Abstract
TET enzymes mediate the conversion of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which is enriched in brain, and its ultimate demethylation. However, the influence of TET and 5hmC on gene transcription in brain remains elusive. We demonstrate that TET1 is downregulated in nucleus accumbens (NAc), a key brain reward structure, by repeated cocaine administration, which enhances behavioral responses to cocaine. We then identify 5hmC induction in both putative enhancers and coding regions of genes that have pivotal roles in drug addiction. Such induction of 5hmC, which is induced similarly upon TET1 knockdown alone, correlates with increased expression of these genes as well as with their alternative splicing in response to cocaine administration. Additionally, 5hmC alterations at certain loci persist for at least one month after cocaine exposure. Together, these findings reveal a novel epigenetic mechanism of cocaine action, and provide new insight into how 5hmC regulates transcription in brain in vivo.
DNA methylation is an epigenetic mechanism where methyl groups are covalently coupled to the C-5 position of cytosine (5-methylcytosine or 5mC) predominantly at CpG dinucleotides1. It is generally accepted that DNA methylation, compared to readily reversible histone modifications, is highly stable and mediates long-term gene silencing. This presents an appealing mechanism for long-lasting transcriptional regulation underlying neural plasticity associated with learning and memory as well as neuropsychiatric disorders. However, accumulating evidence indicates that DNA methylation in brain is reversible2–6, which suggests the existence of DNA demethylation machinery. Though several candidates have been proposed as DNA demethylases7 in mammals, a major breakthrough came in 2009, when ten-eleven translocation protein 1 (TET1) was recognized to convert 5mC to 5-hydroxymethylcytosine (5hmC)8,9. Soon after, two other TET family members, TET2 and TET3, were shown to have 5mC hydroxylase activity10,11. It was further revealed that all three TETs successively oxidize 5mC to 5hmC, 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) in order12,13. As TET proteins directly convert 5mC, oxidized 5mC raises a plausible DNA demethylation mechanism that has become a focus of intensive interest. In fact, studies indicated a number of pathways leading to 5hmC mediated DNA demethylation14–16. For example, TET catalyzed 5hmC conversion into 5fC and 5caC can be efficiently removed from DNA by thymine-DNA glycosylase (TDG). Subsequent repair of the resulting abasic site via base excision repair (BER) can generate an unmethylated cytosine. Alternatively, deamination followed by TDG and BER pathways can also remove 5hmC and replace with unmethylated cytosine17.
We have just started to understand the distribution and function of these novel forms of DNA epigenetic modifications (5hmC, 5fC, 5caC) and their catalyzing TET enzymes in the genome, which are essential in a range of biological processes such as embryonic development, stem cell function, and cancer formation14–16. Of note, 5hmC is most abundant in brain compared with other organs8,18–20, suggesting an important role in neural function21. Indeed, emerging evidence indicates that TET1 regulates active hippocampal DNA demethylation17,22,23, cognition24, and learning and memory22,23,25. However, the influence of TET proteins and 5hmC on the regulation of gene transcription in brain and their role in brain disorders remain largely unknown.
In this study, we set out to determine whether TET proteins and 5hmC are involved in the epigenetic regulation of cocaine action. Repeated cocaine exposure induces persistent changes in gene expression within the nucleus accumbens (NAc)26, a key brain structure of the reward circuitry, and epigenetic mechanisms play important roles in this process as well as in downstream neural and behavioral plasticity27,28. It has been shown that cocaine can alter the expression of several histone-modifying enzymes and DNA methyltransferases in NAc29–32. Here, we show decreased expression of TET1, but not TET2 or TET3, in NAc after repeated cocaine administration. By use of viral manipulations, we establish that decreased TET1 serves to enhance behavioral responses to the drug. In concert with TET1 downregulation, repeated cocaine increases the enrichment of 5hmC at a large subset of genes in NAc that are involved in drug addiction. Such changes in 5hmC are concentrated at both putative enhancers and gene bodies, and correlate with increased expression of these genes as well as with their alternative splicing. Importantly, we show that cocaine-induced increases in 5hmC at representative loci in NAc are mimicked upon Tet1 knockdown alone. In sum, our findings not only advance our understanding of the epigenetic mechanisms involved in cocaine action, but also provide fundamentally new insight into how 5hmC regulates gene expression in the brain.
RESULTS
Repeated cocaine decreases TET1 in NAc to enhance behavioral responses
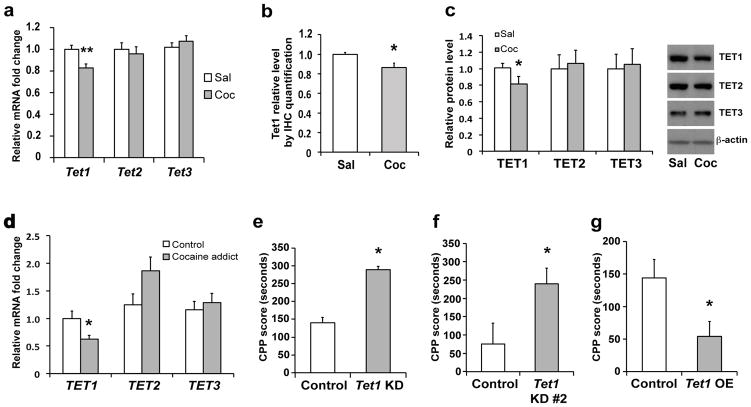
We first examined Tet mRNA regulation in mouse NAc 24 hr after repeated cocaine exposure. Quantitative PCR (qPCR) revealed that, among all three known Tet family members, only Tet1 was down-regulated, with no significant change in Tet2 or Tet3 (Fig. 1a). Quantitative Western blotting and immunohistochemistry revealed a concomitant decrease in TET1 protein, but not TET2 or TET3, in NAc after repeated cocaine (Figs. 1b, 1c, Supplementary Fig. 1a). Importantly, we found a ~40% decrease in TET1 mRNA in the NAc of human cocaine addicts examined postmortem (Fig. 1c). These data demonstrate that TET1 expression is subject to dynamic regulation in the adult brain and implicate TET1 in cocaine action.
Figure 1. TET1 expression is decreased in NAc after repeated cocaine and negatively regulates behavioral responses to cocaine.
a, qPCR analysis shows relative transcription of Tet1, Tet2, and Tet3 mRNA in NAc 24 hr after repeated cocaine (Tet1: P=0.002, t(34)=3.321, n=18/group; Tet2: t(34)=0.498, n=18/group; Tet3: t(46)=0.861, n=24/group). b, Quantification of TET1 in NAc by immunohistochemistry 24 hr after repeated cocaine (P=0.035, t(22)=2.245, n=6/group, ). c, Western blot analysis shows a selective decrease in TET1 protein 24 hr after repeated cocaine (TET1: P=0.034, t(38)=1.863; TET2: t(30)=0.525; TET3: t(39)=0.405. n=15–20/group). A representative blot is shown with β-actin as a loading control. d, qPCR analysis shows a selective decrease in TET1 mRNA, but not TET2 or TET3, in NAc of human cocaine addicts (TET1: P=0.020, t(39)=2.426; TET2: t(36)=1.903; TET3: t(36)=0.593. n=19–21/group). e, Conditioned place preference (CPP) for cocaine (7.5 mg/kg body weight) with viral Tet1 shRNA knockdown (KD) construct#1 in NAc (P=0.049, t(25)=2.060, n=15 for control group and 12 for KD). f, CPP for cocaine with viral Tet1 shRNA KD construct#2 in NAc (P=0.031, t(22)=2.303, n=12/group). g, CPP for cocaine with AAV-TET1 overexpression (OE) in NAc (P=0.027, t(23)=2.358, n=14 for control and 11 for OE group). Data are presented as mean ± SEM. * indicates P<0.05, ** indicates P<0.01. Sal, saline; Coc, cocaine.
