Abstract
Liver X receptors (Lxrs) are master regulators of cholesterol catabolism, driving the elimination of cholesterol from the periphery to the lumen of the intestine. Development of pharmacological agents to activate Lxrs has been hindered by synthetic Lxr agonists’ induction of hepatic lipogenesis and hypertriglyceridemia. Elucidating the function of Lxrs in regulating enterocyte lipid handling might identify novel aspects of lipid metabolism that are pharmacologically amenable. We took a genetic approach centered on the single Lxr gene nr1h3 in zebrafish to study the role of Lxr in enterocyte lipid metabolism. Loss of nr1h3 function causes anticipated gene regulatory changes and cholesterol intolerance, collectively reflecting high evolutionary conservation of zebrafish Lxra function. Intestinal nr1h3 activation delays transport of absorbed neutral lipids, with accumulation of neutral lipids in enterocyte cytoplasmic droplets. This delay in transport of ingested neutral lipids protects animals from hypercholesterolemia and hepatic steatosis induced by a high-fat diet. On a gene regulatory level, Lxra induces expression of acsl3a, which encodes acyl-CoA synthetase long-chain family member 3a, a lipid droplet-anchored protein that directs fatty acyl chains into lipids. Forced overexpression of acls3a in enterocytes delays, in part, the appearance of neutral lipids in the vasculature of zebrafish larvae. Activation of Lxr in the intestine cell-autonomously regulates the rate of delivery of absorbed lipids by inducting a temporary lipid intestinal droplet storage depot.
Keywords: acyl-CoA synthetase long-chain family member 3, diet and dietary lipids, fatty acid/transport, electron microscopy, intestine, nuclear receptors/liver X receptor, nutrition, transcription activator-like effector nuclease, zebrafish
Atherosclerotic cardiovascular disease is the leading cause of death worldwide (1). Elevated serum cholesterol is a major causal risk factor for development and progression of atherosclerosis (2). The most widely used treatment for atherosclerosis is statin drugs, which potently inhibit 3-hydroxy-3-methylglutaryl CoA reductase, the rate-limiting enzyme of cholesterol biosynthesis (3). Nevertheless, statins do not fully mitigate cardiovascular risk. Recent epidemiological investigations reveal that postprandial (nonfasting) plasma lipid levels are a major determinant of cardiovascular risk because they reflect the persistence of highly atherogenic remnant lipoprotein particles in the circulation (4). Statin drugs do not modulate the abundance or atherogenicity of remnant lipoprotein particles (5).
Elucidation of the regulatory events controlling the pace of entry of ingested lipids into the blood stream in chylomicrons might afford the opportunity to blunt postprandial hyperlipidemia. Human jejunal biopsies (6), prolonged, label-free lipid droplet imaging of live loops of mouse intestine (7), and fluorescent lipid probe-based visualization of intestinal lipid droplets in zebrafish larvae (8) have revealed that a substantial fraction of absorbed lipids remain in the enterocyte for many hours following a meal. These findings suggest that there is an intrinsic mechanism for delaying, in part, transit of absorbed lipids. The molecular mechanism accounting for this hours-long retention of absorbed lipids in the intestine is not known.
Liver X receptors (Lxrs) are nuclear receptor transcription factors that regulate energy metabolism through engaging specific metabolite substrates such as oxysterols and then altering the expression of diverse, but functionally integrated genes (9, 10). Lxrs are central inducers of cholesterol catabolism (9, 10). Known direct target genes of Lxr transactivation include factors involved in reverse cholesterol transport; lipoprotein modification; cholesterol uptake, absorption, and excretion; FA and triacylglycerol regulation; bile transport; glucose metabolism; immune and inflammatory signaling; and adipocyte homeostasis (9, 10). Two Lxr paralogs have been described in mammals. Lxra (Nr1h3) is mainly expressed in tissues involved in lipid metabolism regulation, whereas Lxrb (Nr1h2) is more widely expressed (9, 10). Lxra activation inhibits cholesterol absorption and promotion of reverse cholesterol transport. In contrast to its role in atheroprotective cholesterol elimination, Lxra upregulates hepatic lipogenic enzymes and increases liver and blood triacylglycerol levels (11–14), both of which are known risk for cardiovascular disease (15). This dichotomous action, driving elimination of cholesterol while triggering liver FA synthesis, has been a major impediment to developing Lxr-based therapeutics. No studies have directly assessed whether Lxra regulates intestinal FA absorption and trafficking.
Zebrafish has emerged as a major model system for studying metabolism (16). Among many other technical advantages, such as external fertilization, rapid organogenesis, and prolonged optical transparency of larvae, retention of the evolutionarily central cholesteryl ester transfer protein (Cetp) gene in zebrafish has allowed investigators to develop dyslipidemia models without difficulty (17–19). Many dyslipidemic animal models lack Cetp, frustrating drug development (20). Similarly, we have optimized a series of serum and tissue metabolite composition assays to study lipid metabolism (21–23). Vital to this study, zebrafish have a single Lxr ortholog (nr1h3) (24–26), which is similar to mammalian Lxra (25). This fortuitous finding allowed us to avoid the neofunctionalization of Lxrb in mammalian neuronal development and brain lipid homeostasis that clouds studies in higher organisms’ Lxr family members (27).
We found that enterocyte-limited Lxra activation causes retention of absorbed neutral lipids in these cells. This delay in delivering lipids to the systemic circulation confers protection from high-fat diet (HFD)-induced hypercholesterolemia and hepatic steatosis. On a molecular level, we found Lxra induces expression of cellular machinery that diverts the absorbed lipids to cytoplasmic lipid droplets and away from chylomicron assembly and secretion. Specifically, the known direct Lxra target gene acyl-CoA synthetase long-chain family member 3 (acsl3a), which encodes a lipid droplet-anchored long-chain acyl-CoA synthetase that is central to the biogenesis and growth of lipid droplets (28–30), is strikingly induced by Lxra-overexpressing enterocytes. Furthermore, overexpression of acsl3a in enterocytes is sufficient to delay the appearance of absorbed lipid in the vasculature. Our study reveals that Lxr sets the rate of delivery of ingested lipids to the systemic circulation by regulating the production of enterocyte lipid droplets.
MATERIALS AND METHODS
Animals
The Institutional Animal Care and Use Committee and the Radiation Safety Committee of the University of Utah approved all studies. Animals were euthanized by tricaine overdose or immersion in ice. The Centralized Zebrafish Animal Resource at the University of Utah maintains the WT WIK strain zebrafish used in this study.
Gene targeting
Targeted gene deletion of the nr1h3 gene was performed using transcription activator-like effector nucleases (TALENs) targeting the second exon of the gene. The TALEN sequences were designed by the University of Utah Mutation Generation and Detection Facility (31). The TALEN target site was identified with the TALEN Targeter program (https://boglab.plp.iastate.edu/node/add/talen). The TALEN monomers were designed for the second exon of nr1h3 (5′-GGCTTTCATTACAATGTGCTGAGCTGTGAGGGCTGTAAAGGTTTCTTCAGACG). Two plasmids containing the right and left TALEN monomers were cloned with the Golden Gate Assembly method into a modified CS2+ backbone vector (pCS2TAL3-lxrTALEN-DD and pCS2TAL3-lxrTALEN-RR). From those resulting plasmids, the 5′ capped mRNA was produced by in vitro transcription (Promega) of the NotI-linearized plasmids; 1 nL (250 pg of each plasmid per nl) of the transcribed product was injected in zebrafish embryos (G0). Mutations were detected in outcrossed G0 offspring (F1 generation) by means of high-resolution melting analysis (HRMA) and were confirmed with Sanger sequencing (32). Mutations were screened with HRMA analysis using the following primers: forward 5′-TCTGGCTTTCATTACAATGTGC and reverse 5′-GCTGCGTCTGAAGAAACCTT. The resultant WT PCR product for each genotype was a 60 bp amplicon. In z101 mutants, the amplicon was 49 bp. In z102 mutants, the amplicon was 82 bp.
