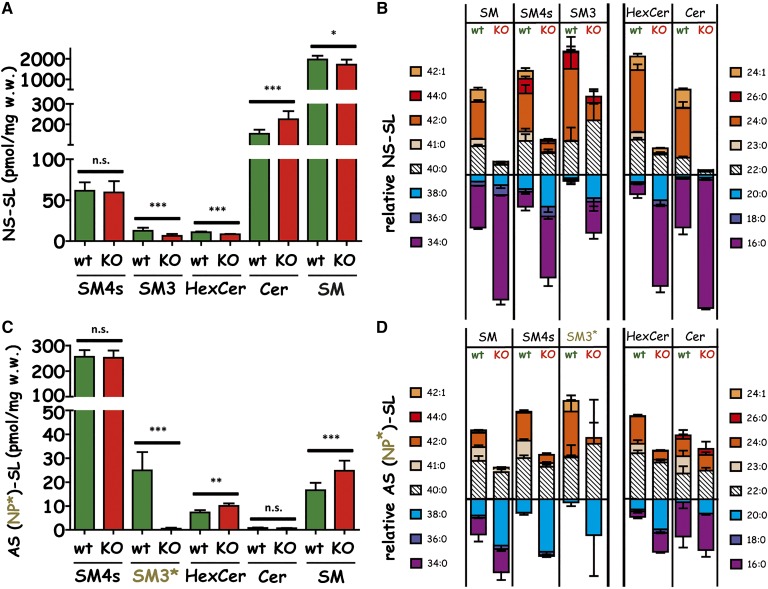
Furthermore, MALDI IMS (Fig. 2) and LC-MS2 [Fig. 5B, D, see also (32)] analyses suggested the majority of sulfatides to contain very long acyl chains (C20 ≤ acyl chain ≤ C26) within their Cer anchor (C40–C44, and in part C38). When analyzing renal sections from CerS2-deficient mice in parallel, we found a strong reduction of C44-sulfatide species together with C42-sulfatides (Figs. 2, 5; including unsaturated species, Fig. 5, supplementary Fig. III), but no complete depletion. Surprisingly, in striking contrast to previously published data for Cers and sphingomyelins (29), C40-sulfatide levels appeared almost unchanged and those of C38-sulfatides even seemed to increase. Similar to the previous results of increasing C34- Cer and C34-sphingomyelin levels in CerS2-deficient kidneys, we observed a compensating increase of C34-species for NS-sulfatides. However, AS-sulfatides and NP-sulfatides failed to compensate in this manner, which resulted in an almost complete depletion of cortical NP-sulfatides (SM3) (Fig. 2, CerS2 KO). Parallel analysis of total renal extracts from wt and CerS2-deficient kidneys corroborated these findings: only a significant reduction of NS-species and a complete depletion of NP-species were observed for the minor compound SM3, whereas the overall levels of other SLs did not alter or even increased slightly (Fig. 5). Intriguingly, compensation was achieved not only by increased expression of C34/C16-species, as had been initially observed for sphingomyelins and Cers, but substantially by C38-species. AS-sulfatides, especially, compensated almost exclusively with C38-species (Fig. 5).
Fig. 5.
Quantification of SLs in the kidneys of 11-week-old female CerS2-deficient and control mice. For NS-SL (A), no difference in the total SM4s concentration is detected. B: The loss of species with C22–C26 fatty acid chain length is compensated via increased concentration of species with C16–C20 fatty acid chain length. C: The total amount of AS-SM4s shows the same behavior (D), while for these molecules no species with palmitic acid are detected (n = 3; n.s. = not significant; *P < 0.05; **P < 0.01; ***P < 0.001).
Most strikingly, sulfatides with the largest Cer anchor (C44) were highly restricted to the papillae (Fig. 2, NS-SM4s and NS-SM3 of wt). Here, roughly every third NS-sulfatide, whether SM4s or SM3, contained this C44-backbone (45% of NS-sulfatides and 43% of all sulfatides within the papillae, as determined by LC-MS2). In total renal extracts, these species contributed only 10% to all NS-sulfatides and made up less than 2% of all renal sulfatides (Fig. 5). Hence, C44-sulfatides were roughly 20-fold enriched over all sulfatides in the papillae, which reflects the papillae making up a very small amount of the total kidney by weight. Analyzing 6-week-old females (supplementary Fig. IV) or 6-week-old males (supplementary Fig. V) did not alter these profiles.
