SUMMARY
Protein interactions form a network whose structure drives cellular function and whose organization informs biological inquiry. Using high-throughput affinity-purification mass spectrometry, we identify interacting partners for 2,594 human proteins in HEK293T cells. The resulting network (BioPlex) contains 23,744 interactions among 7,668 proteins with 86% previously undocumented. BioPlex accurately depicts known complexes, attaining 80-100% coverage for most CORUM complexes. The network readily subdivides into communities that correspond to complexes or clusters of functionally related proteins. More generally, network architecture reflects cellular localization, biological process, and molecular function, enabling functional characterization of thousands of proteins. Network structure also reveals associations among thousands of protein domains, suggesting a basis for examining structurally-related proteins. Finally, BioPlex, in combination with other approaches can be used to reveal interactions of biological or clinical significance. For example, mutations in the membrane protein VAPB implicated in familial Amyotrophic Lateral Sclerosis perturb a defined community of interactors.
INTRODUCTION
A central goal in cell biology is to describe the molecular processes that drive cellular function. While these are genomically encoded, they are executed by the proteome. The proteome can be viewed as constellations of interacting protein modules organized into signal transduction networks, molecular machines, and organelles. However, our knowledge of proteome architecture is fragmentary, as is our conception of how protein interconnectivity is influenced by genetic and cellular variation.
Our understanding of mammalian proteome structure has emerged from 5 strategies. First, focused biochemical studies have revealed stable macromolecular complexes. Second, affinity purification of epitope-tagged proteins followed by mass spectrometry (AP-MS) has identified proteins associated with baits from many protein families, including deubiquitinating enzymes (Sowa et al., 2009), histone deacetylases (Joshi et al., 2013), and chaperones (Taipale et al., 2014). Third, complementary approaches involving either target-specific antibodies for immunoprecipitation (IP)-MS or correlation profiling of soluble protein assemblies have identified many complexes (Havugimana et al., 2012; Malovannaya et al., 2011). Fourth, yeast two-hybrid (Y2H) analysis of ~14,000 human open reading frames (ORFs) has identified binary protein interactions (Rolland et al., 2014). Finally, several databases archive protein interactions from literature (Franceschini et al., 2013; Licata et al., 2012; Ruepp et al., 2009; Stark et al., 2011; Warde-Farley et al., 2010). While these repositories allow in silico interaction network construction, many literature interactions are context-dependent and the stringency of criteria used to identify interactions varies across studies. Thus, databases vary in content and quality.
Given this perspective, remaining challenges concern mapping globally the human protein interactions within a single cell type in a physiological context, and understanding how network architecture depends upon genetic and physiological variation. These challenges reflect 1) the myriad genes, isoforms, and modification states encoded by the human genome, 2) the low abundance of many proteins, which limits detection, 3) many transient interactions that complicate signaling network mapping, and 4) the prevalence of membrane proteins, which often require specialized methods for purification. While no single approach can address all challenges, several attributes of AP-MS will facilitate delivery of a “first-pass” global human interactome. One advantage is its exquisite sensitivity. In addition, unlike binary methods, AP-MS determines components within multi-protein complexes. AP-MS has previously mapped a substantial fraction of yeast (Gavin et al., 2002; Ho et al., 2002; Krogan et al., 2006) and Drosophila (Guruharsha et al., 2011) interactions.
Here, we report AP-MS analysis of 2,594 baits to produce a human interaction map spanning 23,744 interactions among 7,668 proteins. While we detected many known interactions, validating the methodology, most have not been reported, reflecting targeting of many understudied proteins and highlighting AP-MS sensitivity. In addition, we identified 354 communities representing known and previously unidentified complexes. Moreover, integration of protein domain and localization information revealed enrichment of domains within sub-networks and highly correlated localization within complexes, while suggesting biological roles for proteins of unknown function. Finally, we merge isobaric labeling with AP-MS to quantify how genetic variation alters interactions of VAPB variants associated with familial Amyotrophic Lateral Sclerosis (ALS), revealing mutation-specific loss and gain of interactions. Ultimately, BioPlex unveils both individual protein functions and global proteome organization.
RESULTS
Achieving Rapid and Reliable AP-MS
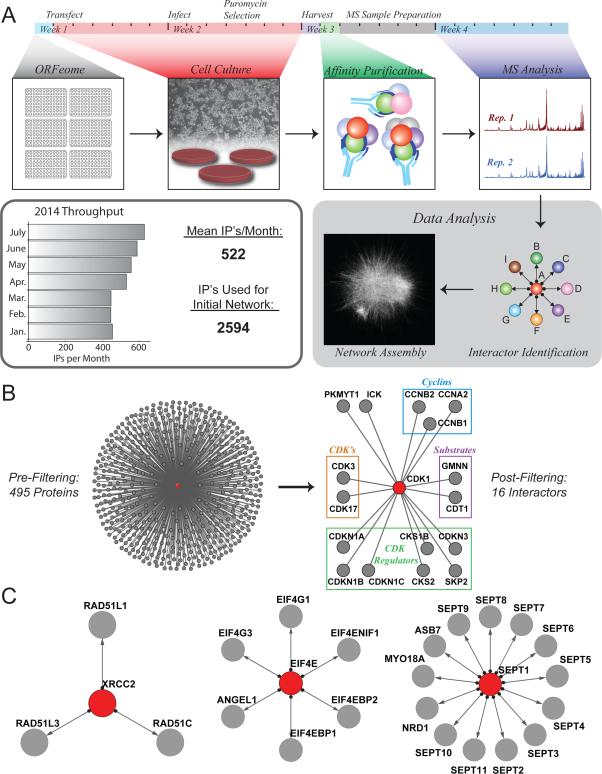
Globally applying AP-MS requires both high capacity and rigorous quality control. As depicted in Figure 1A and described in the Extended Experimental Procedures, we have initiated high-throughput lentiviral expression and AP-MS profiling of C-terminally FLAG-HA-tagged baits from 13,000 protein-coding open reading frames within the sequence-validated Human ORFEOME collection v. 8.1 (Yang et al., 2011). Using this system, single biological replicates of up to 600 baits have been expressed in HEK293T cells, immuno-purified, and analyzed via mass spectrometry in technical duplicate each month. Baits have been processed in batches corresponding to 96-well plates within the ORFEOME, selected randomly from the library. Peptides and proteins were identified and filtered within each IP to a 1% FDR, with additional filters to control network FDR (Extended Experimental Procedures). Given the scale of this endeavor, a paramount concern is data integrity. In addition to clone validation and quality control during the wet-lab pipeline, automated evaluation of MS performance, comparison of LC-MS technical duplicates, automated validation of bait protein detection, and inclusion of positive (RAB11B) and negative (GFP) controls ensured consistent data quality (Figure S1A and Extended Experimental Procedures). While analysis of all protein-coding genes in the Human ORFEOME is ongoing, we focus here on the first 2594 AP-MS experiments (Table S1).
Figure 1. High-Throughput Interaction Mapping via AP-MS.
(A) AP-MS platform: 1) A lentiviral library of 13,000 FLAG-HA-tagged ORFs was constructed from the Human ORFEOME; 2) 293T cells were infected and expanded under puromycin selection; 3) Baits and preys were immuno-purified; 4) tryptic digests were analyzed in technical duplicate by LC-MS; 5) Proteins were identified and specific interactors found; 6) Interactions were assembled to model the human interactome. Up to 600 AP-MS experiments may be completed per month.
(B) CompPASS-Plus extracts 16 interactors for bait CDK1 from a background of nearly 500 proteins.
