Abstract
Phospholipids serve as central structural components in cellular membranes and as potent mediators in numerous signaling pathways. There are six main classes of naturally occurring phospholipids distinguished by their distinct polar head groups that contain many unique molecular species with distinct fatty acid composition. Phospholipid molecular species are often expressed as isobaric species that are denoted by the phospholipid class and the total number of carbon atoms and double bonds contained in the esterified fatty acyl groups (e.g., phosphatidylcholine 34:2). Techniques to separate these molecules exist, and each has positive and negative attributes. Hydrophilic interaction liquid chromatography uses polar bonded silica to separate lipids by polar head group but not by specific molecular species. Reversed phase (RP) chromatography can separate by fatty acyl chain composition but not by polar head group. Herein we describe a new strategy called differential ion mobility spectrometry (DMS), which separates phospholipid classes by their polar head group. Combining DMS with current LC methods enhances phospholipid separation by increasing resolution, specificity, and signal-to-noise ratio. Additional application of specialized information-dependent acquisition methodologies along with RP chromatography allows full isobaric resolution, identification, and compositional characterization of specific phospholipids at the molecular level.
Keywords: lipid profiling, glycerophospholipids, lipid metabolism
The modern field of lipidomics began with the advent of ESI-MS [for review, see (1–3)]. This “soft” ionization technique generates charged molecules (i.e., for m/z < 1,000 Da) without significant analyte decomposition (4). The technique also enables the coupling of HPLC to MS, effectively converting liquid-phase molecules into gas-phase ions. Another leap in technology was the introduction of the triple quadrupole mass spectrometer (5), which when combined with ESI, enables facile, sensitive, and specific analyses of complex lipid mixtures (6, 7). Using ESI-MS, semitargeted scan modes, which exploit unique ion fragmentation patterns during MS/MS experiments, were developed that are presently used to isolate categories and classes of lipids without the need for complex LC separation, a technique termed “shotgun” lipidomics (8). There are several technical limitations of this approach including restricted detection of minor lipid components due to instrumental constraints of the dynamic range and discrimination against low-abundant and/or poorly ionizable lipids. Furthermore, ion suppression and difficulty in differentiating lipids with overlapping fragmentation patterns underline the importance of chromatographic separation prior to MS analysis. A practical alternative to a global lipidomics methodology is a targeted lipidomics approach that utilizes extraction and analysis protocols that are optimized for each lipid category and class (9), providing an effective tool to identify individual lipids in cells (10) and plasma (11) and to quantify molecular species relevant to health and disease (12).
The evolution of lipidomics has closely coincided with the development of new technology, which has allowed for this complex array of molecules to be mined at the category, class, and molecular species levels. For example, early lipid studies relied on basic chromatographic techniques including TLC, HPLC, and GC to characterize the categories and classes of lipids in biological extracts (13, 14). Unfortunately, detailed analyses of fatty acid composition of complex lipids were limited to percentage composition, and the separation process itself often resulted in alteration of the lipid profile (15). As the analytical techniques used for lipidomics have improved, the staggering complexity of the lipidome has emerged (16, 17). Although the actual number of structurally distinct lipids is not known, there may be hundreds of thousands of different lipid molecular species in a given biological extract, all found in a narrow mass range (∼700 Da). This all but ensures isobaric interferences (i.e., multiple species will present as ions with the same integer m/z value) to be the rule rather than the exception. This complex mix of isobars and structural isomers presents significant difficulties in data acquisition and analysis.
The development of two analytical techniques has attempted to address the challenges of isobaric/isomeric lipid populations: 1) HPLC separation and 2) high-resolution, accurate mass MS. However, these techniques have their own inherent analytical problems. Considering the diversity of lipid chemical structures, it has proved impossible to develop LC methods that effectively resolve all lipid categories, classes, and molecular species in a single run, and the time required to develop LC methods is not trivial. High-resolution MS offers the impressive ability to resolve lipids far beyond the capabilities of a triple quadrupole instrument (18), but there are many different lipid species that are iso-elemental and cannot be resolved by high-resolution MS alone. Additionally, no instrument currently available provides high-resolution isolation of target precursor ions. As a consequence, multiple near-isobaric precursor ions are selected for subsequent MS/MS analysis, which makes accurate interpretation of product ion spectra very difficult, especially for low-abundant species. What has been lacking in the field of lipidomics is an orthogonal means of selection that does not necessarily require extensive chromatographic separation or MS with high resolving power or mass accuracy to separate and clearly identify individual lipid molecular species.
Herein we report the use of differential mobility spectrometry (DMS) to enable accurate lipid molecular species identification and quantitation (19, 20). To date, several examples of DMS separation power have been applied to mixtures of tautomers (21), stereoisomers (22), and structural isomers (23); however, this is the first report to apply the technique to lipid profiling. DMS separates ions based on differences in their mobilities during the high- and low-field portions of an applied asymmetric waveform (24). DMS can also separate ions based on chemical effects such as differences in the ions’ dipole moments and the ions clustering with added volatile molecules (termed modifiers), which are related to the chemical environment surrounding the molecule’s charge site (24, 25). DMS was used with and without LC separation to select ionized membrane phospholipid classes prior to mass analysis by a hybrid quadrupole-linear ion trap mass spectrometer. The strengths and weaknesses of different analytical strategies are compared, and the effects DMS contributes to the analysis are discussed. This additional degree of freedom provided by DMS in the analysis of lipids enables “clean” product ion spectra by eliminating interclass isobaric interference during precursor ion selection and dramatically improves the quality of acquired lipidomic data.
