Abstract
Time-of-flight secondary ion mass spectrometry (ToF-SIMS) imaging has been used for the direct analysis of single intact Xenopus laevis embryo surfaces, locating multiple lipids during fertilization and the early embryo development stages with subcellular lateral resolution (∼4 μm). The method avoids the complicated sample preparation for lipid analysis of the embryos, which requires selective chemical extraction of a pool of samples and chromatographic separation, while preserving the spatial distribution of biological species. The results show ToF-SIMS is capable of profiling multiple components (e.g., glycerophosphocholine, SM, cholesterol, vitamin E, diacylglycerol, and triacylglycerol) in a single X. laevis embryo. We observe lipid remodeling during fertilization and early embryo development via time course sampling. The study also reveals the lipid distribution on the gamete fusion site. The methodology used in the study opens the possibility of studying developmental biology using high resolution imaging MS and of understanding the functional role of the biological molecules.
Keywords: oocyte, embryo, time-of-flight secondary ion mass spectrometry, mass spectrometry imaging
The location and identification of lipids is essential to understand their role in biological processes. Their distribution in a cell membrane, nucleus, or other organelles is responsible for their function in inter- and intra-cellular communication and signaling (1, 2). To date, lipid profiling of Xenopus laevis embryos has been accomplished using a selective organic extraction methodology followed by GC-MS/LC-MS. Comprehensive work on the membrane composition of the X. laevis oocyte has been reported by Hill et al. (3). Lipid species in the mass range m/z 400–950 were detected. The plasma membrane is rich in glycerophosphocholine (GPCho), SM, and glycerophosphoinositol (GPIns), as well as cholesterol, comprising approximately 20% of the total lipid extraction. Huang, Liang, and Kam (4) examined the fatty acid composition of egg yolk in three anuran species. Koek et al. (5) analyzed approximately 10% of the cytoplasm in a single X. laevis oocyte using GC/MS. The detected compounds include organic acids, fatty acids (saturated, unsaturated, and hydroxy fatty acids ranging from C8 to C28), amino acids, alcohols, and sugars. These techniques, based on solvent extraction, do not provide any information on spatial localization beyond the fraction which was extracted; however, they offer qualitative and quantitative information about the composition of the cytoplasm with respect to key lipid compounds (6).
A current challenge in lipid biology studies is determining the spatial location of multiple lipids while retaining compositional information. Techniques such as scanning electron microscopy and confocal microscopy have been applied to image the frog egg and embryo to investigate the mechanisms of biological processes and embryo development. Monroy and Baccetti (7) first revealed the outer surface of the plasma membrane in X. laevis eggs by means of scanning electron microscopy. The architecture of the membrane exhibits dramatic changes before and after fertilization and as the fertilized egg moves toward the two-cell embryo. The large hemispherical protrusions on the unfertilized egg gradually become smooth and the microvilli withdraw along with completion of the fertilization and first cleavage. However, these previous studies were either focused on lipid component identification or on surface morphology after complicated sample preparation protocols. Immunofluorescence has revealed cortical and subcortical actin filaments and surface microvilli in late stage and highlighted certain organelles within X. laevis oocytes (8, 9), but this method has been questioned because of the possible interference of the fluorescent tags on in vivo function (10–12).
Mass spectrometric imaging is capable of detecting lipid species in intact biological cells and tissue, and providing lateral resolution down to the micron scale (13–15). Ferreira et al. (16) used MALDI-MS of single and intact embryos or oocytes from humans, cattle, sheep, and fish. The characteristic lipid (represented by GPCho, SM, and TAG) profiles were obtained with high mass resolution. However, due to the lateral resolution of the applied methodology (typically >50 μm), limited spatial distribution can be obtained. Time-of-flight secondary ion mass spectrometry (ToF-SIMS) is another major technique for mass spectrometric imaging of low-medium mass species, offering much higher spatial resolution. The technique involves scanning the sample surface with a finely focused energetic [kiloelectron volts (keV)] primary ion beam (for example C60+ or Au3+) and recording the ejected secondary ions as a function of their m/z and point of origin on the sample to form a “chemical map”. The introduction of polyatomic primary ion beams such as C60+ has facilitated the analysis of molecular species beyond the sample surface allowing complex chemical distributions to be studied in both two and three dimensions (14, 17). Previously, we demonstrated the first ToF-SIMS 3D chemical imaging of a biological cell using the X. laevis oocyte as a model system (18). A C60+ ion beam was used to demonstrate biomolecular depth profiling in the frog oocyte. The spatial distribution of cholesterol (m/z 369) and other lipids at m/z 540–570 and m/z 800–1,000 were revealed to a depth of ∼50 μm beneath the oocyte surface. This study demonstrated proof-of-principle, but practical implementation of high resolution 3D imaging using ToF-SIMS was at that time restricted by existing instrumentation.
We have recently reported on the development and application of a new generation of ToF-SIMS instrumentation, the J105 3D chemical imager (Ionoptika Ltd., Southampton, UK), the details of which are described elsewhere (15). The instrument utilizes a continuous (direct current) primary ion beam and buncher-ToF mass analyzer. This mode of operation offers high mass resolution without sacrificing lateral resolution, improved duty cycle and independence of mass resolution, and mass accuracy from sample topography. These properties are well-suited for studying biological events in topographically challenging samples, including the intact frog embryo. The aim of the current study was to gain new insights into the 2D and 3D arrangement of lipids and other important biomolecules during normal development of X. laevis embryos.
MATERIALS AND METHODS
Obtaining gametes
X. laevis were bred in the Faculty of Life Science at the University of Manchester. The gametes were obtained and fertilization was performed using standard procedures (19). The operations were conducted in conformity with the Public Health Service policy on Humane Care and Use of Laboratory Animals, incorporated in the Institute for Laboratory Animal Research Guide for Care and Use of Laboratory Animals. X. laevis females were preprimed subcutaneously 3–5 days before use with 50 units of pregnant mare serum gonadotropin. Twelve to eighteen hours before use, the animals were injected subcutaneously with 500 units of human chorionic gonadotrophin and placed at 16–18°C overnight in individual tanks. The next day, animals were transferred to tanks containing 5,000 ml 1× Marc’s modified Ringer’s solution (MMR) [1,000 mM NaCl, 20 mM KCl, 10 mM MgCl2, 20 mM CaCl2, 50 mM HEPES = 10× stock solution (pH 7.5), autoclaved before use and diluted accordingly]. Except for HEPES, which was purchased from Biomol, the other chemicals were obtained from Sigma, BioReagent). The laid eggs were then collected in MMR with a wide-bore pipette. Male frogs were euthanized by lethal injection of 400 μl of 40% ethyl 3-aminobenzoate methanesulfonate (MS-222, Sigma, 98%). The testis were then dissected and stored in L-15 cell culture media (Sigma) at 4°C for up to 1 week. Fertilization was then performed by passing a piece of dilacerated testis over the oocytes for 2 min.
Total lipid extraction
A lipid extraction from X. laevis zygotes was initially analyzed to explore the capability of the J105 ToF-SIMS instrument to distinguish the lipid species in a complicated chemical environment and to provide reference spectra for subsequent imaging studies. Methyl tertiary butyl ether (MTBE) extraction of lipids from X. laevis zygotes 10 min after fertilization was employed as follows (20). Ten zygotes 10 min postinsemination of the same batch were washed by HPLC water (Sigma-Aldrich) three times and aspirated to remove all the liquid. Subsequently, the cells were homogenized in a glass homogenizer and transferred into a glass tube with a Teflon-lined cap, the remaining residue was washed with 1.5 ml methanol (Sigma-Aldrich, CHROMASOLV® Plus, for HPLC, ≥99.9%) and mixed with the homogenized zygotes into the glass tube. MTBE(5 ml) (Sigma-Aldrich, CHROMASOLV® Plus, for HPLC, ≥99.9%) was then added into the mixture followed by incubation for 1 h at room temperature on a shaker. Phase separation was induced by adding 1.25 ml MS-grade water. After 10 min of incubation at room temperature, the sample was centrifuged at 1,000 g for 10 min. The upper phase was collected, and the lower phase was re-extracted with 2 ml of the solvent mixture (VMTBE/methanol = 10:3). The total organic phases were divided into 10 aliquots and dried in a vacuum centrifuge for approximately 40 min. To speed up sample drying, 20 μl of MS-grade methanol was added to the organic phase individually after 25 min of centrifugation. The lipid extract was dissolved into 500 μl mixed solvent (Vchloroform:methanol:water = 60:30:4.5) and stored at −20°C before analysis. Prior to analysis, 20 μl solution of lipid extract acquired via the above procedure was pipetted onto a silicon wafer and air-dried. Once dried, the samples were transferred immediately into the ToF-SIMS instrument.