(C) Interaction maps for baits XRCC2, EIF4E, and SEPT1 (red). Nearly all interactions have been previously described. Interactors were identified from backgrounds of 487, 778, and 749 proteins, respectively.
Global Protein Interaction Mapping via CompPASS-Plus
Our work employs CompPASS, which has identified high-confidence interacting proteins (HCIP's) for up to ~100 baits (Sowa et al., 2009). CompPASS quantifies enrichment of each protein in each IP, relative to other unrelated AP-MS datasets, based on abundance, detection frequency, and reproducibility. HCIP's are identified using the Normalized Weighted D-Score (NWD-Score) and Z-Score (Figure S2A).
To improve performance across thousands of AP-MS datasets, we developed CompPASS-Plus, a Naïve Bayes classifier that recognizes HCIP's using several features (Figure S2A). In addition to CompPASS scores, features measure batch variations, overall spectral counts, unique peptide counts, and protein detection frequency. Shannon entropy quantifies a protein's consistency of detection across technical duplicate LC-MS analyses, which removes inconsistent protein identifications and minimizes LC carry-over artifacts (Figure S2B). Because CompPASS-Plus must distinguish HCIP's from background proteins and incorrect protein identifications, proteins are sorted into 3 classes (Figure S2C). Since the few (0.05%) incorrect protein identifications that survive filtering earn NWD- and Z-Scores that most closely resemble those of HCIP's, sorting incorrect protein identifications separately improves accuracy. To train CompPASS-Plus, incorrect protein identifications were modeled using decoy proteins that survived peptide- and protein-level FDR filtering, while HCIP's were modeled using bait-prey pairs reported in STRING (Franceschini et al., 2013) or GeneMania (Warde-Farley et al., 2010); all other bait-prey pairs modeled non-specific background. To minimize over-fitting, each AP-MS experiment was scored using a classifier trained excluding data from its own IP and other IPs on the same plate (Figure S2D). CompPASS-Plus assigns each bait-prey pair in every AP-MS experiment three scores reflecting the probability that it corresponds to a wrong identification, a background protein, or an HCIP. Bait-prey pairs that receive an interaction score of at least 0.75 are considered HCIP's.
CompPASS-Plus effectively distinguishes interactions from background. When AP-MS data were aligned with high-confidence interactions from CORUM (Ruepp et al., 2009), high-scoring bait-prey pairs were most frequently confirmed (Figure S2E). Similarly, over 87% of decoy proteins were classified as incorrect identifications (Figure S2F). When used to classify known true- and false-positive interactions across 30 biological replicate positive and negative control AP-MS experiments, CompPASS-Plus identified known interactions with high sensitivity and specificity (Figure S2G).
The utility of CompPASS-Plus to identify HCIPs from individual AP-MS experiments is depicted in Figure 1B-C. From 495 proteins detected with CDK1, only 16 remained after filtering, and all but 2 (ICK, PKMYT1) are known CDK1 associated proteins or substrates. Notably, all HCIP's for XRCC2 and EIF4E are known (Figure 1C): XRCC2 recovers the entire BCDX2 complex (Masson et al., 2001), while EIF4E binds the EIF4F complex, EIF4E-binding proteins 1 and 2, and its known interactor ANGEL1 (Gosselin et al., 2013). Similarly, 11 of 13 HCIPs for filamentous SEPT1 GTPase are either related SEPT proteins or known SEPT1 interactors. Other examples are highlighted in Figure S1B/C, comparing BioPlex with interactions reported by Y2H (Rolland et al., 2014) or in a previous AP-MS study of DUBs (Sowa et al., 2009).
An AP-MS Map of the Human Interactome
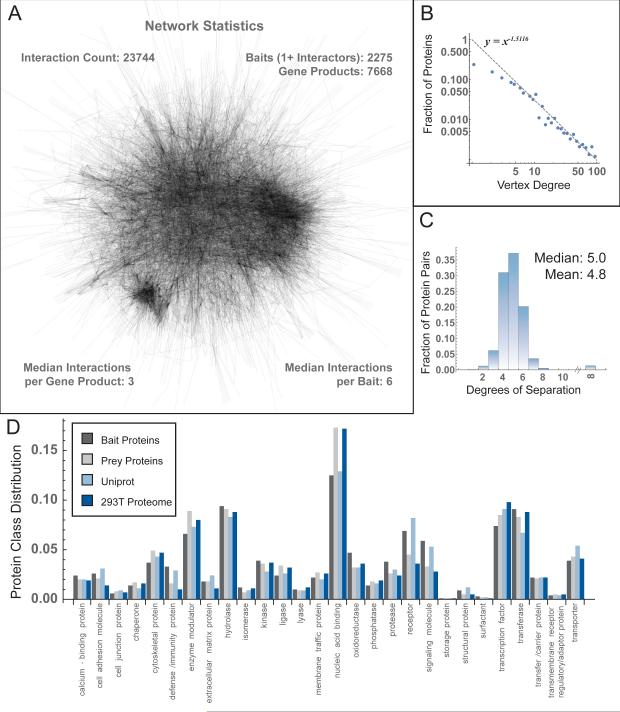
Although each AP-MS experiment identifies proteins associated with one bait, when all are combined, the interactions form a model of the interactome. This network, whose largest component is depicted in Figure 2A, includes 23,744 interactions connecting 7,668 gene products (Table S2). We call this network BioPlex (Biophysical interactions of ORFEOME-derived complexes) and provide a graphical viewer (Figure S3). Of 2,594 baits, 319 were not found to interact with any other proteins in 293T cells under basal conditions as C-terminally tagged proteins (Table S1). The median bait interacted with 6 other proteins, while the median gene product (including preys as well as baits) participated in 3 interactions, suggesting that Bioplex underestimates interactions of proteins not yet targeted for AP-MS.
Figure 2. BioPlex Network Properties.
(A) BioPlex. The largest component is depicted.
(B) Vertex degree distribution of all proteins in BioPlex. The dashed line represents the best fit power law (see Extended Experimental Procedures).
(C) Histogram depicting the number of degrees separating all protein pairs.
(D) Functional classifications of baits and preys assigned by PANTHER and compared against the functional distributions of our HEK293T proteome and the entire human UniProt proteome.
Although the median interaction count for each protein is low, significant variability is observed and the degree distribution is skewed by proteins with many interacting partners, reflecting coverage of proteins participating in large assemblies. When plotted in log-log space (Figure 2B), the fraction of proteins observed across the range of vertex degrees follows a linear trend for degrees above 4 that is consistent with power law behavior typical of scale-free networks (Barabasi and Albert, 1999) and has been observed in protein interaction and metabolic networks (Vidal et al., 2011). In addition, fewer proteins than expected participate in very few (i.e. less than 5) interactions, as indicated by the deviation from power law behavior for low interaction counts. More than 98% of protein pairs connect to each other, most within 5 or fewer degrees of separation (Figure 2C).
To accurately model the interactome, BioPlex should sample proteins evenly across functional categories. Functional classification using PANTHER (Mi et al., 2012) in conjunction with all UniProt proteins (Magrane and Consortium, 2011) revealed a distribution of baits and preys that closely matched UniProt functional categories (Figure 2D). Another consideration is the influence of the AP-MS system on the interaction network. Since baits have been expressed in HEK293T cells, we profiled the 293T proteome, identifying 10,326 proteins and assigning each to abundance percentiles (Figure S4A). HEK293T proteome functional classification mimicked UniProt and most closely resembled the distribution observed for preys (Figure 2D). We then mapped the 293T proteome onto BioPlex. As expected, 90% of preys were also detected in 293T cells via total proteome analysis (Figure S4B); the remaining 10% were only detected via AP-MS, suggesting that their abundance was below detection limits without enrichment. In contrast, only 60% of baits were detected in our 293T proteome (Figure S4B). This is not surprising since bait selection was unbiased and 293T cells only express a fraction of ORFEOME-encoded proteins. Baits were drawn evenly from across native 293T expression levels. In contrast, preys were enriched in the upper third of the abundance range, with proteins in the bottom 25% under-represented (Figure S4C). Though bait levels vary across 2594 IP's, most were detected at moderate levels (Figure S4D). Furthermore, bait abundances and interaction counts were uncorrelated (Figure S4E). Although the extent of bait over-expression is difficult to judge and varies across IP's, previous experimentation has shown that over-expression has little effect on identification of true interacting partners (Sowa et al., 2009). Overall, notwithstanding idiosyncrasies of the 293T expression system, the resulting network constitutes a representative cross-section of the proteome.