In papillae, Cers and GSLs, including sulfatides, are highly enriched in C20-sphingosine content
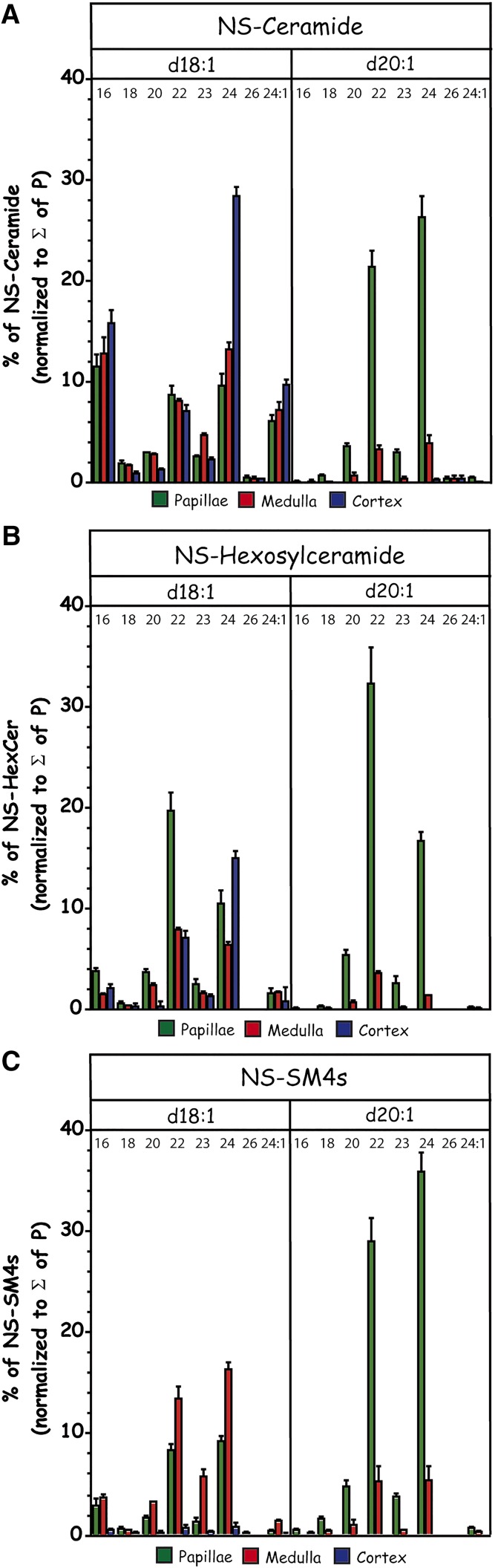
Regarding the C44-Cer anchor of papillar sulfatides, we initially thought of a combination of the most prominent sphingoid base C18-sphingosine linked to a rather rare C26-acyl chain (cerotic acid). Astonishingly, other SLs, i.e., sphingomyelins and especially the sulfatide precursor Cers and HexCers [in kidneys basically galactosylceramide (GalCer)] with a C18-sphingosine did not contain significant amounts of this C44-backbone in any renal region [Fig. 6A (Cer), Fig. 6B (HexCer), data for sphingomyelin not shown]. Therefore, we thought of an alternative structure consisting of an elongated C20-sphingosine linked to the prominent C24-fatty acid (lignoceric acid). Indeed, screening for Cers and HexCers with a C20-sphingosine moiety using the C20-sphingosine-specific MS2 transition to m/z 292 revealed high concentrations of C20-sphingosine containing Cers and HexCers, especially in combination with C22- (behenic acid) and C24-acyl (lignoceric acid) moieties within the papillae (Fig. 6A, B).
Fig. 6.
Cer-anchor compositions differentiating C18- and C20-sphingosine residues of renal Cers (A), HexCers (B) (mainly GalCer), and SM4s sulfatides (C) with a NS-backbone from extracts of cortex (blue), medulla (red), and papillae (green) as determined by LC-(ESI)MS2 (positive ESI). C18-sphingosine (d18:1) and C20-sphingosine (d20:1) containing SLs were detected by their transition to m/z 264 and to m/z 292, respectively. For saturated N-linked acyl chains, only the number of carbon atoms, i.e., 16, 18, 20, 22, 23, 24, and 26, is annotated, whereas for nervonic acid, the number of double bonds is indicated behind the colon (i.e., 24:1) in addition. Sulfatides were detected in positive ESI with their ISD to HexCer and subsequent transition to either m/z 264 or to m/z 292, respectively.
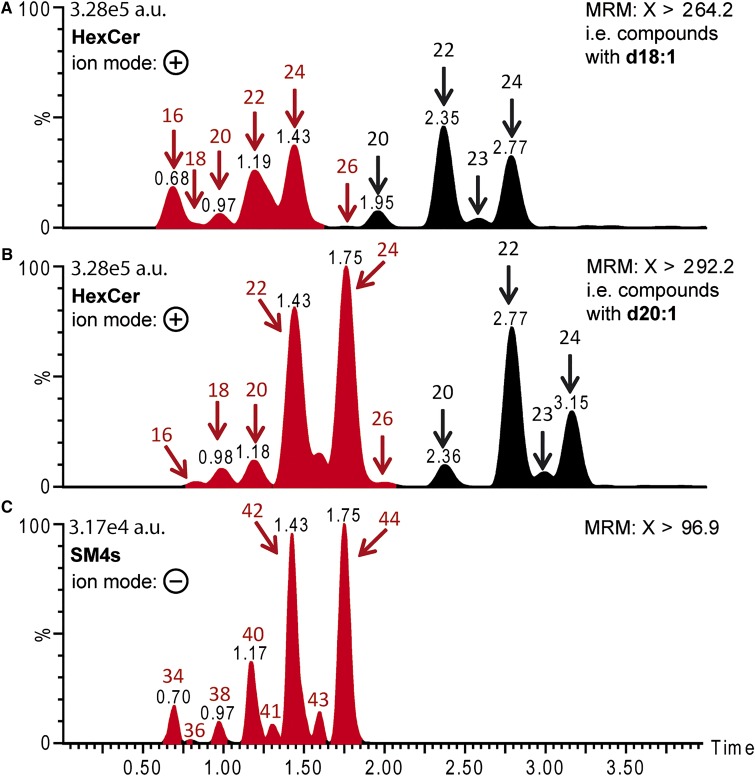
As we had quantified sulfatides by LC-MS2 in the negative mode using the MS2 transition to m/z −97, which reflects the sulfate group, we initially did not obtain any information about the composition of the Cer anchor. Furthermore, only AS-sulfatides, but not NS-sulfatides, produce minor product ions in ESI-MS2 that are indicative of the incorporated sphingoid base [(33) and supplementary Fig. II in (32)]. Surprisingly, every HexCer (likely GalCer, which is the precursor of SM4s) showed two instead of one peak in the SRM chromatograms on the LC-MS2 system. These transitions were obtained in positive ESI mode and the transition is indicative for the incorporated sphingoid base. For each of these transitions, the second peak with higher retention time reflected the expectations of the standard HexCer retention times, whereas the fast eluting first peak reflected exactly the retention times of the corresponding sulfatides, when measured with the identical gradient by LC-MS2 (Fig. 7). Hence, with a certain probability, protonated sulfatides undergo in source decay (ISD) in positive ESI conditions and lose the sulfate group. Then they behave exactly like their biosynthetic precursor GalCer and are detected with the transitions specific for the group of GalCers (HexCers). We had already earlier observed an ISD for Cers and HexCers, which resulted especially in water loss (Fig. 8). The advantage of the observed ISD for sulfatides yielding mainly HexCer in positive mode was that these transitions are specific for the size of the sphingoid base. Hence, positive mode detection allowed sphingoid base-dependent sulfatide quantification demonstrating only marginal signals for sulfatides with a C26-acyl chain, but very prominent signals for C20-sphingosine-containing sulfatides in extracts of the renal papillae. The latter appeared especially in combination with C22- and C24-acyl chains. These profiles were very similar to those observed for Cers and HexCers (Fig. 6C, 7).