Sampling the X. laevis embryos
After fertilization, the eggs were divided into two groups. One group was dejellied 7 min after the start of fertilization by immersing the eggs in 2% l-cysteine [Sigma, BioUltra, ≥98.5% room temperature (RT)] in 0.1× MMR (pH ∼7.5–8.0, with 10 M NaOH) and gently agitated by hand for 2 min. The l-cysteine solution was then decanted and the eggs washed with 0.1× MMR three times within 1 min. The eggs were left in fresh 0.1× MMR. From this time point, the fertilized embryos were collected using a pipette every 10 min until 40 min postfertilization, and immediately fixed with MEMFA [4% (v/v) formaldehyde solution: 2 mM EGTA, 1 mM MgSO4, 20 mM MOPS, 1.23 × 103 mM CH2O; all chemicals were from Sigma, EGTA for molecular biology; MgSO4, BioReagent; formaldehyde, for molecular biology 36.5–38% in H2O] for 1.5 h on a shaker and then washed with 1× PBS (Sigma, BioReagent, without calcium chloride and magnesium chloride, 10× concentration, diluted accordingly) four times and stored in 1× PBS in the refrigerator (21). Thus, the zygotes, 10, 20, 30, and 40 min after the fertilization, were prepared. The other group was dejellied using the same procedure as the first group at 40 min postinsemination, and left in 0.1× MMR at room temperature for further cleavage. The healthy embryos at each cleavage stage were selected, and followed by fixation using MEMFA and washing using 1× PBS, as with the first group, to prepare the embryos at 2 cell stage, 4 cell stage, 8 cell stage, 16 cell stage, and so on, at different cleavage stages. Because the eggs fertilized in vitro at the same time develop almost synchronically (22), the embryos of different cell stages could be collected at certain time intervals, using optical microscopy to confirm the developmental stage.
Prior to ToF-SIMS analysis, the fixed samples were quickly washed with HPLC water three times within 60 s to remove buffer. This was possible without disrupting the integrity of the system due to the resistance of the cell membrane to the osmotic pressure (23, 24). The cells were then loaded onto the SIMS sample holder with the animal hemisphere upwards. After aspirating the liquid around the cells, the samples were plunge-frozen into liquid nitrogen (LN2), followed by freeze-drying (Heto Drywinner, Thermo Electron Corporation) for 6 h. Under the protection of argon gas, the dried samples were then inserted into the ToF-SIMS instrument.
ToF-SIMS analysis
Mass spectra and images were acquired in positive ion mode with a J105 3D chemical imager (Ionoptika Ltd.) using a 40 keV C60 primary ion source operating in direct current mode. The zygote 10 min postfertilization and embryos at different development stages were analyzed. In this work, the C60+ beam was operated at a modest 4 μm focus to match the pixel size of data acquisition, allowing the entire sample to be imaged in ∼45 min. 3D analysis was performed by acquiring sequential image frames with increasing primary ion fluence, thereby revealing the lateral chemical distribution as a function of depth. The mass spectral images were produced using Analyze software (Ionoptika Ltd.). Further principal components analysis (PCA) was performed using MATLAB R2009a (MathWorks Inc., Natick, MA) for better contrast or 3D visualization (analysis routines were developed in-house). The assignment of SIMS ions in the following discussion to lipid classes and specific lipids is consistent with published literature but remains putative at this stage. Further analysis by MS/MS and use of authentic standards would increase confidence in lipid identification.
RESULTS AND DISCUSSION
ToF-SIMS analysis of lipid extracts from zygotes
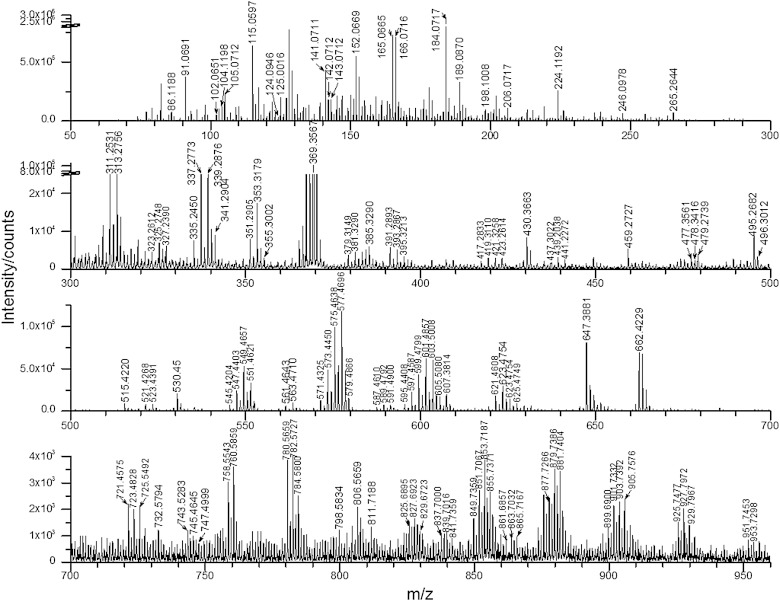
The ToF-SIMS analysis of extracted lipids from the zygotes was performed to provide reference spectra for the single embryo analysis. The positive ion mass spectrum is shown in Fig. 1. The assignment of major peaks based on published literature using MS is listed in Table 1 (25). The mass accuracy of all listed peaks is better than 10 ppm. All the m/z values are rounded to one decimal place in the following description.
Fig. 1.
Positive ion mode ToF-SIMS spectrum of a lipid extraction from X. laevis zygote 10 min after fertilization. Spectral dose density was 1 × 1013 ions/cm2 using a 40 keV C60+ beam. Data acquired with mass resolution m/Δm ∼5,000.
TABLE 1.
