Abstract
The objective of the study was to overview the role of genetic research in fostering translational studies of craniofacial diseases of dental interest. Background information is presented to illustrate influences affecting genetic research studies of Mendelian diseases. Genetic studies of amelogenesis imperfecta, dentinogenesis imperfecta, hereditary gingival fibromatosis and Papillon Lefèvre syndrome are reviewed. Findings are presented to illustrate how translational applications of clinical and basic research may improve clinical care. Clinical and basic science research has identified specific genes and mutations etiologically responsible for amelogenesis imperfecta, dentinogenesis imperfecta, hereditary gingival fibromatosis and Papillon Lefèvre syndrome. These findings are enabling researchers to understand how specific genetic alterations perturb normal growth and development of dental tissues. Identification of the genetic basis of these conditions is enabling clinicians and researchers to more fully understand the etiology and clinical consequences of these diseases of dental importance. Findings from genetic studies of dental diseases provide a basis for diagnostic genetic testing and development of therapeutic intervention strategies directed at the underlying disease etiology. These studies are advancing our understanding of the development of dental tissues in health and disease. The dental community must consider how to incorporate these developments into effective disease prevention paradigms to facilitate the diagnosis and treatment of individuals with genetic diseases.
Keywords: dental, diagnosis, genetic diseases, mutation, treatment
Introduction
Of the more than 7000 inherited human disorders, 30–40% display some oral, dental or craniofacial manifestation (1). Certain conditions, such as amelogenesis imperfecta, may affect only mineralized tooth tissues, while others such as hereditary gingival fibromatosis may affect only the gingiva (2, 3). Many other genetic diseases, such as Papillon-Lefèvre syndrome and tricho-dentoosseous syndrome (OMIM190320), affect both oral and extraoral tissues (1). While oral and dental manifestations for some of these diseases are well characterized, they have not been systematically characterized for many genetic diseases. Unique dental and oral findings are often identified when time is taken to carefully assess the oral cavity (1, 4, 5). Many of these genetic conditions are transmitted within families as Mendelian traits, suggesting that the cause of these conditions may be attributed to genetic alteration of a gene of major effect. With current research approaches it is possible identify genes responsible for these diseases.
Identifying disease genes has been greatly facilitated by advances in genetic research methodologies. As a result, the field of genetics has been transformed into a clinically applied discipline with broad implications for oral medicine (6). While many individual technological advances and discoveries have contributed to the ultimate success of applied genetics research, the successful completion of the human genome project revolutionized genetics research. Collective efforts of an international array of researchers affiliated with academia, private industry and governments integrated efforts in many disciplines to successfully complete the HGP goals (6, 7).
Application of genomic information and technologies has fundamentally altered the way we study human diseases. By applying an ever increasing array of research tools, researchers are able to characterize gene function in health and disease. A vibrant ‘Bench to Bedside’ clinical research paradigm has emerged, designed to speed translation of promising laboratory discoveries to improve diagnosis and treatment of diseases. Identifying these etiologic genetic alterations provides a first step to enable cellular and molecular studies to characterize how genes function and to understand how alterations of normal gene functions cause disease. The goal of understanding the genetic basis of disease is to facilitate the diagnosis and treatment of disease conditions (Fig. 1). Development of treatments directed at correcting specific biologic defects will permit clinicians to move from a ‘one size fits all’ treatment approach to a personalized treatment paradigm.
Fig. 1.

Clinical implications of understanding the genetic basis of disease.
A first step in identifying the genetic basis of disease has traditionally employed gene mapping strategies. Studying affected and unaffected members of families segregating a genetic disease, researchers employ genetic linkage and genetic association strategies to determine the chromosomal location of disease causing gene(s). Gene mapping studies are often the first step to identifying disease causing genes, permitting their role in normal and abnormal development to be understood. These approaches have identified thousands of disease causing gene mutations, including many that affect dental, oral and craniofacial structures (1), several of which are illustrated below.