MATERIALS AND METHODS
Chemicals
Methylene chloride, methanol, n-propanol, acetonitrile, and isopropanol (HPLC grade or better) were obtained from Thermo Fisher Scientific (Pittsburgh, PA). Ammonium acetate and human serum were obtained from Sigma Chemical Co. (St. Louis, MO). Lipid standards, including phosphatidylcholine (PC) (16:0/18:1); phosphatidylethanolamine (PE) (16:0/18:1); phosphatidylglycerol (PG) (16:0/18:1); phosphatidylinositol (PI) (16:0/18:1); phosphatidylserine (PS) (16:0/18:1); phosphatidic acid (PA) (16:0/18:1); lysophosphatidylcholine (LPC) (18:0); lysophosphatidylethanolamine (LPE) (18:0); lysophosphatidylglycerol (16:0); lysophosphatidylinositol (16:0); lysophosphatidylserine (18:0); lysophosphatidic acid (16:0); SM (d18:1/16:0); ceramide (Cer) (16:0); and class-specific lipid extracts from egg (PC, PE, PG, PA, SM, and Cer), soy (PI and LPC), and brain (PS) were obtained from Avanti Polar Lipid Co. (Alabaster, AL).
Sample preparation
Purified phospholipid extract stock was prepared by mixing the individual phospholipid classes to a final concentration of 10 µg/ml each in methanol-methylene chloride (50:50; v/v) containing 5 mM ammonium acetate. For LC and DMS analysis, the stock was further diluted to 1 µg/ml in appropriate analysis buffer. Pooled human serum was extracted using the method of Bligh and Dyer (26). Dried lipid extracts were redissolved in the same solvent as the purified lipid extracts. For HPLC ESI-MS/MS analysis of lipids, samples were dissolved in 100% HPLC solvent A.
ESI-MS/MS analyses
Lipid samples were analyzed using a QTRAP® 6500 LC/MS/MS system (AB SCIEX, Redwood Shores, CA), which is a hybrid quadrupole-linear ion trap mass spectrometer. Source parameters (e.g., temperatures, gas flows, etc.) were optimized using a mixture of phospholipid standards that were tee-infused with a syringe pump into the flow of an Acquity UPLC (ultra performance liquid chromatography) system (Waters, Milford, MA) delivering the sample at a final flow rate of 500 µl/min. Nontargeted analysis was carried out using the enhanced MS (EMS) mode, which uses the linear ion trap functionality of the instrument to perform the equivalent of an MS scan with higher resolution and better sensitivity. In other experiments, semitargeted scans were used to focus on the different phospholipid classes including precursor ion scan (PIS) and neutral loss scan (NLS) modes (i.e., “shotgun” lipidomics). For phospholipid analyses, different scan modes and ion polarities were used to monitor all six classes optimally in a multiplexed fashion: PC, PIS m/z 184 Da (+); PE, NLS 141 Da (+); PS, NLS 185 Da (+); PA, NLS 98 Da (+); PG, NLS 172 Da (+); and PI, PIS m/z 241 Da (−). The sphingolipid classes Cer, PIS m/z 264 (+); and SM, PIS m/z 184 (+) were also included in the analysis.
Infusion-DMS method
The mass spectrometer was equipped with SelexIONTM DMS technology. The DMS-based experiments were carried out using 1.5% 1-propanol as the DMS chemical modifier. Operating conditions were optimized using flow injection of lipid extract mixture and were determined as follows: DMS cell temperature = 150°C, chemical modifier = 1-propanol, separation voltage (SV) = 4,000 V (+) or 3,750 (−), DMS offset = 3.0 V, and nitrogen resolving gas = 37 psi. The lipids were dissolved in methanol-methylene chloride (50:50; v/v) containing 5 mM ammonium acetate and infused at a rate of 10 μl per minute for 4 min. The individual phospholipid classes were separated by ramping the compensation voltage (COV). The COV is a direct current voltage that stabilizes the path of selected molecules through the DMS cell. The molecular ions were recorded by EMS scan with a duty cycle of 0.15 s.
LC method I: HILIC-UPLC
Chromatography was performed with an Acquity UPLC system (Waters Corporation) operating at a flow rate of 0.5 ml/min, a column oven temperature maintained at 40°C, an autosampler maintained at 4°C, and 10 µl sample injections. A Kinetix hydrophilic interaction liquid chromatography (HILIC) column (1.7 µm, 2.1 mm × 100 mm) (Phenomenex, Torrance, CA) was used using a binary solvent gradient consisting of acetonitrile-water (95:5; v/v) with 10 mM ammonium acetate (buffer A) and acetonitrile-water (50:50; v/v) with 10 mM ammonium acetate (buffer B). The gradient ran from 0% to 9% B for 6 min, increasing to 30% for 5.5 min. After each chromatographic run, the LC column was reequilibrated prior to the next injection.
LC method II: RP-UPLC
Chromatography was performed with an Acquity UPLC system operating at a flow rate of 0.3 ml/min, a column oven temperature maintained at 40°C, an autosampler maintained at 4°C, and 10 µl sample injections. A CSH C18 column (1.7 um, 2.1 mm × 100 mm) (Waters Corporation) was used using a binary solvent gradient consisting of water-acetonitrile (60:40; v/v) with 10 mM ammonium acetate (buffer A) and isopropanol-acetonitrile (50:50; v/v) with 10 mM ammonium acetate (buffer B). An isocratic gradient was run at 30% B for 9 min. After each chromatographic run, the LC column was reequilibrated prior to the next injection.