Characteristic molecular ions and fragments of common lipids in ToF-SIMS spectra
| +SIMS | ||||
| m/z | Proposed Fragments | Species | Reference | |
| GPCho | 86.1188 | [C5H12N]+ | Fragment | (18, 26) |
| 104.1198 | [C5H14NO]+ | Fragment | (27) | |
| 125.0016 | [C2H6PO4]+ | Fragment | (28) | |
| 166.0716 | [C5H13NPO3]+ | Fragment | (18, 26) | |
| 184.0717 | [C5H15NPO4]+ | Fragment | (18, 26) | |
| 206.0717 | [C5H14NPO4Na]+ | Fragment | (18, 26) | |
| 224.1192 | [C8H19NPO4]+ | Fragment | (28) | |
| 246.0978 | [C8H18NPO4Na]+ | Fragment | (27) | |
| 459.2727 | [C23H40O7P]+ C20:3 monoglycerolphosphate | Fragment | (29) | |
| 478.3416 | [C9H18NPO6-C15H31]+ C16:0 monoacylglycerophosphocholines | Fragment | (29, 30) | |
| 496.3012 | [C9H20NPO7-C15H31]+ C16:0 monoacylglycerophosphocholines | Fragment | (29, 30) | |
| 521.4268 | [C33H61O4]+ C30:1 | Fragment | (29) | |
| 523.4391 | [C33H63O4]+ C30:0 | Fragment | (29) | |
| 721.4575 | [C39H71O7PK]+ C34:3 | [M+K-TMA]+ | (31) | |
| 723.4828 | [C39H73O7PK]+ 34:2 | [M+K-TMA]+ | (31) | |
| 725.5492 | [C39H75O7PK]+ C34:1 | [M+K-TMA]+ | (28, 31) | |
| 732.5794 | [C10H19NPO8-C30H60]+ C32:1 | [M+H]+ | (3) | |
| 758.5543 | [C10H19NPO8-C32H62]+ C34:2 | [M+H]+ | (3) | |
| 760.5859 | [C10H19NPO8-C32H64]+ C34:1 | [M+H]+ | (3) | |
| 780.5659 | [C42H80NPO8Na]+ C34:2 | [M+Na]+ | (28) | |
| 782.5727 | [C42H82NPO8Na]+ C34:1 | [M+Na]+ | (28) | |
| 784.5800 | [C42H84NPO8Na]+ C34:0 | [M+Na]+ | (28) | |
| 798.5834 | [C42H82NPO8K]+ C34:1 | [M+K]+ | (28) | |
| 806.5659 | [C10H19NPO8-C36H62]+ C38:6 | [M+H]+ | (3) | |
| GPEtn | 124.0946 | [C2H7NPO3]+ | Fragment | (18, 26) |
| 142.0712 | [C2H9NPO4]+ | Fragment | (18, 26) | |
| 743.5283 | [C7H12NPO8-C34H66]+ C36:2 | [M+H]+ | (3) | |
| 745.4645 | [C7H12NPO8-C34H68]+ C36:1 | [M+H]+ | (3) | |
| 811.7188 | [C7H12NPO8-C39H74]+ C41:3 | [M+H]+ | (3) | |
| GPGro | 198.1008 | [C5H11PO6]+ | Fragment | |
| 747.4999 | [C8H12PO10-C32H64]+ C34:1 | [M+H]+ | (3) | |
| GPSer | 185.0894 | [C3H8NPO6]+ | Fragment | (18, 26) |
| Sphingolipids | 86.1188 | [C5H12N]+ | Fragment | (18, 26) |
| 102.0651 | [C5H12NO]+ | Fragment | (27) | |
| 104.1198 | [C5H14NO]+ | Fragment | (27) | |
| 166.0716 | [C5H13NPO3]+ | Fragment | (18, 26) | |
| 184.0717 | [C5H15NPO4]+ | Fragment | (18, 26) | |
| 206.0717 | [C5H14NPO4Na]+ | Fragment | (18, 26) | |
| 265.2644 | [C18H35N]+ | Fragment | (29) | |
| 725.5492 | [C39H80N2PO6Na]+ C34:1 | [M+Na]+ | (28, 31) | |
| Sterol lipids, cholesterol | 369.3567 | [C27H45]+ | [M+H-H2O] + | (18) |
| 385.3290 | [C27H45O]+ | [M-H] + | (18) | |
| Prenol lipids, vitamin E | 430.3663 | [C29H50O2]+ | [M] + | (31) |
| MAG | 311.2531 | [C19H35O3]+ C16:1 | All the ions listed here could be fragments from phospholipids, DAG, and TAG, or could be [M+H-OH]+ from MAG. | (31) |
| 313.2756 | [C19H37O3]+ C16:0 | (31) | ||
| 323.2612 | [C20H35O3]+ C17:2 | (32) | ||
| 325.2748 | [C20H37O3]+ C17:1 | (32) | ||
| 327.2948 | [C20H39O3]+ C17:0 | (32) | ||
| 335.2450 | [C21H35O3]+ C18:3 | (32) | ||
| 337.2773 | [C21H37O3]+ C18:2 | (31) | ||
| 339.2876 | [C21H39O3]+ C18:1 | (31) | ||
| 341.2904 | [C21H41O3]+ C18:0 | (31) | ||
| 351.2905 | [C22H39O3]+ C19:2 | (32) | ||
| 353.3179 | [C22H41O3]+ C19:1 | (32) | ||
| 355.3002 | [C22H43O3]+ C19:0 | (32) | ||
| 391.2893 | [C25H43O3]+ C22:2 | (32) | ||
| 393.2867 | [C25H45O3]+ C22:1 | (32) | ||
| 395.3213 | [C25H47O3]+ C22:0 | (32) | ||
| DAG | 515.4220 | [C33H55O4]+ C30:0 | All the ions listed here could be fragments from phospholipids and TAG, or could be [M+H-OH]+ from DAG. | (31) |
| 519.4109 | [C33H59O4]+ C30:2 | (31) | ||
| 545.4204 | [C35H61O4]+ C32:3 | (31) | ||
| 547.4403 | [C35H63O4]+ C32:2 | (31) | ||
| 549.4657 | [C35H65O4]+ C32:1 | (31) | ||
| 551.4621 | [C35H67O4]+ C32:0 | (31) | ||
| 561.4643 | [C36H65O4]+ C33:2 | (31) | ||
| 563.4710 | [C36H67O4]+ C33:1 | (31) | ||
| 571.4325 | [C37H63O4]+ C34:4 | (31) | ||
| 573.4450 | [C37H65O4]+ C34:3 | (31) | ||
| 575.4638 | [C37H67O4]+ C34:2 | (31) | ||
| 577.4696 | [C37H69O4]+ C34:1 | (31) | ||
| 579.4866 | [C37H71O4]+ C34:0 | (31) | ||
| 587.4610 | [C38H67O4]+ C35:3 | (31) | ||
| 589.4792 | [C38H69O4]+ C35:2 | (31) | ||
| 591.4900 | [C38H71O4]+ C35:1 | (31) | ||
| 595.4408 | [C39H63O4]+ C36:6 | (31) | ||
| 597.4587 | [C39H65O4]+ C36:5 | (31) | ||
| 599.4799 | [C39H67O4]+ C36:4 | (31) | ||
| 601.4857 | [C39H69O4]+ C36:3 | (31) | ||
| 603.5008 | [C39H71O4]+ C36:2 | (31) | ||
| 605.5080 | [C39H73O4]+ C36:1 | (31) | ||
| 607.3814 | [C39H75O4]+ C36:0 | (31) | ||
| 621.4608 | [C41H65O4]+ C38:7 | (31) | ||
| 623.4754 | [C41H67O4]+ C38:6 | (31) | ||
| 625.4749 | [C41H69O4]+ C38:5 | (31) | ||
| TAG | 825.6895 | [C53H93O6]+ C50:5 | All the ions listed here are [M+H]+ or [M+Na]+. | (31) |
| 827.6923 | [C53H95O6]+ C50:4 | (31) | ||
| 829.6723 | [C53H97O6]+ C50:3 | (31) | ||
| 837.7000 | [C54H93O6]+ C51:6 | (31) | ||
| 839.7016 | [C54H95O6]+ C51:5 | (31) | ||
| 849.7359 | [C54H97O6]+ C51:4 | (31) | ||
| 851.7067 | [C55H95O6]+ C52:6 | (31) | ||
| 853.7187 | [C55H97O6]+ C52:5/[C53H98O6 Na]+ C50:2 | (31) | ||
| 855.7371 | [C55H99O6]+ C52:4/[C53H100O6Na]+ C50:1 | (31) | ||
| 861.6857 | [C55H105O6]+ C52:0 | (31) | ||
| 863.7032 | [C56H95O6]+ C53:7 | (31) | ||
| 865.7167 | [C56H97O6]+ C53:6 | (31) | ||
| 867.7572 | [C56H99O6]+ C53:5 | (31) | ||
| 877.7266 | [C55H98O6Na]+ C52:4/[C57H97O6]+ C54:2 | (31) | ||
| 879.7386 | [C55H100O6Na]+ C52:3/[C57H99O6]+ C54:3 | (31) | ||
| 881.7404 | [C55H102O6Na]+ C52:2/[C57H101O6]+ C54:4 | (31) | ||
| 899.6900 | [C59H95O6]+ C56:10 | (33)a | ||
| 901.7332 | [C59H97O6]+ C56:9 | (33)a | ||
| 903.7392 | [C59H99O6]+ C56:8 | (33)a | ||
| 905.7576 | [C59H101O6]+ C56:7 | (33)a | ||
| 925.7477 | [C59H99O6Na]+ C56:8 | (33)a | ||
| 927.7972 | [C59H101O6Na]+ C56:7 | (33)a | ||
| 929.7967 | [C59H103O6Na]+ C56:6 | (33)a | ||
| 951.7453 | [C61H100O6Na]+ C58:4 | (33)a | ||
| 953.7298 | [C61H102O6Na]+ C58:4 | (33)a | ||
Adapted from (25).