Fig. 7.
Comparison of retention times of peaks observed in extracted ion chromatograms (EICs) recording the transitions of C18- and C20-sphingosine-containing HexCers in positive ESI with EICs recording the transitions of sulfatides in negative ESI. A renal papilla lipid extract was separated on UPLC and detected by SRMs. C18- and C20-sphingosine-containing HexCers were recorded with the transitions of the precursor ions [HexCer + H]+ and [HexCer − H2O + H]+ with the acyl chains C16:0, C18:0, C20:0, C22:0, C23:0, C24:0, and C26:0 to the product ion [C18-sphingosine – 2H2O + H]+ (m/z 264.2) (upper spectrum) or to the product ion [C20-sphingosine – 2H2O + H]+ (m/z 292.2) (middle spectrum), respectively. Bottom spectrum: EIC of the identical sample using the identical gradient in negative ESI for the transition of the precursor ions [SM4s – H]− with the NS-Cer backbones C34, C36, C38, C40, C41, C42, C43, and C44 to the product ion [HSO4]− (m/z −97).
Fig. 8.
Gas phase reactions of Cers (A), HexCers (GalCer, but glucosylceramide behaves identically) (B), and sulfatides SM4s (C) in positive ESI during ESI-MS2 measurements. ISD appears during ionization (probably by ion acceleration in an environment of substantial residual pressure) prior to the ions entering the first MS analyzing the first quadrupole, whereas CID occurs in the second quadrupole (using argon) prior to the second MS analyzing the third quadrupole.
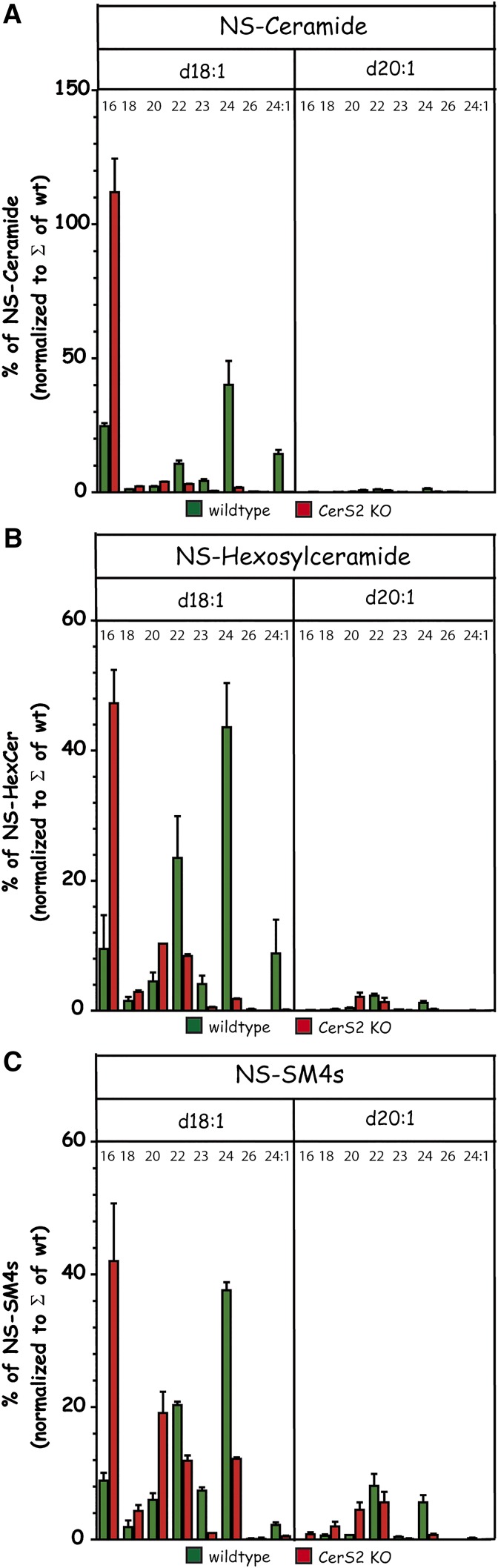
With this information and using ISD, we reanalyzed the composition of Cers, HexCers, and SM4s sulfatides in total extracts of wt and CerS2-deficient kidneys. Indeed, C20-sphingosine-containing SLs were minor components of total renal extracts. These data also confirmed a significant compensation of sulfatides with acyl chains longer than C22 by those with C20-acyl chains in CerS2-deficient kidneys (Fig. 9).
Fig. 9.
Comparison of renal C18- and C20-sphingosine-containing NS-Cers (A), NS-HexCers (B) (mainly GalCer), and NS-SM4s sulfatides (C) in wt and CerS2-deficient mice as determined by LC-(ESI)MS2 (positive ESI). C18-sphingosine (d18:1)- and C20-sphingosine (d20:1)-containing SLs were detected by their transition to m/z 264 and to m/z 292, respectively. For saturated N-linked acyl chains, only the number of carbon atoms, i.e., 16, 18, 20, 22, 23, 24, and 26, is annotated, whereas for nervonic acid, the number of double bonds is indicated behind the colon (i.e., 24:1) in addition. Sulfatides were detected in positive ESI with their ISD to HexCer and subsequent transition to either m/z 264 or to m/z 292, respectively.