Amelogenesis imperfecta
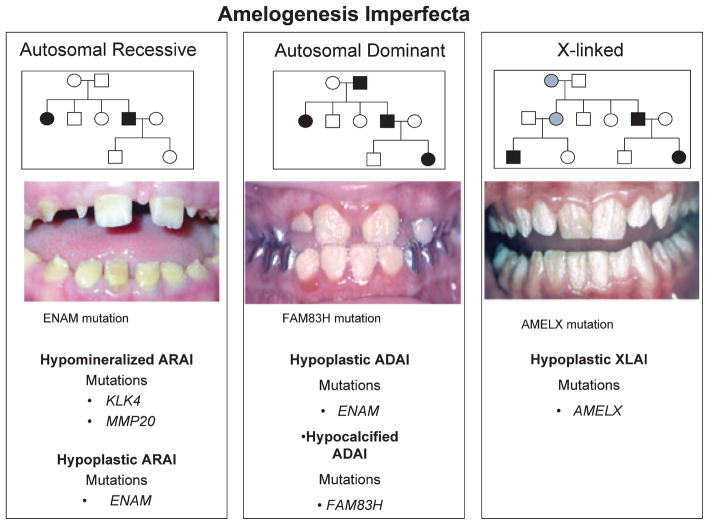
The amelogenesis imperfecta (AI) are a genetically and phenotypically heterogeneous group of disorders that affect the quality or quantity of enamel (2, 8). AI phenotypes are classified as hypoplastic, hypomaturation and hypocalcified. A total of 14 types have been recognized historically based upon phenotype and mode of inheritance. Mutations in five genes have been associated with non-syndromic AI: amelogenin (AMELX), enamelin (ENAM), family with sequence similarity, member H (FAM83H), kallikrein 4 (KLK4), and matrix metalloproteinase 20 (MMP20), improving AI nosology and increasing our understanding of enamel development in health and disease (Fig. 2) (2, 8–10). Mutations of AMELX, which encodes the predominant enamel protein amelogenin, are associated with X-linked forms of AI. Mutations of the major enamel proteases, KLK4 and MMP20, are associated with autosomal recessive hypomaturation AI (8). Characterizations of disease associated clinical phenotypes are critical to genetic studies, facilitating a better understanding of genotype-phenotype correlations. For example allelic ENAM mutations have been identified in both autosomal dominant and recessive hypoplastic AI (2, 8, 10), and a gene dosage effect has been demonstrated for certain ENAM mutations (11). Mutations of the FAM83H gene are responsible for the most common form of AI in North America, autosomal dominant hypocalcified AI, (Fig. 2) (12). Despite these advances, it is clear that not all AI causing genes have been identified.
Fig. 2.
Amelogenesis imperfecta summary. Genes for which mutations have been identified for each of the Mendelian modes of transmission are illustrated. Clinical photographs illustrating representative examples for each form of Mendelian transmission for AI are shown. The genetic mutation responsible for each case is indicated directly below each photograph. ARAI, autosomal recessive AI; ADAI, autosomal dominant AI; XLAI, X-linked AI).
Dentinogenesis imperfecta
Dentin defects are broadly classified into the dentinogenesis imperfectas (DIs, types I–III) and dentin dysplasias (DDs, types -I and -II) (13). In DI, the underlying dentin is defective and soft. Although the overlying enamel is structurally normal, it fractures easily, exposing the underling dentin. DI can be an isolated finding due to mutations in the dentin sialophosphoprotein (DSPP) gene or as part of a syndrome. Syndromic DI can be a feature of osteogenesis imperfecta, a disorder of brittle bones, that results from mutations in the genes encoding the type-1 collagens: collagen 1A1 (COL1A1) and collagen 1A2 (COL1A2) (2, 13). In Dentin Dysplasia (DD), the permanent dentition often appears clinically normal, but radiographically the pulp chambers may be obliterated by abnormal dentin. In DD type I, all teeth are normal in shape, form and color, but have short roots, contributing to premature exfoliation. Periapical radiolucencies are frequently noted. Molecular genetic studies have not been performed for DD type I. In DD type II, the deciduous teeth are often clinically indistinguishable from DI. Radiographically, pulp cavities show thistletube deformity and commonly have pulp stones. Mutations in DSPP have been found to cause DD type II. Until recently it was not possible to sequence the entire coding region of DSPP due to a highly repetitive region of the gene. Now that this region can be sequenced, it appears that the majority of DSPP mutations occur in this repetitive region (Table 1) (14). The ability to completely sequence the DSPP gene has important implications for genetic testing for DI conditions. As the entire DSPP gene can now be sequenced, when DSSP coding region mutations are not found, the COL1A1 and COL1A2 genes can be evaluated for disease causing mutations. We have identified cases of apparently isolated (non-syndromic) DI, in which we have excluded DSPP mutations, and subsequently identified COL1A1 or COL1A2 mutations. These may reflect very mild cases of OI/DI, where DI is the only clinically apparent phenotypic finding. The ability to determine the genetic basis of DI may have important clinical implications for some families. In such cases, it may be important for families to be aware of individuals who have DI due to collagen type-1 mutations, and for which the clinical expression of disease may vary with some cases presenting only DI, while other cases may show more significant clinical findings affecting bone and vasculature tissues. The ability to determine the etiology of DI in an individual can impact their access to dental care. Definitive genetic testing may be important for individuals to obtain state and/or third party insurance coverage to help with dental care.