Represents the species assigned according to the LIPID MAPS structure database.
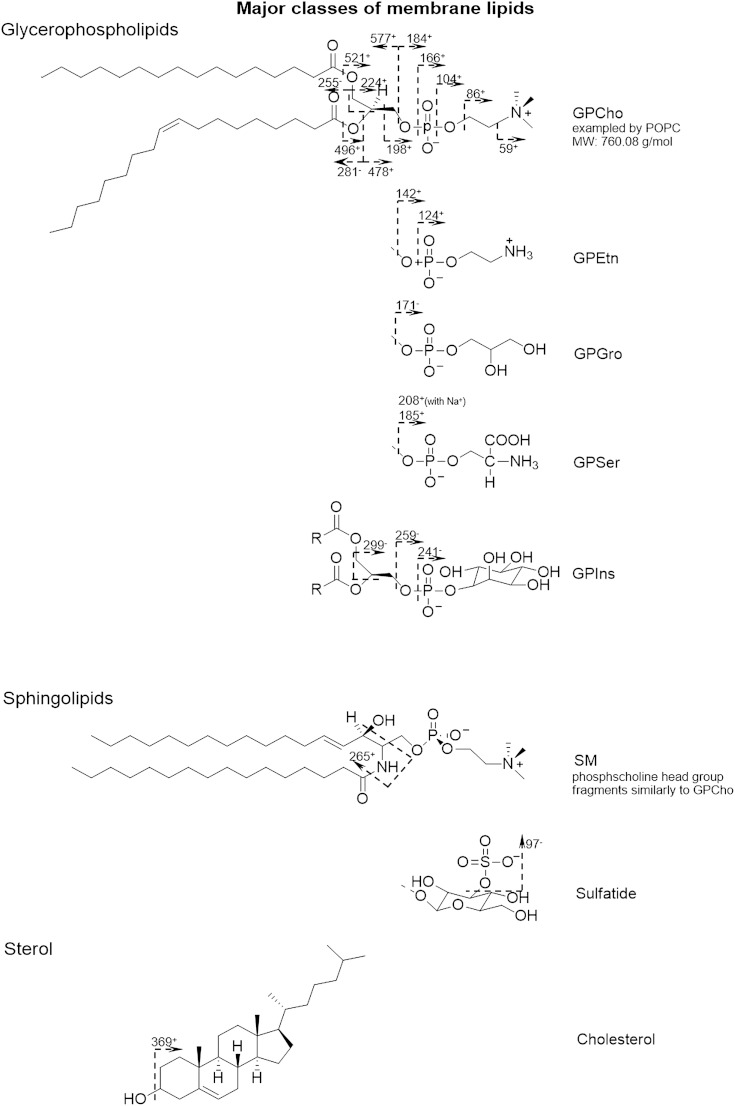
The major lipid classes in the lipid extraction were detected in a single run. The characteristic fragments, as well as the molecular ions, were used to assign the lipids. GPCho produced intense fragments at m/z 86.1, 104.1, 125.0, 166.1, 184.1, and 224.1 from the phosphocholine headgroup. The glycerol tail of GPCho was mainly fragmented to monoglycerol phosphate at m/z 459.3 (C20:3), various monoacylglycerol (MAG) and diacylglycerol (DAG) species, and potassiated DAG phosphate at m/z 721.5 (C34:3), 723.5 (C34:2), and 725.5 (C34:1). Sphingolipids, including SM, shared common fragments derived from the phosphocholine headgroup with GPCho. However, instead of two fatty acyl groups bonded to a glycerol backbone, SM had a fatty acyl group bonded to a basic nitrogen leading to the unique ion at m/z 265.2 [C18H35N]+, loss of phosphocholine head group, and neutral loss of the fatty acid as a ketene. There were some less abundant lipids, such as glycerophosphoethanolamine (GPEtn) and glycerophosphoserine (GPSer). GPEtn had characteristic peaks at m/z 124.1 and 142.1. Some common fragments were expected due to phospholipids having the same structure of glycerol backbone and phosphate group, e.g., m/z 125.0 [C2H6PO4]+ and m/z 143.1 [C2H8O5P]+. Schematic fragmentation paths are shown in Fig. 2 (25). Several phospholipid species were detected as molecular ions such as 32:1, 34:2, 34:1, 38:4, and 38:6 GPCho; 36:2, 36:1, and 41:3 GPEtn; and sodiated 34:1 SM. The results are consistent with the observations of Hill et al. (3). Cholesterol had the characteristic fragments at m/z 369.4 [M+H-H2O]+ and m/z 385.4 [M-H]+. The molecular ion of vitamin E was observed at m/z 430.4. The assignment of various DAG and TAG species in the region m/z >500 benefited from the available mass resolution (m/Δm 5,000). DAG could be the degradation product of TAG and phospholipids, as opposed to ion beam-induced changes to the structure, however, DAG type lipid fragments do not normally arise from protonated phosphocholine-containing species, as during fragmentation the charge stays on the head group. Fragmentation of the Na and K adducts could also lead to the DAG peaks (34). Considering the abundant DAG and TAG in the frog egg yolk (22), these two lipids are listed individually in Table 1.
Fig. 2.
Fragmentation of the major classes of lipids in biological cell membranes. Adapted from (25).
ToF-SIMS analysis of a lipid extraction of X. laevis zygotes demonstrates that the technique is capable of detecting the major classes of lipid species within complex mixtures without prior chromatographic separation. The characteristic peaks of GPEtn, GPCho, GPSer, glycerophosphoglycerol (GPGro), SM, cholesterol, vitamin E, and DAG are identified and can be used as reference signals to locate the lipid species on the real zygote surface. However, as noted above, without MS/MS verification, these assignments could be uncertain due to possible isobaric interference. Our caution has been demonstrated by listing possible alternative assignments in Table 1. For example, m/z 725.5 could be potassiated DAG phosphate or sodiated SM 34:1.
Monitoring the lipid distribution on the animal hemisphere of zygotes sampled shortly after fertilization
The fertilization of eggs is initiated with sperm entry in the “animal” hemisphere. For X. laevis, studies have indicated a significantly higher number of spermatozoa binding to the animal hemisphere (35, 36). The first visible sign of egg activation is the movement of pigment granules toward the animal pole (termed cortical contraction), which occurs approximately 4 min after fertilization (37). Our aim was to investigate the biological phenomenon on the surface of the egg shortly after the fertilization. The zygotes fixed at 10 min postfertilization were analyzed using a 4 μm-focused 40 keV C60+ primary ion beam to visualize any local differences of surface chemistry.