LC-DMS method
For the LC-DMS experiments, the analytes within each lipid class were monitored using EMS scans with predetermined optimal COV settings. The COV values were optimized for each phospholipid class during infusion-DMS by ramping the COV at increments of 0.1 V once the other parameters were set (e.g., COV for PC in positive mode is 0.3V). The optimal COV values for the 14 phospholipid classes are listed in supplementary Table I. To generate a total ion current (TIC), the MS experiment consisted of 14 EMS scans using the optimal COV values. Each duty cycle was 1.9 s, and the EMS scans were repeated over the 10 min chromatography period. For information-dependent acquisition (IDA) experiments, the analytes within each lipid class were monitored using their optimal scan types [e.g., (+) NLS 141 for PE] with predetermined COV settings.
RESULTS
HILIC-UPLC MS
The resolution of lipids prior to MS analysis minimizes the isobaric interference among different lipid categories and classes. Because the MS/MS spectra for lipids are information poor (as compared with the information-rich spectra obtained from product ion analysis of peptides), it is difficult to identify lipids in a complex mixture qualitatively without some type of separation. HPLC has long been the preferred method to resolve lipid molecular species. Thus, initial experiments were aimed at establishing a baseline set of results to demonstrate the ability of UPLC-ESI-MS/MS to resolve and identify phospholipids and Cers (Fig. 1) and to provide a data set that can be used to compare the separation efficacy of conventional LC methods with the performance of the DMS. Using LC method I, phospholipid classes and Cer were separated with good resolution. The MS method used to analyze the lipid mixture (Fig. 1A) and extracted human serum (Fig. 1B) was developed based on the traditional shotgun lipidomic methodology to identify lipid classes based on specific precursor fragment ions and neutral losses associated with the different lipid classes.
Fig. 1.
Separation of phospholipid classes by HILIC-UPLC. Multiplex analysis of purified phospholipids (A) and human serum (B) using PIS and NLS in positive and negative ion mode. Top panel shows HILIC chromatographic separation and mass spectrometric detection of phospholipid/sphingolipid classes by either precursor m/z 184, 241, 264 or neutral loss m/z 141, 172, 185, 98. Middle panel shows NLS m/z 141 in positive ion mode specific for PE and LPE. Bottom panel shows summed mass spectral (MS) data from NLS m/z 141 in positive ion mode shown indicated in gray above. Inset: magnification from m/z 762 to m/z 774.
The top panel of Fig. 1 shows HILIC chromatographic separation and mass spectrometric detection of phospholipid classes in a standard mixture of purified phospholipids (left) or human serum (right) by either PISs for m/z 184, 241, and 264 or NLSs for m/z 141, 172, 185, and 98, which collectively correspond to PC, PI, and Cer and PE, PG, PS, and PA, respectively, in the positive ion mode. The middle set of panels shows the results for the phospholipid class PE using an NLS of 141 Da in the positive ion mode. The bottom set of panels shows the extracted spectrum for PE and LPE for the standard mixture (left) and human serum (right), with a magnified view of a subset of individual PE lipids within all detectable molecular species.
DMS MS
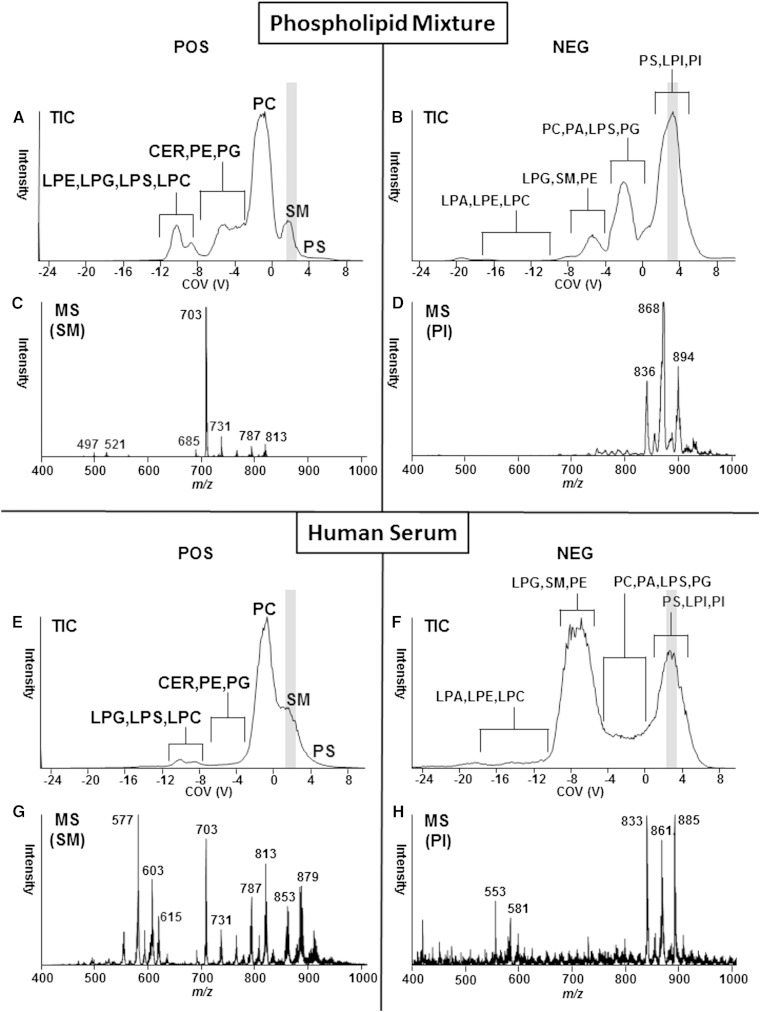
To assess the ability of DMS to resolve lipid classes within a complex lipid mixture, a phospholipid lipid standard mixture was infused and resolved by DMS and analyzed by EMS in the positive ion mode (Fig. 2A) and the negative ion mode (Fig. 2B). Ramping the COV during an EMS experiment resulted in the separation of lipids by class that can be visualized by extracting the spectrum at the appropriate COV value that is specific for each phospholipid class and Cer (Fig. 2C, D). Supplementary Table I shows the optimal COV values for each phospholipid class, as determined using lipid standards. These data show that DMS is capable of resolving a mixture of lipid standards; however, the demonstration of effective resolution using a complex lipid extract is needed to establish the technique for biologically derived samples with complex matrices. Figure 2E–H demonstrates the resolving power of DMS on human serum lipid extracts. Like the defined standard samples, DMS resolves the phospholipid and sphingolipid classes in human serum extract, demonstrating the resolution power of DMS even in samples with complex matrices. The resolution is dependent on the polarity of the experiment, with better separation between phospholipid classes achieved in the negative ion mode, whereas the positive ion mode is better suited for the separation of the lipid classes Cer, SM, and PC.