Transgenesis
Tg(fabp2:EGFP-nr1h3)z103, Tg(actb2:EGFP-nr1h3)z104, and Tg(fabp2:EGFP-acsl3a)z105 transgenic lines were prepared in WIK animals using the Tol2 transposase (33). Our breeding strategy led to the standard incorporation of a single transgene copy: half of the progeny from an outcross of each founder animal showed transmission of enhanced green fluorescent protein (EGFP) fluorescence. The following primers were used to add the recombination sites to nr1h3 and acsl3a sequences and to recombine them into the pDONRP2R-P3 plasmid: lxra forward 5′-ggggacagctttcttgtacaaagtggCGGCAGAAGTGAAACAGGAGATTCTCA, lxra reverse 5′-ggggacaactttgtataataaagttgTCATTCGTGAACATCCCAAATTT, acsl3a forward 5′-ggggacagctttcttgtacaaagtggGGAGGCTGACGCAGGATTTGAGTC, and acsl3a reverse 5′-ggggacaactttgtataataaagttgTTACTTGGCTCCGTACATCCTCTC. The 4.5 kb fabp2 promoter was amplified from genomic DNA, as previously described (34). The recombination sites were added with the following primers to recombine the promoter into the pDONRP4-P1R plasmid: forward 5′-ggggacaactttgtatagaaaagttgAAACCTTTAGTTTGGGTTTTTCAGA and reverse 5′-ggggactgcttttttgtacaaacttgGATGATGACAGACTGTTGTGTGATC. The rest of the plasmids for the transgenic constructs were kindly donated by the Kwan lab at the University of Utah (http://tol2kit.genetics.utah.edu).
Immunoblot analysis
Homogenates of 30 larvae [5 days post-fertilization (dpf)] were prepared in 150 µl of RIPA Buffer (Sigma) containing a protease inhibitor cocktail (Roche Complete Mini) with a micropestle. Forty micrograms of protein were separated in a 15% sodium dodecyl sulfate-denaturing polyacrylamide gel and transferred to a polyvinylidene difluoride membrane. Membranes were incubated with antibodies against EGFP (rabbit, A-11122, Life Technologies, Carlsbad, CA), β-tubulin (Btub or Tubb rabbit HRP conjugated, Abcam ab6046), and Lxra (goat, Abcam ab24362). Secondary HRP-conjugated antibodies for rabbit (goat anti-rabbit IgG-HRP) and goat (donkey anti-goat IgG-HRP) were obtained from Santa Cruz Biotechnology.
Larvae high-fat feeding experiments and Oil Red O staining
A 5% w/v dried egg yolk (Sigma) suspension was prepared in embryo water (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4, pH 7.4) as previously described (8, 21). Ten 6 dpf larvae were distributed per well in 6-well plates (Nest Biotech Co. Ltd.), and the egg yolk suspension was added (5 ml per well). After 6 h, 3 wells per experimental group were rinsed with embryo water to remove uningested lipids. After the rinses, the larvae were fixed in 4% paraformaldehyde (PFA)-PBS for 1 h at room temperature. For the following time points (24, 48, and 72 h), larvae were kept in clean 6-well plates until the fixation in 4% PFA-PBS. After all larvae were collected and fixed, they were stained with Oil Red O (ORO) for 1 h as described previously (21). After staining, larvae were rinsed in PBS, and lipid staining was observed with an AF6000 microscope (Leica Microsystems). Representative pictures were acquired with an attached DFC319FX camera.
Defined diet preparation
Experimental diets were formulated based on previously described zebrafish diets (35). All the ingredients listed in supplementary Table I were homogenized in a mixer (KitchenAid Artisan Stand Mixers), and the resulting dough was air dried for 48 h. After drying, the diets were ground with a coffee grinder, sieved into convenient pellet size (400–600 μm), and stored at −20°C. Animals were fed these two diets for 3 weeks (juvenile experiments) or 2 months (adult experiments). Proximate composition analyses from the diets were performed by the Mississippi State University Chemical Laboratory. For all adult studies, 5 female and 5 male 3 months post-fertilization (mpf) zebrafish were distributed in 3 liter tanks. The animals were fed twice daily with automatic feeders (Auto Fish feeder Petco, San Diego, CA). After 2 months of eating these diets, animals were euthanized, and their intestines and livers were dissected for further analysis described in the following sections.
The high-cholesterol (4% w/w) diet was prepared as previously described (19). Commercial flakes (TetraMin Tropical Flakes, Blacksburg, VA) were soaked in a cholesterol-diethyl ether solution, and the flakes were left to dry overnight. For all adult studies, 5 female and 5 male 3 mpf zebrafish were distributed in 3 liter tanks. The animals were fed twice daily with automatic feeders for 21 days.
Blood and tissue lipid composition analysis
Blood was collected by cardiac puncture with glass capillaries attached to a microinjector (Microinjection Systems, Harvard System). Blood was diluted in 0.2 ml tubes with 20 µl of PBS-EDTA. After centrifugation at maximum speed for 5 min, plasma was collected. Tissues were homogenized in lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100) by sonication. Protein concentration was determined with the BCA protein assay reagent (Thermo Scientific). The triacylglycerol and total cholesterol levels in the blood or tissue homogenates were analyzed with colorimetric assay kits (Spinreact, Mexico), with normalization to protein content.
Oral gavage
Zebrafish (3 mpf) fed a regular diet (twice a day brine shrimp, housing conditions) were fasted overnight and then gavaged with 0.2 µCi of [3H]triolein dissolved to a final volume of 2 μl in a 5:1:1 (v/v) flaxseed oil-cod liver oil-sunflower oil mixture that emulated the lipid mixture from the experimental diets (36). After the gavage, animals were kept in separate tanks (400 ml) at 28°C. Twenty-four hours postgavage, the animals were euthanized, and the intestines were dissected, cleaned with PBS, and digested overnight at 37°C in 200 μl of 1 N NaOH. The protein content of a dilution (1/10 in distilled water) of digested homogenates was measured by BCA protein assay reagent (Thermo Scientific), and the remainder of the homogenate was mixed with 5 ml of scintillation liquid for counting 3H disintegrations in a Beckman L5-5000 instrument.
FA utilization
4,4-Difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid (BODIPY FL C12, Life Technologies) and 1-palmitoyl-2-(dipyrrometheneboron difluoride)undecanoyl-sn-glycero-3-phosphocholine (TopFluorPC, Avanti Polar Lipids) were diluted in the 5% dried egg yolk suspension to a final concentration of 20 ng/ml. Larvae (6 dpf) were fed the suspension for 6 h, anesthetized, rinsed, and sonicated in chloroform-methanol to perform a lipid extraction by Folch method (37). After extraction, lipids were resuspended in 200 µl of chloroform, and a 10 µl aliquot was run on a TLC plate (TLC silica gel 60, Merck Millipore). The standards for triacylglycerol, diacylglycerol, and phosphatidylcholine were synthesized as previously described (8). A two-solvent system was used to separate phospholipids from nonpolar lipids: the first solvent mixture was run until the middle of the plate (chloroform-methanol-acetic acid-water, 50:37.5:3.5:2, v/v), and the second solvent mixture was run to 2 cm from the end of the plate (hexane-diethyl ether-acetic acid, 70:30:1, v/v). Once the plate was completely dry, the fluorescent bands were detected with a blue fluorescence laser (Typhoon Trio variable mode imager, Amersham Bioscience). Fluorescent bands for each lipid class were normalized to the total fluorescence.
Electron microscopy
The anterior half of the intestine from 5 mpf animals was fixed in 2.5% PFA and 1% glutaraldehyde in 100 mM sodium cacodylate (pH 7.4), and counterstained with OsO4. Electron microscopy was performed using JEOL JEM-1400 Plus transmission electron microscope at the University of Utah’s Electron Microscopy Core facility.
Cell culture
Caco2 cells were purchased from the American Type Cell Collection, and were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium supplemented with 4.5 mg/dl glucose, 10% fetal bovine serum, 1% l-glutamine, 1% penicillin/streptomycin, and 1% lipid (38). Cells were seeded onto Transwell membranes (24 mm diameter inserts, 3 µm pore size, Corning Incorporated Costar, Corning, NY) in 6-well plates and grown to confluence for 21 days prior to treatment with vehicle (DMSO) or GW-3965. Cells were either fixed and stained with ORO or subjected to lysis and RNA extraction after 24 h of treatment.
Quantitative PCR analysis
RNA was extracted from the intestines of 5 mpf animals fed a control diet or HFD for 2 months with the Qiagen RNeasy MiniKit. cDNA was synthesized from 1 µg of intestinal RNA using the Vitro cDNA synthesis kit (Life Technologies). Primers (0.5 μM), 1/40 of the cDNA synthesis reaction, and SYBR Green PCR mix (Life Technologies) were combined in a total volume of 20 μl. The quantitative PCR primer sequences for target genes and the reference gene are shown in supplementary Table II. Reactions were performed in an Applied Biosystems 7900HT Detection System in duplicate, and the fluorescence data acquired during the extension phase were normalized to rplp0 by the delta-delta method (39). The primers for human ACSL3 were 5′-CCCCTGAAACTGGTCTGGTG and 5′-TCCGCCTGGTAATGTGTTTTAA (40).