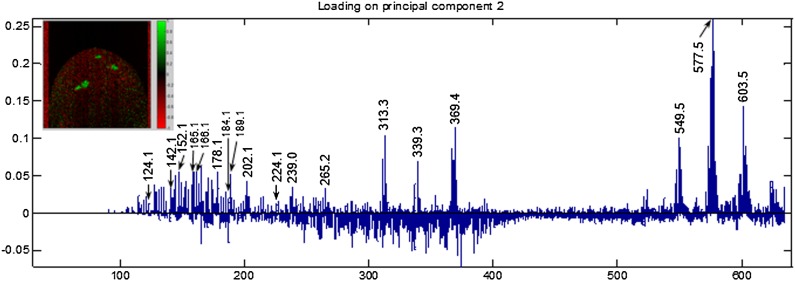
Image data acquired from the animal hemisphere of the zygote was binned to 0.1 Da to facilitate data processing. PCA was used to identify the significant differences in the image, and positive loaded peaks in PC2 (Fig. 3) guided the selection of m/z values to generate selected ion images as in Fig. 4 (25).
Fig. 3.
PCA loading plot of ToF-SIMS data of single X. laevis zygote 10 min postinsemination. Inset is Principal Component 2 scores image which shows that positive loadings (shown in green) contribute to the possible fusion site of the gametes on the animal hemisphere of the zygote. ToF-SIMS signals associated with the possible fusion site on the animal pole of the zygote are labeled in the positive loading in PC2. Adapted from (25).
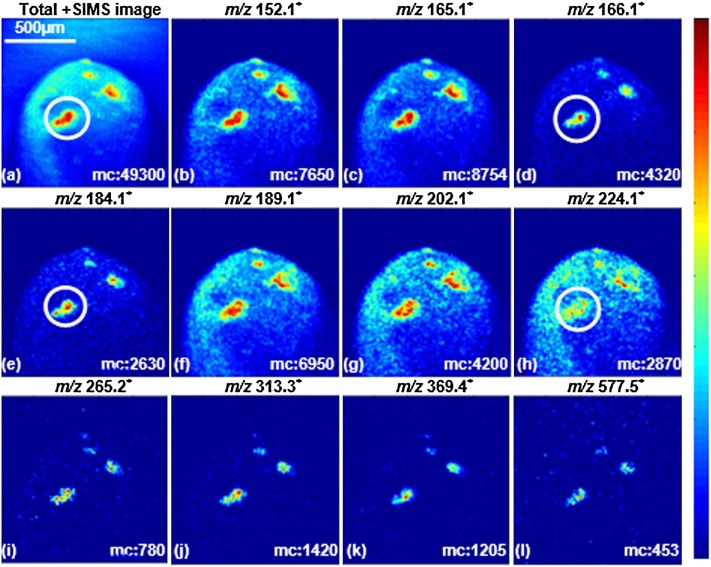
Fig. 4.
Total ion and selected ion images (positive mode ToF-SIMS) of the animal hemisphere of X. laevis zygote 10 min postinsemination. Spectral dose density is 5 × 1013 ions/cm2 using a 4 μm-focused 40 keV C60+ beam over 1,200 × 1,200 μm2 with 256 × 256 pixels. The surface region marked with a white circle is the proposed egg-sperm fusion site. The m/z 165.1 is a common glycerophospholipid fragment; m/z 166.1 and 184.1, the fragments from GPCho and SM; m/z 224.1, the fragment from GPCho; m/z 265.2, the characteristic peak of SM; m/z 313.3, MAG [C19H37O3]+ C16:0; m/z 369.4, cholesterol fragment; m/z 577.5, DAG [C37H69O4]+ C34:1; m/z 152.1, 189.1 and 202.1 are unassigned ions. The color scale corresponds to the relative ion intensity and the maximum count (mc) per pixel is displayed in each image frame. Adapted from (25).
The overall morphology of the zygote is clearly seen in the total ion image shown in Fig. 4a. Several features with distinctive chemical gradients are scattered across the zygote surface, the same phenomenon has not been seen in the unfertilized egg surface (data not shown). This indicates that these circular features could be the sperm entry sites. The abundant biomolecules residing within these features are mapped across the animal side of the zygote as in the diagnostic secondary ion images, revealing the varying distribution patterns for different lipids. The common fragment m/z 165.1 shared by all glycerophospholipids is spread across the feature areas; m/z 166.1 and 184.1 from GPCho and/or SM condense in the middle of the features particularly in one area marked with a white circle in Fig. 4d, e; GPGro at m/z 189.1 is spread more widely across the features; an unidentified ion at m/z 202.1 and GPCho ion at m/z 224.1 appear to circle the features in Fig. 4g, h, while another SM fragment at m/z 265.2 (Fig. 4i) is strongly localized within the distribution of m/z 224.1. The other components, such as MAG at m/z 313.3, cholesterol at m/z 369.4, and possibly DAG at m/z 577.5, are concentrated in the center of the features as shown in Fig. 4j–l.
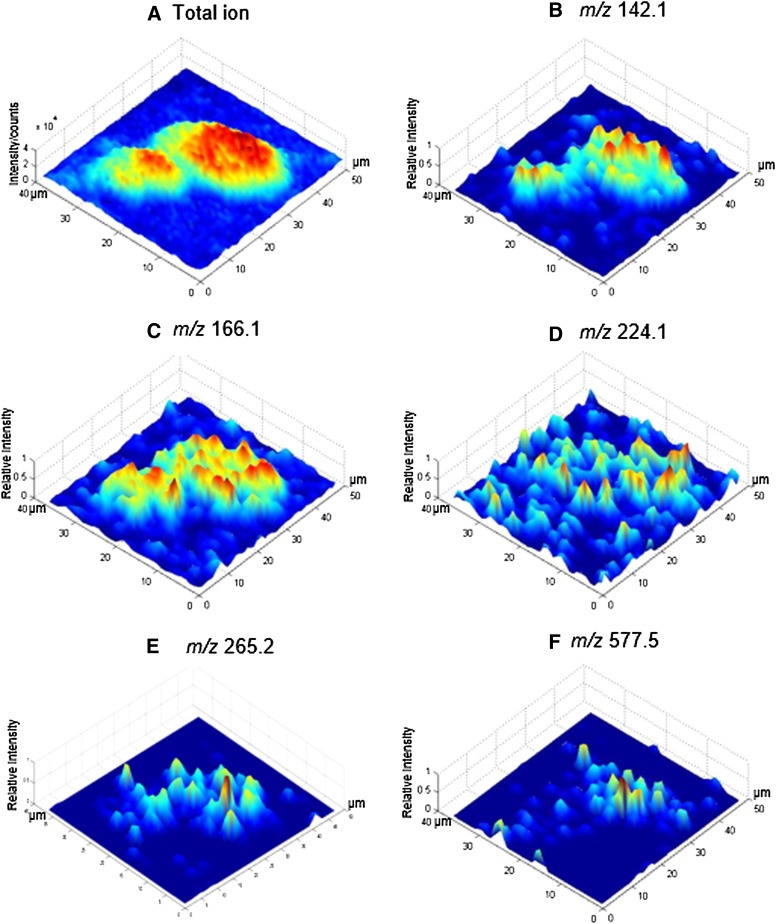
The 3D intensity maps of selected ions are further formed by normalizing the intensity of selected ions on each pixel to the maximum intensity of selected ions across the fusion site marked with the white circle in Fig. 4a. Peaks at m/z 142.1 (Fig. 5b) from GPEtn and m/z 577.5 (Fig. 5f) from DAG reach higher intensity in the center of the fusion site, whereas m/z 224.1 (Fig. 5d) from GPCho shows more uniform intensity extending to the outer region of the fusion site; m/z 265.2 (Fig. 5e) from SM is more tightly distributed at the fusion center, as with possible DAG. Combining the information from Fig. 4 and Fig. 5, the pattern of intensity changes across the region of interest, implying different distribution patterns of the selected biomolecules across the feature areas. GPCho is in the outer circle of the fusion site, GPEtn and SM are distributed more toward the fusion center, and DAG and cholesterol are most abundant in the center.
Fig. 5.