Table 1.
Dentin sialophosphoprotein (DSPP) mutations reported for non-syndromic dentinogenesis imperfecta and dentin dysplasia
| Location | cDNA* | Protein† | Diagnosis‡ |
|---|---|---|---|
| Exon 2 | c.16T > G | p.Y6D | DD-II |
| Exon 2 | c.44C > T | p.A15V | DGI-II |
| Exon 2 | c.49C > A | p.P17T | DGI-II |
| Exon 2 | c.49C > T | p.P17S | DGI-II |
| Intron 2 | c.52-3C > G | p.V18_Q45del | DGI-II |
| Intron 2 | c.52-3C > A | p.V18_Q45del | DGI-II |
| Exon 3 | c.52G > T | p.V18_Q45del or p.V18F | DGI-II |
| DGI-III | |||
| DGI-III | |||
| DGI-II | |||
| Exon 3 | c.133C > T | p.V18_Q45del or p.Q45X | DGI-II |
| Intron 3 | c.135 + 1G > A | p.V18_Q45del | DGI-II |
| Intron 3 | c.135 + 1G > T | p.V18_Q45del | DGI-II |
| Exon 5 | c.1870_1873 delTCAG | p.S624TfsX687 | DD-II |
| Exon 5 | c.1918_1921 delTCAG | p.S640TfsX671 | DD-II |
| Exon 5 | c.2040delC | p.S680fsX1313 | DGI-II |
| Exon 5 | c.2272delA | p.S758AfsX554 | DGI-II |
| Exon 5 | c.2525delG | p.S842TfsX471 | DGI-II |
| Exon 5 | c.2593delA | p.S865fsX1313 | DGI-II |
| Exon 5 | c.2684delG | p.S895fsX1313 | DGI-II |
| Exon 5 | c.3141delC | p.S1047fsX223 | DD-II |
| Exon 5 | c.3438delC | p.D1146fsX1313 | DGI-II |
| Exon 5 | c.3546_3550 delTAGCAinsG | p.D1182fsX1312 | DGI-II |
Hereditary gingival fibromatosis
Gingival overgrowth, a benign enlargement of the gingiva, may occur as an isolated clinical finding (hereditary gingival fibromatosis; HGF), in association with additional clinical anomalies as part of a syndrome (syndromic gingival fibromatosis) or as a side effect of certain medications (drug induced gingival overgrowth) (3, 15). There is a genetic basis to the etiology of all three forms of gingival overgrowth. HGF is most commonly transmitted as an autosomal dominant trait. The condition may be moderate or severe in its clinical presentation, with gingival tissues covering the crowns of teeth, preventing normal eruption and leading to malocclusion, difficulty eating and speaking and esthetic concerns. Genetic studies of families segregating HGF have determined that the condition is genetically heterogeneous, with at least four different genes mapped (16–19). Studies of a large Brazilian family segregating HGF localized the responsible gene to chromosome 2 and identified a son of sevenless-1 (SOS-1) gene mutation as etiologic (16). A deletion mutation introduces a premature stop codon in the SOS-1 coding sequence. This mutation creates a truncated SOS-1 protein which is more active than wild-type SOS-1 protein. SOS-1 is functionally important in signal transduction, controlling whether cells grow, differentiate or divide (OMIM182530) (1). SOS-1 is expressed by many cell types in many tissues. An intriguing question is why this disruption of such an important gene only affects the gingiva, leading to gingival overgrowth. Immunohistochemistry studies demonstrate that SOS-1 expression is much higher in gingiva than other types of skin. This highlights the uniqueness of many oral tissues and structures including enamel, dentin and gingiva. Dental tissues are unique in part due to unique patterns of gene expression.
Studies of gingiva from individuals with HGF show increased amounts of collagen, increased numbers of fibroblasts, and increased rates of fibroblast proliferation (20). Studies also demonstrate specific changes in fibroblast signal transduction associated with the SOS-1 mutation in HGF patients (21). Mutant SOS-1 can translocate to the plasma membrane without growth factor stimulation, leading to sustained activation of RAS/MAPK signaling. The resulting increase in the magnitude and duration of ERK signaling and increased phosphorylation of certain proteins in the nucleus is associated with the up-regulation of cell cycle regulators and transcription factors which promote cell cycle progression from the G(1) to S phase. These findings illustrate the mechanism by which mutant SOS-1 causes HGF.