ACSL activity
Individual intestines from 4 mpf animals fed a control diet were homogenized by sonication in 200 µl of a buffer containing 250 mM sucrose, 10 mM Tris (pH 7.4), 1 mM EDTA, 1 mM dithiothreitol, and protease inhibitor cocktail (Roche). Homogenates were centrifuged at 16,000 rpm at 4°C for 30 min. The clear supernatant was collected, and the protein concentration was measured with BCA protein assay reagent (Thermo Scientific). Acsl activity in 10 µg of protein was measured as previously described (41). The reaction was performed at 28°C for 5 min in the presence of 175 mM Tris-HCl, pH 7.4, 8 mM MgCl2, 5 mM dithiothreitol, 10 mM ATP, 250 µM CoA, and 3 µCi of [3H]oleic acid (specific activity 15 Ci/mmol) dissolved in 0.5 mM Triton X-100, and 10 µM EDTA. The reaction was stopped with the addition of 1 ml isopropanol-heptane-1 M H2SO4 (40:10:1, v/v). After two washes with heptane, the 3H disintegrations in the recovered [3H]oleolyl-CoA were counted in a Beckman L5-5000 TD liquid scintillation counter.
Ex vivo FA oxidation
FA oxidation was measured in tissue homogenates as previously described (42). Homogenates from two pools of zebrafish intestines per genotype (n = 6 intestines per pool) were incubated for 2 h at 28°C in reaction buffer (100 mM sucrose, 10 mM Tris-HCl, 5 mM KH2PO4, 0.2 mM EDTA, 0.3% FA-free BSA, 80 mM KCl, 1 mM MgCl2, 2 mM l-carnitine, 0.1 mM malate, 0.05 mM CoA, 200 µM oleate, pH 8.0) containing [14C]oleate (0.4 µCi per sample). The generated CO2 was trapped with filter paper soaked in hyamine hydroxide. The acid-soluble metabolites where separated from the unoxidized FAs by adding 1 M perchloric acid. The CO2 trapped and the acid-soluble metabolite fraction were mixed with 5 ml of scintillation liquid (Ultima Gold, PerkinElmer), and the 14C disintegrations were counted in a Beckman L5-5000 TD instrument. Oxidation values were normalized per milligram of protein.
Intestinal transit assay
Intestinal transit was monitored as previously described (43). Larvae were fed powdered dry food (30 micron Hatchfry, Argent Labs) from 5 dpf until 7 dpf. At 7 dpf, the animals were fed a mix of powder and a nondigestible fluorescence tracer for 1 h: 300 µl of 2 µm yellow-green FluoroSpheres carboxylate-modified microspheres (Life Technologies) and 200 mg of larval powder. After 1 h, the animals were transferred to clean plates, and animals with intestines that were full of ingested material were selected for serial imaging at 6 and 24 h. The animals were screened in a Leica AF6000 microscope (Leica Microsystems), and the position of the remaining fluorescent ingested material was scored.
Microsomal triglyceride transfer protein activity assay
Microsomal triglyceride transfer protein (Mtp) activity was measured in adult zebrafish anterior intestines with a commercial, fluorescence-based kit according to the manufacturer’s instructions (44) (Roar Biomedical, New York, NY). Tissues were homogenized by sonication in 100 µl of assay buffer (150 mM NaCl, 1 mM EDTA, 10 mM Tris, pH 7.4). The assay was performed adding 50 µg of protein per sample, 5 µl of donor particle, and 5 µl of acceptor particle and incubated for 21 h at 28°C. The fluorescence was measured at different time points (3, 6, and 21 h) at the excitation wavelength of 465 nm and emission wavelength of 538 nm using a SpectraMax M5e microplate reader (Molecular Devices).
Enterocytes isolation and FA uptake analysis
Enterocyte isolation was performed as previously described in other fish species (45) with minor modifications. WT and Tg(fabp2:EGFP-nr1h3) 2 mpf animals were fasted overnight. The animals were anesthetized, and the intestines (anterior and middle intestines) were dissected and rinsed in PBS. The intestines from 20 animals per group were minced in Hanks’ solution (Sigma H8264) with two scalpels. After the tissue was minced, the tissue was digested in collagenase (Sigma C5138). After 15 min, the digested tissue was filtered through 300 µm (Pentair Industrial, Milwaukee, WI), 100 µm, 70 µm, and 40 µm (Cell strainers, BD Falcon, San Jose, CA) meshes to remove ∼90% of the goblet cells present. After the filtrations, the cell suspension was centrifuged at 80 g for 10 min to obtain the enterocyte pellet. After two washes with Hanks’ medium, the enterocytes were resuspended in 1.5 ml of Hanks’-BSA. The cells were counted with a Neubauer chamber, and the concentration was adjusted to have 1 × 106 cells/ml. After the isolation, 100 µl of cell suspension was added in a 96-well plate (Flat bottom, Greiner CELLSTAR 96-well plates, No. 655180), and 0.5 µl of BODIPY FL C12 solution was added (20 ng/ml in Hanks’ buffer). The plate was incubated at 28°C in a shaker for 15 or 30 min. After the incubation time points, the cells were transferred to 1.5 ml tubes and centrifuged for 5 min at maximum speed to pellet the cells, and the buffer was removed. After two washes with Hanks’ solution, cells were lysed with 100 µl of lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100) by sonication. Protein content was measured from 5 µl of the homogenate by BCA protein assay reagent (Thermo Scientific), and the remaining lysate was used to perform a lipid extraction. After the lipid extraction, lipids were resuspended in 100 µl of chloroform and run on a TLC plate. Fluorescence bands were detected with a blue light laser (Typhoon Trio variable mode imager, Amersham Bioscience). Cell viability was also measured with an LDH Cytotoxicity Assay kit (Cayman Chemical Co., Ann Arbor, MI) after 2 and 4 h of incubation time.
Body fat composition analysis
Adult 5 mpf animals were anesthetized, rinsed, dried, and weighed. They were then homogenized in chloroform-methanol to perform a lipid extraction. After extraction, the mass of lipids was divided by body mass.
Statistical analysis
Statistical analyses were performed using SigmaStat software. Data are presented as means ± standard error of the mean. Unless explicitly stated otherwise, differences with the WT group were evaluated using Student’s t-test or ANOVA (followed by Tukey’s test for multiple comparisons). A significance of P < 0.05 was applied to all statistical tests performed.
RESULTS
Generation of Lxra-null mutant zebrafish
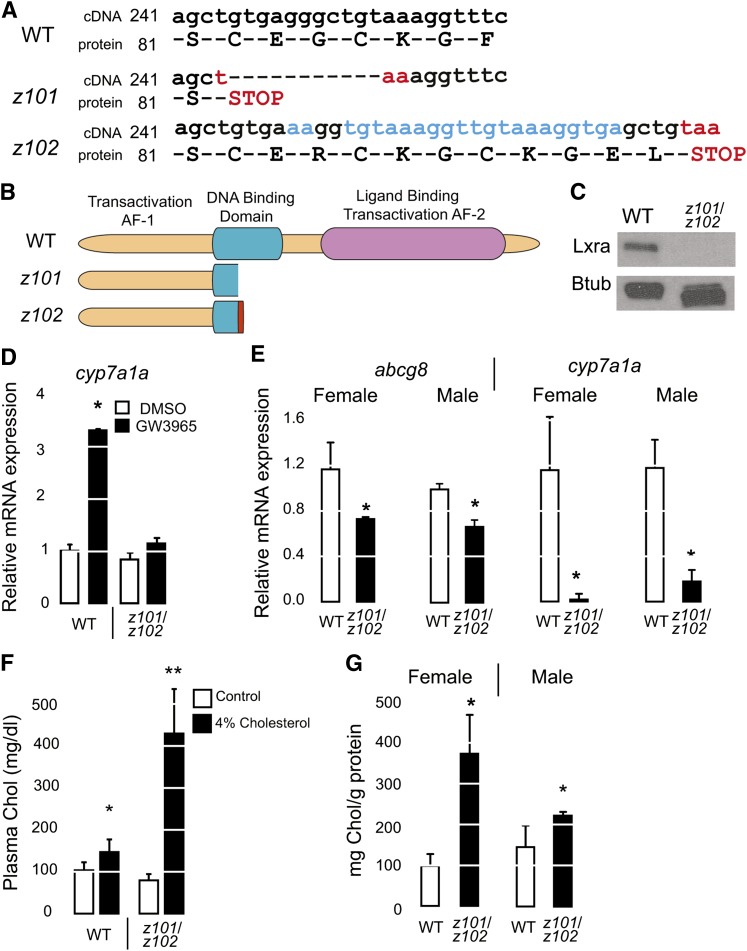
We prepared two nr1h3 deletion mutants using TALENs targeting the second exon of the gene (31). The first nr1h3 mutation (hereafter, strain z101) has an 11 bp deletion causing an in-frame stop codon truncating the protein in the DNA binding domain. The second nr1h3 mutation (hereafter, strain z102) has a 22 bp insertion that also causes a premature stop codon (Fig. 1A). Both of these mutations cause a premature truncation in the DNA binding domain (Fig. 1B). Heterozygous carriers of both mutations (nr1h3+/z101 and nr1h3+/z102) were viable and fertile and transmitted the mutations in the germ line in the anticipated Mendelian ratios. Transheterozygous (nr1h3z101/z102) mutants showed complete loss of Lxra immunoreactivity with an antibody raised against the C-terminal portion of the protein (Fig. 1C). When treated with GW3965, nr1h3z101/z102 mutant larvae did not show induction of the Lxra target gene cyp7a1a, whereas WT larvae did (Fig. 1D). In livers from adult animals fed a high-cholesterol diet, abcg8 and cyp7a1a transcript abundance was lower in nr1h3z101/z102 animals (Fig. 1E). Similar to Lxra−/−;Lxrb−/− mice, nr1h3z101/z102 mutants were cholesterol intolerant, with striking hypercholesterolemia and hepatic cholesterol accumulation in the liver when fed high-cholesterol diets (Fig. 1F, G). These gene expression and dietary studies revealing cholesterol intolerance confirmed that nr1h3z101/z102 are Lxra-null mutants.