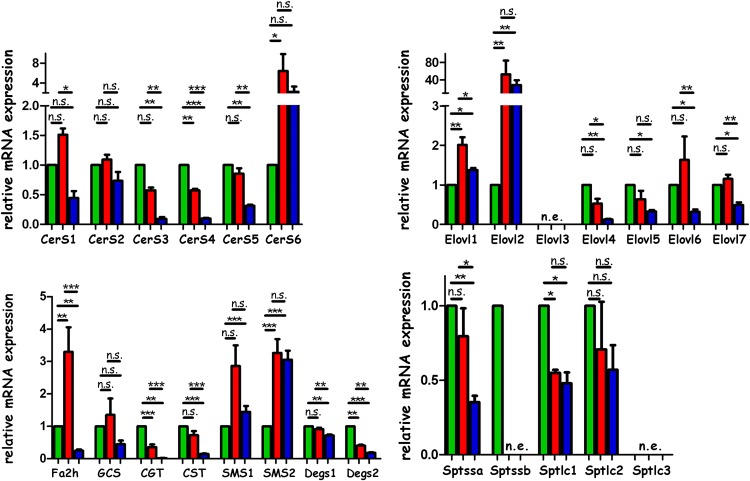
Region-specific mRNA expression of enzymes involved in SL biosynthesis
Local variations in the expression and activity of enzymes involved in GSL anabolism might be responsible for the observed local different sulfatide structures. Therefore, we sought to investigate regional mRNA expressions of the corresponding (iso-)enzymes.
SL biosynthesis starts with the activity of SPT, a heterodimer consisting of two of the three subunits serine palmitoyl-CoA transferase long chain (Sptlc)1, Sptlc2, and Sptlc3 (38). Sptlc3 appears to synthesize shorter C16-sphingoid bases (16), which were not present in renal sulfatides. Expression of Sptlc1 and of Sptlc2 was observed in all renal regions, whereas that of Sptlc3 was not detectable (Fig. 10, supplementary Fig. VI).
Fig. 10.
Quantitative real-time PCR-based analyses of mRNA expression of enzymes involved in the anabolic pathway of sulfatides in renal papillae (green), medulla (red), and cortex (blue). Expression values were calculated by the 2−ΔΔCt method and are represented relative to papillary expression. Significance was calculated using one-way ANOVA followed by post hoc Tukey’s test. Error bars represent mean ± SEM (n = 3).
The Sptlc-heterodimer is further regulated by the serine palmitoyl-CoA transferase small subunits (Sptss)a and Sptssb. Sptssa supports the production of C18-sphingoid bases and Sptssb guides the complex toward C20-sphingoid base production (15). We observed Sptssa in all renal regions with highest expression in the papillae and cortex. Sptssb expression was low and could only be detected in the papillae (Fig. 10, supplementary Fig. VI), which is in line with the particular presence of C20-SLs in the papillae.
For the synthesis of higher SLs, activated fatty acids are required. It is currently assumed that fatty acids are α-hydroxylated by fatty acid 2-hydroxylase (Fa2h) before being incorporated into Cers, resulting in renal AS-sulfatides. Fa2h expression was highest in the medulla and papillae and lowest in the cortex (Fig. 10, supplementary Fig. VI), correlating with the high medullary AS-sulfatide levels.
SLs contain a high degree of very long chain fatty acids which requires the action of special elongases, Elovl1–7. Except for Elovl3, all Elovl-mRNAs were detected in the three renal compartments. Elovl7 and Elovl1 can elongate saturated acyl chains from C16/18 up to C24/C28 (Elovl7 also elongates polyunsaturated fatty acids) (8, 11, 39), and saturated fatty acids are especially found in SLs (40). Both Elovl1 and Elovl7 were present in all regions, with Elovl1 concentrating in the cortex and Elovl7 being mostly expressed in the papillae and medulla (Fig. 10, supplementary Fig. VI).
The condensation of sphingoid bases with activated fatty acids is catalyzed by one of the six CerSs in mice. CerS2 (preferring C22- and C24-acyl-CoAs) was the most highly expressed, followed by CerS4 (preferring C20-acyl-CoA), CerS5, and CerS6 (both preferring C14- and C16-acyl-CoAs), whereas CerS1 (preferring C18-acyl-CoA) and CerS3 (being quite promiscuous) were almost undetectable (supplementary Fig. VI). Out of the prominent CerSs, only CerS4 concentrated significantly in the papillae over the other regions with the lowest levels found in the cortex.
Subsequent to the condensation of sphingoid bases with acyl chains via the CerSs, the desaturases, gene of dihydroceramid-Δ4-desaturase (Degs)1 and Degs2, hydroxylate the C4-position, yielding phytosphingosine, and Degs1 further dehydrates this base to finally produce sphingosines with a trans-Δ4 double bond (41–43). Analysis of Degs1 and Degs2 reflected the highest expression of Degs2 in the papillae, followed by the medulla, and lowest in the cortex, which contradicts the cortex-specific expression of phytosphingosine-containing NP-sulfatides.
Reducing acyl chain length in the Cer anchor of sulfatides by about two carbon units does not affect urinary pH
Inspired by our latest findings (6) that: i) papillar sulfatides are important for the renal ammonia exchange process, ii) urinary pH decreased in mice with renal sulfatide deficiency, and iii) strong reduction of the characteristic very long chain sulfatides especially in the papillae of CerS2-deficient mice; we monitored the urine pH of CerS2-deficient mice over 24 h of diet. No significant differences compared with the wt were observed, however (wt, pH = 5.99 ± 0.38; CerS2-KO, pH = 6.04 ± 0,31; n = 14).