To investigate directly whether altered levels of TET1 in NAc regulate behavioral responses to cocaine, we performed stereotaxic viral manipulations to express Tet1-shRNA to knockdown TET1 selectively in the adult NAc (Supplementary Figs. 2a, 2b). We first confirmed the accuracy of viral injection and integrity of brain structure after the surgeries (Supplementary Fig. 1b). We then analyzed the mice in an unbiased cocaine conditioned place preference paradigm, which provides an indirect measure of drug reward. Tet1 knockdown with either of two independent shRNA constructs robustly enhanced cocaine place conditioning (Figs. 1e, 1f). Conversely, we utilized a previously validated AAV-TET1 vector to express TET117 in NAc (Supplementary Fig. 2c) and found significantly decreased cocaine preference (Fig. 1g), which indicates that Tet1 in the NAc is sufficient to suppress cocaine reward. Together, these data suggest that TET1 normally functions to negatively regulate cocaine reward and that cocaine-induced suppression of TET1 in NAc contributes to enhanced drug sensitivity.
5hmC profiling in NAc after repeated cocaine administration
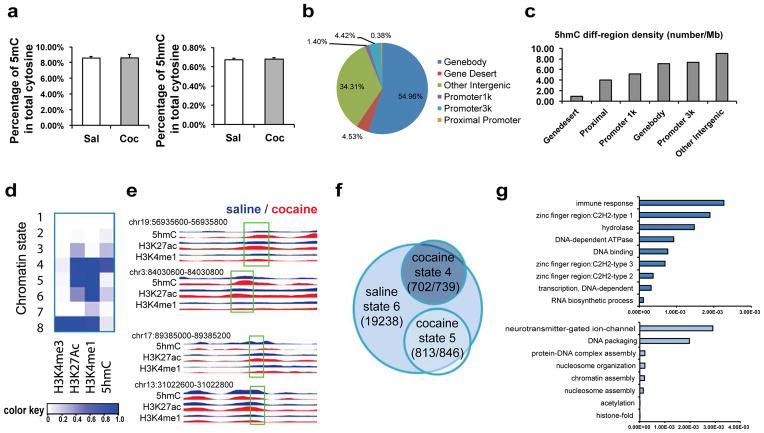
As TET1 oxidizes 5mC into 5hmC, we next investigated cocaine regulation of these DNA modifications in NAc. First, total levels of 5mC and 5hmC were quantified by liquid chromatography–electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS)5,33. Neither 5mC nor 5hmC demonstrated a significant global change as a percentage of total cytosine in NAc after repeated cocaine (Fig. 2a). This suggests that any alterations in 5hmC may be locus specific. Alternatively, given the ~2-fold greater abundance of Tet2 and Tet3 mRNAs in NAc (Supplementary Fig. 3), it is conceivable that they compensate for TET1 downregulation despite their lack of regulation by cocaine (Figs. 1a, 1c). To test these possibilities, we obtained genome-wide mapping of 5hmC by performing a selective chemical labeling method for 5hmC capture followed by deep sequencing18,21. This methodology has proven to be an accurate, selective, and comprehensive approach to studying 5hmC18, and we obtained high quality sequencing results in our analysis of NAc (Supplementary Table 1).
Figure 2. Repeated cocaine induces 5hmC alterations in NAc.
a, Quantitative analysis of 5mC and 5hmC using LC-ESI-MS/MS. Global 5mC and 5hmC content is expressed as the percentage in the total cytosine pool (5mC: t(16)=0.018, 5hmC: t(16)=0.295. n=9/group). Data are presented as mean ± SEM. b, Genomic distribution of sites that show cocaine-induced changes in 5hmC enrichment (n=3/group). Gene deserts are any regions that do not contain gene annotations and are at least 1 Mb in length. Other intergenic regions refer to those not otherwise classified as any category displayed in the chart. c, 5hmC differential region density is presented as the number of cocaine-induced differential regions per Mb sequence. d, “Chromatin state” is generated is generated by ChromHMM40 to define combinatorial patterns of H3K4me3 (a promoter mark), H3K27ac and H3K4me1 (enhancer marks), and 5hmC in NAc. Eight distinct states are demonstrated, and each has a different combination of chromatin mark enrichment. Color key reflects the emission probabilities of a hidden Markov model, which denotes the frequencies of chromatin mark presences. e, Representative tracks of 5hmC, H3K27ac, and H4K4me1 in saline (blue) or cocaine (red) illustrate a chromatin state switch from state 6 in saline to state 4 in cocaine (top two samples) or to state 5 in cocaine (bottom two samples). The regions that display enrichment changes in 5hmC and H3K27ac are highlighted with a box and denoted with the corresponding genomic coordinates. Y-axis reflects normalized read counts that are set to the same scale for saline and cocaine. f, The Venn diagram demonstrates that the vast majority of cocaine-specific putative enhancer genes (702 out of 739 for state 4 and 813 out of 846 for state 5) have a chromatin state switch at the putative enhancer site from state 6 to state 4 or 5 after repeated cocaine. g. List of gene ontology terms that are most significantly enriched in cocaine-specific state 4 putative enhancer genes (top) and cocaine-specific state 5 putative enhancer genes (bottom).
In total, we identified 208,801 5hmC peak regions from NAc of saline-treated control mice and 226,185 peaks from cocaine-treated mice. Though the gross chromosomal distribution of 5hmC peak regions was equivalent between saline- and cocaine-treated conditions, we found that sex chromosomes have extremely low 5hmC enrichment (Supplementary Fig. 4a). We also identified 11,511 regions that displayed differential levels of 5hmC after repeated cocaine (Supplementary Table 2), which appears to be evenly distributed across all autosomes (Supplementary Fig. 4b). These 5hmC differential regions were heavily distributed in gene bodies (~55%) and intergenic regions (~34%) by calculating the percentage of differential region counts across all genomic features (Fig. 2b, Supplementary Fig. 4c). In addition, we measured the density of these 5hmC alterations and revealed intergenic regions, gene promoters, and gene bodies as among the top categories (Fig. 2c). Hence, we speculated that 5hmC might play functional roles in both intergenic as well as genic regions in cocaine action.
5hmC dynamics at putative enhancers
We first characterized 5hmC dynamics at putative enhancer regions. Enhancers are DNA regulatory elements that play critical roles in gene expression. Many of them reside long distances away from gene promoters in either upstream or downstream regions. It is estimated that hundreds of thousands of enhancers exist in the genome, vastly outnumbering the ~20,000 coding genes. Recently, it was shown that enhancers in the mammalian genome are associated with characteristic histone modification patterns34, for example, H3K27ac and H3K4me1 co-binding has been widely recognized as a signature of active enhancers that engage in transcription regulation34–38. In accordance with these studies of several tissues, to define putative enhancers in NAc, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for H3K4me1 and H3K27ac (Supplementary Table 1). We also included a previously reported ChIP-seq dataset for H3K4me339, which is highly enriched at promoters, to exclude our predicted enhancers from promoters.