Fig. 1.
Generation of nr1h3z101/z102 Lxra mutants. A: cDNA and protein sequence of nr1h3 mutations generated with TALENs. B: Illustrations of the truncated Lxr proteins predicted by the mutations. C: Immunoblot analysis showing complete loss of Lxra protein expression in z101/z102 transheterozygous animals. D: cyp7a1a expression in 18 dpf larvae incubated with GW3965 (n = 3 pools of 3 larvae each). A significant increase was observed only in WT larvae treated with the agonist (P < 0.05). E: mRNA expression of abcg8 and cyp7a1a in livers of adults fed a high-cholesterol diet (n = 3 to 5). Asterisks (*) indicate P < 0.05 for each genotype compared with WT. F, G: Plasma and hepatic cholesterol (Chol) was measured in adults fed a high-cholesterol diet at the conclusion of the experiment (n = 3 to 5). Asterisks (*) indicate P < 0.05 compared with WT. Btub, β-tubulin.
Generation of enterocyte-limited Lxra-overexpressing zebrafish
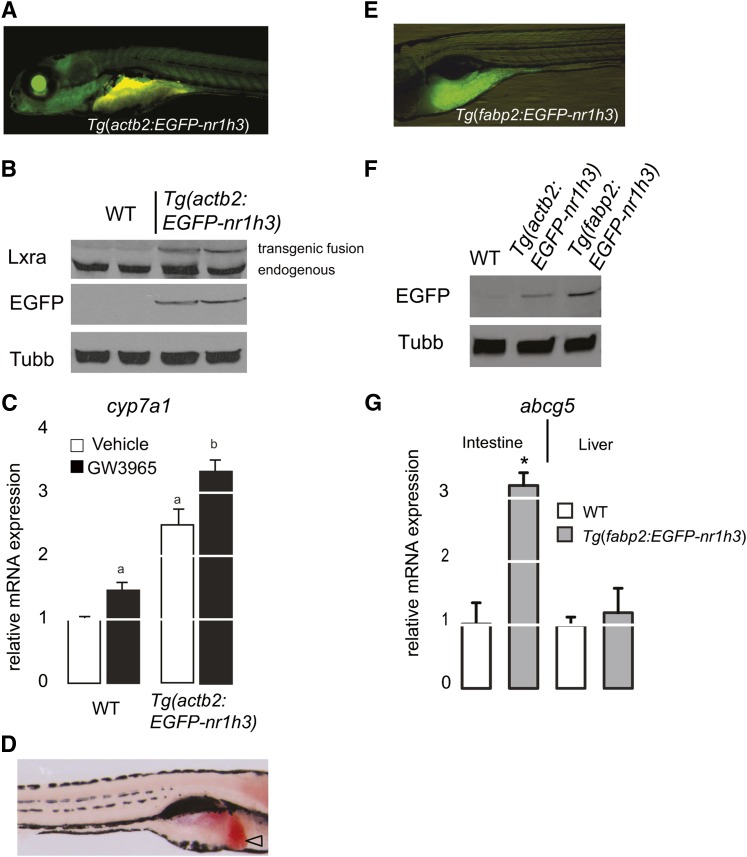
We prepared a cDNA encoding an EGFP-Lxra fusion protein that would allow us to visualize the transgene product with fluorescent imaging and immunoblotting. First established with the Glucocorticoid receptor, N-terminal tags do not appear to alter the subcellular localization or ligand-dependent transactivation of nuclear receptor target genes (46). The stable Tg(actb2:EGFP-nr1h3) transgenic line was prepared using Tol2 transposase-mediated transgenesis (33) and showed broad EGFP reporter was expression (Fig. 2A). We analyzed EGFP-Lxra protein expression in the progeny of two independent transgenic founders; equal EGFP-Lxra fusion protein expression was detected in both transgenic cohorts with both anti-Lxra and anti-EGFP antibodies, confirming desired transgene product expression (Fig. 2B). These immunoblot analyses of two separate founders minimized our concern for positional or transgene copy number effects in working with this construct. Likewise, the transgenic protein’s expression did not alter the much higher abundance of the endogenous Lxra protein. This is a concern because Lxra is a retinoid X receptor (Rxr) heterodimerization partner, and very high expression of one such partner may titrate the available Rxr protein that would otherwise be in complex with other partners (47).
Fig. 2.
Generation of Lxra transgenic zebrafish. A: A 5 dpf Tg(actb2:EGFP-nr1h3) larva photographed in the left lateral view with a GFP long-pass filter shows broad transgene expression. The yellow autofluorescence of the intestinal lumen. B: Immunoblot analyses with anti-Lxra and anti-EGFP antibodies demonstrate that the fusion protein is expressed to equivalent levels in transgenic animals, and the presence of the transgenic fusion protein does not alter the abundance of the endogenous Lxra protein. Anti-Tubb antibody was used as a loading control. C: cyp7a1a expression in 18 dpf larvae incubated with vehicle (0.1% DMSO) or GW3965 (200 nM) for 24 h (n = 3 pools of 3 larvae each). Values that do not share a common letter are significantly different (P < 0.05) from one another. D: All 6 dpf Tg(actb2:EGFP-nr1h3) larvae show hepatic steatosis (open arrowhead) when fixed and stained with ORO. Note, this larva is photographed in the right lateral view. E: EGFP expression in a 6 dpf Tg(fabp2:EGFP-nr1h3) larva is restricted to the anterior intestine. Note this left lateral view is higher magnification than in A. F: Western blot analysis of EGFP-Lxra fusion proteins in extracts prepared from WT, Tg(fabp2:EGFP-nr1h3), and Tg(actb2:EGFP-nr1h3). G: abcg5 expression in intestine and liver of 42 dpf WT and Tg(fabp2:EGFP-nr1h3) (f2) juveniles (n = 4). Asterisk (*) indicates significant differences with the WT group (P < 0.05).
Next, we addressed whether the global overexpression of EGFP-Lxra would increase transactivation of a known Lxra target gene. Specifically, we measured cyp7a1 mRNA abundance in WT and Tg(actb2:EGFP-nr1h3) transgenic larvae in the presence or absence of the potent Lxra synthetic agonist GW3965. Whereas WT larvae treated with agonist showed a 50% increase in cyp7a1 transcript abundance, Tg(actb2:EGFP-nr1h3) transgenic larvae showed a 250% increase in the absence of exogenous ligand and a 350% increase in the presence of exogenous ligand (Fig. 2C). Finally, we fixed and stained never-fed 6 dpf Tg(actb2:EGFP-nr1h3) larvae with the neutral lipid dye ORO to assess whether these animals have increased liver lipid accumulation, a phenotype that would be expected in an organism with overly active hepatic Lxra (11, 12). Tg(actb2:EGFP-nr1h3) had hepatic steatosis at 6 dpf (Fig. 2D). Collectively, these preliminary studies show that the EGFP-Lxra fusion protein we prepared is functional.