Findings of SOS-1 related HGF illustrate clinical implications of genetic studies of diseases. Genetic studies of families segregating HGF enabled investigators to identify the etiologic gene responsible for the condition, providing a basis for genetic testing. Identification of the responsible gene provided a starting point to understand how the SOS-1 gene mutation causes gingival fibromatosis. This provides an understanding of the biological systems involved in development of HGF, and provides a rationale basis to consider development of treatment intervention strategies. Ultimately this kind of information increases our fundamental understanding of gingival development and may help develop tissue engineering strategies designed to repair damaged gingiva.
While the SOS-1 gene mutation is etiologic for some cases of HGF, other cases of gingival fibromatosis are due to other types of genetic changes including chromosomal anomalies (3). Genetic studies of additional HGF families clearly indicate genetic heterogeneity. This means that mutations of additional genes can also cause HGF. As additional HGF causing genes are identified and their mechanism of action understood, researchers will develop a better understanding of gingival development that should facilitate development of better strategies to treat gingival diseases.
Papillon-Lefèvre syndrome
Papillon-Lefèvre syndrome (PLS) is characterized by plamar-plantar hyperkeratosis and severe, early onset periodontitis (OMIM245000) (1, 22). Affected individuals manifest severe gingival inflammation soon after the primary teeth erupt. Severe periodontitis develops with destruction of the periodontium. The primary teeth are prematurely exfoliated, and the gingival inflammation resolves until eruption of the secondary dentition, when the process of inflammation and destruction of the periodontium begins again. PLS is transmitted an autosomal recessive trait. Genetic studies localized the gene for PLS to chromosome 11, and subsequently cathepsin-C (CTSC) gene mutations were determined to be etiologic (22). CTSC is a proteinase, which functions to remove dipeptides from the terminus of protein substrates. CTSC is expressed in many tissues and cells, particularly neutrophils. CTSC is also highly expressed in the gingiva and skin on the palms and soles. More than 70 different CTSC mutations have been identified. These mutations span the entire gene and are found in highly conserved regions of the protein. Conservation of amino acids across species indicates that the conserved amino acids are functionally important. Specific mutations in highly conserved regions of cathepsin-C disrupt the enzymatic function of the protein (23, 24).
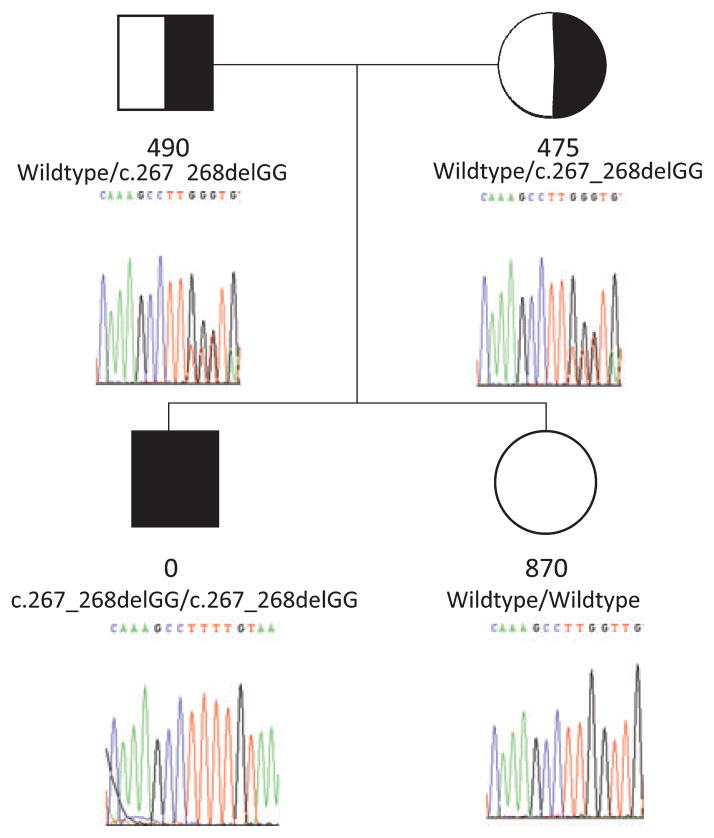
As cathepsin-C mutations associated with PLS cause the CTSC protein to lose its enzymatic activity, genetic testing for PLS may be performed by direct sequencing of the CTSC gene or by determining CTSC enzyme activity (Fig. 3) (25). Basic research has determined that cathepsin-C activity is needed to activate several important enzymes produced by neutrophils, called neutrophil derived serine proteases (NSPs). The NSPs cathepsin-G, elastase and proteinase 3 are secreted in an inactive, proenzyme form. Active cathepsin-C is needed to cleave the NSP propeptides and activate them. Neutrophils from PLS patients with CTSC mutations do not have active cathepsin-C and cannot activate the NSPs (26). Additional studies have determined that one of the biological molecules released at sites of inflammation is the chemokine macrophage inflammatory protein-1 alpha (MIP-1α). Inflammatory factors including microbial lipopolysaccharides (LPS) and the proinflammatory cytokine interleukin -1β that are associated with microbial plaque and inflammation in the gingival crevice induce