When we plot our ChIP-seq data of H3K4me1 and H3K27ac over putative mouse brain enhancers derived from the ENCODE database (http://genome.ucsc.edu/ENCODE/), both display enriched binding at enhancer regions (Supplementary Fig. 5). This analysis suggests good antibody specificity and sequencing quality. Likewise, 5hmC showed enrichment at these putative enhancer regions genome-wide. We then performed a study on the combination of these epigenetic modifications —‘or chromatin states’34,38, by using ChromHMM38,40. We applied combinatorial patterns of H3K4me1 and H3K27ac, together with 5hmC, to define several distinct chromatin states (see Online Methods, Fig. 2d)38,40. We first excluded state 8 from our putative enhancer analysis because of its high levels of H3K4me3, which reflect promoter presence (state 8 in Fig. 2d). We then focused on the chromatin states that are characterized by enhancer marks H3K4me1 and H3K27ac, as well as 5hmC (states 4, 5, and 6 in Fig. 2d). To further validate our definition of putative enhancers by H3K4me1 and H3K27ac ChIP-seq through ChromHMM, we applied a second algorithm (RFECS, Random Forest based Enhancer identification from Chromatin States) that reportedly accurately predicts enhancers based solely on H3K4me1 and H3K27ac ChIP-seq34. We found that the enhancer predictions from the two algorithms have >70% overlap, which provides validity for our enhancer predication approach. We next performed qChIP to validate H3K4me1 and H3K27ac enrichment at multiple predicted putative enhancer sites (Supplementary Fig. 6). Importantly, these putative enhancer sites exhibited various degrees of enrichment of P300 (Supplementary Fig. 6), a transcription co-activator that also marks enhancer regions. These findings thus further support our method of identifying putative enhancers.
Our analyses revealed robust regulation of these various chromatin states at putative enhancer regions in NAc by cocaine. A subgroup of putative enhancers displayed increased prevalence of H3K27ac together with either increased 5hmC (switch from state 6 to state 4) or decreased 5hmC (switch from state 6 to state 5) after repeated cocaine (Figs. 2d, 2e). As enhancers can regulate expression of neighboring protein-coding genes41, we assigned these putative enhancers to the nearest genes to connect them to plausible targets (see Discussion for limitations of this approach). We recognized 739 and 846 cocaine-specific chromatin state 4 and state 5 putative enhancer genes, respectively (Supplementary Table 3). The vast majority (>90%) of these genes’ enhancers were under state 6 at saline condition (Fig. 2f). This analysis indicates robust 5hmC dynamics —a gain in 5hmC in switching from state 6 to state 4 or a loss of 5hmC in switching from state 6 to state 5—after cocaine. Gene ontology (GO) analysis of cocaine-specific state 4 and state 5 putative enhancer genes reveals enrichment in several interesting categories (Fig. 2g, Supplementary Table 4). For example, cocaine-specific state 4 putative enhancer genes include C2H2 zinc finger proteins that bind to methylated DNA42 and immune genes that are implicated in neuronal plasticity43. In contrast, cocaine-specific state 5 putative enhancer genes are mainly clustered in neurotransmitters as well as many chromatin assembly genes, both of which are important for neural plasticity and memory44. As transcription factors are increasingly recognized to bind to enhancers as part of their regulatory roles, we performed a motif analysis of the cocaine-specific state 4 and state 5 putative enhancer regions and found significant enrichment of a handful of candidate transcription factors (Supplementary Table 5). Some of these same transcription factors were deduced from our recent analysis of cocaine regulation of histone modifications and alternative splicing39, which suggests follow-up studies.
5hmC positively correlates with alternative splicing
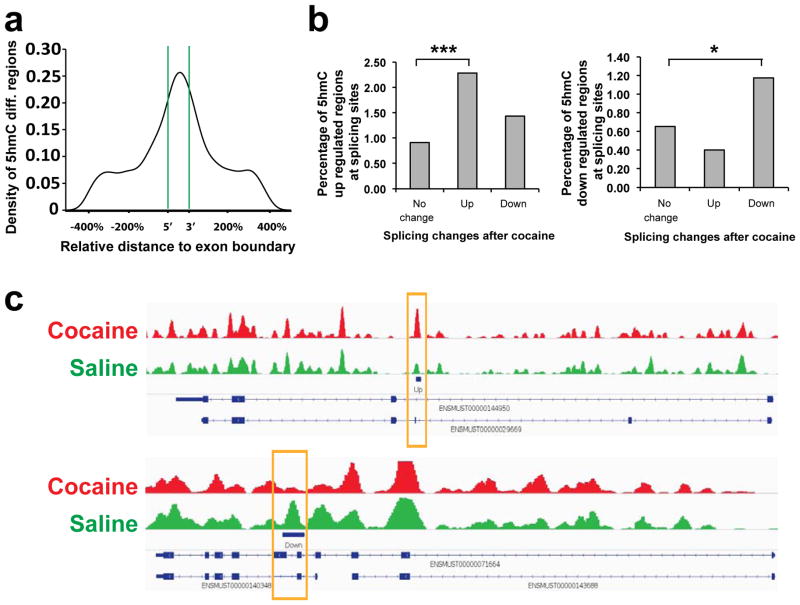
Next, we explored the influence of altered 5hmC enrichment at coding regions of NAc in response to repeated cocaine. We identified significant enrichment of cocaine-induced changes in 5hmC at flanking exon boundaries (Fig. 3a), which implicates 5hmC in mRNA alternative splicing. Furthermore, we overlaid our 5hmC profiling with RNA-seq analysis of NAc 24 hr after repeated cocaine or saline treatment, the same time point used for our 5hmC experiments. We found significant enrichment of 5hmC-increased regions (35 regions) at splicing sites of up-regulated splicing isoforms (1488 sites), as well as significant enrichment of 5hmC-decreased regions (18 regions) at splicing sites of down-regulated isoforms (1536 sites; Fig. 3b, 3c). To our knowledge, this is the first documentation of detailed alternative splicing sites coupled with 5hmC changes in brain (Supplementary Table 6). Together, our findings support the involvement of 5hmC at exonic regions in alternative RNA splicing in response to cocaine.
Figure 3. 5hmC alterations positively correlate with alternative splicing regulation.
a. 5hmC differential region density plot at exon regions. The X-axis represents an exon at the center with flanking regions that are of the size of 400% of the central exon. Green lines illustrate exon boundaries. b, Percentage enrichment of up-regulated 5hmC regions (left) or down-regulated 5hmC regions (right) at splicing sites of increased (up), decreased (down), and not significantly changed (no change) splicing isoforms in response to cocaine. (Fisher’s exact test; ***= 4.36E-06, *=0.022). c, Representative 5hmC tracks in saline (green) and cocaine (red) conditions that correlate with altered expression of a splicing isoform. A yellow box highlights the altered 5hmC after repeated cocaine. In the upper sample, increased 5hmC is associated with upregulation of the splicing isoform (transcript ID:ENSMUST00000029669, RNAseq log2 fold change=1.079, Q-value=0.0085). In the lower sample, decreased 5hmC is associated with downregulation of the splicing isoform (transcript ID:ENSMUST00000071664, RNAseq log2 fold change=-0.507, Q-value=0.0005).