BioPlex Recapitulates Known Complexes and Reveals Thousands of New Interactions
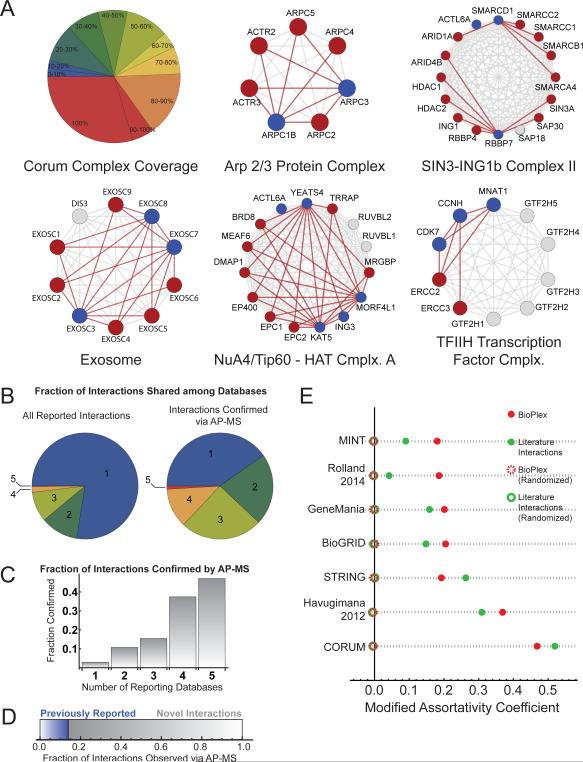
To assess its accuracy and completeness, we superimposed BioPlex onto high-confidence CORUM complexes detected via low-throughput methods, attaining high overlap. When at least two constituent proteins were selected as baits, more than 25% of CORUM complexes were perfectly recapitulated by AP-MS (Figure 3A). Similarly, over 1/3 of complexes achieved at least 90% coverage, while 1/2 were 80% complete and nearly 3/4 attained at least 50% coverage. Proteins within a complex are often highly interconnected, as for the Arp 2/3 and NuA4/Tip60-HAT complexes and the Exosome. In contrast, RBBP7 belongs to a large complex assembled from two smaller complexes, each of which has been separately isolated by AP-MS: HDAC1/2-ING1-SIN3A-SAP30-SAP18-RBBP4 (Complex I) and ARID1A-ARID4B-SMARCD1-SMARCC1-SMARC2-SMARCB1-SMARCA4 (Complex II) (Kuzmichev et al., 2002). Our RBBP7 analysis identified all components of Complex I except SAP18, possibly reflecting its small size (153 amino acids), as well as a single component of Complex II (ARID4B). In contrast, SMARCD1 associated with 6 of 8 subunits of the Complex II SMARC/ARID sub-complex (Figure 3A). This may reflect relatively weaker interactions among the sub-assemblies. Likewise, the CDK Activating Kinase (CAK) complex CDK7-CCNH-MNAT1 is a multifunctional protein kinase that is involved in both CDK activation and in transcription-coupled repair through the TFIIH complex. While the core kinase complex and its interactions with 3 TFIIH complex components (ERCC2, ERCC3, and GTF2H1) were detected using CDK7, CCNH, and MNAT1 as baits, several TFIIH components were not detected (Figure 3A). This incomplete TFIIH complex identification with tagged CAK could reflect stringent washing or the relative abundance of CAK complexes versus TFIIH complexes. To understand why many TFIIH complex members were absent, we examined AP-MS data targeting GTF2H3 that was acquired via our pipeline while this manuscript was in preparation. Encouragingly, we detected interactions with GTF2H1, GTF2H2, GTF2H4, and ERCC3, attaining nearly complete coverage of this complex (data not shown). Overlap with CORUM will continue to improve as the network grows to include analysis of most human proteins.
Figure 3. Evaluation of AP-MS Protein Interactions.
(A) AP-MS interactions superimposed onto CORUM complexes. The pie chart depicts the fraction of complexes achieving the indicated coverage in BioPlex. Only complexes containing two or more baits were considered. Five representative CORUM complexes: baits are colored blue, while preys are red and proteins not observed by AP-MS are gray. Interactions among CORUM complex members are gray while interactions confirmed by AP-MS are red.
(B) Physical protein interactions reported in BioGrid, CORUM, GeneMania, STRING, and MINT were merged. Left: overlap among databases. Right: overlap among databases for interactions confirmed by AP-MS.
(C) Fraction of database interactions confirmed by AP-MS as a function of the number of supporting database reports. The composite interaction database was filtered to include only interactions connecting one of 2594 baits with proteins observed as baits or preys in the interaction network.
(D) 86% of AP-MS interactions have not been reported in the databases listed above.
(E) Pairwise comparisons of BioPlex with published interaction networks were performed, using graph assortativity to quantify preferential interaction in cases of shared localization among proteins detected in both networks. Literature datasets included BioGRID, CORUM, GeneMania, STRING, and MINT, as well as interactions recently reported via yeast-two-hybrid (Rolland et al., 2014) and LC-MS correlation profiling (Havugimana et al., 2012). Each analysis was repeated with randomized localizations as a control.
To enable more extensive comparison, we compiled all human physical interactions reported in CORUM, BioGRID (Stark et al., 2011), GeneMania, STRING, and MINT (Licata et al., 2012). The latter 4 databases accept a variety of evidence in support of protein interactions, including high-throughput techniques, and thus contain many more interactions at correspondingly higher false-positive rates. Only physical interactions supported by direct experimental evidence were included; interactions due to co-expression, co-localization, text-mining, or predictions were excluded. Inter-database overlap was limited, with fewer than 25% of interactions reported by multiple databases (Figure 3B). Notwithstanding such narrow agreement, interactions seen in BioPlex were more frequently found in multiple databases (Figure 3B). This suggests that BioPlex preferentially overlaps with the most frequently reported interactions. While interactions found in all 5 databases were confirmed by AP-MS nearly 50% of the time and more than 35% of interactions reported by 4 databases were confirmed, only 2% of interactions unique to just one database were observed (Figure 3C).
Approximately 84% of the 23,744 BioPlex interactions have not been reported (Figure 3D), reflecting sampling of many “pioneer” proteins and increased analytical depth afforded by new instrumentation. For further validation, we compared reported subcellular localizations of interacting protein pairs as an indirect measure of plausibility. While co-localization cannot guarantee physical association, interacting protein pairs must at least partially co-localize. In contrast, false-positive interacting proteins would localize randomly with respect to each other. To assess co-localization, we mapped UniProt subcellular localizations onto BioPlex. The tendency for co-localized proteins to interact is measured by graph assortativity (Extended Experimental Procedures). Positive assortativities indicate preferential interactions among proteins in the same compartment, while values near zero imply random interactions.