MIP-1α expression by epithelial cells lining the gingival sulcus (27). Active NSPs are needed to regulate MIP-1α activity. Since PLS patients do not have active NSPs, they cannot regulate MIP-1α activity. Incorporating findings from several studies that have characterized the function of normal and mutant CTSC, we developed a model that may explain, in part, the mechanism by which CTSC mutations are etiologic for severe periodontitis. Severe periodontal destruction seen in PLS may result from a localized dysregulation of the inflammatory response (27). In this pathogenesis model, microbial plaque in the gingival crevice produces proinflammatory molecules such as microbial LPS that trigger a localized inflammatory response. Production of additional proinflammatory molecules is chemotactic for PMNs which invade the gingival crevice and the surrounding periodontium. MIP-1α, one of the many proinflammatory cytokines and chemokines produced at sites of inflammation, is a powerful chemoattractant for the continued influx of PMN. Normally, MIP-1α is cleaved and inactivated by the NSPs. As PLS patients can not produce active CTSC they cannot activate the NSPs and therefore cannot inactivate MIP-1α. Consequently, the recruitment and activation of additional PMNs cannot be regulated at sites of periodontal inflammation. Accordingly, PMNs continue to infiltrate the periodontium, enhancing the proinflammatory cascade, leading to the destruction of periodontal soft tissues and bone. Cathepsin-C mutations have also been associated with pre-pubertal periodontitis and aggressive periodontitis, indicating a role in other forms of periodontitis (28, 29).
Fig. 3.
PLS pedigree illustrating phenotype, CTSC genotype and CTSC enzyme activity. Filled symbols indicate individuals with a clinical diagnosis of PLS. Half shaded and open symbols are carriers and non carriers, respectively, based upon biochemical and/or mutational analyses. The CTSC protease activity (μmol/min/mg protein) in leukocytes is given below each individual. Mutated CTSC alleles associated with PLS have no enzyme activity. Heterozygote individuals demonstrate approximately 50% enzyme activity levels found in individuals with 2 normal CTSC alleles. The affected individual is homozygous for a c.267_268delGG mutation.
Clinical implications and applications
A primary focus of biomedical research is directed at utilizing genetic information and technology to improve our understanding of genetic diseases to improve clinical care. Table 2 illustrates several of the numerous studies that have successfully identified the genetic basis of diseases of dental importance. It is important to consider how we can use genetic information to help diagnose and treat patients (Fig. 1). Identifying the genetic basis of a disease enables clinicians and scientists to understand the genetic underpinnings of disease and facilitate development of diagnostic tests. Diagnostic testing may be based on identification of mutations affecting specific genes such as those listed in Table 2. In addition to DNA based testing, other types of tests may be applied, such as biochemical testing for CTSC activity in PLS (Fig. 3) (25), and cytogenetic testing to identify chromosomal anomalies associated with syndromic forms of HGF such as Zimmerman-Laband syndrome (OMIM135500) (1). The ability to correctly diagnose a clinical disease also permits development of better nosology. Being able to identify individuals with similar forms of disease permits evaluation of treatments in individuals with shared disease etiology (Fig. 1). Ultimately an important goal is to develop proactive, preventative treatments, whereby susceptible individuals can be identified before the onset of disease, and intervention strategies employed to prevent development of clinical disease.
Table 2.