5hmC dynamics correlate with cocaine-induced transcriptional regulation
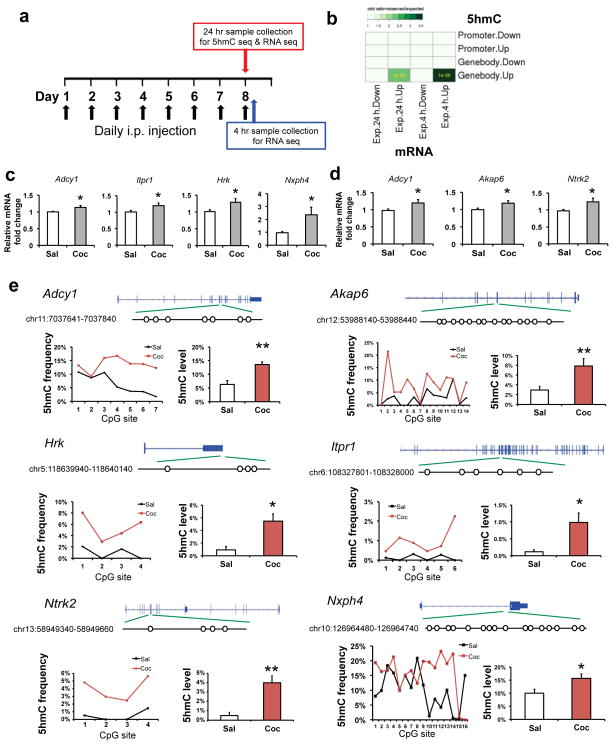
To complement our transcriptome analysis at 24 hr of withdrawal from repeated cocaine treatment, we generated another RNA-seq dataset from NAc, which was obtained 4 hr after a subsequent challenge dose of cocaine (Fig. 4a). This new dataset better reflects altered inducibility of genes in contrast to the 24 hr dataset which reflects longer-lasting or steady-state changes in gene expression. Interestingly, cocaine-induced increases in 5hmC at gene bodies correlated significantly with increased gene expression after 24 hr of withdrawal from cocaine exposure (odds ratio=observation/expectation is 2.5, P-value=4e-03, 24 out of 356 up-regulated genes displayed increased 5hmC in gene body regions, Fig. 4b), and this overlap is even more significant with genes that are induced by a cocaine challenge (odds ratio=3.5, P-value is 1e-06, Fig. 4b; odd ratio=~1, P>0.05 for all other cross comparisons). 31 out of 261 genes up-regulated at 4 hr showed a pre-existing increase in 5hmC enrichment (Table 1). GO analysis indicated that these overlapping genes were concentrated in certain meaningful categories that have pivotal roles in drug addiction, such as long-term plasticity, synaptic transmission, and glutamate neurotransmitter (Table 1, Supplementary Fig. 7). These data indicate that, after repeated exposure to cocaine, increased 5hmC in NAc correlates with both steady-state gene induction as well as with gene inducibility in response to a subsequent cocaine challenge. Of note, however, the enhancer changes did not correlate genome-wide with gene expression (see Discussion).
Figure 4. Gene body 5hmC alterations correlate with gene transcription changes after repeated cocaine.
a, Schematic drawing of the experimental timeline of 24 hr 5hmC-seq and RNA-seq (24 hr after a course of repeated cocaine treatment), and 4 hr RNA-seq (4 hr after a subsequent cocaine challenge). b, Heatmap showing significant overlaps between 5hmC alterations at gene bodies after repeated cocaine (24 hr) and gene expression change (at 24 hr or 4 hr). Color code represents odds ratio (observed/expected). Only significant P-values are shown in grids. c, qPCR validation confirms RNA-seq findings of increased expression of Adcy1 (P=0.010, t(22)=2.815), Itpr1 (P=0.043, t(22)=2.151), Hrk (P=0.031, t(18)=2.345), and Nxph4 (P=0.041, t(21)=2.177) 24 hr after repeated cocaine (c) and Adcy1 (P=0.047, t(18)=2.133), Akap6 (P=0.046, t(18)=2.146), and Ntrk2 (P=0.043, t(18)=2.180) 4 hr after a subsequent challenge (d). N=10–12/group. e, oxBS-seq validates 5hmC differential changes in Adcy1(P=0.005, t(6)=4.34), Akap6 (P=0.004, t(13)=3.549), Hrk (P=0.011, t(6)=3.641), Itpr1 (P=0.034, t(5)=2.888), Ntrk2 (P=0.005, t(3)=7.692), and Nxph4 (P=0.034, t(15)=2.324). Schematic gene structures are illustrated on top. Each differential locus is enlarged with open circles denoting CpG sites. Genomic coordinate of the differential locus is also shown. A summary of 5hmC frequency at each CpG site is demonstrated in a line chart and the mean 5hmC levels of individual alleles is displayed in a bar graph (n=2–4 biological replicates per condition). All data are presented as mean ± SEM. * indicates P<0.05, ** indicates P<0.01. Sal, saline; Coc, cocaine.
Table 1. List of genes that show overlap of cocaine-induced changes by 5hmC-seq and RNA-seq.
The table lists those genes that show in increase both in 5hmc at gene bodies and in RNA expression at 4 or 24 hr.
| 24hr 5hmC up and 24 hr mRNA up overlapping genes | ||
|---|---|---|
| gene | gene ID | representative gene ontology terms |
| 6330403A02Rik | ENSMUSG00000053963 | membrane |
| Adcy1 | ENSMUSG00000020431 | adenyl nucleotide binding; learning or memory; Behavior; Long-term potentiation;Gap junction; memory;Calcium signaling pathway; GnRH signaling pathway |
| Atp2b1 | ENSMUSG00000019943 | calcium ion transport;phosphoprotein; ATP binding; brain development; aging |
| Crtac1 | ENSMUSG00000042401 | calcium ion binding |
| Etl4 | ENSMUSG00000036617 | glycoprotein; embryonic skeletal system development; glycosylation site:O-linked (GlcNAc); phosphoprotein |
| Fam49b | ENSMUSG00000022378 | breast cancer? |
| Foxo1 | ENSMUSG00000044167 | phosphoprotein; nerve growth factor receptor signaling; transcription regulation |
| Hrk | ENSMUSG00000046607 | neronal death; apoptosis |
| Hs3st4 | ENSMUSG00000078591 | sulfotransferase activity |
| Itpr1 | ENSMUSG00000030102 | Long-term potentiation; synapse; Long-term potentiation; metal ion transmembrane transporter activity; cellular ion homeostasis; calcium ion binding; phosphoprotein; GnRH signaling |
| Kremen1 | ENSMUSG00000020393 | membrane; glycoprotein; nervous system development; Wnt signaling pathway |
| Lars2 | ENSMUSG00000035202 | ATP binding; gene expression; splicing; translation fidelity. |
| Mapk10 | ENSMUSG00000046709 | GnRH signaling pathway; phosphoprotein; ATP binding; Reelin Signaling in Neurons; Interleukin signaling; GDNF signaling |
| Mbp | ENSMUSG00000041607 | transmission of nerve impulse; cellular ion homeostasis; phosphoprotein |
| Ndrg3 | ENSMUSG00000027634 | phosphoprotein; negative regulation of cell growth; positive regulation of Ras protein signaling |
| Nxph4 | ENSMUSG00000040258 | neuropeptide signaling pathway |
| Pcsk5 | ENSMUSG00000024713 | glycoprotein; cytoplasmic membrane-bounded vesicle;embryonic skeletal system development; nerve growth factor receptor signaling pathway |
| Rgnef | ENSMUSG00000021662 | plasma membrane; phosphoprotein; central nervous system neuron axonogenesis |
| Satb2 | ENSMUSG00000038331 | embryonic skeletal system development;phosphoprotein |
| Scube1 | ENSMUSG00000016763 | calcium ion binding; glycoprotein |