As an indirect validation, we calculated assortativities for several databases and interaction datasets and compared each with BioPlex (Figure 3E). Because localizations of proteins in each dataset varied considerably, pairwise comparisons with BioPlex were performed focusing on interactions that connected proteins in both networks. Assortativities quantify each network's tendency to connect these shared proteins according to subcellular localization. Pairwise analyses were also repeated with randomized subcellular localizations. While assortativities in each pairwise comparison varied due to differences in the extent of biological characterization and quality of subcellular localization information available for shared proteins, all networks exhibited non-random interactions. Proteins included in both CORUM and BioPlex were well-characterized and predominantly nuclear or cytoplasmic, attaining the highest coefficients. In contrast, comparisons that included higher proportions of less studied proteins exhibited reduced, though highly significant, preferences for connecting proteins with shared localization. Overall, BioPlex compared favorably with published interaction networks: in 5 of 7 cases, BioPlex showed a greater tendency to connect co-localized proteins; in the remaining 2 cases, differences were small and reflected in part interactions missing from BioPlex because neither protein has been targeted for AP-MS. This analysis suggests that the BioPlex network is at minimum comparable in quality to previously published interaction data, a conclusion that extends to the many unreported interactions it contains.
BioPlex Community Structure Reveals the Interactome Functional Organization
While several BioPlex complexes have been highlighted by comparison with CORUM, complexes may also be revealed from network topology alone as clusters of highly interconnected proteins. Since no prior knowledge is required, community detection algorithms may associate new proteins with known complexes and identify unknown complexes. We have employed a two-stage strategy to map BioPlex community structure, using clique percolation (Palla et al., 2005) to identify 256 communities that were further subdivided via modularity-based clustering (Newman, 2004) into 354 communities and sub-communities (Table S3).
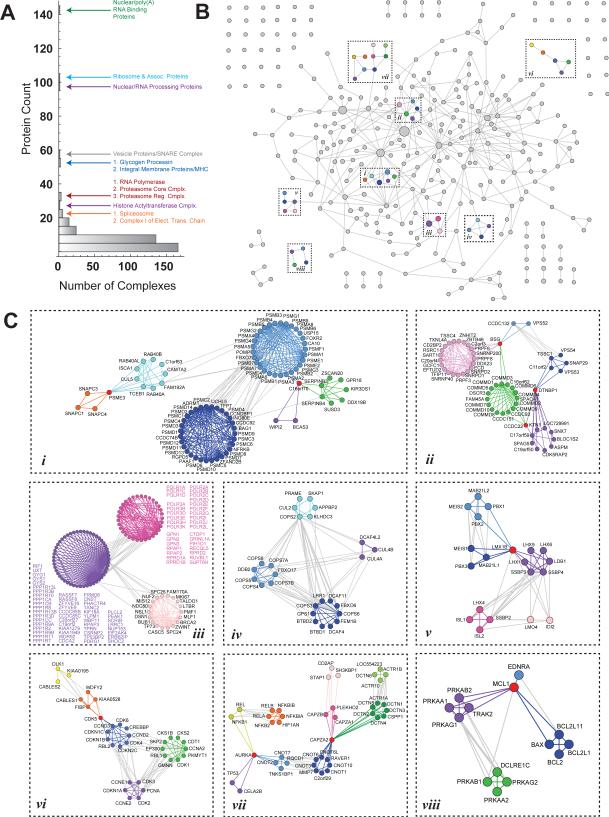
Community size varied from 2 to 140 proteins, though most included fewer than 20 members (Figure 4A). Most communities encompassed related proteins, as 84% were enriched for at least one GO term or Pfam domain (Figure S5A). Moreover, many communities match known complexes, forming a network that depicts interactome organization (Figure 4B). Subsets of this network have been enlarged (Figure 4C) to reveal underlying protein interactions.
Figure 4. Community Analysis of the AP-MS Interactome.
(A) BioPlex communities: 256 initial communities were identified via 3-clique percolation and sub-divided via modularity-based clustering, resulting in 354 communities.
(B) Network of communities. Gray circles represent 354 BioPlex communities; circle size is proportional to the number of member proteins. Gray edges connect communities that share a common protein or were initially classified as a single community and were sub-divided via modularity-based clustering. Labeled boxes highlight communities expanded in part C.
(C) Each box depicts communities highlighted in part B at resolution sufficient to observe individual interactions. Proteins are grouped by community membership; communities that initially clustered together and subsequently split are rendered in similar hues; proteins shared among clusters are red. Interactions that span multiple communities are gray while interactions among members of a community share the color of that cluster.
The power of community detection to retrieve known complexes is exemplified by Figure 4Ci. While the proteasome emerged as one large community following clique percolation, further modularity-based clustering subdivided the proteasome into 2 clusters corresponding largely to its catalytic particle (primarily PSMA and PSMB subunits) and regulatory particle (primarily PSMC, PSMD and UCHL5 subunits), as well as a CUL5-TCEB1-RAB40 ubiquitin ligase complex which links with the proteasome via the alternative cap protein PSME3 and FAM192A. Chaperones that assist proteasome assembly (e.g. PSMG1/2, POMP) and other regulators (PAAF1 and FOXO7) also cluster with the proteasome, along with proteins (e.g. ZFAND2B, CCDC74B, C16orf70) that have no known proteasomal connections.
A unique feature of clique percolation is that it allows proteins to be shared among multiple communities. Such proteins often physically connect or coordinate distinct cellular activities. Alternatively, specificity factors for enzymes involved in diverse regulatory processes may be shared with multiple communities. For example, the community in Figure 4Ciii contains a cluster of protein phosphatase catalytic and regulatory subunits that connects to a highly interconnected RNA Polymerase cluster as well as a cluster of kinetochore components. Phosphorylation controls both RNA polymerase and kinetochore function and linkage to phosphatase components may reveal common regulatory factors and mechanisms for distinct target complexes. Further examples include CDK (Figure 4Cvi), vesicle function (Figure 4Cii), LIM domain and homeobox transcription factor (Figure 4Cv), and cullin ring ubiquitin ligase/signalosome (Figure 4Civ) communities.
Many communities are united by shared traits that cause member proteins to preferentially associate (Table S3; Figure S5A). Some match known complexes, such as the Mediator (Clusters 11a and 11b) and 43S translation pre-initiation complexes (Clusters 25a and 25b). Indeed, 33 known Mediator subunits separated into 2 sub-complexes, and was devoid of non-Mediator Complex connections (Figure S5B). Similarly, Cluster 73 contains the FTS1-HOOK1,2,3 protein complex accompanied by TNFSF13B, whose association was until now unknown. Such associations can reveal much about unknown proteins: the little-known protein C11orf74 associates with 5 members of the Intraciliary Transport Particle A (Cluster 74). Not only do its neighbors share biological functions and domains, but all have been implicated in related ciliary disorders (cranioectodermal dysplasia 1-4, short-rib thoracic dysplasia 9). Finally, community structure can reflect functional subtleties. While Clusters 6a, 6b, and 6c uniformly regulate histone acetylation, each comprises a different acetyltransferase complex and targets a distinct histone subset.
Protein Interactions Reveal Associations among Functional Domains
Many proteins may be decomposed into domains, or self-contained modules that have independently evolved to perform specific functions. Domains often recur in the proteome, performing related functions within many proteins. Although thousands of domains have been identified, the functions of many are unknown. Since many domains mediate interactions by binding complementary structures within other proteins, analyzing interactions of their parent proteins may provide functional insights. Such efforts would complement previous attempts to characterize known or predicted domain interactions (Boxem et al., 2008). We have mapped Pfam (Finn et al., 2014) domains to each protein in BioPlex and identified domain pairs whose parent proteins interact with unusual frequency (Figure 5A). While co-occurring domains do not necessarily interact directly and may instead relate indirectly through other protein features or shared function, these associations nevertheless can provide insights into each domain's unique biology.