Next, we prepared the stable transgenic line Tg(fabp2:EGFP-nr1h3), which expresses the EGFP-Lxra fusion protein under the control of the enterocyte-limited fatty acid binding protein 2 (fabp2) promoter (Fig. 2E). The EGFP-Lxra fusion protein was detected by immunoblotting protein extracts from larvae with an anti-GFP antibody; as expected, its abundance was less than that seen in extracts prepared from Tg(actb2:EGFP-nr1h3) larvae (Fig. 2F). Increased transcriptional activation of Lxra target genes was confirmed by measuring abcg5 transcript abundance. We observed increased abcg5 expression in the intestine, but not in the liver of adult Tg(fabp2:EGFP-nr1h3) animals (Fig. 2G). These gene expression profiles confirm that we successfully prepared an enterocyte-limited Lxra overexpressing line that shows anticipated induction of known target genes.
Intestinal overexpression of Lxra delays lipid absorption
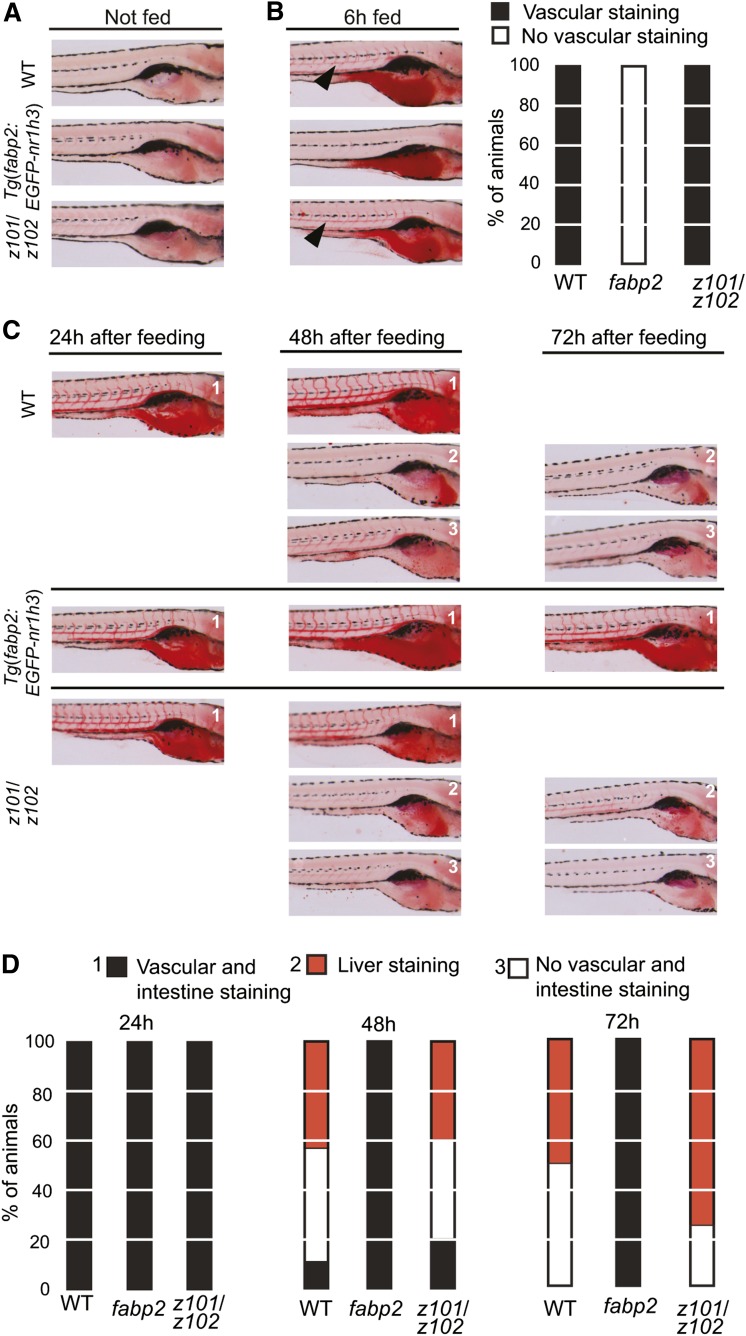
We previously established that 6 dpf larvae show scant lipid staining, and that following a 6 h high-fat meal, the intestine, liver, and vasculature show prominent staining with ORO; this staining correlates with whole-carcass triacylglycerol levels (21). Furthermore, complete clearance of ingested lipids (resolution of intestinal, hepatic, and vascular staining) occurs in the majority of WT larvae over the next 18 h. We fed 6 dpf WT, Tg(fabp2:EGFP-nr1h3) transgenic, and nr1h3z101/z102 mutants a high-fat meal for 6 h and assessed whole-body lipid stores with ORO staining. While all three cohorts showed no vascular, liver, or intestinal staining prior to feeding (Fig. 3A), only WT and nr1h3z101/z102 mutant larvae showed vascular lipid staining at the conclusion of the meal (Fig. 3B). This absence of vascular lipid staining in Tg(fabp2:EGFP-nr1h3) transgenic larvae after 6 h of feeding suggested a delay in the absorption of ingested lipids or a delay in the transport of absorbed lipids into the circulation. To address these possibilities, we placed fed larvae in fresh medium lacking food and scored with ORO staining 24 h after the meal. At this point, larvae of all three genotypes showed intestinal and vasculature staining (Fig. 3C, D). When examined 48 and 72 h following the meal, WT and nr1h3z101/z102 larvae showed near-complete digestion and use of lipids, as reflected by fewer animals showing vascular, intestinal, or liver lipid staining (Fig. 3C, D). In contrast, Tg(fabp2:EGFP-nr1h3) transgenic larvae had persisting intestinal and vascular lipid staining. Importantly, the transit of nondigestible fluorescence beads included in a meal (43) was similar among all three genotypes (supplementary Fig. I). Thus, in larvae, overexpression of Lxra in the intestine delays enterocyte trafficking of ingested lipids but does not alter intestinal transit of ingested materials.
Fig. 3.
Delayed lipid absorption in Tg(fabp2:EGFP-nr1h3) transgenic zebrafish larvae. A, B: Larvae were fasted or fed a high-fat meal for 6 h, and fixed and stained with ORO (left) and scored for location of lipids (B, right). C: Other fed animals were placed in clean embryo water and fixed and stained 24, 48, and 72 h later. D: Staining patterns were quantified (n = 25 to 28 per genotype and time point). Black arrow heads indicate lipid staining in vasculature in WT and nr1h3z101/z102 larvae. In B and D, “fabp2” refers to Tg(fabp2:EGFP-nr1h3).
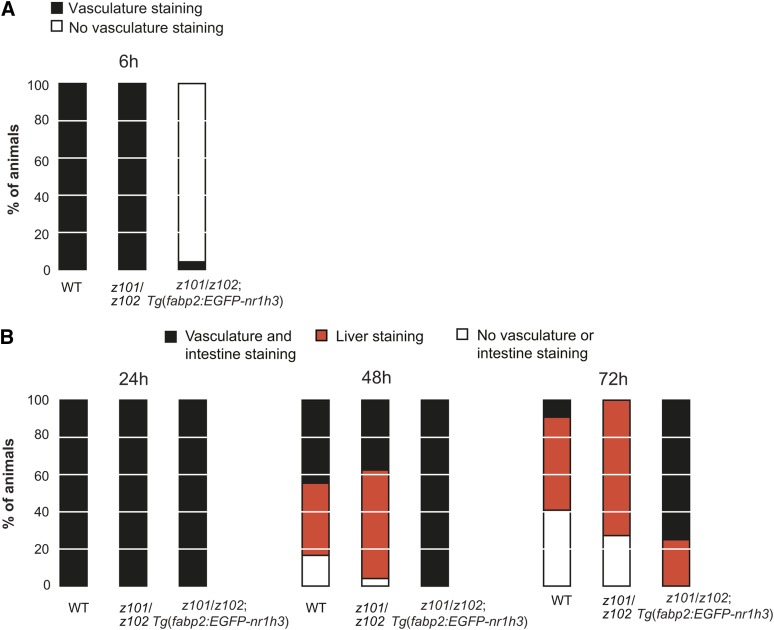
To establish that intestinal overexpression of Lxra in the intestine is sufficient to delay the transport of absorbed lipids even in the absence of normal Lxra expression in all other organs, we crossed Tg(fabp2:EGFP-nr1h3) to nr1h3z101/z102 mutants and characterized the fate of ingested lipids in larvae. Corroborating our results on the WT Lxra background, forced intestinal expression of Lxra in global Lxra mutant animals delayed the appearance of lipids in the vasculature of larvae fed a high-fat meal (Fig. 4A). Furthermore, nr1h3z101/z102; Tg(fabp2:EGFP-nr1h3) larvae showed a delay in complete digestion of the ingested lipids (Fig. 4B). Collectively, these results indicate a cell-autonomous role for Lxra in regulating the fate of absorbed lipids.
Fig. 4.