3D intensity map of total ion (A) and selected ions (B–F) across the proposed fusion site identified in Fig 4a. Selected peak assignment is as follows: m/z 142.1, fragment from GPEtn (B); m/z 166.1, fragment from GPCho and SM (C); m/z 224.1, fragment of GPCho (D); m/z 265.2, fragment of SM (E); and m/z 577.5, DAG [C37H69O4]+ C34:1 (F).
Membrane fusion is a profound biological process likely involving lipid rearrangement and possible synthesis. As DAG could be derived from TAG and phospholipids, a similar distribution pattern of DAG at m/z 577.5 was expected as phospholipid fragments at m/z 166.1 and 224.1, however these ions show different locations in Fig. 4d, h, i. It was also thought that the cleavage of phosphatidylinositol 4,5-bisphosphate into DAG during the fertilization was involved in the egg activation (38). Therefore, it is likely that the DAG ion detected in the fusion site reflects the biological process rather than beam-induced fragmentation. Considering that DAG can be rapidly phosphorylated to phosphatidic acid (GPA), which is a critical step in phospholipid biosynthesis (39, 40), it is speculated that in the center of the fusion site phospholipase-mediated hydrolysis of SM and phosphatidylinositol 4,5-bisphosphate occurs to generate possible DAG (41), which quickly builds new lipids such as GPEtn and GPCho to stop the dissolution of the membrane induced by the sperm binding. GPEtn is able to form highly curved structures that could be used to seal a space inside the fusion center for the release of the sperm nucleus. Most synthesized GPCho is transported away from the center to form the outer layer of the fusion site. This proposed model involving lipid synthesis and rearrangement can explain the distribution pattern of selected ions. For example, DAG and SM have high relative intensity exclusively in the fusion center; GPEtn only exists in the fusion center and GPCho is lower in the center and higher away from the center.
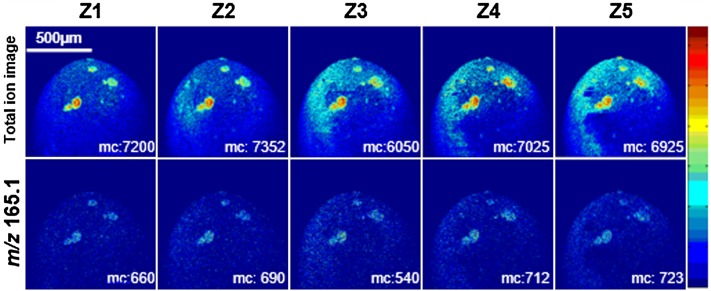
To investigate the fusion progression beneath the surface, depth profiling ToF-SIMS analysis was further performed on the zygote. The distribution of total ion intensity and an image of m/z 165.1 are shown for different depth layers (Z1 to Z5) in Fig. 6. This reveals that the fusion sites penetrate the cell membrane and expand with ring-like structures over the depth of the analysis (∼100 nm in total, based on the estimated etch rate using the 40 keV C60+ beam). At this point, it is difficult to identify which fusion site is truly the point of sperm entry. Amphibians contain two groups of mechanisms exhibiting very different blocks to polyspermy (42). One is a fast electrical block before second sperm-egg fusion by cortical granule exocytosis at the fertilization envelope. Another is a slow process that occurs in the egg cytoplasm after sperm entry, allowing only a single sperm nucleus with a single centrosome to ultimately participate in embryonic development, while the other sperm nuclei and centrosomes degenerate before cleavage (43). It is likely that the slow block occurs in the sampled zygote, as several fusion sites penetrating the X. laevis egg membrane are seen in Fig. 6.
Fig. 6.
Depth profile of X. laevis zygote 10 min postinsemination using dynamic ToF-SIMS imaging (positive mode ToF-SIMS) with a 4 μm-focused 40 keV C60+ beam, showing that the gamete fusion progressed through the surface region. The distribution of total ion signal (top row) and m/z 165.1 (bottom row) is shown as a function of depth (Z1 to Z5, ∼20 to 100 nm total depth). The depth profiling was performed on the animal hemisphere. Starting from the image at the surface (Z1) each image was acquired with an additional primary ion dose density of 1 × 1013 ions/cm2 over an area of 1,200 × 1,200 μm2 (256 × 256 pixels).
Monitoring the lipid distribution on X. laevis embryos through the early development stages
Mitotic cell division, a regulated dynamic biological event, is of such central importance for animal development and associated dynamic processes that the visualization of the mitosis directly in the developing embryo is of great interest. However, only the species featuring a transparent embryo that develops extracorporeally (e.g., teleosts) can be observed noninvasively by optical microscopy (44). With regard to amphibian embryos with almost complete optical opacity, such as the X. laevis embryo, observations are restricted. In this section, ToF-SIMS is employed to monitor the lipid distribution in the ∼10 nm surface region through early embryo development offering an alternative chemically specific method for embryonic study.
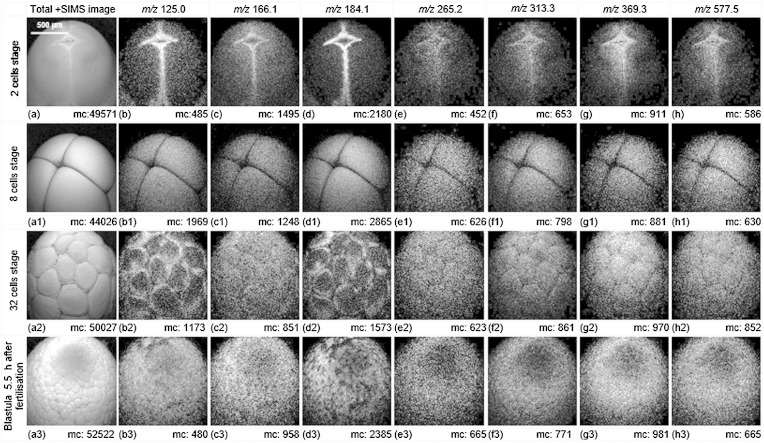
The biomolecular arrangements on the animal hemisphere of embryos at the 2-, 4-, and 32-cell stages and blastulas at 3.3 h and 5.5 h postfertilization are shown in the positive ion mode ToF-SIMS images in Fig. 7 [data previewed from (45)]. During the cleavage, the embryo will not grow, but divides into smaller cells called blastomeres packed within the sphere. This morphological change is clearly reflected in the total ion images in Fig. 7a, a1, a2, a3. The phosphocholine lipid headgroup fragments at m/z 125.0, m/z 166.1, and m/z 184.1 imaged at the 2-cell and 32-cell stages concentrate in the blastomere junctions, and are less intense on the cell surface at these developmental stages as shown in Fig. 7b, b2, c, c2, d, d2. This species is more uniformly distributed on the embryo at the 8-cell stage as in Fig. 7b1, c1, d1. Cholesterol (m/z 369.4) and the possible DAG ion at m/z 577.5 spread on the animal side homogenously as seen in Fig. 7g1, g2, g3, h1, h2, h3, except for the embryo at the 2-cell stage, where these signals too appear to concentrate at the blastomere junction. It is worth mentioning that the MAG ion at m/z 313.3, shown in Fig. 7f, f1, f2,f3, shares a similar distribution pattern to cholesterol and DAG. The cholesterol lends the membrane rigidity and mobility (46), which could be a reason why the cholesterol is abundant on the animal hemisphere to keep the membrane lipids stable and to adapt to fast biomolecular rearrangement during the cell cleavage. However, considering the vacuum environment of the ToF-SIMS instrument, cholesterol migration to the surface of the membrane (47) cannot be discounted. The distribution of the SM ion at m/z 265.2 shows that SM has a high affinity for cholesterol, as it colocates with cholesterol as shown in Fig. 7e–e3 and Fig. 7g–g3. These two biomolecules are able to pack tightly to form microdomains that are thought to function as signaling platforms in order to regulate the localization and interactions of proteins (48). The blastula, 5.5 h postinsemination, appears to show a complementary distribution of phosphocholine to SM, cholesterol, MAG, and DAG (Fig. 7d3–h3); although from the image of the whole embryo, the detailed distribution is uncertain due to the relatively small feature size. To explore the lipid distribution at this development stage would require reanalysis of a smaller region exploiting the high resolution capabilities of the C60+ microprobe for micron-level spatial localization.