DISCUSSION
Using MALDI IMS for the histological analysis of renal sulfatides, we discovered special sulfatides with a C44-Cer anchor as highly concentrated markers of the papillary region. As the papillae contribute by not more than 5% to the total kidney wet weight, these sulfatides make up less than 2% of all sulfatides within total kidney extracts. To reveal the structure of the C44-sulfatides, negative ion mode MALDI-MS2, either directly from tissue sections or from corresponding extracts, was too insensitive to collect relevant specific fragments, which are among the very low abundance product ion fraction recorded by collision-induced dissociation (CID)-MS2 spectra from sulfatides (44). To determine the type of sphingoid base and acyl chain of the Cer anchor of sulfatides, we established a new LC-MS2 method in which sulfatides were recorded indirectly in positive ESI mode as their precursor GalCers. Probably due to the acceleration of ions within the step wave of the Xevo TQ-S instrument at a relatively low vacuum, molecular ions partially undergo low energy CID. This induces ISD of sulfatides to produce protonated GalCers before mass analysis is performed. Hence, sulfatides mimic their precursor lipids, GalCers, which then are specifically detected in SRM mode by their CID transition to sphingoid base-specific fragments. Combining LC with the MS2 detection, sulfatides and their precursor GalCers, which are also present in the renal samples, are easily differentiated by their chromatographic retention times (chromatographic resolution R ≈ 6). To our knowledge, this is the first method to quantify unmodified sulfatides while simultaneously obtaining structural information on their sphingoid base and fatty acid composition. Analysis of papillary extracts by this method revealed the papillary C44-sulfatides to contain C20-sphingosine instead of the otherwise dominant C18-sphingosine. Also sulfatides with a shorter C40- and C42-Cer anchor were substantially built on the bases of C20-sphingosine. In fact, more than 50% of the papillary sulfatides were based on C20-sphingosine, whereas in the medulla, the concentration of C20-sphingosine-containing sulfatides was five times lower, and in the cortex they were undetectable. We also observed a similar gradient of C20-sphingosine-containing sulfatides in human kidneys (supplementary Fig. VII). C20-sphingosine had been reported as a major sphingoid base of bovine renal papillae (45), but the acid hydrolysis used obscured the SLs it was originating from. Interestingly, C20-sphingosine is the major sphingoid base in sulfatide-rich salt (nasal) glands from Eider duck and Herring Gull. Here the sulfatide content correlates with changes of Na+-K+-ATPase activity upon NaCl loading (4). These results were obtained after SL hydrolysis, but the sphingolipid from which the C20-sphingosine originated was not documented. Analyzing intact compounds, we observed a similar concentration gradient for C20-sphingosine-containing Cers and HexCers (most probably GalCers). Initially, these structures were probably not recognized because: i) total renal extracts contain less than 2% of them; ii) common MS2 screenings for Cers and HexCers with the fragment m/z 264 (derived from doubly dehydrated sphingosine) are C18-sphingoid base specific; and iii) there is no difference in mass between sulfatides containing C20-sphingosine and sulfatides with C18-sphingosine and an additional C2H4-unit in the fatty acid. Only for C44-sulfatides, there is no overlap with a C18-sphingosine-containing sulfatide, as cerotic acid (C26:0) is basically not present in sulfatides. Therefore, only these C44-sulfatides reflected a papillae-specific signal in IMS working in MS1 mode. This biological example of differentially located structural isomers also demonstrates that high resolution MS will not always be sufficient for complete compound identification.
However, the simultaneous documentation of sulfatide tissue distributions according to their membrane anchor compositions reveals new aspects in the complexity of cell type-specific pathways, which were not observed by previous immunocytochemical studies (46), as the anti-sulfatide antibodies recognize only the sulfatide head groups (sulfated glycan moieties). Taking quantitative aspects into account, we could document with MALDI IMS systematic distribution differences of sulfatides, which were based exactly on these differences of their Cer anchor moieties. These differences reflect cell-specific enzyme expression/activity variations for sulfatide biosynthesis for which we analyzed mRNA expression patterns. Hence, NS-sulfatides (containing nonhydroxy fatty acids and sphingosines, no matter if SM4s or SM3) with very long acyl chains were synthesized in the papillae, which was reflected by high CerS2-mRNA, Degs1-mRNA, cerebroside galactosyltransferase (Cgt; UDP-galactosyltransferase 8A or UDP-galactose-ceramide-galactosyltransferase)-mRNA, and cerebroside sulfotransferase (Cst)-mRNA, but lower Fa2h-mRNA levels. In addition, the medulla expressed high mRNA levels of Fa2h, which introduces the α-hydroxy group into fatty acids (47, 48). Indeed, the bulk of AS-sulfatides concentrates in this region. In all regions, sulfatides with C18-sphingoid bases were found, which is in line with a general expression of the small a-subunit of the SPT complex (Sptssa). The longer C20-sphingosine, however, concentrated highly in papillary NS-sulfatides, which was reflected by the presence of the Sptssb-mRNA (encoding the small b-subunit) in the papillae, but not in the medulla and cortex. When incorporated into the SPT complex, the b-subunit had been demonstrated recently to trigger the production of C20-sphingosine, whereas the a-subunit leads to the production of C18-sphingoid bases (15). Even in the papillae, Sptssb-mRNA levels were quite low as compared with those of the a-subunit. For a detailed study, protein levels of the two subunits would have to be compared. In the cortex, in line with minor Cst-mRNA levels, overall sulfatide levels were low, but NP-sulfatides expressing phytosphingosine were exclusively detected here. This was not reflected by the relatively minor cortical mRNA levels of Degs2, which is needed for phytosphingosine synthesis (42). As only special minor subcortical regions contained these sulfatides, it may be reasoned that the corresponding cell-specific mRNA levels are not represented correctly by the average cortical mRNA levels. Furthermore, mRNA levels may not directly correlate with enzyme activities and always have to be discussed with caution.
Mice are often taken as model organisms for human diseases, which works only if the corresponding molecules facilitate the same functions in both organisms. One prerequisite for this is a similar local expression. Therefore, we investigated the local sulfatide anchor composition in the papillae, medulla, and cortex of two human cases. Here also, the highest expression of NS-sulfatides was found in the papillae. More interestingly, C20-sphingosine-containing sulfatides were also restricted to the papillae in human kidney. Furthermore, they contained, basically, only very long acyl chains like NS-sulfatides of mouse papillae (supplementary Fig. VII). This similarity triggers speculations on special papillary sulfatide function in mammals that might depend on these particular Cer anchors.