Intestinal overexpression of nr1h3 in nr1h3z101/z102 mutants is sufficient to delay transport of ingested lipids. A: WT, nr1h3z101/z102, and nr1h3z101/z102; Tg(fabp2:EGFP-nr1h3) larvae were fed a high-fat meal for 6 h and then fixed and stained with ORO. B: After the 6 h meal, animals were placed in clean embryo water and fixed and stained 24, 48, and 72 h later. The staining patterns were quantified (n = 19–20 per genotype and time point).
Lxra activation in the intestine delays absorption of FAs by promoting cytoplasmic lipid droplet formation
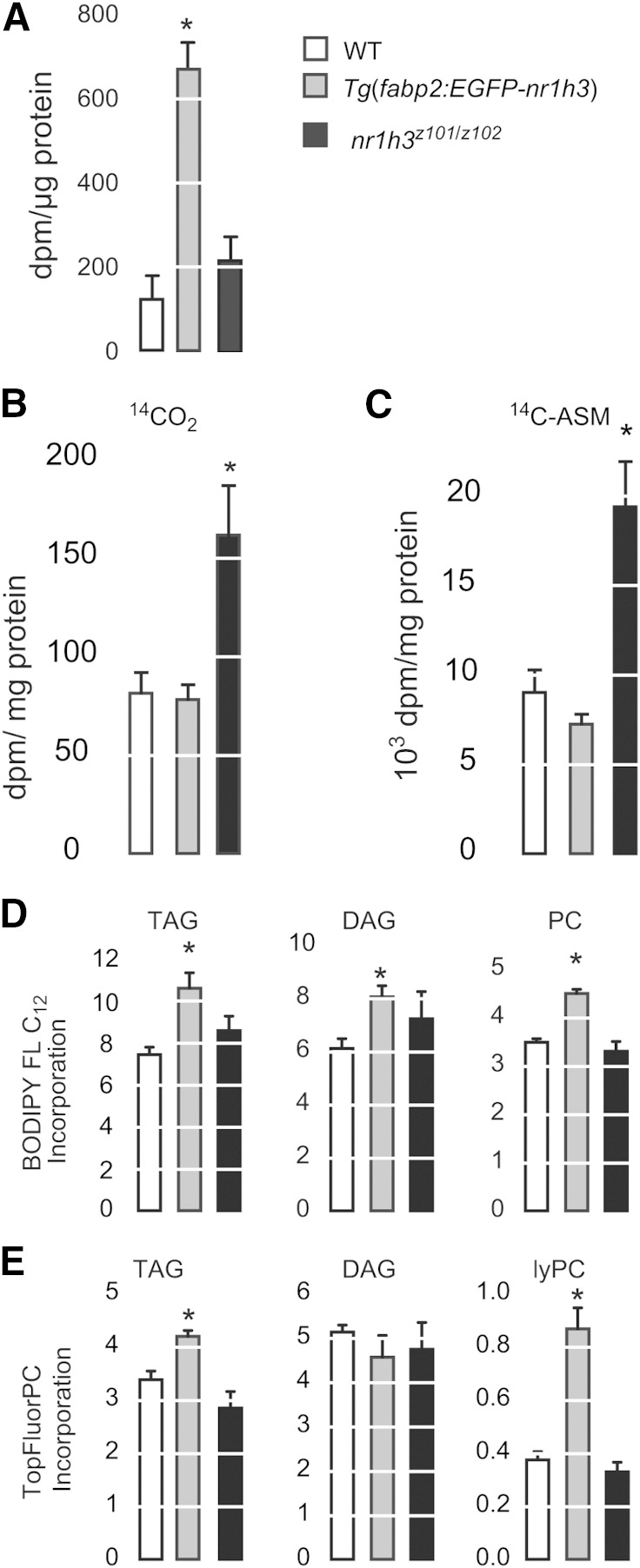
To probe the mechanism accounting for the delayed appearance of neutral lipids in the vasculatures of Tg(fabp2:EGFP-nr1h3) animals, we gavaged 3 mpf adults with [3H]triolein and measured tracer signal in the dissected and washed intestine 24 h later (Fig. 5A). Tg(fabp2:EGFP-nr1h3) transgenic adults showed greatest tracer signal in intestines following the gavage, suggesting that intestinal storage of absorbed lipids accounts for the delayed appearance of neutral lipids in the vasculature in these animals. While intestinal retention of radiotracer in Tg(fabp2:EGFP-nr1h3) transgenic animals was gratifying, this single labeling method does not account for lipid pool size or rates of tracer oxidation. Thus, we measured FA oxidation in isolated intestines. Tg(fabp2:EGFP-nr1h3) and WT intestines showed equal rates of 14CO2 and 14C-labeled acid-soluble metabolite production when incubated with 14C-oleate. (Fig. 5B, C). This result indicates that decreased FA oxidation in Tg(fabp2:EGFP-nr1h3) does not account for the persistence of absorbed lipids in the form of cytoplasmic lipid droplets. Indeed, nr1h3z101/z102 intestines showed nearly double the rates of CO2 and acid-soluble metabolite production.
Fig. 5.
Retentions of ingested FAs in Tg(fabp2:EGFP-nr1h3) transgenic adult intestines. A: Three mpf males were gavaged with 2 µl of oil containing 0.2 µCi of [3H]triolein. Twenty-four hours postgavage, intestines were harvested, and the remaining 3H was quantified. * P < 0.05 for each genotype compared with WT (n = 4). B, C: Ex vivo production of 14CO2 and 14C -acid-soluble metabolites (ASM) by intestinal homogenates incubated with 14C-oleate. * P < 0.05 for each genotype compared with WT (n = 6 intestines per pool, and the results represent the mean of two pools per genotype). D, E: Larvae (6 dpf) were fed an emulsion of 5% dried egg yolk containing a fluorescence FA (BODIPY FL C12) or fluorescent phospholipid (TopFluorPC). After 6 h, the larvae were rinsed, the lipids were extracted, and the incorporation of the fluorescent tracers into the different lipid classes was quantified. * P < 0.05 for each genotype compared with WT (n = 3 pools of 30 larvae each). DAG, diacylglycerol; lyPC, lysophosphatidylcholine; PC, phosphatidylcholine; TAG, triacylglycerol.
While decreased FA oxidation as a potential explanation for retention of absorbed lipids in the intestines of Tg(fabp2:EGFP-nr1h3) animals was ruled out, we wished to further explore the role of Lxra in regulating absorbed FA handling. To this end, we performed experiments with two fluorescent medium-chain FA analogs: a 12-carbon FA analog, BODIPY FL C12, and a phospholipid analog containing an 11-carbon FA analog side chain. These analogs have been previously used in zebrafish to visualize lipid trafficking (8) and allowed us to circumvent the pool size quandary: owing to their medium-chain lengths (11 and 12 carbons plus fluorophore), these FA analogs can traverse the intestine without having to be incorporated into neutral lipids that are carried as chylomicron cargo. Tg(fabp2:EGFP-nr1h3) larvae fed a high-fat meal spiked with BODIPY FL C12 showed greater incorporation of the FA analog in triacylglycerol, diacylglycerol, and phosphatidylcholine (Fig. 5D). A similar pattern was observed in Tg(fabp2:EGFP-nr1h3) larvae after the administration of TopFluorPC (Fig. 5E). In total, these experiments indicate that intestine-limited activation of Lxr promotes incorporation of fatty acyl chains into neutral and phospholipids that are then stored in the enterocyte.