Figure 5. Detection of Associations Including Domains of Unknown Function.
(A) After mapping Pfam domains onto BioPlex, the numbers of proteins containing each domain were tallied. Numbers of interactions that linked one domain with another were also determined. Contingency tables were then populated relating observation of one domain with the other, and Fisher's Exact Test determined the likelihood of a non-random association between the two domains. This process was repeated for all domain pairs and p-values were adjusted for multiple hypothesis testing (Benjamini and Hochberg, 1995).
(B) Heat map depicting significant domain associations involving domains of unknown function (highlighted in red). Blue boxes label domain pairs that associate at a 1% FDR. Labels indicate domain associations highlighted in C-H.
(C-H) Interaction networks corresponding to associated domain pairs. Blue vertices match proteins that contain the indicated domains of unknown function, while red vertices indicate proteins containing the associated domain; gray nodes match neither domain. P-values were determined by Fisher's Exact Test with multiple testing correction. Schematics depicting the domain structure of each central protein are displayed below each network.
Across BioPlex, 2,968 domain pairs associated at a 1% FDR (Table S4; Figure S6A,B). While many associations describe known relationships among familiar domains, most link domains with no known connections. As expected, proteins with Protein Kinase domains frequently interact with proteins bearing Cyclin N/C domains, reflecting cyclin-dependent kinases. In addition, synaptobrevin associates with SNARE and septin-containing proteins co-occur. Similarly, TRiC chaperones containing Cpn60 TCP1 domains associate with proteins containing WD40 domains whose folding they facilitate (Figure S6C) (Spiess et al., 2004). Finally, as expected, SCAN-domain-containing proteins self-associate (Figure S6D), and frequently assort with KRAB and zf-C2H2 domains.
Among unreported domain associations are a subset that relate domains of unknown function with other Pfam domains, including 14-3-3, UBX, SCAN, and WD40 domains (Figure 5B). Proteins bearing domains of unknown function provide context for reported domain associations (Figures 5C-5H). For example, the DUF2045 domain-containing protein KIAA0930 is surrounded by 14-3-3 proteins (Figure 5C), raising the possibility that KIAA0930 and the DUF2045 domain may participate in intracellular signaling. Similarly, DUF1162 proteins VPS13A and VPS13C interact with proteins containing UBX domains (Figure 5D). Each domain of unknown function presented in Figures 5C-5H is unaccompanied within its parent proteins, ruling out influence of other domains.
BioPlex Aids Protein Subcellular Localization Prediction
Because proteins must exist in close proximity to physically interact, the interactome necessarily reflects the subcellular localizations of its proteins. Thus, we mapped subcellular localization data from UniProt onto BioPlex and calculated the enrichment of each subcellular compartment among each protein's first and second degree neighbors (Table S5). While enrichment could indicate that a protein associates with a compartment in multiple ways, the simplest interpretation is that a protein at least partially localizes to that compartment. Figure S7 displays predicted localizations for several proteins and complexes. Panel A depicts predicted localizations for proteasome subunits and the sub-network surrounding PSMA1 to illustrate its most likely localizations. As expected, essentially all proteasome subunits are found in both nucleus and cytoplasm; the only exception is PSME3, the alternative regulatory cap protein that clustered separately from the rest of the proteasome (Figure 4Ci). Indeed, previous experiments have demonstrated that unlike other proteasome subunits, PSME3 localizes specifically in the nucleus (Wojcik et al., 1998). Other panels depict localizations for CDK's and related proteins (B), Complex I of the mitochondrial Electron Transport Chain (C), and the nuclear Mediator Complex (D), which are all consistent with their known localization.
BioPlex Enables Characterization of Unknown Proteins
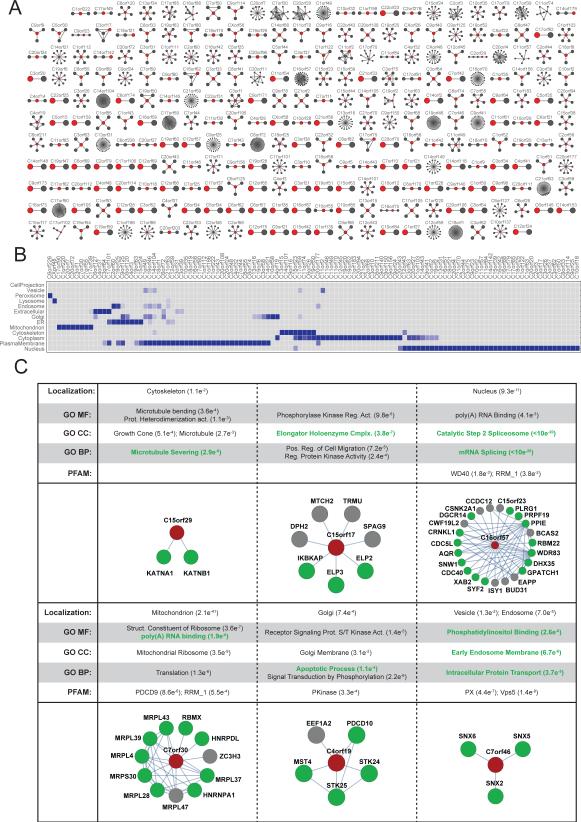
A motivation for unbiased mapping of the human interactome is to explore the biology of uncharacterized proteins. BioPlex contains interactions for 271 unstudied open reading frames (Figure 6A) that suggest functions and localizations of these proteins. As described, 117 open reading frames were assigned to at least one subcellular location at a 1% FDR (Table S5; Figure 6B). Of 8 open reading frames predicted by AP-MS to localize to mitochondria, 6 are known or suspected mitochondrial proteins (UniProt and/or www.mitocarta.org) and 2 (C1orf220, C17orf39) have not been assigned any localization. For these 2 proteins among many others, their positions within the AP-MS interaction network provide insights into their primary subcellular localizations.
Figure 6. BioPlex Topology Aids Unknown Protein Characterization.
(A) Network maps displaying interacting partners (gray) for 271 proteins assigned generic names based on chromosome and open reading frame position (red). Most are uncharacterized.
(B) Expected subcellular localizations for 117 of 271 uncharacterized ORFs, based on localization enrichment among the protein's primary and secondary neighbors.
(C) Networks surrounding 6 uncharacterized proteins and listing enriched GO terms, Pfam domains, and subcellular localizations. P-values were determined by Fisher's Exact Test with multiple testing correction. Red nodes: open reading frames; green nodes: neighboring proteins that match the enriched term highlighted with green text.
To highlight the information encoded by an uncharacterized protein's position in the AP-MS network, we provide several examples in Figure 6C. Inferences about each ORF's specific interactions, biological function and localization are summarized (Table S6). Three of these proteins have been characterized, affording an opportunity to evaluate network predictions. In each case, agreement was excellent. C15orf29 was found to interact with proteins KATNA1 and KATNB1, which associate with the cytoskeleton and participate in microtubule severing. Recently, C15orf29 was renamed KATNBL1, and may govern KATNA1 activity. Similarly, C16orf57 has been renamed USB1 and identified as a nuclear protein that participates in RNA splicing and mRNA processing (Mroczek et al., 2012). Furthermore, C7orf30 is thought to act as a silencing factor for the mitochondrial ribosome (Fung et al., 2013). In contrast, little is known about C15orf17, C4orf19, and C7orf46. As these proteins show, BioPlex is a roadmap for exploring the uncharted expanses of the proteome and illuminating the dark corners of cell biology.