Summary of genetic diseases with genes identified as etiologic for each
| Clinical disease | Etiologic Gene/OMIM* | Chromosomal location of gene |
|---|---|---|
| Amelogenesis imperfecta | Enamelin (ENAM ) OMIM606585 | 4q21 |
| Kallikrein 4 (KLK4) OMIM603767 | 19q13.4 | |
| Matrix Metalloproteinase 20 (MMP20) OMIM604629 | 11q22.3-q23 | |
| Family with sequence similarity, member H (FAM83H) OMIM611927 | 8q24.3 | |
| Amelogenin (AMELX) OMIM300391 | Xp22.3-p22.1 | |
| Dentinogenesis imperfecta | Dentin sialophosphoprotein (DSPP) OMIM125485 | 4q21.3 |
| Osteogenesis imperfecta/dentinogenesis imperfecta | Collagen type1 A1 (COL1A1) OMIM120150 | 17q21.31-q22 |
| Osteogenesis imperfecta/dentinogenesis imperfecta | Collagen type1 A2 (COL1A2) OMIM120160 | 7q22.1 |
| Papillon Lefèvre syndrome | Cathepsin C (CTSC ) OMIM602365 | 11q14.1-q14.3 |
| Hereditary gingival fibromatosis | Son of sevenless-1 (SOS-1) OMIM182530 | 2p22-p21 |
Online Mendelian Inheritance in man catalogue number (1).
Genetic diseases from a public health perspective
As the genetic etiologies of diseases are increasingly understood, dentists must consider how this information can be used to improve patient care. Treatment of genetic diseases poses unique challenges to traditional treatment paradigms. For millennia, infectious diseases were responsible for significant human morbidity and mortality. Through research efforts, the etiology of significant infectious diseases affecting the US population, such as smallpox, diphtheria, measles and paralytic poliomyelitis, were identified, and etiology based treatment strategies developed. Once the infectious agents for these diseases were identified, and vaccines developed, the number of cases in the USA decreased dramatically. As a consequence, by the late 1990s the morbidity and mortality associated with these conditions virtually disappeared in the USA.
From a public health perspective, treatment of infectious diseases can be broadly considered at three levels of intervention [See Fig. 2; Hart et al. 2000 (30)]. To illustrate an infectious disease prevention paradigm, consider a more recent public health challenge, human immunodeficiency virus (HIV) infection. Untreated, individuals infected with HIV usually develop the clinical disease acquired immune deficiency syndrome (AIDS). Untreated individuals with AIDS develop complications including opportunistic infections. Public health efforts for infectious diseases are based upon identification of the etiologic infectious agent, and prevention of individuals becoming infected. Primary prevention is directed at preventing individuals becoming infected. For individuals already infected, treatment is directed at secondary prevention, controlling the onset of clinical disease, such as with antiretroviral treatments. Before such treatments were available, treatment was directed at tertiary prevention, controlling complications of the clinical disease. Ultimately successful treatment is based on the ability to identify and understand the etiologic basis of a disease. For infectious diseases, treatment prevention strategies are directed at preventing infection: by avoiding infection or by enabling the host to effectively overcome the infectious challenge such as with vaccines. When a disease is not understood, palliative care and tertiary prevention are usually the only treatment possible. As the disease is understood, secondary prevention can be directed at controlling clinical disease. Primary prevention is the most effective, but is predicated on understanding the etiologic basis of disease (30).
Disease prevention paradigm for a Mendelian genetic disease
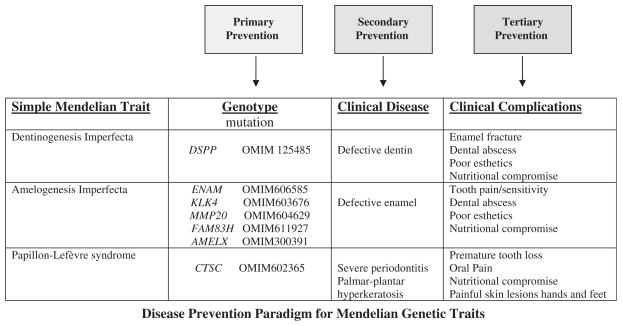
When considering genetic diseases, we must reconsider prevention strategies, as it is not usually possible to prevent individuals inheriting disease genes. Figure 4 illustrates the case for several of the Mendelian diseases we have discussed. Improving treatment beyond tertiary prevention will depend upon our ability to correctly diagnose the etiologic form of disease and to understand each disease sufficiently to permit identification of biologic targets for treatment to overcome the primary genetic defect (30).
Fig. 4.
Disease prevention paradigm for Mendelian diseases. Mendelian diseases reviewed in text are listed in the left column. Specific gene(s) for which etiologic mutations have been identified for each condition are listed in the second column, with the associated Online Mendelian Inheritance in Man (OMIM) catalogue number (1). Third column lists primary pathologic findings associated with mutations of each gene. Fourth column lists clinical complications that commonly result due to the underlying disease pathology. Possible points of treatment intervention (primary, secondary and tertiary) are indicated above.