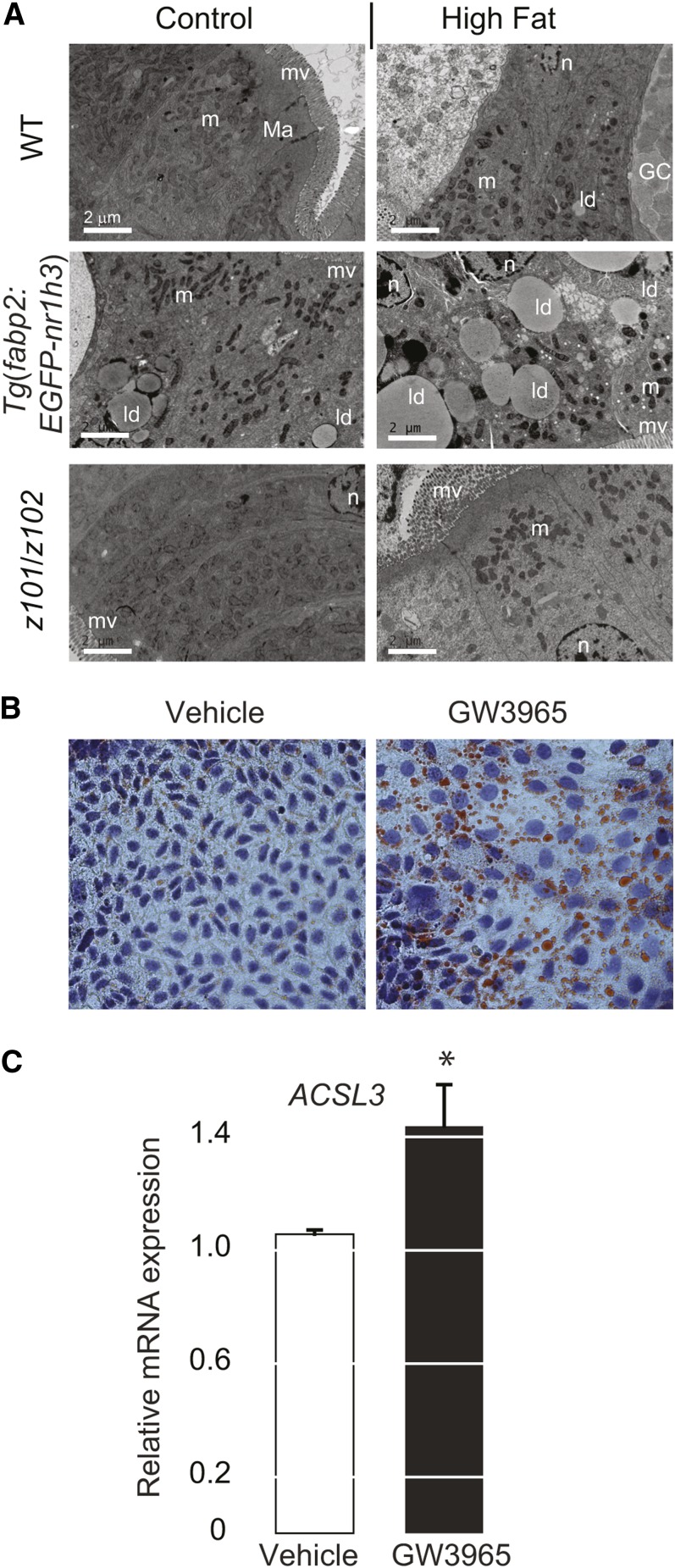
Next, we fed adult animals defined diets containing normal (control) or excessive (high) fat for 2 months and then performed ultrastructural analysis of adult zebrafish intestines. After a 24 h fast, the intestines of WT and nr1h3z101/z102 mutant animals had scant lipid content as revealed by transmission electron microscopy. Moreover, cytoplasmic lipid droplets were seen in Tg(fabp2:EGFP-nr1h3) enterocytes, with the largest and most numerous lipid droplets appearing in animals fed HFDs (Fig. 6A). Demonstrating that the Lxr effect on intestinal lipid handling is conserved across evolution, a 24 h treatment of the polarized human enterocyte line Caco2 with GW3965 caused accumulation of cytoplasmic lipid droplets (Fig. 6B). These structural results confirm the radiotracer and fluorescent tracer findings and indicate that Lxra activation promotes intestinal neutral lipid storage.
Fig. 6.
Intestinal activation of Lxra induces cytoplasmic lipid droplet formation. A: Electron microscopy of anterior intestine sections showing lipid droplet accumulation in Tg(fabp2:EGFP-nr1h3) transgenic animals] 24 h postfeeding. GC, goblet cells; ld, lipid droplets; m, mitochondria; Ma, macula adherens or desmosome; mv, microvilli; n, nuclei. B, C: Caco2 cells were culture in the presence or absence (vehicle, DMSO 0.1% v/v) of 2 μM GW3965 for 24 h and then either fixed and stained with ORO (and hematoxylin counterstain of nuclei) or subjected to RNA extraction for quantification of ACSL3 transcript. * P < 0.01 in a two-sided Student’s t-test.
Intestinal lipid accumulation might reflect impaired chylomicron assembly and secretion (48). To test this possibility, we assayed the activity of the β lipoprotein packaging enzyme Mtp. When fed either diet, Tg(fabp2:EGFP-nr1h3) animals had the highest intestinal Mtp activity, excluding the possibility that impaired Mtp activity accounted for the intestinal lipid storage seen in these animals (supplementary Fig. II). This increased Mtp activity in the Tg(fabp2:EGFP-nr1h3) animals’ intestines corresponded to the upregulation in mtp transcript abundance (Table 1). This experiment does not exclude a post-Mtp defect in nascent chylomicron assembly; however, the major regulatory step in halting β lipoprotein assembly is endoplasmic reticulum-associated degradation of Apob that fails to be lipidated by Mtp (49).
TABLE 1.
Intestinal gene expression profiles of juvenile animals after control diet and HFD
| Control Diet | HFD | ||||||
| Gene | Protein | WT | Tg(fabp2: EGFP-nr1h3) | nr1h3z101/z102 | WT | Tg(fabp2: EGFP-nr1h3) | nr1h3z101/z102 |
| FA CoA activation | |||||||
| acsl3a | Acyl-CoA synthetase long-chain family member 3a | 1.04 ± 0.11a | 17.88 ± 5.48b | 0.68 ± 0.12a | 1.17 ± 0.12a | 26.53 ± 4.03b | 0.5 ± 0.21c |
| acsl3b | Acyl-CoA synthetase long-chain family member 3b | 1.19 ± 0.11 | 1.34 ± 0.10 | 0.47 ± 0.07 | 1.66 ± 0.22a | 1.91 ± 0.09a | 0.63 ± 0.07b |
| acsl5 | Acyl-CoA synthetase long-chain family member 5 | 1.05 ± 0.11 | 1.09 ± 0.04 | 1.11 ± 0.12 | 2.32 ± 0.31 | 2.28 ± 0.24 | 2.58 ± 0.43 |
| FA transport | |||||||
| cd36 | CD36 antigen | 1.02 ± 0.08a | 0.41 ± 0.04b | 1.05 ± 0.17a | 1.26 ± 0.17a | 0.32 ± 0.08b | 0.80 ± 0.15a |
| slc27a4 | Solute carrier family 27, member 4 | 1.01 ± 0.03 | 0.81 ± 0.12 | 1.16 ± 0.12 | 1.19 ± 0.19a | 0.73 ± 0.03b | 2.05 ± 0.45c |
| Lipoprotein assembly | |||||||
| apoa1a | Apolipoprotein A-Ia | 1.01 ± 0.10 | 0.65 ± 0.08 | 1.08 ± 0.07 | 12.30 ± 0.84a | 3.55 ± 0.92b | 6.29 ± 1.32b |
| apoa1b | Apolipoprotein A-Ib | 1.20 ± 0.42 | 1.35 ± 0.57 | 2.57 ± 1.25 | 0.37 ± 0.08a | 5.84 ± 1.73b | 1.90 ± 0.43ab |
| apoa4 | Apolipoprotein A-IV | 1.02 ± 0.12 | 0.60 ± 1.56 | 2.73 ± 0.89 | 7.68 ± 1.24 | 6.54 ± 0.36 | 8.90 ± 1.17 |
| apoba | Apolipoprotein Ba | 1.33 ± 0.44 | 1.17 ± 0.30 | 1.68 ± 0.62 | 2.40 ± 0.93 | 4.52 ± 2.08 | 2.97 ± 0.65 |
| apobb | Apolipoprotein Bb | 1.01 ± 0.08a | 0.49 ± 0.06c | 1.58 ± 0.22b | 2.25 ± 0.24a | 1.20 ± 0.06b | 2.37 ± 0.27a |
| dgat2 | Diacylglycerol O-acyltransferase 2 | 1.04 ± 0.11a | 0.26 ± 0.04b | 2.39 ± 0.18c | 1.68 ± 0.23a | 0.54 ± 0.17b | 3.57 ± 0.30c |
| mtp | Microsomal triglyceride transfer protein | 1.01 ± 0.08a | 1.84 ± 0.33b | 1.24 ± 0.18a | 3.36 ± 0.47a | 6.55 ± 0.87b | 3.76 ± 0.59a |
Results for each gene were analyzed by one-way ANOVA, followed by Tukey’s test. Values that do not share a common letter are significantly different (P < 0.05). n = 5–6.
Next, we tested the possibility that an Lxra-driven increase in FA uptake in enterocytes might simply overwhelm the ability of these cells to package FAs into chylomicrons, diverting them passively into cytoplasmic lipid droplets (50). In isolated enterocytes, less BODIPY FL C12 was absorbed when Lxra was overexpressed (supplementary Fig. III). One limitation of this type of ex vivo experiment is that it is impossible to distinguish between apical and basolateral uptake. Nevertheless, this decrease in FA uptake by Tg(fabp2:EGFP-nr1h3) enterocytes is supported at a molecular level in the downregulation of cd36 and slc27a4 transcripts, both encoding proteins involved in enterocyte uptake of luminal FAs (Table 1), although the extent of their contribution to FA uptake remains a matter of contention (51). Taken together, these studies in larvae, adults, and isolated enterocytes indicate that overexpression of Lxra in the intestine drives storage of absorbed FAs in cytoplasmic lipid droplets. Neither defective chylomicron assembly nor excessive FA uptake by enterocytes appears to be the cause of Lxr-driven intestinal storage.