While recently the involvement of renal sulfatides for papillary ammonia transport could be demonstrated in vivo (6), functional reasons for such diverse sulfatide anchor expressions within different renal cell types remain elusive. As Stettner et al. (6) discussed sulfatides of the papillae to regulate ammonia homeostasis, we hypothesized that the abundant very long acyl chains of papillary sulfatides may contribute to these sulfatide functions. Therefore, we investigated CerS2-deficient mice that had lost most of the very long acyl chains in neutral SLs (29), but could not observe similar differences in urinary pH as had been documented in sulfatide-deficient kidneys. Detailed analysis revealed no changes in the overall sulfatide levels, only the minor population of cortical NP-sulfatides were absent. As expected from previous investigations of Cers, sphingomyelins, and HexCers in CerS2-deficient mice (29, 49), sulfatides with C41- up to C44-acyl moieties were significantly decreased, however, those with C40-acyl moieties did not change significantly. In contrast to NS-ceramides and NS-sphingomyelins, sulfatides and SLs with AS-moieties compensated C41- to C44-SLs significantly with C20-acyl chains. This may reflect a compensatory role of CerS4, which preferentially incorporates C22-acyl chains, but may also use C20-CoA (13, 50). Besides in the papillae, CerS4 is especially expressed in the medulla of control kidneys, where AS-sulfatides concentrate. An additional upregulation of CerS4-mRNA levels in mutant tissue cannot be excluded, and may further contribute to the rather unchanged levels of C22-sulfatides. However, CerS4 principally could also use C24-acyl-CoAs. Why this potential is not utilized in vivo remains to be investigated. A compensatory role by CerS3 in mutant mice is rather unlikely due to the very low mRNA levels detected. Furthermore, especially with CerS3, we would have expected a higher capacity to compensate for C24- than for C22-sulfatides, and especially for C20-sulfatides (12).
In summary, IMS is a unique and valuable tool to dissect local tissue lipid changes that may occur upon metabolic disease, metabolic stimulus, or in corresponding model organisms. By that, it reveals a new level of complexity of cell type-specific lipid expression patterns which hint at the individual fine tuning of cell functions. To dissect the physiological functions of lipids in defined cell populations, cell type-specific mouse models have been and will be developed. The phenotype of fine tuning defects, however, cannot be expected to be obvious, and revealing it will often remain difficult. Nevertheless, the precise evaluation of cell type-specific mouse models of lipid metabolism again will depend on location-dependent lipid data which can be generated by IMS, but no other MS technique.
Supplementary Material
Acknowledgments
The authors thank Benita von Tümpling-Radosta and Ulrike Rothermel, German Cancer Research Center (DKFZ) Heidelberg, for excellent technical assistance, as well as Björn Meyer, University of Applied Sciences Mannheim, for critical comments.
Footnotes
Abbreviations:
- 9-AA
- 9-aminoacridine
- AS
- ceramide anchor with an alpha-hydroxy acyl chain and a sphingosine
- Cer
- ceramide
- CerS
- ceramide synthase
- Cgt
- cerebroside galactosyltransferase (UDP-galactosyltransferase 8A or UDP-galactose-ceramide-galactosyltransferase)
- CID
- collision-induced dissociation
- Cst
- cerebroside sulfotransferase
- Degs
- gene of dihydroceramid-Δ4-desaturase
- Des
- dihydroceramid-Δ4-desaturase
- Elovl
- elongation of very long chain fatty acids
- Fa2h
- fatty acid 2-hydroxylase
- GalCer
- galactosylceramide
- GSL
- glycosphingolipid
- HexCer
- hexosylceramide
- IMS
- imaging MS
- ISD
- in source decay
- NS
- ceramide anchor with a nonhydroxy acyl chain and a sphingosine
- NP
- ceramide anchor with a nonhydroxy acyl chain and a phytosphingosine
- SL
- sphingolipid
- SM3
- lactosylceramide II3-sulfate
- SM4s
- galactosylceramide I3-sulfate
- SPT
- serine palmitoyl-CoA transferase
- Sptlc
- serine palmitoyl-CoA transferase long chain
- Sptss
- serine palmitoyl-CoA transferase small subunit
- SRM
- single reaction monitoring
- UPLC
- ultra-performance LC
- wt
- wild-type
This work was supported by allocations to R.S. and H-J.G. within a joint grant (Zentren für Angewandte Forschung an Hochschulen: Applied Biomedical Mass Spectrometry, “ZAFH ABIMAS”) from ZO IV by the Landesstiftung Baden-Württemberg and the Europäischer Fonds für regionale Entwicklung (EFRE) to C.H. Work in the Bonn laboratory with the CerS2-deficient mice was supported by the German Research Foundation (SFB 645,B2) to K.W.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of text, seven figures, and one table.
REFERENCES
- 1.Ishizuka I. 1997. Chemistry and functional distribution of sulfoglycolipids. Prog. Lipid Res. 36: 245–319. [DOI] [PubMed] [Google Scholar]
- 2.Raff M. C., Fields K. L., Hakomori S. I., Mirsky R., Pruss R. M., Winter J. 1979. Cell-type-specific markers for distinguishing and studying neurons and the major classes of glial cells in culture. Brain Res. 174: 283–308. [DOI] [PubMed] [Google Scholar]
- 3.Vos J. P., Lopes-Cardozo M., Gadella B. M. 1994. Metabolic and functional aspects of sulfogalactolipids. Biochim. Biophys. Acta. 1211: 125–149. [DOI] [PubMed] [Google Scholar]
- 4.Karlsson K. A., Samuelsson B. E., Steen G. O. 1974. The lipid composition and Na+-K+-dependent adenosine-triphosphatase activity of the salt (nasal) gland of eider duck and herring gull. A role for sulphatides in sodium-ion transport. Eur. J. Biochem. 46: 243–258. [DOI] [PubMed] [Google Scholar]
- 5.Zalc B., Helwig J. J., Ghandour M. S., Sarlieve L. 1978. Sulfatide in the kidney: how is this lipid involved in sodium chloride transport? FEBS Lett. 92: 92–96. [DOI] [PubMed] [Google Scholar]