| Slc24a2 | ENSMUSG00000037996 | memory; metal ion transmembrane transporter activity; cellular ion homeostasis; long term synaptic depression |
| Spock2 | ENSMUSG00000058297 | calcium ion binding; phosphoprotein; glycoprotein; synapse assembly |
| Sv2b | ENSMUSG00000053025 | synapse; regulation of neurotransmitter levels; synaptic vesicle; transmission of nerve impulse; neurotransmitter secretion |
| Syn1 | ENSMUSG00000037217 | transmission of nerve impulse; neurotransmitter secretion; ATP binding |
| 24hr 5hmC up and 4 hr mRNA up overlapping genes | ||
|---|---|---|
| gene | gene_id | representative gene ontology terms |
| Adcy1 | ENSMUSG00000020431 | adenyl nucleotide binding; learning or memory; Behavior; Long-term potentiation;Gap junction; memory;Calcium signaling pathway; GnRH signaling pathway |
| Akap6 | ENSMUSG00000061603 | Spectrin/alpha-actinin; kinase; endomembrane system; nuclear envelope; positive regulation of cell growth |
| Arhgap32 | ENSMUSG00000041444 | dendritic spine; cell membrane; phosphoprotein; dendrite; neuron projection; lipid binding; postsynaptic density; synapse |
| Atp1a3 | ENSMUSG00000040907 | locomotory behavior; learning; cellular chemical homeostasis; ion homeostasis; atp-binding; visual learning; visual behavior |
| Birc6 | ENSMUSG00000024073 | apoptosis; protein ubiquitination |
| Cacna1e | ENSMUSG00000004110 | transport; calcium channel; di-, tri-valent inorganic cation transport; Calcium signaling pathway; behavioral response to pain; fear response |
| Camk2a | ENSMUSG00000024617 | regulation of synaptic plasticity; autophosphorylation; phosphoprotein; GnRH signaling pathway; ATP binding; Transcription factor CREB signaling; neurotransmitter secretion |
| Cyld | ENSMUSG00000036712 | phosphoprotein; central nervous system morphogenesis; apoptosis |
| D10Bwg1379e | ENSMUSG00000019852 | membrane; phosphoprotein |
| Daam2 | ENSMUSG00000040260 | cytoskeletal protein binding; Wnt signaling pathway; coiled coil; actin binding; Wnt signaling pathway |
| Dst | ENSMUSG00000026131 | axonogenesis; calcium ion binding; cytoskeleton organization; intermediate filament-based process; regulation of microtubule polymerization or depolymerization; coiled coil |
| Gabrb1 | ENSMUSG00000029212 | synapse; postsynaptic cell membrane; cell junction; ligand-gated channel activity |
| Gpr158 | ENSMUSG00000045967 | phosphoprotein; G-protein coupled receptor signaling pathway |
| Gpr26 | ENSMUSG00000040125 | G-protein coupled receptor signaling pathway |
| Gria2 | ENSMUSG00000033981 | dendrite; neuron projection; neurotransmitter receptor; glutamate receptor activity; synaptic transmission; transmission of nerve impulse; Long-term depression; Long-term potentiation |
| Grin2a | ENSMUSG00000059003 | synapse; memory; synaptic transmission;regulation of transmission of nerve impulse; Calcium signaling pathway; Glutamate receptor-related; learning or memory;behavior |
| Grin2b | ENSMUSG00000030209 | synapse; memory; synaptic transmission;regulation of transmission of nerve impulse; Calcium signaling pathway; Glutamate receptor-related; learning or memory;behavior |
| Hipk2 | ENSMUSG00000061436 | locomotory behavior; phosphoprotein; atp-binding;regulation of transcription |
| Ina | ENSMUSG00000034336 | coiled coil; cytoskeleton; phosphoprotein; nervous system development |
| Itpr1 | ENSMUSG00000030102 | Long-term potentiation; synapse; Long-term potentiation; metal ion transmembrane transporter activity; cellular ion homeostasis; calcium ion binding; phosphoprotein; GnRH signaling |
| Kif1a | ENSMUSG00000014602 | axon guidance; anterograde synaptic vesicle transport; cytoskeleton; atp-binding; coiled coil |
| Lars2 | ENSMUSG00000035202 | ATP binding; gene expression; splicing; translation fidelity |
| Mtap1b | ENSMUSG00000052727 | induction of synaptic plasticity; phosphoprotein; microtubule-based process; cytoskeleton |
| Ntrk2 | ENSMUSG00000055254 | behavior; phosphoprotein; dendrite; atp-binding; regulation of synaptic transmission; regulation of neurotransmitter secretion; regulation of transmission of nerve impulse; leaning and memory |
| Ppp1r16b | ENSMUSG00000037754 | coiled coil; signal transduction |
| Prkcb | ENSMUSG00000052889 | serine/threonine-specific protein kinase; Long-term potentiation; calcium ion transport; Synaptic Long Term Depression; GnRH signaling pathway; NFkB signaling |
| Setd7 | ENSMUSG00000037111 | histone lysine methylation |
| Sipa1l1 | ENSMUSG00000042700 | coiled coil; regulation of synaptic plasticity; ephrin receptor signaling; Rap GTPase signaling |
| Syne1 | ENSMUSG00000019769 | actin binding; brain development; dendrite morphogenesis |
| Tmtc1 | ENSMUSG00000030306 | membrane |
| Unc13c | ENSMUSG00000062151 | coiled coil; synapse; cell junction; transmission of nerve impulse; synaptic transmission |
To validate our findings, we carried out qPCR to measure gene expression and oxBS-seq to quantify 5hmC alterations45. We first confirmed induction of several genes at the 24 hr time point (Adcy1, Itpr1, Hrk, Nsph4, Fig. 4c) or 4 hr after a cocaine challenge (Adcy1, Akap6, Ntrk2, Fig. 4d) that are consistent with our RNA-seq findings. We then chose two genomic regions from our 5hmC-seq analysis for validation with oxBS-seq (Supplementary Fig. 8). This experiment confirmed significant induction of 5hmC at each of these genes in an independent cohort of animals (Fig. 4e).
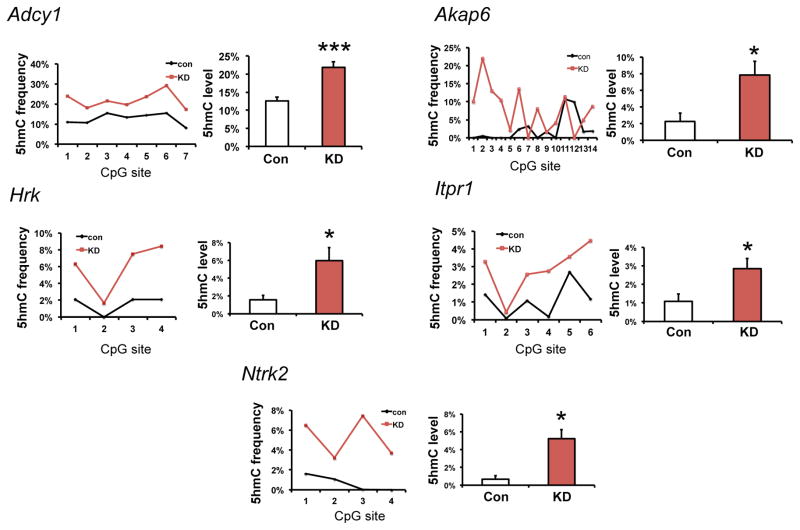
To establish a causal role for the cocaine-induced downregulation of TET1 in NAc in mediating the increased enrichment of 5hmC at gene targets, we investigated whether TET1 knockdown in NAc by itself in cocaine naïve mice is able to mimic the 5hmC changes seen after cocaine. Indeed, for the six loci that we studied, five displayed robust induction of 5hmC upon TET1 knockdown (Fig. 5).