Fig. 2.
Separation of phospholipid and sphingolipid classes by DMS. DMS of purified phospholipids/sphingolipids in positive mode and negative mode. Top panels (A, B) show total ion current of DMS separation and MS detection using EMS and the trap function. COV voltage was ramped from −25 to 10. Bottom panels show summed mass spectral data from COV 1.7 to 2.3 specific for SM (C) or COV 2.4 to 3.8 specific for PI (D) indicated in gray above. DMS of human serum in positive mode or negative mode. Top panels (E, F) show total ion current of DMS separation and MS detection using the trap function. COV was ramped from −25 to 10. Bottom panels show summed mass spectral data from COV 1.7 to 2.3 specific for SM (G) or COV 2.4 to 3.8 specific for PI (H) indicated in gray above.
The addition of the DMS to the HILIC LC/MS separation strategy shows improved sensitivity and selectivity for some of the phospholipids as evidenced in the improved signal-to-noise ratio in the HILIC chromatograms (Fig. 3A, B). While the overall multiple reaction monitoring (MRM) signal levels decreased upon application of the DMS to this workflow, the ultimate outcome is the reduction of chemical noise levels from endogenous interferences (27, 28); MRMs are traditionally blind to the presence of such closely related species. In biological samples such as serum, there is increased background noise that contributes to a loss in sensitivity and selectivity for HILIC LC/MS. However, by including DMS as a filter to reduce background noise, the net sensitivity may be increased as shown in Fig. 3.
Fig. 3.
Application of DMS combined with HILIC-UPLC to enhance sensitivity. HILIC-based chromatographic separation of lipids extracted from human serum in positive ion mode (left panels) or negative ion mode (right panels) without DMS (A) or with DMS (B). Selected COV values were specific for each phospholipid class.
Application of DMS to reduce background interference
Initial experiments to evaluate the use of DMS and UPLC together focused on reducing the general noise level in the spectrum obtained during PIS and NLS analyses. Appropriate precursor ion and neutral loss experiments were combined in a single MS experiment, and human serum extract was resolved using LC method I. Figure 3A shows the TIC in the positive and negative ion modes using LC-MS alone. When the same experiment was performed using the DMS, there was a dramatic reduction in background noise (Fig. 3B). These results suggest that concomitant product ion analysis could be greatly improved using DMS in addition to LC by the reduction of interfering isobars, especially in untargeted discovery experiments such as data-dependent and data-independent acquisitions (27, 28).
In order to determine whether the application of DMS causes any loss of detectable lipid species or leads to any changes in phospholipid lipid profiles, we compared the mass spectra of serum phospholipids recorded using either HILIC separation alone with those obtained with the combination of HILIC and DMS (supplementary Fig. I). While the mass spectra for several phospholipid classes look very similar, some differences related to relative intensities between molecular species were noted. The cause of this needs to be further investigated, and additional work will be needed to fully evaluate the quantitative potential of DMS.
RP-UPLC and enhancement with DMS
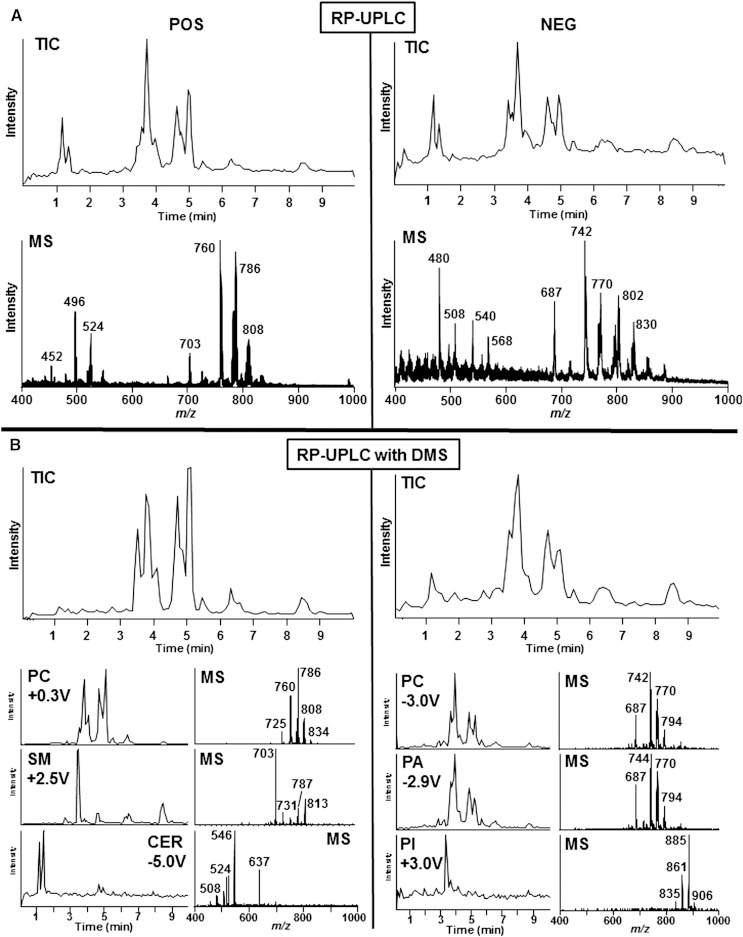
The SelexIONTM unit can function equally well either with infusion or with LC-based sample input strategies. Considering the huge diversity of lipid molecular species present in a biological sample, neither strategy alone is sufficient to resolve isobaric interference during MS analysis. To test the concept of DMS as an orthogonal separation strategy to LC resolution, experiments were designed to separate phospholipid mixtures based on the fatty acid composition and fatty acid chain length using reversed phase (RP) chromatography (method II) and concomitantly resolve eluting lipids based on lipid class using DMS (Fig. 4). It was anticipated that combining both methods would increase the resolution of the lipidomics experiment to better identify the nature of individual molecular species in complex biological samples. In the first set of experiments, human serum lipid extracts were analyzed by ESI-MS in the positive and negative polarities using the nonspecific EMS scan mode (Fig. 4A, upper panels). Because RP