- 6.Stettner P., Bourgeois S., Marsching C., Traykova-Brauch M., Porubsky S., Nordström V., Hopf C., Koesters R., Sandhoff R., Wiegandt H., et al. 2013. Sulfatides are required for renal adaptation to chronic metabolic acidosis. Proc. Natl. Acad. Sci. USA. 110: 9998–10003. [Erratum. 2013. Proc. Natl. Acad. Sci. USA. 110: 14813.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niimura Y., Nagai K. 2008. Metabolic responses of sulfatide and related glycolipids in Madin-Darby canine kidney (MDCK) cells under osmotic stresses. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 149: 161–167. [DOI] [PubMed] [Google Scholar]
- 8.Guillou H., Zadravec D., Martin P. G., Jacobsson A. 2010. The key roles of elongases and desaturases in mammalian fatty acid metabolism: Insights from transgenic mice. Prog. Lipid Res. 49: 186–199. [DOI] [PubMed] [Google Scholar]
- 9.Mizutani Y., Kihara A., Chiba H., Tojo H., Igarashi Y. 2008. 2-Hydroxy-ceramide synthesis by ceramide synthase family: enzymatic basis for the preference of FA chain length. J. Lipid Res. 49: 2356–2364. [DOI] [PubMed] [Google Scholar]
- 10.Mizutani Y., Kihara A., Igarashi Y. 2006. LASS3 (longevity assurance homologue 3) is a mainly testis-specific (dihydro)ceramide synthase with relatively broad substrate specificity. Biochem. J. 398: 531–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sassa T., Ohno Y., Suzuki S., Nomura T., Nishioka C., Kashiwagi T., Hirayama T., Akiyama M., Taguchi R., Shimizu H., et al. 2013. Impaired epidermal permeability barrier in mice lacking elovl1, the gene responsible for very-long-chain fatty acid production. Mol. Cell. Biol. 33: 2787–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jennemann R., Rabionet M., Gorgas K., Epstein S., Dalpke A., Rothermel U., Bayerle A., van der Hoeven F., Imgrund S., Kirsch J., et al. 2012. Loss of ceramide synthase 3 causes lethal skin barrier disruption. Hum. Mol. Genet. 21: 586–608. [DOI] [PubMed] [Google Scholar]
- 13.Tidhar R., Futerman A. H. 2013. The complexity of sphingolipid biosynthesis in the endoplasmic reticulum. Biochim. Biophys. Acta. 1833: 2511–2518. [DOI] [PubMed] [Google Scholar]
- 14.Russo S. B., Tidhar R., Futerman A. H., Cowart L. A. 2013. Myristate-derived d16:0 sphingolipids constitute a cardiac sphingolipid pool with distinct synthetic routes and functional properties. J. Biol. Chem. 288: 13397–13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harmon J. M., Bacikova D., Gable K., Gupta S. D., Han G., Sengupta N., Somashekarappa N., Dunn T. M. 2013. Topological and functional characterization of the ssSPTs, small activating subunits of serine palmitoyltransferase. J. Biol. Chem. 288: 10144–10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hornemann T., Penno A., Rutti M. F., Ernst D., Kivrak-Pfiffner F., Rohrer L., von Eckardstein A. 2009. The SPTLC3 subunit of serine palmitoyltransferase generates short chain sphingoid bases. J. Biol. Chem. 284: 26322–26330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kremser C., Klemm A. L., van Uelft M., Imgrund S., Ginkel C., Hartmann D., Willecke K. 2013. Cell-type-specific expression pattern of ceramide synthase 2 protein in mouse tissues. Histochem. Cell Biol. 140: 533–547. [DOI] [PubMed] [Google Scholar]
- 18.Sandhoff R. 2010. Very long chain sphingolipids: tissue expression, function and synthesis. FEBS Lett. 584: 1907–1913. [DOI] [PubMed] [Google Scholar]
- 19.Mizutani Y., Mitsutake S., Tsuji K., Kihara A., Igarashi Y. 2009. Ceramide biosynthesis in keratinocyte and its role in skin function. Biochimie. 91: 784–790. [DOI] [PubMed] [Google Scholar]
- 20.Rabionet M., van der Spoel A. C., Chuang C. C., von Tumpling-Radosta B., Litjens M., Bouwmeester D., Hellbusch C. C., Korner C., Wiegandt H., Gorgas K., et al. 2008. Male germ cells require polyenoic sphingolipids with complex glycosylation for completion of meiosis: a link to ceramide synthase-3. J. Biol. Chem. 283: 13357–13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rabionet M., Gorgas K., Sandhoff R. 2014. Ceramide synthesis in the epidermis. Biochim. Biophys. Acta. 1841: 422–434. [DOI] [PubMed] [Google Scholar]
- 22.Poulos A., Johnson D. W., Beckman K., White I. G., Easton C. 1987. Occurrence of unusual molecular species of sphingomyelin containing 28–34-carbon polyenoic fatty acids in ram spermatozoa. Biochem. J. 248: 961–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandhoff R., Geyer R., Jennemann R., Paret C., Kiss E., Yamashita T., Gorgas K., Sijmonsma T. P., Iwamori M., Finaz C., et al. 2005. Novel class of glycosphingolipids involved in male fertility. J. Biol. Chem. 280: 27310–27318. [DOI] [PubMed] [Google Scholar]
- 24.Jakobsson A., Westerberg R., Jacobsson A. 2006. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog. Lipid Res. 45: 237–249. [DOI] [PubMed] [Google Scholar]
- 25.Li W., Sandhoff R., Kono M., Zerfas P., Hoffmann V., Char-Hoa Ding B., Proia R. L., Deng C. 2007. Depletion of ceramides with very long chain fatty acids causes defective skin permeability barrier function, and neonatal lethality in ELOVL4 deficient mice. Int. J. Biol. Sci. 3: 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mandal M. N., Ambasudhan R., Wong P. W., Gage P. J., Sieving P. A., Ayyagari R. 2004. Characterization of mouse orthologue of ELOVL4: genomic organization and spatial and temporal expression. Genomics. 83: 626–635. [DOI] [PubMed] [Google Scholar]
- 27.Zadravec D., Tvrdik P., Guillou H., Haslam R., Kobayashi T., Napier J. A., Capecchi M. R., Jacobsson A. 2011. ELOVL2 controls the level of n-6 28:5 and 30:5 fatty acids in testis, a prerequisite for male fertility and sperm maturation in mice. J. Lipid Res. 52: 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandhoff R., Hepbildikler S. T., Jennemann R., Geyer R., Gieselmann V., Proia R. L., Wiegandt H., Grone H. J. 2002. Kidney sulfatides in mouse models of inherited glycosphingolipid disorders: determination by nano-electrospray ionization tandem mass spectrometry. J. Biol. Chem. 277: 20386–20398. [DOI] [PubMed] [Google Scholar]