Figure 5. Tet1 knockdown in NAc of cocaine naïve mice induces 5hmC at cocaine-regulated loci.
oxBS-seq demonstrates increased 5hmC at Adcy1 (P=0.0002, t(6)=8.017), Akap6 (P=0.019, t(13)=2.684), Hrk (P=0.022, t(3)=4.356), Itpr1 (P=0.011, t(5)=3.945), and Ntrk2 (P=0.027, t(3)=4.082) from NAc of naïve animals that received viral Tet1 shRNA knockdown as in Fig. 1e. A summary of 5hmC frequency at each CpG site is demonstrated in a line chart and the mean 5hmC level of individual alleles is displayed in a bar graph (n=2–4 biological replicates per condition). All data are presented as mean ± SEM. * indicates P<0.05, ** indicates P<0.01. Con, control shRNA; KD, knockdown with Tet1 shRNA.
Repeated cocaine-induced 5hmC regulation can be long-lasting
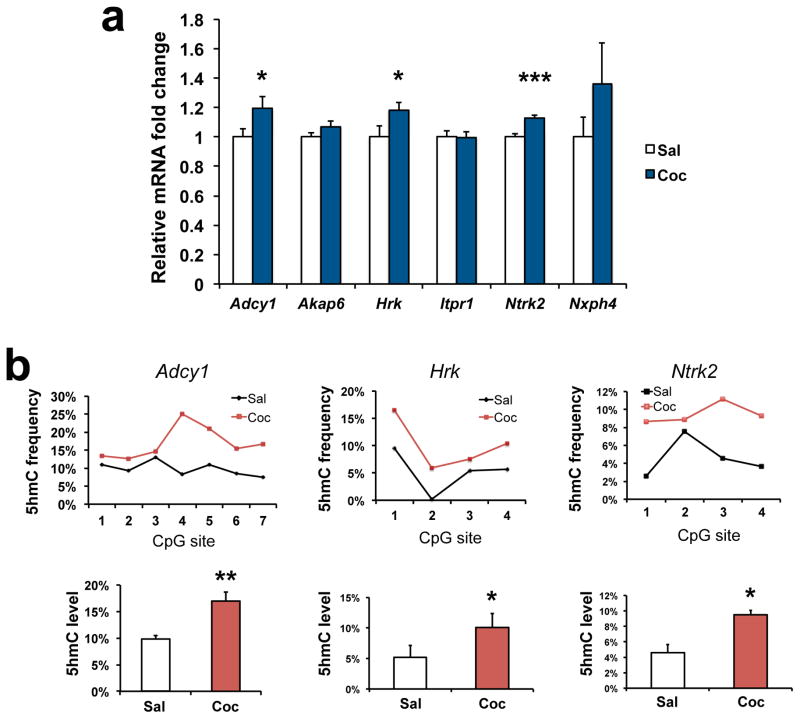
5hmC is generally considered a transient intermediate state between 5mC and 5fC/5caC that ultimately leads to unmethylated cytosine. However, 5hmC exists at much more abundant levels—particularly in brain—compared with 5fC or 5caC. Hence, we speculated that 5hmC might also serve as a stable epigenetic mark and tested if some of the cocaine-induced changes in 5hmC persist long after cocaine exposure. Among the genes we studied (Fig. 4), we demonsrated sustained induction of 3 of them (Adcy1, Hrk, and Ntrk2) (Fig. 6) one month after repeated cocaine treatment, and this induction was associated with prolonged increases in 5hmC (Fig. 6), effects not seen for the other genes (data not shown). These findings indicate that 5hmC can be a persisting epigenetic mark associated with prolonged transcriptional activation. As Adcy1 and Ntrk2 play important roles in drug addiction46–48, the sustained induction of 5hmC might contribute to the persistent, strong actions of cocaine in brain.
Figure 6. Long-lasting induction of 5hmC at particular loci after cocaine.
a, qPCR shows increased expression of Adcy1 (P=0.043, t(32)=2.098), Hrk (P=0.044, t(30)=1.517), and Ntrk2 (P=7.04E-05, t(30)=4.607) one month after repeated cocaine exposure, with no persisting change observed for Akap6 (t(30)=1.517), Itpr1(t(30)=0,071), or Nxph4 (t(30)=1.308). N=16–17/group. Data are displayed as mean ± SEM. b, oxBS-seq demonstrates concomitant increase of 5hmC at genes with persistent transcriptional induction (Adcy1: P=0.012, t(6)=3.567; Hrk: P=0.018, t(3)=4.688; Ntrk2: P=0.027, t(3)=4.050). The results are displayed in the same manner as in Fig. 5. N=2–3 biological replicates/condition. Data are presented as mean ± SEM. * indicates P<0.05, ** indicates P<0.01. Sal, saline; Coc, cocaine.
DISCUSSION
Results of the present study reveal previously unappreciated roles for TET enzymes and 5hmC in the molecular and behavioral effects of cocaine. We demonstrate selective downregulation of TET1 in NAc in response to chronic cocaine administration, which serves to promote behavioral responses to the drug. We then go on to link such TET1 downregulation to alterations of 5hmC at putative enhancer regions and gene bodies, changes associated with altered gene expression and alternative splicing. Together, the studies reveal new epigenetic mechanisms involved in cocaine action.
Though most studies of 5mC have focused on gene promoters, our finding that 5hmC alterations in gene bodies are associated with altered gene transcription is consistent with previous observations of 5hmC enrichment at gene bodies in stem cells49 and during neural development21. While the exonic enrichment of 5hmC is suggestive of a role in pre-mRNA alternative splicing50, our study is the first to directly associate regulation of 5hmC at alternative splicing sites with altered splicing. We recently demonstrated that cocaine induces an order of magnitude more changes in alternative splicing than changes in total transcript levels39. The present results thus implicate 5hmC in this prominent form of transcriptional regulation. Interestingly, MeCP2, a protein that binds to 5mC, has been shown to regulate drug addiction51,52. Recent work also revealed the direct binding of MeCP2 to 5hmC53. In concert with previous reports of MeCP2’s regulation of alternative splicing54, it will be interesting to explore whether MeCP2 interacts with 5hmC changes at splicing sites in NAc in the context of cocaine.