chromatography resolves lipids based on their hydrophobicity, which is a quality that is directly related to the fatty acyl chain lengths of each complex lipid, lipids are not resolved by class based on their defining polar head groups as is the case with normal phase chromatography. The TIC data in Fig. 4A demonstrate reasonable resolution in a relatively short chromatographic run (∼10 min). The bottom panels of Fig. 4A display the mass spectra (0–10 min) for the corresponding TIC panels. Note that there is no class identification among the many peaks in the spectrum. Figure 4B shows data from experiments designed identically to those in Fig. 4A except that the DMS was active and COV values for each lipid class and subclass of interest were used (supplementary Table I) to identify the lipid category and class of lipids eluting from the column.
Fig. 4.
Application of DMS combined with RP-UPLC to enhance selectivity. A: RP-UPLC separation of lipids extracted from human serum in positive ion mode (left panels) or negative ion mode (right panels) without DMS (A) or with DMS (B). Top panel of A shows total ion current of RP chromatogram by EMS scan. Bottom panel shows mass spectral data of EMS scan. Top panel of B shows total ion current of RP chromatograms by EMS scan with application of COV values specific for each phospholipid class (sum of 14 scans). Bottom panel shows three selected EMS scans with COV voltages of 0.3, 2.5, and −5.0 specific for PC, SM, and Cer in positive ion mode or −3.0, −2.9, and 3.0 for PC, PA, and PI in negative ion mode. Also shown are mass spectral data.
The TIC panels in Fig. 4B differ from those in Fig. 4A because the DMS only allows certain lipid classes to enter the mass spectrometer, selecting only those lipids with the appropriate COV values. Figure 4B (lower panels) shows the respective extracted spectra for PC, SM, and Cer (positive ion mode) and for PC, PS, and PA (negative ion mode) from human serum extract using COV values specific for each lipid class. Because a nonspecific scan mode was used (i.e., EMS), the results show the ability of this analytical strategy to separate lipid classes by DMS while concomitantly separating the molecules based on their hydrophobicity by LC. Thus, DMS acts as an orthogonal, mass-independent means of lipid class identification, greatly simplifying interpretation of the mass spectra. Data acquired in the positive ion mode used COV values of +0.3, +2.5, and −5.0, specific for PC, SM, and Cer, respectively, and in the negative ion mode used COV values of −3.0, −2.9, and +3.0 to isolate PC, PA, and PI, respectively.
Improved qualitative analysis using DMS
Characterization of lipid molecular species is dependent on clear, high-quality product ion spectra to identify the individual fatty acids esterified to the glycerol backbone of complex lipids. Due to the extensive isobaric overlap of lipid species within the lipidome and the relatively low resolution of precursor ion selection prior to MS/MS (by either triple quadrupole instruments or high resolution instruments), it is commonplace for multiple lipid species sharing the same nominal mass to be isolated for MS/MS analysis even when LC is used. To determine whether DMS can improve the quality of product ion spectra generated during an IDA discovery experiment, human serum extract was separated using LC method II, and lipids were analyzed by MS without DMS (Fig. 5A) and with DMS (Fig. 5B). For the IDA experiment, the survey scan was set to a neutral loss of 141 Da (Fig. 5, top panel), wherein eluting PE molecular species triggered a product ion scan. Although the survey scan is specific for PE, when the instrument switches to MS/MS mode, any molecule with the same nominal mass will fragment and appear in the product ion spectrum. By setting the DMS to a COV value specific for PE, only PE will pass through the DMS, effectively improving the clarity of the product ion spectrum.
Fig. 5.
Combination of RP-UPLC and DMS using IDA reveals molecular species. Top panels show RP-UPLC separation of PE using NLS m/z 141 without DMS (A) or with DMS (B). COV of −4.2 specific for PE was selected. Middle panels show extracted m/z of 744.5 from NLS m/z 141. Bottom panels show enhanced product ion (EPI) scans of the areas indicated in gray above. Inset: magnification from m/z 250 to m/z 325. Of note, the NLSs were performed in the positive ion mode (+); the IDA was performed in the negative ion mode (−).