Fig. 7.
Total ion and selected ion images (positive mode ToF-SIMS) of the animal hemisphere of X. laevis embryos in different cleavage stages using dynamic ToF-SIMS imaging. The 40 keV C60+ beam primary ion dose density was 2 × 1013 ions/cm2 over a 1,200 × 1,200 μm2 area with 256 × 256 pixels. The biomolecular arrangements on the animal hemisphere of embryos at 2 cells, 4 cells, 32 cells, and blastula 5.5 h postinsemination are shown. During the cleavage, the embryo does not grow but divides into smaller cells (blastomeres) packed on the surface of the sphere. The maximum counts (mc) can be read on the ion images. The m/z 165.1, common fragment from glycerophospholipids; GPCho and SM share the fragments at m/z 125.0, 166.1, 184.1, and m/z 265.2 is unique to SM; m/z 313.3, MAG [C19H37O3]+ C16:0; m/z 369.4, cholesterol fragment; m/z 577.5, DAG [C37H69O4]+ C34:1 or the fragment from phospholipid. [Data previewed from (45)].
CONCLUSION AND OUTLOOK
Single embryo analysis using ToF-SIMS shows the possibility of spatiotemporal chemical mapping of lipids on an embryo surface with high mass resolution and high spatial resolution. The sample preparation is simple without requiring lipid extraction and retains the pristine chemistry on the cell membrane.
The biological process of fertilization of the X. laevis egg is visualized by ToF-SIMS imaging to gain insight into the possible mechanism of egg-sperm fusion on the membrane. Lipid profiling in the possible egg-sperm fusion sites shows DAG is located almost exclusively in the center along with lower intensity of SM; GPEtn is mainly in the fusion center; and GPCho is more intense in surrounding regions of fusion center. Cholesterol is evenly distributed over a wider range than DAG, possibly to stabilize the newly formed membrane. The results are consistent with the following lipid synthesis pathway. The phospholipase-mediated hydrolysis of SM-generated DAG in the fusion center swiftly builds new lipids such as GPEtn and GPCho. GPEtn then forms the highly curved structure in order to seal a space inside the fusion center for the release of the sperm nucleus. Most synthesized GPCho is transported away from the center to form the outer layer of the fusion site.
For the first time, intact developing embryos have been imaged using ToF-SIMS with relatively simple sample preparation and without considering the embryonic pigment effect as encountered in the optical microscopic approach. Studies such as this may serve as a starting point to further explore the role of lipid distributions in the mechanisms of biological processes in single cells on the micron and submicron scale, which is currently beyond other techniques.
Acknowledgments
The authors are grateful to Dr. Arno Christian Gutleb and Prof. Nicholas Winograd for the creative discussions, and to Dr. Jimmy Moore for assistance with image processing.
Footnotes
Abbreviations:
- DAG
- diacylglycerol
- GPCho
- glycerophosphocholine
- GPEtn
- glycerophosphoethanolamine
- GPGro
- glycerophosphoglycerol
- GPIns
- glycerophosphoinositol
- GPSer
- glycerophosphoserine
- MAG
- monoacylglycerol
- MMR
- Marc’s modified Ringer’s solution
- MTBE
- methyl tertiary butyl ether
- PCA
- principal components analysis
- ToF-SIMS
- time-of-flight secondary ion mass spectrometry
This work was funded by a studentship to H.T. from the Analytical Trust Fund of the Royal Society of Chemistry. The development and use of the J105 ToF-SIMS instrument was funded by the UK Engineering and Physical Sciences Research Council (EPSRC) under grant EP/G045623.
REFERENCES
- 1.Andreyev A. Y., Fahy E., Guan Z., Kelly S., Li X., McDonald J. G., Milne S., Myers D., Park H., Ryan A., et al. 2010. Subcellular organelle lipidomics in TLR-4-activated macrophages. J. Lipid Res. 51: 2785–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quehenberger O., Armando A. M., Brown A. H., Milne S. B., Myers D. S., Merrill A. H., Bandyopadhyay S., Jones K. N., Kelly S., Shaner R. L., et al. 2010. Lipidomics reveals a remarkable diversity of lipids in human plasma. J. Lipid Res. 51: 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill W. G., Southern N. M., MacIver B., Potter E., Apodaca G., Smith C. P., Zeidel M. L. 2005. Isolation and characterization of the Xenopus oocyte plasma membrane: a new method for studying activity of water and solute transporters. Am. J. Physiol. Renal Physiol. 289: F217–F224. [DOI] [PubMed] [Google Scholar]
- 4.Huang C. H., Liang M. F., Kam Y. C. 2003. The fatty acid composition of oophagous tadpoles (Chirixalus eiffingeri) fed conspecific or chicken egg yolk. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 135: 329–336. [DOI] [PubMed] [Google Scholar]
- 5.Koek M. M., Bakels F., Engel W., van den Maagdenberg A., Ferrari M. D., Coulier L., Hankemeier T. 2010. Metabolic profiling of ultrasmall sample volumes with GC/MS: from microliter to nanoliter samples. Anal. Chem. 82: 156–162. [DOI] [PubMed] [Google Scholar]
- 6.Touboul D., Brunelle A., Laprévote O. 2011. Mass spectrometry imaging: towards a lipid microscope? Biochimie. 93: 113–119. [DOI] [PubMed] [Google Scholar]
- 7.Monroy A., Baccetti B. 1975. Morphological changes of the surface of the egg of Xenopus laevis in the course of development. I. Fertilization and early cleavage. J. Ultrastruct. Res. 50: 131–142. [DOI] [PubMed] [Google Scholar]
- 8.Colombo R., Benedusi P., Valle G. 1981. Actin in Xenopus development: indirect immunofluorescence study of actin localization. Differentiation. 20: 45–51. [DOI] [PubMed] [Google Scholar]
- 9.Chow R. L., Elinson R. P. 1993. Local alteration of cortical actin in Xenopus eggs by the fertilizing sperm. Mol. Reprod. Dev. 35: 69–75. [DOI] [PubMed] [Google Scholar]
- 10.Ramírez-Zacarías J. L., Castro-Muñozledo F., Kuri-Harcuch W. 1992. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry. 97: 493–497. [DOI] [PubMed] [Google Scholar]
- 11.Rogers W. J., Prichard J. W., Hu Y. L., Olson P. R., Benckart D. H., Kramer C. M., Vido D. A., Reichek N. 2000. Characterization of signal properties in atherosclerotic plaque components by intravascular MRI. Arterioscler. Thromb. Vasc. Biol. 20: 1824–1830. [DOI] [PubMed] [Google Scholar]
- 12.Wenk M. R. 2005. The emerging field of lipidomics. Nat. Rev. Drug Discov. 4: 594–610. [DOI] [PubMed] [Google Scholar]
- 13.Nygren H., Hagenhoff B., Malmberg P., Nilsson M., Richter K. 2007. Bioimaging TOF-SIMS: High resolution 3D Imaging of single cells. Microsc. Res. Tech. 70: 969–974. [DOI] [PubMed] [Google Scholar]
- 14.Fletcher J. S., Lockyer N. P., Vickerman J. C. 2011. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom. Rev. 30: 142–174. [DOI] [PubMed] [Google Scholar]