- 29.Imgrund S., Hartmann D., Farwanah H., Eckhardt M., Sandhoff R., Degen J., Gieselmann V., Sandhoff K., Willecke K. 2009. Adult ceramide synthase 2 (CERS2)-deficient mice exhibit myelin sheath defects, cerebellar degeneration, and hepatocarcinomas. J. Biol. Chem. 284: 33549–33560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folch J., Lees M., Sloane-Stanley G. H. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 31.Zhang X., Nakajima T., Kamijo Y., Li G., Hu R., Kannagi R., Kyogashima M., Aoyama T., Hara A. 2009. Acute kidney injury induced by protein-overload nephropathy down-regulates gene expression of hepatic cerebroside sulfotransferase in mice, resulting in reduction of liver and serum sulfatides. Biochem. Biophys. Res. Commun. 390: 1382–1388. [DOI] [PubMed] [Google Scholar]
- 32.Marsching C., Jennemann R., Heilig R., Gröne H. J., Hopf C., Sandhoff R. 2014. Quantitative imaging mass spectrometry of renal sulfatides: validation by classical mass spectrometric methods. J. Lipid Res. 55: 2343–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsu F. F., Turk J. 2004. Studies on sulfatides by quadrupole ion-trap mass spectrometry with electrospray ionization: structural characterization and the fragmentation processes that include an unusual internal galactose residue loss and the classical charge-remote fragmentation. J. Am. Soc. Mass Spectrom. 15: 536–546. [DOI] [PubMed] [Google Scholar]
- 34.Jennemann R., Sandhoff R., Langbein L., Kaden S., Rothermel U., Gallala H., Sandhoff K., Wiegandt H., Grone H. J. 2007. Integrity and barrier function of the epidermis critically depend on glucosylceramide synthesis. J. Biol. Chem. 282: 3083–3094. [DOI] [PubMed] [Google Scholar]
- 35.Livak K. J., Schmittgen T. D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 36.Marsching C., Eckhardt M., Grone H. J., Sandhoff R., Hopf C. 2011. Imaging of complex sulfatides SM3 and SB1a in mouse kidney using MALDI-TOF/TOF mass spectrometry. Anal. Bioanal. Chem. 401: 53–64. [DOI] [PubMed] [Google Scholar]
- 37.Grove K. J., Voziyan P. A., Spraggins J. M., Wang S., Paueksakon P., Harris R. C., Hudson B. G., Caprioli R. M. 2014. Diabetic nephropathy induces alterations in the glomerular and tubule lipid profiles. J. Lipid Res. 55: 1375–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowther J., Naismith J. H., Dunn T. M., Campopiano D. J. 2012. Structural, mechanistic and regulatory studies of serine palmitoyltransferase. Biochem. Soc. Trans. 40: 547–554. [DOI] [PubMed] [Google Scholar]
- 39.Naganuma T., Sato Y., Sassa T., Ohno Y., Kihara A. 2011. Biochemical characterization of the very long-chain fatty acid elongase ELOVL7. FEBS Lett. 585: 3337–3341. [DOI] [PubMed] [Google Scholar]
- 40.Kolter T., Sandhoff K. 1999. Sphingolipids—their metabolic pathways and the pathobiochemistry of neurodegenerative diseases. Angew. Chem. Int. Ed. Engl. 38: 1532–1568. [DOI] [PubMed] [Google Scholar]
- 41.Fabrias G., Munoz-Olaya J., Cingolani F., Signorelli P., Casas J., Gagliostro V., Ghidoni R. 2012. Dihydroceramide desaturase and dihydrosphingolipids: debutant players in the sphingolipid arena. Prog. Lipid Res. 51: 82–94. [DOI] [PubMed] [Google Scholar]
- 42.Omae F., Miyazaki M., Enomoto A., Suzuki M., Suzuki Y., Suzuki A. 2004. DES2 protein is responsible for phytoceramide biosynthesis in the mouse small intestine. Biochem. J. 379: 687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michel C., van Echten-Deckert G., Rother J., Sandhoff K., Wang E., Merrill A. H., Jr 1997. Characterization of ceramide synthesis. A dihydroceramide desaturase introduces the 4,5-trans-double bond of sphingosine at the level of dihydroceramide. J. Biol. Chem. 272: 22432–22437. [DOI] [PubMed] [Google Scholar]
- 44.Hsu F. F., Bohrer A., Turk J. 1998. Electrospray ionization tandem mass spectrometric analysis of sulfatide. Determination of fragmentation patterns and characterization of molecular species expressed in brain and in pancreatic islets. Biochim. Biophys. Acta. 1392: 202–216. [DOI] [PubMed] [Google Scholar]
- 45.Karlsson K. A., Samuelsson B. E., Steen G. O. 1968. Structure and function of sphingolipids. 1. Differences in sphingolipid long-chain base pattern between kidney cortex, medulla, and papillae. Acta Chem. Scand. 22: 1361–1363. [DOI] [PubMed] [Google Scholar]
- 46.Trick D., Decker J., Groene H. J., Schulze M., Wiegandt H. 1999. Regional expression of sulfatides in rat kidney: immunohistochemical staining by use of monospecific polyclonal antibodies. Histochem. Cell Biol. 111: 143–151. [DOI] [PubMed] [Google Scholar]
- 47.Alderson N. L., Rembiesa B. M., Walla M. D., Bielawska A., Bielawski J., Hama H. 2004. The human FA2H gene encodes a fatty acid 2-hydroxylase. J. Biol. Chem. 279: 48562–48568. [DOI] [PubMed] [Google Scholar]
- 48.Eckhardt M., Yaghootfam A., Fewou S. N., Zoller I., Gieselmann V. 2005. A mammalian fatty acid hydroxylase responsible for the formation of alpha-hydroxylated galactosylceramide in myelin. Biochem. J. 388: 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pewzner-Jung Y., Brenner O., Braun S., Laviad E. L., Ben-Dor S., Feldmesser E., Horn-Saban S., Amann-Zalcenstein D., Raanan C., Berkutzki T., et al. 2010. A critical role for ceramide synthase 2 in liver homeostasis: II. Insights into molecular changes leading to hepatopathy. J. Biol. Chem. 285: 10911–10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mizutani Y., Kihara A., Igarashi Y. 2005. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 390: 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.