Figure 5A, top panel shows the survey scan using the semitargeted NLS 141 Da experiment specific for PE without DMS. The LC strategy used a RP column, so PE molecular species elute from the column based on their acyl chain lengths. Figure 5A (middle panel) shows the extracted ion chromatogram for m/z 744.5 from the NLS 141 Da (+), which is the appropriate mass for PE (36:2). In the absence of DMS, the RP-UPLC separation based on hydrophobicity is incomplete and cannot be further resolved. The event-triggered MS/MS scan results in a mass spectrum that indicates the presence of interfering molecules, presumably derived from other chromatographically overlapping lipid categories. Figure 5A (bottom panel) shows multiple fragments associated with fatty acids (i.e., m/z 255, palmitic acid; m/z 281, oleic acid; m/z 279, linoleic acid; m/z 283, stearic acid; and m/z 307, eicosadienoic acid). Thus, the mass spectrum shows a fragmentation pattern that does not allow unequivocal identification of the molecular species and undermines accuracy in quantitation. However, the application of DMS to the chromatographically resolved sample maintains the specificity of the semitargeted scan during MS/MS that is defined by the COV. As shown in Fig. 5B, bottom panel, when DMS is used with a COV specific for PE (COV = −4.2), only two fatty acid peaks are observed, stearic acid (18:0) and linoleic acid (18:2), as well as fragment for LPE (18:0). Note, the MS/MS spectra were acquired in the negative ion mode versus the survey scan (i.e., NLS 141), which was run in the positive ion mode. During the IDA run, masses are adjusted to reflect different polarities; thus, in the middle panels, the extracted ion chromatograms (XICs) reflect the positive ion mass (m/z 744.5), and in the bottom panels, the product ion spectra were acquired from the appropriate negative ion mass, m/z 742.4. Thus, the product ion spectrum is clarified, and the fragment ions fully support the identification of the molecule as PE (18:0/18:2) and can be used to accurately quantitate the molecule.
DISCUSSION
A major difficulty encountered in the analysis of lipids by MS is related to the extensive isobaric or near-isobaric overlap of different lipid molecular species within a sample. For example, at a nominal mass of m/z 762.4 with a mass tolerance of ± 0.1, there are at least 25 different lipid molecular species, some of which are iso-elemental, meaning they have the same molecular formula but are different chemical species, each with its own biological function.4 The problem of isobaric and near-isobaric interference is magnified during product ion analysis wherein a precursor ion is selected for fragmentation. The isolation window is much wider than the instrument tolerance (isolation widths vary between instrument types but range between 0.4 and 1.2 amu), so the number of molecules that are funneled to the collision cell can be dramatically higher (∼2503). This issue is the fundamental challenge in the identification and quantification of lipids by MS.
Various strategies have been devised to address lipid isobaric interference. The most common method is online separation by HPLC. Using chromatography columns to separate lipids by category, class, and/or fatty acyl chain length greatly simplifies acquired mass spectra to improve identification and quantitation (11). However, due to the wide diversity in chemical structures and physical properties among lipids, it is challenging to develop a single HPLC method to resolve all lipids in a complex, biological lipid extract. The introduction of UPLC and the use of tightly packed columns have greatly improved the resolution of complex lipid mixtures. In these studies, UPLC resolved lipids into their respective categories and classes using an HILIC column in a short chromatographic time frame (Figs. 1 and 3), and the use of an RP column resolved the same lipids by their respective fatty acyl chain lengths (Figs. 4 and 5). These methods provide reasonable resolution, but even with chromatographic separation, lipid isobars still coelute, as demonstrated in Fig. 5A, making qualitative identification problematic. The balance between resolving lipid classes based on the polarity of their head groups versus resolving lipids based on their hydrophobicity contributed by the fatty acyl chains inherently diminishes lipid coverage in a profiling experiment. An orthogonal approach using DMS to separate lipids by category and class while simultaneously resolving lipids by chromatography offers one solution to this difficult problem.
The chromatographic separation of lipid extracts was enhanced with the use of DMS coupled with MS to allow orthogonal separation of components in the analyzed mixtures without a significant increase in the analysis time, yielding a set of more specific, less convolved data resulting in confident species identification and improved detection of low-level components within a matrix. This new workflow adds a new dimension to the collected data and, thus, improves the quality of the results and confidence in the identification of lipid species.
The ability of DMS to resolve lipid isobars lies in its ability to separate molecules based on their chemical properties (24, 25, 29) rather than size and shape alone, factors that are the basis for separation by ion mobility in drift tubes (19). The length of the DMS cell is short (3 cm) compared with more traditional ion mobility drift tubes, as the resolution lies in the separation of molecules based on their differential mobilities under oscillating high and low electrical fields. The chemical effects are enhanced in the presence of chemical modifiers, such as the 1-propanol used in these studies. Under low electrical fields, the chemical modifier clusters around the ions, affecting their shape, size, and chemical properties, resulting in a specific mobility through the cell. Under high electrical fields, the modifier declusters from the ion. Hence, the net effect of the modifier is enhancement of the differential mobility under the oscillating electrical field (25, 29). This technique differs from traditional ion mobility spectrometry wherein molecules are separated by their cross-sectional area, which is related to their shape and size (and hence molecular weight), rather than chemical effects (30).
The power of DMS to resolve lipid categories and classes is most prominently exemplified during multiplexed NLS analysis (Fig. 5). Triple quadrupole instruments, by the nature of their design and configuration, are capable of semitargeted scans that isolate specific classes of lipids, depending on its specific structural motifs. For example, PS can be isolated by looking for neutral losses of 185 Da in the positive ion mode. This transition is specific to the loss of the phosphoserine head group, and the resultant spectrum displays only PS molecular species. However, during product ion analysis of precursor ions identified in the NLS, the specificity is lost, and any molecule with the same nominal mass as the target precursor will fragment and contribute to the product ion spectrum. As a consequence, it can be a challenge to determine which fragments are associated with a specific precursor ion. Furthermore, considering that there are a limited number of common fatty acids, and hence a limited number of unique fragments, among all complex lipids, multiple different precursor ions can generate some of the same fatty acid fragments. This not only confounds identification but can significantly affect quantitative rigor due to isobaric MRM transitions.