Functional validation of the VAPA/VAPB Interaction Sub-network
Perhaps the greatest value of BioPlex is the potential of its interactions to inspire hypothesis-driven research into under-explored areas of biological inquiry. To demonstrate this application while further validating BioPlex, we examined a sub-network involving VAPB, previously found mutated in familial ALS, and the related protein VAPA.
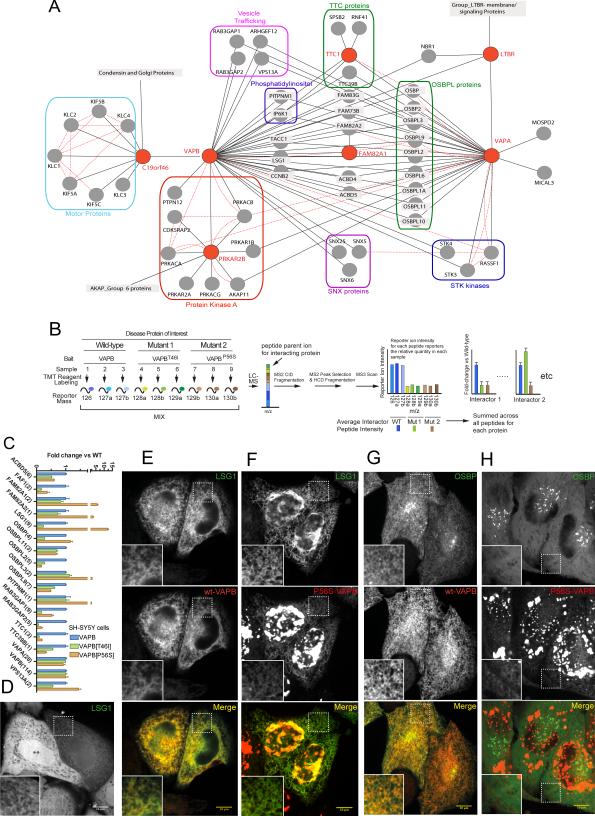
VAPA and B are endoplasmic reticulum (ER)-localized transmembrane proteins that anchor proteins implicated in lipid dynamics, primarily lipid transfer proteins (Lev et al., 2008). Lipid-transfer proteins, such as oxysterol-binding proteins (OSBP's) and other VAPA/B associated proteins, contain FFAT motifs that interact with the cytoplasmic MSP domain of VAPA/B (Kaiser et al., 2005). VAPB and several interacting partners were found in BioPlex, and VAPA was analyzed using the high-throughput pipeline during manuscript preparation, yielding a highly interconnected network (Figure 7A). As expected, VAPA and VAPB associated with several OSBPs (Mesmin et al., 2013a), and other proteins linked with membrane traffic or signaling, several of which were seen reciprocally.
Figure 7. Quantitative Interaction Proteomics of the VAPB Network Reveals Differential Interactions for VAPB Variants Associated with ALS.
(A) BioPlex interaction network for VAPA, VAPB and associated proteins. Dotted red lines: interactions reported by BioGRID; solid black lines: BioPlex interactions.
(B,C) Overview of our TMT approach for examining how ALS-associated mutations in VAPB affect interaction partners. VAPB and its variants were stably expressed in SH-SY5Y cells as FLAG-HA-tagged fusion proteins and subjected to AP-MS. Triplicate purifications were digested with trypsin prior to reaction with isobaric tag reagents. TMT reporter ion intensities were extracted from MS3 spectra and combined to determine changes in association of individual proteins with VAPB (panel B). Relative intensities for VAPB and VAPB mutant interacting proteins are shown in panel C. Error bars represent mean +/− standard error. Numbers of peptides quantified for each protein are listed in parentheses.
(D) Confocal image of HeLa cells expressing EGFP-LSG1 alone. When expressed at low/moderate level (cell with the single white asterisk) LSG1 localizes in a reticular compartment reminiscent of the ER (high magnification in the inset printed at higher contrast). When expressed at very high level (cell with two black asterisks) a diffuse cytosolic distribution prevails possibly due to saturation of ER binding sites. Scale bar: 10 μm.
(E,F) Fluorescence images of HeLa cells co-expressing EGFP-LSG1 with mCherry-tagged VAPB variants. (E) Upon co-expression of wild type VAPB, LSG1 is recruited to the ER. (F) VAPBP56S forms aggregates and only weakly localizes to the ER. Even in these cells LSG1 colocalizes with VAPB, although preferentially with the small pool of VAPBP56S which retains a reticular (ER) distribution.
(G,H) Fluorescence images of HeLa cells coexpressing EGFP-OSBP with mCherry-tagged VAPB variants. (G) When co-expressed with VAPBWT, OSBP is recruited to the ER. (H) OSBP has a diffuse cytosolic distribution when coexpressed with VAPBP56S, which lacks a functional FFAT binding site.
To validate the interactions found in 293T cells and to understand how patient mutations in VAPB affect individual associations, we stably expressed VAPB and its mutants (VAPBT46I and VAPBP56S) in SH-SY5Y cells and performed quantitative AP-MS using Tandem Mass Tagging (TMT) (see EXTENDED EXPERIMENTAL PROCEDURES and Figure 7B). Biological triplicate AP-MS complexes from all 3 variants were subjected to 9-plex TMT with reporter ion quantification by LC-MS3-based mass spectrometry. Many interactors identified in 293T cells also associated with wild-type VAPB in SH-SY5Y cells (Figure 7C). Normalized reporter ion intensities revealed proteins that displayed altered VAPB association with specific mutants (Figure 7C), including increased association of the VAPBP56S mutant with FAM82A2 (also called PTPIP51) as recently reported (Stoica et al., 2014). Moreover, while previous studies indicated that FAF1 associated equivalently with VAPBWT and VAPBP56S in vitro (Baron et al., 2014), our in vivo results indicate that FAF1 associates more weakly with VAPBT46I and VAPBP56S relative to VAPBWT, consistent with interaction via the FFAT motif (Figure 7C).
To further validate enhanced association with LSG1 and reduced association of OSBP with VAPBP56S (Figure 7C) we examined localization of EGFP-LSG1 and OSBP-EGFP with or without expression of VAPBWT or VAPBP56S in HeLa cells. Importantly, VAPBP56S is known to intrinsically aggregate. In cells not expressing exogenous VAP proteins, EGFP-LSG1 displayed reticular ER localization, and therefore binding to VAP, when expressed at low level, and a predominant cytosolic diffuse distribution at high expression as seen previously with OSBP (Mesmin et al., 2013b) (Figure 7D). This cytosolic distribution likely reflects VAP binding site saturation in the ER membrane. Accordingly, VAPBWT over-expression strongly increased the association of EGFP-LSG1 and OSBP with the ER (Figure 7E,G). In contrast, expression of aggregation-prone VAPBP56S led to recruitment of EGFP-LSG1, but not of OSBP, into the aggregates (Figure 7F,H). Additionally, a pool of LSG1, but not of OSBP, co-localized with the small VAPBP56S pool that remained diffusely distributed throughout the ER (Figure 7G,H). These observations confirm that the P56S mutation within the MSP domain of VAPB reduced FFAT motif-dependent interactions, while enhancing interactions with LSG1, which does not contain an FFAT motif. These data demonstrate the potential for BioPlex to inform and enhance focused study of individual proteins.