We also identified numerous cocaine-induced changes in 5hmC at putative enhancer sites in NAc. Using H3K4me1 and H3K27ac as tentative marks of active enhancers34–38, we studied 5hmC dynamics at enhancer regions in response to cocaine. Although enhancers have long been recognized for their regulatory importance, the lack of common sequence features makes them difficult to identify. Our study indicates that 5hmC is enriched at enhancer regions in NAc and that such enrichment is regulated by cocaine. By assigning cocaine-regulated putative enhancers to the nearest genes to probe for potential enhancer targets, we found that the gene targets are enriched in interesting GO categories. However, we failed to detect a correlation between these regulated enhancers and our RNA-seq transcriptome profiling, emphasizing that the degree to which modifications at enhancer sites control dynamic regulation of gene expression in brain remains uncertain. Moreover, while defining enhancer-regulated targets by proximity alone has been a fruitful approach35, we must emphasize its limitations. Many enhancers bypass hundreds of kilobases of interspersed sequence on the linear genome to interact with distant targets via chromosomal looping. Accordingly, studying the three-dimensional structure of chromatin can provide a precise prediction of enhancer target genes55. However, studies of higher order genome architecture are just beginning to be applied to the brain and major advances are needed before they can be applied to a microdissected brain region such as the NAc. Another means by which enhancers influence transcription is by directing the expression of a group of non-coding RNAs termed enhancer RNAs (eRNAs)56. These eRNAs are particularly sensitive to neural activity. However, many of the eRNAs are nonpolyadenylated57 which is beyond the detection range of our poly-A selection based Illumina RNA-seq protocol (see Online Method for details). Moreover, given the cell-type specific feature of enhancers36, enhancer-driven transcriptional regulation likely occurs in only a subtype of cells, which is difficult to examine in our current heterogeneous tissue dissections, and is impossible to isolate in sufficient quantities for more selective analysis. Functional implications of putative enhancers are often confirmed in in vitro reporter assays, although this approach is limited for brain since cultured cells—even cultured neurons—do not always reflect modes of transcriptional regulation seen in brain in vivo. Continued technology development55,58,61 should improve our ability to study enhancer regulation in NAc in addiction models in future investigations. Nevertheless, despite these limitations, our putative identification of enhancer regions identified numerous genes whose expression in NAc is associated with altered 5hmC enrichment. Our analysis also identified several transcription factor motifs at cocaine-regulated putative enhancer regions, which is consistent with the emerging role of transcription factors in non-coding enhancer regions35. These findings illustrate the importance of overlaying analyses of dynamic enhancer sites with genome-wide mapping of essential transcription factors in addition models.
Our finding that cocaine-induced 5hmC enrichment in NAc positively correlates with increased gene expression is consistent with previous reports53. 5hmC may promote such transcription activation through dissociation of 5mC binding protein repressors or recruitment of novel effector proteins16. It has been proposed that a decrease in TET1 leads to less 5mC conversion to 5hmC, and thus to promoter hypermethylation and transcription repression22–24. Our demonstration that TET1 downregulation leads to significantly overlap between increased 5hmC and gene induction after cocaine appears to counter to this view. There are several possible explanations. 1) The previous studies focused on gene promoters, whereas the significant alterations in 5hmC that we identified occur at gene bodies, and it is known that DNA modifications exert different effects at these distinct regions59. 2) None of the earlier studies compared 5hmC landscapes with genome-wide transcriptome profiling, as we do here, and instead utilized candidate gene approaches. 3) Although 5mC oxidation patterns were not examined in some studies22,23, TET1 knockout mice did display elevated 5hmC at selected genes24, suggesting that TET1 deficiency can cause increased 5hmC at least at certain loci. 4) If TET1 downregulation causes universal DNA hypermethylation, it is hard to explain why there is a global decrease in 5mC when TET1 is repressed in mouse hippocampus in the context of seizures23. In fact, TET1 has been implicated in both transactivation and repression49,60. Within TET1 depleted ES cells, more TET1 targets were upregulated than downregulated. It is hypothesized that this TET1 effect might reflect its normal coupling with repressor complexes49,60. Moreover, how 5hmC conversion into 5fC/5caC is affected by TET1 deficiency is unknown. An interruption of this process after TET1 loss might contribute to 5hmC accumulation. Importantly, we show that TET1 knockdown in NAc caused similar induction of 5hmC (Fig. 5) as observed after cocaine exposure, which supports a causal role of TET1 downregulation in 5hmC induction at the genomic loci we investigated. However, the lack of a global DNA methylation change (Fig. 2a), the observation that TET1 knockdown did not mimic all cocaine-triggered 5hmC increases in NAc, and possible non-enzymatic activities of TET123 suggest several directions for future research. In addition, it would be interesting to explore a possible role for TET2 and TET3 in cocaine action, even though they are not regulated in NAc by cocaine, given their relative abundance in this brain region (Supplementary Fig. 3). Nevertheless, our data show that TET1 has at least some unique targets in NAc that are not compensated by TET2 or TET3 and that demonstrate 5hmC induction in response to cocaine.
It has been challenging to correlate DNA epigenetic modifications and gene expression regulation genome-wide in brain under several experimental paradigms. This is possibly because: 1) available methodologies are not sufficiently sensitive to capture minor changes of both simultaneously; 2) most studies have focused on a subset of genomic regions (e.g., promoters) and might have missed important regulation elsewhere; 3) sodium bisulfite sequencing technology used to date for DNA methylation analysis has not differentiated 5hmC and 5mC, which can exert opposite effects on transcription; and 4) a change in DNA methylation can reflect not only an alteration in steady-state gene expression but also a gene’s poising for future transcription. Our study addresses several of these questions by showing, genome-wide in a discrete region of adult brain, that dynamic regulation of 5hmC correlates with several features of gene expression, including alternative splicing of primary transcripts, alterations in steady-state levels of expression as well as future inducibility, and that some of the 5hmC changes are long-lasting.
In summary, this study establishes 5hmC as a novel epigenetic mark in cocaine action. By mapping 5hmC in NAc in response to repeated cocaine exposure, we identified numerous target genes that now warrant investigation for their role in cocaine action. That some of the genes identified have previously established roles in cocaine addiction46–48 supports the importance of examining the novel targets. A prominent feature of drug addiction is that, once formed, addicts can have life-long behavioral abnormalities, which implicates stable brain changes in this process26. Our findings thus present 5hmC as one plausible underpinning mechanism. The next important steps are to carry out more sophisticated behavioral experiments of drug addiction, such as self-administration, to probe a role of 5hmC at particular genomic loci in NAc in the development and maintenance of addiction. Such work has the potential of generating new means of interrupting this process for therapeutic goals.
METHODS
Methods and any associated references are available in the online version of the paper.
Supplementary Material
Supplementary Figure 1. Representative immunohistochemistry images.
Supplementary Figure 2. Validation of viral-mediated Tet1 shRNA knockdown and overexpression.
Supplementary Figure 3. Relative mRNA expression of Tets in adult mouse nucleus accumbens.
Supplementary Figure 4. Distribution of 5hmC-enriched regions in NAc of saline- and cocaine-treated mice.
Supplementary Figure 5. Selective enrichment of H3K4me1, H3K27ac, and 5hmC at enhancer regions.
Supplementary Figure 6. qChIP validation of putative enhancers.
Supplementary Figure 7. Gene ontology of genes that show cocaine-induced changes by both 5hmC-seq and RNA-seq.
Supplementary Figure 8. oxBS-seq methodology validation.
Supplementary Table 1. Sequencing data summary: quality control analysis.
Supplementary Table 2. Gene regions that display cocaine-induced changes in 5hmC.
Supplementary Table 3. Genes whose putative nearby enhancers show cocaine-induced switches to chromatin states 4 or 5.
Supplementary Table 4. Gene ontology of genes whose putative nearby enhancers show cocaine-induced switches to chromatin states 4 or 5.
Supplementary Table 5. Motif analysis of putative enhancer genes.