Figure 5 demonstrates that even with reasonable chromatography, lipid isobars from different lipid classes do coelute. The clarity of the product ion spectrum in Fig. 5B obtained using the DMS to isolate PE molecular species makes identification of the molecule straightforward. The PE-specific COV setting ensures that only PE molecules enter the MS, and, thus, the issue of low resolution precursor isolation is no longer relevant. The diagnostic fatty acid anions can be directly attributed to a single precursor with a defined total number of carbons and double bonds, which enables facile determination of the fatty acid distribution among the positional isobars and simultaneously provides quantitative insight into their relative concentrations. For example, PC 36:2 (m/z 786.6) may have multiple isomers, including PC (18:1/18:1) or (18:0/18:2); these molecules can be identified in a sample based on the presence of the specific fatty acid fragments, and their relative concentrations can be assessed by integrating the area under the peak for these fragments only if interfering isobaric peaks are eliminated from the spectrum. These results demonstrate the power of including DMS in lipid separation strategies: DMS enables facile class identification and, in combination with RP chromatography, which resolves the lipids based on their chain lengths, results in the identification of lipids at the level of molecular species in a single analytical run.
The ability of DMS to resolve lipid mixtures can be applied to shotgun lipidomics via infusion as well. Indeed, one of the major criticisms of the shotgun approach has been the difficulty to deconvolute the mass spectra at the precursor ion and product ion levels. With DMS, however, lipids can be isolated with category- and class-specific COV values (supplementary Table I) prior to MS analysis, thereby greatly reducing the complexity of the sample and analysis. Figure 2 demonstrates the ability of DMS to resolve lipids during an infusion experiment. Although not the central focus of this study, the possibility of DMS to improve shotgun lipidomic analysis is intriguing. The resolution of human serum extract using RP-UPLC in conjunction with DMS demonstrates the orthogonality of the method. RP separation and product ion analysis in the negative ion mode does not allow for class identification. However, concomitantly using lipid class-specific COV values enables class identification, fatty acid composition, and baseline noise reduction to improve quantitative rigor by effectively increasing the signal-to-noise ratio of analytes (Fig. 4).
The COV values given in supplementary Table I remain constant over time provided conditions within the DMS cell are not changed. A test of the stability of the COV voltages over an extended time period showed no significant fluctuation over 4 days (not shown).5 Another consideration regarding COV values is the in-source formation of adducts. PA, an anionic lipid, readily forms chloride and acetate adducts. Care should be taken to select a mobile phase or infusion solvent that favors the formation of one adduct, if any, because each adduct has its own unique COV value.
The use of DMS as an independent, orthogonal separation tool during LC ESI-MS/MS analysis of lipids significantly improves lipidomic analyses. Specifically, DMS in combination with RP-UPLC enhances the qualitative aspects of lipid analysis by providing an additional separation element that complements the separation physics underlying the hydrophobic interactions of RP chromatography. As exemplified in Fig. 5, in combination with RP-UPLC, DMS enhances the qualitative aspects of lipid analysis and enables the resolution of isobaric species into individual molecular species that would not be possible with chromatography alone. In this report, we have not addressed the quantitative aspects of DMS but show its resolution potential as a supplementary technique to LC to resolve targets of interest in case of ambiguity. Lipid classes can be readily identified owing to each class having a unique COV value among the two different instrument polarities; the instrument baseline noise is reduced, thereby increasing the signal-to-noise ratio; and the resolution of lipids provided by the column remains unchanged. The cost of additional resolution, as is the case for all mass spectrometric measurement, is a decrease of three to five times in the base sensitivity of the instrument. However, because the chemical noise is reduced, the effects on sensitivity appear to be lipid class-specific and must be tested with authentic standards during assay development and validation. Of note, because DMS dramatically reduces background noise interferences, especially when working with biological samples, the overall detection limits generally improve. Overall, DMS significantly enhances the resolution power of lipids compared with HPLC alone. The clarity it provides to qualitative analyses makes DMS an attractive technology to MS-based studies in lipidomics.
Supplementary Material
Footnotes
Abbreviations:
- Cer
- ceramide
- COV
- compensation voltage
- DMS
- differential ion mobility spectrometry
- EMS
- enhanced mass spectrometry
- HILIC
- hydrophilic interaction liquid chromatography
- IDA
- information-dependent acquisition
- LPE
- lysophosphatidylethanolamine
- MRM
- multiple reaction monitoring
- NLS
- neutral loss scan
- PA
- phosphatidic acid
- PC
- phosphatidylcholine
- PE
- phosphatidylethanolamine
- PG
- phosphatidylglycerol
- PI
- phosphatidylinositol
- PIS
- precursor ion scan
- PS
- phosphatidylserine
- RP
- reversed phase
- TIC
- total ion current
- UPLC
- ultra performance liquid chromatography
This work was supported by LIPID MAPS Glue Grant U54 GM069338. For research use only. Not for use in diagnostic procedures. The trademarks mentioned herein are the property of AB Sciex Pte. Ltd. or their respective owners. AB SCIEX is being used under license.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one table and one figure.