Clinical relevance
Clinical and basic research has identified the genetic basis for many diseases of dental importance. Findings from these studies provide a basis for improved diagnosis, classification and treatment of diseases. The dental community must consider how to incorporate these developments into effective disease prevention paradigms to facilitate the diagnosis and treatment of individuals with genetic diseases.
Acknowledgments
The authors were supported by the Intramural Research Programs of the National Institute of Dental and Craniofacial Research (Z01-DE000711) and the National Human Genome Research Institute of the National Institutes of Health, Bethesda, MD. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Dental and Craniofacial Research, the National Human Genome Research Institute or the National Institutes of Health.
Contributor Information
T.C. Hart, Human Craniofacial Genetics Section, Skeletal and Craniofacial Diseases Branch, National Institute of Dental and Craniofacial Research, National Institutes of Health, Bethesda, MD, USA
P.S. Hart, Office of the Clinical Director, National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA
References
- 1.Online Mendelian Inheritance in Man, OMIM (TM) McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University; Baltimore, MD: National Center for Biotechnology Information, National Library of Medicine; Bethesda, MD: Oct 15, 2008. World Wide Web URL: http://www.ncbi.nlm.nih.gov/omim/ [Google Scholar]
- 2.Bailleul-Forestier I, Molla M, Verloes A, Berdal A. The genetic basis of inherited anomalies of the teeth. Part 1: clinical and molecular aspects of non-syndromic dental disorders. Eur J Med Genet. 2008;51:273–91. doi: 10.1016/j.ejmg.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Hart TC, Pallos D, Bozzo L, Cortelli J. Evidence of genetic heterogeneity for hereditary gingival fibromatosis. J Dent Res. 2000;79:1758–64. doi: 10.1177/00220345000790100501. [DOI] [PubMed] [Google Scholar]
- 4.Gorlin RJ, Cohen MM, Hennekam RCM. Syndromes of the Head and Neck. 4. New York: Oxford University Press; 2001. p. 1332. [Google Scholar]
- 5.Domingo DL, Freeman AF, Davis J, Puck JM, Tianxia W, Holland SM, et al. Novel intraoral phenotypes in hyperimmunoglobulin-E syndrome. Oral Dis. 2008;14:73–81. doi: 10.1111/j.1601-0825.2007.01363.x. [DOI] [PubMed] [Google Scholar]
- 6.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–47. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 7.Collins FS, Morgan M, Patrinos A. The Human Genome Project: lessons from large-scale biology. Science. 2003;300:286–90. doi: 10.1126/science.1084564. [DOI] [PubMed] [Google Scholar]
- 8.Wright JT. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet A. 2006;140:2547–55. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, et al. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am J Hum Genet. 2008;82:489–94. doi: 10.1016/j.ajhg.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- 11.Ozdemir D, Hart PS, Firatli E, Aren G, Ryu OH, Hart TC. Phenotype of ENAM mutations is dosage-dependent. J Dent Res. 2005;84:1036–41. doi: 10.1177/154405910508401113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wright JT, Simmons D, Frazier-Bowers S, Crawford P, Sangwoo TH, Hart PS, et al. Phenotypic variation of autosomal dominant hypocalcified amelogenesis imperfecta associated with multiple FAM83H mutations. J Dent Res. 2009;88:356–60. doi: 10.1177/0022034509333822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart PS, Hart TC. Disorders of human dentin. Cells Tissues Organs. 2007;186:70–7. doi: 10.1159/000102682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKnight D, Hart PS, Hart TC, Hartsfield J, Wilson A, Wright JT, et al. A comprehensive analysis of normal variation and disease-causing mutations in the human DSPP gene. Hum Mutat. 2008;29:1392–404. doi: 10.1002/humu.20783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trackman PC, Kantarci A. Connective tissue metabolism and gingival overgrowth. Crit Rev Oral Biol Med. 2004;15:165–75. doi: 10.1177/154411130401500305. [DOI] [PubMed] [Google Scholar]