Supplementary Table 6. Genes that show overlap in cocaine regulation of 5hmC and alternative splicing.
Supplementary Table 7. List of primers used.
Acknowledgments
We thank A. Chess for critical comments, and O. Jabado and M. Mahajan from the Mount Sinai Genomics Core for technical support. This work was supported by grants from the National Institute on Drug Abuse (E.J.N.) and National Institutes of Health (P.J., P.C., G.F., H.S.).
Footnotes
DATA ACCESS
All new ChIP-seq and RNA-seq data have been deposited into the Gene Expression Omnibus with accession number GSE63749. Datasets of 24 hr RNA-seq and H3K4me1 and H3K4me3 ChIP-seq were previously deposited with accession number GSE42811.
Note: Supplementary information is available in the online version of the paper.
AUTHOR CONTRICUTIONS
The studies were conceived and designed by E.J.N., P.J., and J.F. J.F. performed RNA-seq and ChIP-seq. K.E.S., Y.L., and J.F. performed 5hmC capture and sequencing. L.S., N.S., and J.F. performed bioinformatic analyses. J.F. and J.H. performed oxBS-seq. J.F., V.V., B.M.L., V.S., and I.M. performed qPCR analyses. V.V., D.E., T.C., and J.F. performed immunohistochemistry. J.F., V.V., D.F., J.K., and E.R. performed stereotaxic surgeries and behavioral assays. T.L., K.F.F., and G.F. contributed LC-ESI-MS/MS data. C.Z. and H.S. provided AAV-Tet1shRNA and AAV-TET1 viruses. M.E.C. performed Western blotting. G.T. contributed human samples. D.F., B.L., B.M.L., V.S., and P.K. also helped to prepare the samples and collect the data. The paper was written by J.F. and E.J.N. and was edited by other authors.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 2.Weaver IC, et al. Epigenetic programming by maternal behavior. Nature neuroscience. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 3.Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 4.Ma DK, et al. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng J, et al. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nature neuroscience. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo JU, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nature neuroscience. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 8.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He YF, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ito S, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu H, Zhang Y. Mechanisms and functions of Tet protein-mediated 5-methylcytosine oxidation. Genes & development. 2011;25:2436–2452. doi: 10.1101/gad.179184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Branco MR, Ficz G, Reik W. Uncovering the role of 5-hydroxymethylcytosine in the epigenome. Nature reviews. 2012;13:7–13. doi: 10.1038/nrg3080. [DOI] [PubMed] [Google Scholar]
- 16.Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14:341–356. doi: 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song CX, et al. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Globisch D, et al. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One. 2010;5:e15367. doi: 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic acids research. 2010;38:e181. doi: 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szulwach KE, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nature neuroscience. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudenko A, et al. Tet1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaas GA, et al. TET1 Controls CNS 5-Methylcytosine Hydroxylation, Active DNA Demethylation, Gene Transcription, and Memory Formation. Neuron. 2013;79:1086–1093. doi: 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang RR, et al. Tet1 regulates adult hippocampal neurogenesis and cognition. Cell stem cell. 2013;13:237–245. doi: 10.1016/j.stem.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li X, et al. Neocortical Tet3-mediated accumulation of 5-hydroxymethylcytosine promotes rapid behavioral adaptation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7120–7125. doi: 10.1073/pnas.1318906111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nature reviews. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- 27.Feng J, Nestler EJ. Epigenetic mechanisms of drug addiction. Current opinion in neurobiology. 2013;23(4):521–528. doi: 10.1016/j.conb.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogge GA, Wood MA. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology. 2013;38:94–110. doi: 10.1038/npp.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 30.Renthal W, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nature neuroscience. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le T, Kim KP, Fan G, Faull KF. A sensitive mass spectrometry method for simultaneous quantification of DNA methylation and hydroxymethylation levels in biological samples. Anal Biochem. 2011;412:203–209. doi: 10.1016/j.ab.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajagopal N, et al. RFECS: a random-forest based algorithm for enhancer identification from chromatin state. PLoS computational biology. 2013;9:e1002968. doi: 10.1371/journal.pcbi.1002968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malik AN, et al. Genome-wide identification and characterization of functional neuronal activity-dependent enhancers. Nature neuroscience. 2014;17:1330–1339. doi: 10.1038/nn.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst J, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng J, et al. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orom UA, Shiekhattar R. Long Noncoding RNAs Usher In a New Era in the Biology of Enhancers. Cell. 2013;154:1190–1193. doi: 10.1016/j.cell.2013.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasai N, Nakao M, Defossez PA. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic acids research. 2010;38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huh GS, et al. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vogel-Ciernia A, et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nature neuroscience. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Booth MJ, et al. Quantitative sequencing of 5-methylcytosine and 5-hydroxymethylcytosine at single-base resolution. Science. 2012;336:934–937. doi: 10.1126/science.1220671. [DOI] [PubMed] [Google Scholar]
- 46.Zachariou V, et al. Distinct roles of adenylyl cyclases 1 and 8 in opiate dependence: behavioral, electrophysiological, and molecular studies. Biological psychiatry. 2008;63:1013–1021. doi: 10.1016/j.biopsych.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Graham DL, et al. Tropomyosin-related kinase B in the mesolimbic dopamine system: region-specific effects on cocaine reward. Biological psychiatry. 2009;65:696–701. doi: 10.1016/j.biopsych.2008.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobo MK, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–390. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–348. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khare T, et al. 5-hmC in the brain is abundant in synaptic genes and shows differences at the exon-intron boundary. Nature structural & molecular biology. 2012;19:1037–1043. doi: 10.1038/nsmb.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nature neuroscience. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deng JV, et al. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nature neuroscience. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–1430. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Young JI, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell AC, et al. The genome in three dimensions: a new frontier in human brain research. Biological psychiatry. 2014;75:961–969. doi: 10.1016/j.biopsych.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annual review of genetics. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 58.Cruz FC, et al. New technologies for examining the role of neuronal ensembles in drug addiction and fear. Nature reviews. 2013;14:743–754. doi: 10.1038/nrn3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hellman A, Chess A. Gene body-specific methylation on the active X chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 60.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Representative immunohistochemistry images.
Supplementary Figure 2. Validation of viral-mediated Tet1 shRNA knockdown and overexpression.
Supplementary Figure 3. Relative mRNA expression of Tets in adult mouse nucleus accumbens.
Supplementary Figure 4. Distribution of 5hmC-enriched regions in NAc of saline- and cocaine-treated mice.
Supplementary Figure 5. Selective enrichment of H3K4me1, H3K27ac, and 5hmC at enhancer regions.
Supplementary Figure 6. qChIP validation of putative enhancers.
Supplementary Figure 7. Gene ontology of genes that show cocaine-induced changes by both 5hmC-seq and RNA-seq.
Supplementary Figure 8. oxBS-seq methodology validation.
Supplementary Table 1. Sequencing data summary: quality control analysis.
Supplementary Table 2. Gene regions that display cocaine-induced changes in 5hmC.
Supplementary Table 3. Genes whose putative nearby enhancers show cocaine-induced switches to chromatin states 4 or 5.
Supplementary Table 4. Gene ontology of genes whose putative nearby enhancers show cocaine-induced switches to chromatin states 4 or 5.
Supplementary Table 5. Motif analysis of putative enhancer genes.
Supplementary Table 6. Genes that show overlap in cocaine regulation of 5hmC and alternative splicing.
Supplementary Table 7. List of primers used.