DISCUSSION
High-Throughput AP-MS Complements other Global Interaction Mapping Approaches
To date, most near global studies of human protein interactions have relied upon Y2H, which is amenable to automation and measures binary, and often direct, interactions. However, as emphasized through BioPlex community analysis, many interactions involve large protein assemblies whose detection is facilitated by AP-MS, and may not be detectable as binary complexes due to complex structural interactions. An example is the Mediator Complex. AP-MS using 5 subunits (MED7, MED19, CDK8, CDK19, and CCNC) identified nearly all Mediator subunits, with each bait capturing 23 - 37 subunits (Figure S5B, Table S2). In contrast, analysis of baits CCNC, MED4, MED18, MED20, MED25, MED28, and MED30 with a Y2H library containing 16 Mediator subunits yielded 24 partners, only one of which was a known Mediator subunit (Rolland et al., 2014). This reflects that Mediator architecture involves co-assembly of multiple subunits rather than modular assembly most easily captured via Y2H. The ability of AP-MS to pinpoint primary and secondary interactions, while simultaneously recognizing independent complexes with shared components, is the cornerstone of our large-scale network construction.
Network structures for multi-functional and dynamic protein associations will emerge as a larger fraction of the proteome is sampled as baits. Surprisingly, 86% of BioPlex interactions have not been reported. This reflects: 1) interrogation of many poorly studied proteins; 2) AP-MS sensitivity, enabling detection of low abundance proteins; 3) co-associated protein identification as complex members. These factors are exemplified by the SH2-SH3 domain containing protein NCK2. We identified 46 HCIPs for NCK2 (Table S2), 31 of which are reported in STRING or GENEMANIA databases as proteins that interact with NCK2 or NCK2-associated proteins. Of 15 NCK2 associated proteins found in BioPlex, but not in STRING or GENEMANIA, C3orf10 and SHB are known to interact with other NCK2 associated proteins and SEMA6A, KIAA1522, PEAK1, and SH3PXD2B contain proline-rich sequences of the type known to associate with SH2 domains or other adaptor elements. Thus, AP-MS provides a complementary approach to binary interaction measurements for understanding interactome connectivity.
Experimental Challenges of Global Interaction Mapping via AP-MS
Although AP-MS has proven reliable for interaction mapping, technical factors have important implications for BioPlex: 1) Some proteins require an intact C-terminus for proper complex assembly and may not provide reliable interacting partners when C-terminally tagged. Conceivably, some of the 322 baits that produced no interacting partners are affected by C-terminal tagging. 2) Some baits may be toxic upon expression in HEK293T cells. Since >93% of bait-expressing cell lines have survived to harvest, only a small fraction of baits targeted thus far are toxic. 3) 28% of proteins encoded by ORFEOME 8.1 do not represent the longest open reading frame in GenBank, which could affect BioPlex in unpredictable ways. 4) While bait expression levels vary, there was no correlation between bait abundance and HCIP count (Figure S4D, E) (Sowa et al., 2009). In addition, protein associations generally confirm known bait localizations (Figure S7; Table S5). Together, these findings suggest that lentiviral expression has not unduly biased our network. 5) Up to 1/3 of the genome encodes membrane proteins. Previous studies have suggested that extraction conditions can substantially affect membrane protein complex recovery (Babu et al., 2012). For several well-studied membrane proteins, our pipeline captured largely intact membrane protein complexes, including components of Complex I of the electron transport chain (Figure S7C) and the VAPB complex (Figure 7A). However, for unstudied trans-membrane proteins, further studies will be required to determine whether extraction conditions allow retrieval of intact complexes.
An additional consideration is that BioPlex incorporates only one biological replicate for each bait. While technical replicate LC-MS analyses address some sources of experimental error and many quality control measures have been implemented, variability in expression and affinity purification remain. Such effects would be best addressed with biological replicates, though our project scope has rendered this impractical. Nevertheless, as our network has grown to include AP-MS experiments targeting much of the human proteome, complexes have been purified multiple times as each subunit is targeted via AP-MS. These distinct affinity purifications tend to reinforce each other (e.g. Figure 3A, Figure 4C, Figure 7A). In such cases, the confidence of our final network is higher than a single IP of an individual bait would be.
BioPlex Accurately Models the Human Interactome
Notwithstanding challenges, BioPlex appears to accurately model the human interactome. The accuracy of CompPASS-Plus is highlighted by its performance on known true- and false-positives (Figure S2F,G). Furthermore, the reliability of the resulting interaction network is most apparent from interactions of well-studied baits (Figure 1B,C) and from the extensive overlap observed with CORUM (Figures S2E and 3A). Additionally, subcellular co-localization analysis suggests that AP-MS interactions achieve accuracies equal or higher than previous studies and databases (Figure 3E).
Further evidence of BioPlex quality emerges from overall network architecture. When subdivided into communities (Figures 4 and S5), 85% reflect functional or structural properties of their constituent proteins and many correspond to known complexes, including Mediator and Histone Acetyltransferase Complexes (Figure S5B) as well as RNA Pol II, the Signalsome, and CNOT and COMM complexes (Figure 4C), among others (Table S3). Network structure largely distinguishes proteasome core and regulatory subunits (Figure 4C). More generally, patterns of domain association (Figures S6 and 5; Table S4) and subcellular localization (Figure S7; Table S5) match expectations. Many previously unreported interactions involve understudied proteins whose properties may be revealed by overlaying GO classification, Pfam domains, and sub-cellular localization (Figure 6B,C; Tables S5 and S6). By modeling the interactome in its entirety, BioPlex amounts to more than the sum of its several thousand constituent AP-MS experiments.
While BioPlex is drawn from a single cell type, interactions may differ in distinct cell lineages and in response to specific stimuli. Thus, the network is best viewed as a framework that can be used for hypothesis generation and for design and interpretation of targeted studies that address dynamic and genetic underpinnings of individual networks, as illustrated through our quantitative analysis of the VAPB complex (Figure 7C).
Conclusions
Utilizing the human ORFEOME, we have assembled the largest AP-MS network of human interactions. Resulting from AP-MS analysis of more than 10% of human proteins, BioPlex spans over 1/3 of the human proteome and includes nearly 24,000 interactions, most of which have not been described. Viewed individually or in aggregate, these interactions are a valuable resource for both focused and systems-level biological research. The network will also establish a foundation for future efforts to expand interactome coverage and to explore network dynamics and the effect of disease mutations on network architecture.
EXPERIMENTAL PROCEDURES
An overview of experimental procedures is provided below. See Extended Experimental Procedures for details.
Protein Expression and Purification
The sequence-validated Human ORFEOME v. 8.1 (Yang et al., 2011) was used to construct a lentiviral library containing 13,000 ORFs bearing C-terminal FLAG-HA epitopes. Following sequence verification and virus production, HEK293T cells were infected and expanded under puromycin selection. Upon harvest, cell lysates were clarified and baits captured with anti-HA agarose prior to washing and elution with HA peptide. Clones will be distributed through the Dana Farber/Harvard Cancer Center DNA Resource Core (http://dnaseq.med.harvard.edu/).
Mass Spectrometry
After purification, proteins were precipitated with 10% TCA, and digested with trypsin prior to technical duplicate analyses on ThermoFisher Q-Exactive mass spectrometers. Using Sequest (Eng et al., 1994), spectra were searched against human protein sequences from Uniprot (Magrane and Consortium, 2011) and common contaminants. The target-decoy method (Elias and Gygi, 2007) was used to filter each LC-MS dataset to a preliminary 1% protein FDR. Additional filtering controlled the network FDR (Extended Experimental Procedures).
Identification of Interacting Proteins and Network Assembly
An extension of CompPASS (Sowa et al., 2009) called CompPASS-Plus was developed to distinguish interactors from non-specific background and false-positive identifications. Interactions were pooled across AP-MS experiments to assemble BioPlex (Extended Experimental Procedures).