REFERENCES
- 1.Loizides-Mangold U. 2013. On the future of mass-spectrometry-based lipidomics. FEBS J. 280: 2817–2829. [DOI] [PubMed] [Google Scholar]
- 2.Wenk M. R. 2005. The emerging field of lipidomics. Nat. Rev. Drug Discov. 4: 594–610. [DOI] [PubMed] [Google Scholar]
- 3.Dennis E. A., Brown H. A., Deems R. A., Glass C. K., Merrill A. H., Murphy R. C., Raetz R. H., Shaw W., Subramaniam S., Russel D. W., et al. 2005. The LIPID MAPS approach to lipidomics. In Functional Lipidomics. L. Feng and G. Prestwich, editors. CRC Press/Taylor & Francis Group, Boca Raton, FL. 1–15. [Google Scholar]
- 4.Fenn J. B., Mann M., Meng C. K., Wong S. F., Whitehouse C. M. 1989. Electrospray ionization for mass spectrometry of large biomolecules. Science. 246: 64–71. [DOI] [PubMed] [Google Scholar]
- 5.Yost R. A., Enke C. G. 1979. Triple quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal. Chem. 51: 1251–1264. [DOI] [PubMed] [Google Scholar]
- 6.Han X., Gross R. W. 1994. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. USA. 91: 10635–10639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milne S., Ivanova P., Forrester J., Alex Brown H. 2006. Lipidomics: an analysis of cellular lipids by ESI-MS. Methods. 39: 92–103. [DOI] [PubMed] [Google Scholar]
- 8.Han X., Gross R. W. 2003. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 44: 1071–1079. [DOI] [PubMed] [Google Scholar]
- 9.Harkewicz R., Dennis E. A. 2011. Applications of mass spectrometry to lipids and membranes. Annu. Rev. Biochem. 80: 301–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dennis E. A., Deems R. A., Harkewicz R., Quehenberger O., Brown H. A., Milne S. B., Myers D. S., Glass C. K., Hardiman G. T., Reichart D., et al. 2010. A mouse macrophage lipidome. J. Biol. Chem. 285: 39976–39985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quehenberger O., Dennis E. A. 2011. The human plasma lipidome. N. Engl. J. Med. 365: 1812–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cooper M. J., Anders M. W. 1975. High pressure liquid chromatography of fatty acids and lipids. J. Chromatogr. Sci. 13: 407–411. [DOI] [PubMed] [Google Scholar]
- 14.McCluer R. H., Ullman M. D., Jungalwala F. B. 1986. HPLC of glycosphingolipids and phospholipids. Adv. Chromatogr. 25: 309–353. [PubMed] [Google Scholar]
- 15.DeLong C. J., Baker P. R., Samuel M., Cui Z., Thomas M. J. 2001. Molecular species composition of rat liver phospholipids by ESI-MS/MS: the effect of chromatography. J. Lipid Res. 42: 1959–1968. [PubMed] [Google Scholar]
- 16.Shevchenko A., Simons K. 2010. Lipidomics: coming to grips with lipid diversity. Nat. Rev. Mol. Cell Biol. 11: 593–598. [DOI] [PubMed] [Google Scholar]
- 17.Dennis E. A. 2009. Lipidomics joins the omics evolution. Proc. Natl. Acad. Sci. USA. 106: 2089–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwudke D., Hannich J. T., Surendranath V., Grimard V., Moehring T., Burton L., Kurzchalia T., Shevchenko A. 2007. Top-down lipidomic screens by multivariate analysis of high-resolution survey mass spectra. Anal. Chem. 79: 4083–4093. [DOI] [PubMed] [Google Scholar]
- 19.Kanu A. B., Dwivedi P., Tam M., Matz L., Hill H. H., Jr 2008. Ion mobility-mass spectrometry. J. Mass Spectrom. 43: 1–22. [DOI] [PubMed] [Google Scholar]
- 20.Shvartsburg A. A., Isaac G., Leveque N., Smith R. D., Metz T. O. 2011. Separation and classification of lipids using differential ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 22: 1146–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell J. L., Le Blanc J. C., Schneider B. B. 2012. Probing electrospray ionization dynamics using differential mobility spectrometry: the curious case of 4-aminobenzoic acid. Anal. Chem. 84: 7857–7864. [DOI] [PubMed] [Google Scholar]
- 22.Jin W., Jarvis M., Star-Weinstock M., Altemus M. 2013. A sensitive and selective LC-differential mobility-mass spectrometric analysis of allopregnanolone and pregnanolone in human plasma. Anal. Bioanal. Chem. 405: 9497–9508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maccarone A. T., Duldig J., Mitchell T. W., Blanksby S. J., Duchoslav E., Campbell J. L. 2014. Characterization of acyl chain position in unsaturated phosphatidylcholines using differential mobility-mass spectrometry. J. Lipid Res. 55: 1668–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneider B. B., Covey T. R., Coy S. L., Krylov E. V., Nazarov E. G. 2010. Planar differential mobility spectrometer as a pre-filter for atmospheric pressure ionization mass spectrometry. Int. J. Mass Spectrom. 298: 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell J. L., Zhu M., Hopkins W. S. 2014. Ion-molecule clustering in differential mobility spectrometry: lessons learned from tetraalkylammonium cations and their isomers. J. Am. Soc. Mass Spectrom. 25: 1583–1591. [DOI] [PubMed] [Google Scholar]
- 26.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 27.Jasak J., Le Blanc Y., Speer K., Billian P., Schoening R. M. 2012. Analysis of triazole-based metabolites in plant materials using differential mobility spectrometry to improve LC/MS/MS selectivity. J. AOAC Int. 95: 1768–1776. [DOI] [PubMed] [Google Scholar]
- 28.Ray J. A., Kushnir M. M., Yost R. A., Rockwood A. L., Wayne Meikle A. 2014. Performance enhancement in the measurement of 5 endogenous steroids by LC-MS/MS combined with differential ion mobility spectrometry. Clin. Chim. Acta. In press. [DOI] [PubMed] [Google Scholar]
- 29.Schneider B. B., Covey T. R., Coy S. L., Krylov E. V., Nazarov E. G. 2010. Control of chemical effects in the separation process of a differential mobility mass spectrometer system. Eur. J. Mass Spectrom. (Chichester, Eng.). 16: 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kliman M., May J. C., McLean J. A. 2011. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim. Biophys. Acta. 1811: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.