- 15.Fletcher J. S., Rabbani S., Henderson A., Blenkinsopp P., Thompson S. P., Lockyer N. P., Vickerman J. C. 2008. A new dynamic in mass spectral imaging of single biological cells. Anal. Chem. 80: 9058–9064. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira C. R., Saraiva S. A., Catharino R. R., Garcia J. S., Gozzo F. C., Sanvido G. B., Santos L. F., Lo Turco E. G., Pontes J. H., Basso A. C., et al. 2010. Single embryo and oocyte lipid fingerprinting by mass spectrometry. J. Lipid Res. 51: 1218–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher J. S., Lockyer N. P., Vickerman J. C. 2006. C60, Buckminsterfullerene: its impact on biological ToF-SIMS analysis. Surf. Interface Anal. 38: 1393–1400. [Google Scholar]
- 18.Fletcher J. S., Lockyer N. P., Vaidyanathan S., Vickerman J. C. 2007. TOF-SIMS 3D biomolecular imaging of Xenopus laevis oocytes using buckminsterfullerene (C60) primary ions. Anal. Chem. 79: 2199–2206. [DOI] [PubMed] [Google Scholar]
- 19.Strauss B., Adams R. J., Papalopulu N. 2006. A default mechanism of spindle orientation based on cell shape is sufficient to generate cell fate diversity in polarised Xenopus blastomeres. Development. 133: 3883–3893. [DOI] [PubMed] [Google Scholar]
- 20.Matyash V., Liebisch G., Kurzchalia T. V., Shevchenko A., Schwudke D. 2008. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 49: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dykstra M. J., Reuss L. E. 2003. Biological Electron Microscopy: Theory, Techniques, and Troubleshooting. 2nd edition. Springer Netherlands. [Google Scholar]
- 22.Gilbert S. F. 2010. Developmental Biology. 9th edition. Singuer Association. [Google Scholar]
- 23.Berntsson K-E., Haglund B., Lovtrup S. 1965. Osmoregulation in the amphibian egg the influence of calcium. J. Cell. Physiol. 65: 101–112. [DOI] [PubMed] [Google Scholar]
- 24.Kelly S. M., Butler J. P., Macklem P. T. 1995. Control of cell volume in oocytes and eggs from Xenopus laevis. Comp. Biochem. Physiol. A Physiol. 111: 681–691. [DOI] [PubMed] [Google Scholar]
- 25.Tian H. 2011. Visualisation and Profiling of Lipids in Single Biological Cells Using Time-of-Flight Secondary Ion Mass Spectrometry. Ph.D Thesis. University of Manchester, Manchester, UK. [Google Scholar]
- 26.Ostrowski S. G., Szakal C., Kozole J., Roddy T. P., Xu J., Ewing A. G., Winograd N. 2005. Secondary ion MS imaging of lipids in picoliter vials with a buckminsterfullerene ion source. Anal. Chem. 77: 6190–6196. [DOI] [PubMed] [Google Scholar]
- 27.Yang H-J., Ishizaki I., Sanada N., Zaima N., Sugiura Y., Yao I., Ikegami K., Setou M. 2010. Detection of characteristic distributions of phospholipid head groups and fatty acids on neurite surface by time-of-flight secondary ion mass spectrometry. Med. Mol. Morphol. 43: 158–164. [DOI] [PubMed] [Google Scholar]
- 28.Tahallah N., Brunelle A., De La Porte S., Laprévote O. 2008. Lipid mapping in human dystrophic muscle by cluster-time-of-flight secondary ion mass spectrometry imaging. J. Lipid Res. 49: 438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pulfer M., Murphy R. C. 2003. Electrospray mass spectrometry of phospholipids. Mass Spectrom. Rev. 22: 332–364. [DOI] [PubMed] [Google Scholar]
- 30.Löhmann C., Schachmann E., Dandekar T., Villmann C., Becker C. M. 2010. Developmental profiling by mass spectrometry of phosphocholine containing phospholipids in the rat nervous system reveals temporo-spatial gradients. J. Neurochem. 114: 1119–1134. [DOI] [PubMed] [Google Scholar]
- 31.Passarelli M. K., Winograd N. 2011. Lipid imaging with time-of-flight secondary ion mass spectrometry (ToF-SIMS). Biochim. Biophys. Acta. 1811: 976–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalo P. J., Ollilainen V., Rocha J. M., Malcata F. X. 2006. Identification of molecular species of simple lipids by normal phase liquid chromatography–positive electrospray tandem mass spectrometry, and application of developed methods in comprehensive analysis of low erucic acid rapeseed oil lipids. Int. J. Mass Spectrom. 254: 106–121. [Google Scholar]
- 33.LIPID MAPS Lipid Structure Database (LMSD). Accessed April 10, 2014, at http://www.lipidmaps.org/data/structure/LMSDSearch.php?Mode=ProcessClassSearch&LMID=LMGL03.
- 34.Passarelli M. K., Winograd N. 2011. Characterizing in situ glycerophospholipids with SIMS and MALDI methodologies. Surf. Interface Anal. 43: 269–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stein K. K., Primakoff P., Myles D. 2004. Sperm-egg fusion: events at the plasma membrane. J. Cell Sci. 117: 6269–6274. [DOI] [PubMed] [Google Scholar]
- 36.Katagiri C., Yoshizaki N., Kotani M., Kubo H. 1999. Analyses of oviductal pars recta-induced fertilizability of coelomic eggs in Xenopus laevis. Dev. Biol. 210: 269–276. [DOI] [PubMed] [Google Scholar]
- 37.Stewart-Savage J., Grey R. D. 1982. The temporal and spatial relationships between cortical contraction, sperm trail formation, and pronuclear migration in fertilized Xenopus eggs. Dev. Genes Evol. 191: 241–245. [DOI] [PubMed] [Google Scholar]
- 38.Larabell C., Nuccitelli R. 1992. Inositol lipid hydrolysis contributes to the Ca2+ wave in the activating egg of Xenopus laevis. Dev. Biol. 153: 347–355. [DOI] [PubMed] [Google Scholar]
- 39.Luo B., Regier D. S., Prescott S. M., Topham M. K. 2004. Diacylglycerol kinases. Cell. Signal. 16: 983–989. [DOI] [PubMed] [Google Scholar]
- 40.Athenstaedt K., Daum G. 1999. Phosphatidic acid, a key intermediate in lipid metabolism. Eur. J. Biochem. 266: 1–16. [DOI] [PubMed] [Google Scholar]
- 41.Berridge M. J. 1993. Inositol trisphosphate and calcium signaling. Nature. 361: 315–325. [DOI] [PubMed] [Google Scholar]
- 42.Tarin J. J., Cano A. 2000. Fertilization in Protozoa and Metazoan Animals: Cellular and Molecular Aspects. Springer, Berlin. [Google Scholar]
- 43.Glahn D., Nuccitelli R. 2003. Voltage-clamp study of the activation currents and fast block to polyspermy in the egg of Xenopus laevis. Dev. Growth Differ. 45: 187–197. [DOI] [PubMed] [Google Scholar]
- 44.Papan C., Boulat B., Velan S. S., Fraser S. E., Jacobs R. E. 2006. Time-lapse tracing of mitotic cell divisions in the early Xenopus embryo using microscopic MRI. Dev. Dyn. 235: 3059–3062. [DOI] [PubMed] [Google Scholar]
- 45.Large Complex Samples. Accessed April 10, 2014 at http://www.ionoptika.com/j105-features/119-applications/j105-features/144-large-complex-samples.html.
- 46.van Meer G., Voelker D. R., Feigenson G. W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones E. A., Lockyer N. P., Vickerman J. C. 2008. Depth profiling brain tissue sections with a 40 keV C60+ primary ion beam. Anal. Chem. 80: 2125–2132. [DOI] [PubMed] [Google Scholar]
- 48.Milhas D., Clarke C. J., Hannun Y. A. 2010. Sphingomyelin metabolism at the plasma membrane: Implications for bioactive sphingolipids. FEBS Lett. 584: 1887–1894. [DOI] [PMC free article] [PubMed] [Google Scholar]