- 16.Hart TC, Zhang Y, Gorry MC, Hart PS, Cooper M, Marazita ML, et al. A mutation in the SOS1 gene causes hereditary gingival fibromatosis type 1. Am J Hum Genet. 2002;70:943–54. doi: 10.1086/339689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye X, Shi L, Cheng Y, Peng Q, Huang S, Liu J, et al. A novel locus for autosomal dominant hereditary gingival fibromatosis, GINGF3, maps to chromosome 2p22.3–p23. 3. Clin Genet. 2005;68:239–244. doi: 10.1111/j.1399-0004.2005.00488.x. [DOI] [PubMed] [Google Scholar]
- 18.Xiao S, Bu L, Zhu L, Zheng G, Yang M, Qian M, et al. A new locus for hereditary gingival fibromatosis (GINGF2) maps to 5q13–q22. Genomics. 2001;74:180–185. doi: 10.1006/geno.2001.6542. [DOI] [PubMed] [Google Scholar]
- 19.Zhu Y, Zhang W, Huo Z, Zhang Y, Xia Y, Li B, et al. A novel locus for maternally inherited human gingival fibromatosis at chromosome 11p15. Hum Genet. 2007;121:113–123. doi: 10.1007/s00439-006-0283-1. [DOI] [PubMed] [Google Scholar]
- 20.Lee EJ, Jang SI, Pallos D, Kather J, Hart TC. Characterization of fibroblasts with son of sevenless-1 mutation. J Dent Res. 2006;85:1050–1055. doi: 10.1177/154405910608501115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang SI, Lee EJ, Hart PS, Ramaswami M, Pallos D, Hart TC. Germ line gain of function with SOS1 mutation in hereditary gingival fibromatosis. J Biol Chem. 2007;282:20245–20255. doi: 10.1074/jbc.M701609200. [DOI] [PubMed] [Google Scholar]
- 22.Hart TC, Hart PS, Bowden DW, Michalec MD, Callison SA, Walker SJ, et al. Mutations of the cathepsin C gene are responsible for Papillon-Lefevre syndrome. J Med Genet. 1999;36:881–887. [PMC free article] [PubMed] [Google Scholar]
- 23.Hart PS, Zhang Y, Firatli E, Yugur C, Lotfazar M, Michalec MD, et al. Identification of cathepsin C mutations in ethnically diverse Papillon-Lefevre syndrome patients. J Med Genet. 2000;37:927–932. doi: 10.1136/jmg.37.12.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Turk D, Janjić V, Stern I, Podobnik M, Lamba D, Dahl SW, et al. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20:6570–6582. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Hart PS, Moretti AJ, Bouwsman OJ, Fisher EM, Dudlicek L, et al. Biochemical and Mutational Analyses of the Cathepsin C Gene in Three North American Families with Papillon Lefevre Syndrome. Hum Mutat. 2002;20:75–81. doi: 10.1002/humu.9040. [DOI] [PubMed] [Google Scholar]
- 26.de Haar SF, Jansen DC, Schoenmaker T, De Vree H, Everts V, Beertsen W. Loss-of-function mutations in cathepsin C in two families with Papillon-Lefèvre syndrome are associated with deficiency of serine proteinases in PMNs. Hum Mutat. 2004;23:524. doi: 10.1002/humu.9243. [DOI] [PubMed] [Google Scholar]
- 27.Ryu OH, Choi SJ, Linares AM, Song IS, Kim YJ, Jang KT, et al. Gingival epithelial cell expression of macrophage inflammatory protein-1alpha induced by interleukin-1beta and lipopolysaccharide. J Periodontol. 2007;78:1627–1634. doi: 10.1902/jop.2007.070066. [DOI] [PubMed] [Google Scholar]
- 28.Hart TC, Hart PS, Michalec MD, Zhang Y, Marazita ML, Cooper M, et al. Localization of a gene for prepubertal periodontitis to chromosome 11q14 and identification of a cathepsin C gene mutation. J Med Genet. 2000;37:95–101. doi: 10.1136/jmg.37.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noack B, Görgens H, Hempel U, Fanghänel J, Hoffmann T, Ziegler A, et al. Cathepsin C gene variants in aggressive periodontitis. J Dent Res. 2008;87:958–963. doi: 10.1177/154405910808701017. [DOI] [PubMed] [Google Scholar]
- 30.Hart TC, Marazita ML, Wright JT. The impact of molecular genetics on oral health paradigms. Crit Rev Oral Biol Med. 2000;11:26–56. doi: 10.1177/10454411000110010201. [DOI] [PubMed] [Google Scholar]
- 31.McKnight DA, Simmer JP, Hart PS, Hart TC, Fisher LW. Overlapping DSPP mutations cause Dentin Dysplasia and Dentinogenesis Imperfecta. J Dent Res. 2008;87:1108–1111. doi: 10.1177/154405910808701217. [DOI] [PMC free article] [PubMed] [Google Scholar]