Data Accessibility
BioPlex interactions were deposited into BioGRID in September 2014 and are available for download and browsing at gygi.med.harvard.edu/projects/bioplex. All 5,200 RAW files are also available.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Marc Vidal and David Hill for ORFEOME 8.1, and Yasunori Saheki for advice. This work was supported by the NIH (U41 HG006673 to SPG, JWH, and MES; R37NS036251 to PDC) and Biogen (SPG, JWH, and PDC). LPV is supported by a fellowship from Damon Runyon Cancer Research Foundation. JAP is supported by K01DK098285. PDC is an Investigator of the Howard Hughes Medical Institute. MW was supported by the Charles A. King Trust Fellowship. SPG and JWH are consultants for Biogen.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
The study was conceived by SPG and JWH. ELH developed CompPASS-Plus and software for data collection and integration, performed network, domain, and informatic analyses, and oversaw data collection and pipeline quality. LT directed the cell culture and biochemistry pipeline and organized samples for MS analysis. JAP was responsible for all mass spectrometry, including instrument operation, method development, maintenance, and troubleshooting. GC, FG, MPG, RAO, ST, and JS performed DNA and cell line production. RJB and GZ performed protein purifications. BZ and JC performed HEK293T proteomic analysis. BE, DK, RR, MES, and JM provided computational support. VGP and LPV constructed the lentiviral library. MW assisted with localization analyses. AO performed TMT experiments. RD performed cell biological experiments under the direction of PDC. Data interpretation was performed by ELH, TH, RD, PDC, SAT, SPG, and JWH. The data visualization tool was constructed by SPG. The paper was written by ELH, JWH, and SPG and edited by all authors.
REFERENCES
- Babu M, Vlasblom J, Pu S, Guo X, Graham C, Bean BD, Burston HE, Vizeacoumar FJ, Snider J, Phanse S, et al. Interaction landscape of membrane-protein complexes in Saccharomyces cerevisiae. Nature. 2012;489:585–589. doi: 10.1038/nature11354. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Baron Y, Pedrioli PG, Tyagi K, Johnson C, Wood NT, Fountaine D, Wightman M, Alexandru G. VAPB/ALS8 interacts with FFAT-like proteins including the p97 cofactor FAF1 and the ASNA1 ATPase. BMC Biol. 2014;12:39. doi: 10.1186/1741-7007-12-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: a Practical and Powerful Approach to Multiple Testing. J Royal Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, et al. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S, Nishimura T, Sasarman F, Shoubridge EA. The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation. Mol Biol Cell. 2013;24:184–193. doi: 10.1091/mbc.E12-09-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- Gosselin P, Martineau Y, Morales J, Czjzek M, Glippa V, Gauffeny I, Morin E, Le Corguille G, Pyronnet S, Cormier P, et al. Tracking a refined eIF4E-binding motif reveals Angel1 as a new partner of eIF4E. Nucleic Acids Res. 2013;41:7783–7792. doi: 10.1093/nar/gkt569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guruharsha KG, Rual JF, Zhai B, Mintseris J, Vaidya P, Vaidya N, Beekman C, Wong C, Rhee DY, Cenaj O, et al. A protein complex network of Drosophila melanogaster. Cell. 2011;147:690–703. doi: 10.1016/j.cell.2011.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havugimana PC, Hart GT, Nepusz T, Yang H, Turinsky AL, Li Z, Wang PI, Boutz DR, Fong V, Phanse S, et al. A census of human soluble protein complexes. Cell. 2012;150:1068–1081. doi: 10.1016/j.cell.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Joshi P, Greco TM, Guise AJ, Luo Y, Yu F, Nesvizhskii AI, Cristea IM. The functional interactome landscape of the human histone deacetylase family. Mol Syst Biol. 2013;9:672. doi: 10.1038/msb.2013.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT. Structural basis of FFAT motif-mediated ER targeting. Structure. 2005;13:1035–1045. doi: 10.1016/j.str.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lev S, Ben Halevy D, Peretti D, Dahan N. The VAP protein family: from cellular functions to motor neuron disease. Trends Cell Biol. 2008;18:282–290. doi: 10.1016/j.tcb.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, et al. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857–861. doi: 10.1093/nar/gkr930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011. 2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson JY, Tarsounas MC, Stasiak AZ, Stasiak A, Shah R, McIlwraith MJ, Benson FE, West SC. Identification and purification of two distinct complexes containing the five RAD51 paralogs. Genes Dev. 2001;15:3296–3307. doi: 10.1101/gad.947001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Antonny B, Drin G. Insights into the mechanisms of sterol transport between organelles. Cell Mol Life Sci. 2013a;70:3405–3421. doi: 10.1007/s00018-012-1247-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesmin B, Bigay J, Moser von Filseck J, Lacas-Gervais S, Drin G, Antonny B. A four-step cycle driven by PI(4)P hydrolysis directs sterol/PI(4)P exchange by the ER-Golgi tether OSBP. Cell. 2013b;155:830–843. doi: 10.1016/j.cell.2013.09.056. [DOI] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2012;41:D377–386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek S, Krwawicz J, Kutner J, Lazniewski M, Kucinski I, Ginalski K, Dziembowski A. C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3' end modification. Genes Dev. 2012;26:1911–1925. doi: 10.1101/gad.193169.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Fast algorithm for detecting community structure in networks. Phys Rev E. 2004;69 doi: 10.1103/PhysRevE.69.066133. [DOI] [PubMed] [Google Scholar]
- Palla G, Derenyi I, Farkas I, Vicsek T. Uncovering the overlapping community structure of complex networks in nature and society. Nature. 2005;435:814–818. doi: 10.1038/nature03607. [DOI] [PubMed] [Google Scholar]
- Rolland T, Tasan M, Charloteaux B, Pevzner SJ, Zhong Q, Sahni N, Yi S, Lemmens I, Fontanillo C, Mosca R, et al. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes HW. CORUM: the comprehensive resource of mammalian protein complexes--2009. Nucleic Acids Res. 2009;38:D497–501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C, Meyer AS, Reissmann S, Frydman J. Mechanism of the eukaryotic chaperonin: protein folding in the chamber of secrets. Trends Cell Biol. 2004;14:598–604. doi: 10.1016/j.tcb.2004.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoica R, De Vos KJ, Paillusson S, Mueller S, Sancho RM, Lau KF, Vizcay-Barrena G, Lin WL, Xu YF, Lewis J, et al. ER-mitochondria associations are regulated by the VAPB-PTPIP51 interaction and are disrupted by ALS/FTD-associated TDP-43. Nat Commun. 2014;5:3996. doi: 10.1038/ncomms4996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, Larsen B, Choi H, Berger B, Gingras AC, Lindquist S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell. 2011;144:986–998. doi: 10.1016/j.cell.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik C, Tanaka K, Paweletz N, Naab U, Wilk S. Proteasome activator (PA28) subunits alpha, beta, and gamma (Ki antigen) in NT2 neuronal precursor cells and HeLa S3 cells. Eur J Cell Biol. 1998;77:151–160. doi: 10.1016/s0171-9335(98)80083-6. [DOI] [PubMed] [Google Scholar]
- Yang X, Boehm JS, Salehi-Ashtiani K, Hao T, Shen Y, Lubonja R, Thomas SR, Alkan O, Bhimdi T, Green TM, et al. A public genome-scale lentiviral expression library of human ORFs. Nat Methods. 2011;8:659–661. doi: 10.1038/nmeth.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BioPlex interactions were deposited into BioGRID in September 2014 and are available for download and browsing at gygi.med.harvard.edu/projects/bioplex. All 5,200 RAW files are also available.