Enterocyte-limited overexpression of Lxra protects animals from hypercholesterolemia and hepatic steatosis
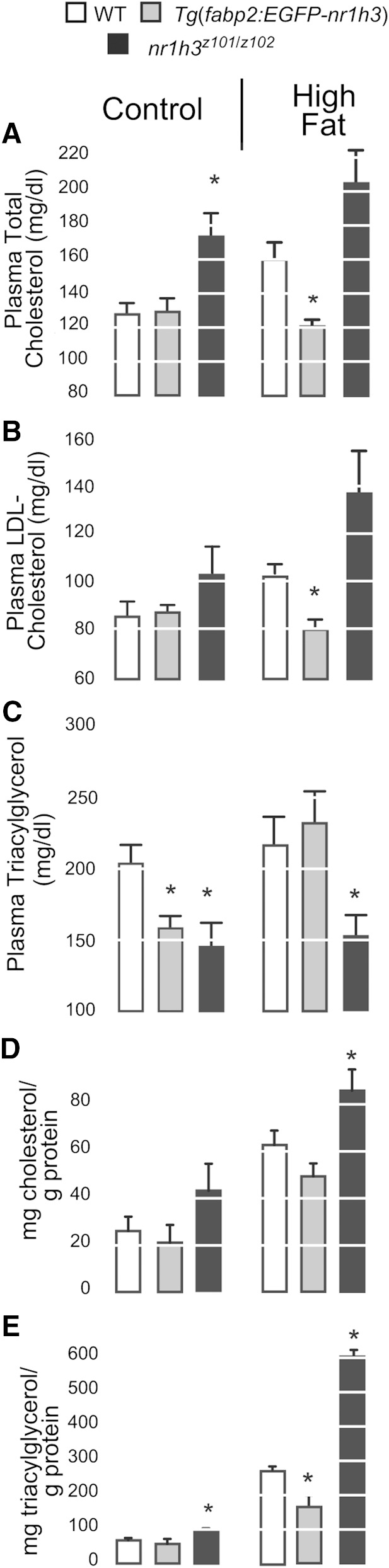
Next, we wished to examine the metabolic consequences of modulating intestinal Lxr activity in response to HFDs. When fed the control diet for 2 months, Tg(fabp2:EGFP-nr1h3) transgenic adults and nr1h3z101/z102 mutants had lower masses at the conclusion of the defined diet feeding period (supplementary Fig. IVA). Tg(fabp2:EGFP-nr1h3) transgenic adults had less body fat at the conclusion of the feeding period (supplementary Fig. IVB). The lower mass of the nr1h3z101/z102 mutants is reminiscent of the lower mass of young Lxra−/−;Lxrab−/− mice (52). Out of concern that these differences in body mass and composition might cloud further analysis, we limited feeding of the HFDs to 2 months. Similar to the effect of high-cholesterol feeding (Fig. 2F), deletion of nr1h3 caused increased plasma total cholesterol (Fig. 7A, B), although the effect was significant only on control diets. Intestine-limited overexpression of Lxr caused decreases in both total and LDL cholesterol when animals were fed either diet (Fig. 7A, B). Again mirroring the phenotype of young Lxra−/−;Lxrb−/− mice (52), nr1h3z101/z102 mutants had lower fasting plasma triacylglycerol on both diets (Fig. 7C). Overexpression of Lxra in the intestine also caused a decrease in fasting plasma triacylglycerol on control diets only (Fig. 7C). On the HFD, nr1h3z101/z102 mutants showed increased hepatic cholesterol (Fig. 7D). Furthermore, nr1h3z101/z102 mutants showed striking, HFD-induced increases in hepatic triacylglycerol, while Tg(fabp2:EGFP-nr1h3) animals were protected from this steatosis (Fig. 7E). In summary, enterocyte-limited overexpression of Lxra protects against diet-induced hypercholesterolemia and hepatic steatosis.
Fig. 7.
Intestinal overexpression of Lxr prevents HFD-induced hypercholesterolemia and hepatic steatosis. A–C: Adult animals were fed HFDs for 2 months. After an overnight fast, plasma total and LDL cholesterol, and triacylglycerol levels were highest in nr1h3z101/z102 mutants and lowest in Tg(fabp2:nr1h3) transgenic animals (n = 5). D, E: Liver cholesterol and triacylglycerol were highest in nr1h3z101/z102 mutants and lowest in Tg(fabp2:nr1h3) transgenic animals (n = 4). * P < 0.05 for each genotype compared with WT.
Lxra promotes lipid accumulation in enterocytes through the upregulation of acsl3a, which encodes a cytoplasmic lipid droplet-anchored enzyme of FA activation
We performed a focused survey of Lxra target genes that could be involved in the intestinal lipid accumulation (Table 1). The acsl3a transcript (orthologous to human ACSL3) showed the highest induction in Lxra-overexpressing animals and reduced expression in Lxra mutants. Similarly, following a short pharmacological treatment with Lxr agonist, Caco2 cells showed a 50% increase in ACSL3 transcript abundance (Fig. 6C). Acyl-CoA synthetases are enzymes that activate FAs for lipid synthesis, catabolism, and phospholipid remodeling (53). There are five subfamilies with diverse isoforms in mammals showing differential substrate FA length specificity (54). The long-chain acyl-CoA synthetases, or Acsls, activate FAs with a chain length of 12 to 20 carbons. Acsl3 is ubiquitously expressed (55). Acsl3 targeting to cytoplasmic lipid droplets is achieved by an N-terminal cytoplasmic lipid droplet anchor motif that is not present in other Acsl family members (28) This enzyme acts throughout lipid droplet biogenesis to activate CoA thioesters for subsequent incorporation into the growing storage depot (29). The induction of the encoding acls3a transcript raised the possibility that this enzyme might serve to activate FAs for incorporation into glycerolipids and cholesteryl esters at the surface of lipid droplets, “channeling” them away from incorporation into chylomicrons (56, 57).
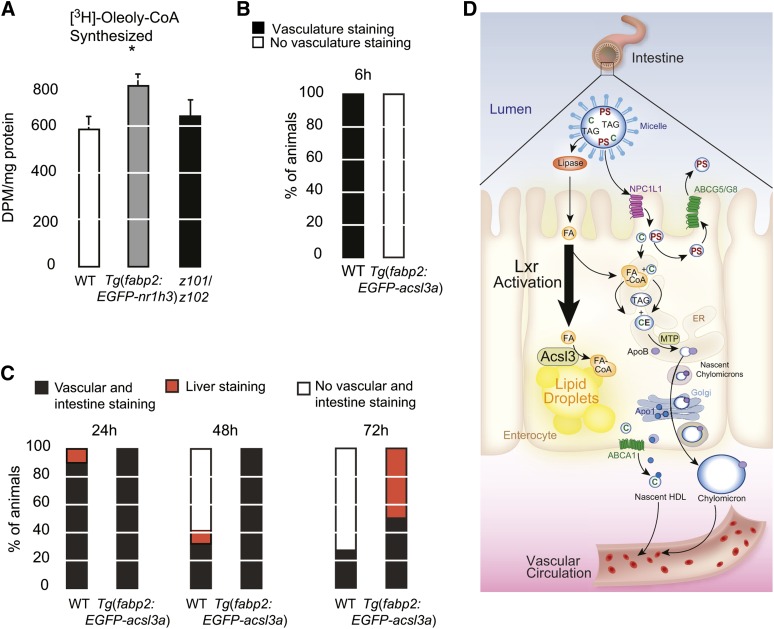
To test our hypothesis, we confirmed that the increase in acslc3a transcript abundance correlated with increased Acsl activity in intestine homogenates from Tg(fabp2:EGFP-nr1h3) animals (Fig. 8A). Because the main Acsl enzymes expressed in intestine are Acsl3 and Acsl5 (58), this assay cannot distinguish between the activities of these two Acsls. Nevertheless, a recent study demonstrated that Acsl3 has a higher substrate affinity for oleic acid than longer-chain FAs (41). Furthermore, neither the paralogous acsl3b transcript nor the acsl5 transcript were induced in Tg(fabp2:EGFP-nr1h3) enterocytes, arguing that Acsl3a activity is uniquely induced by intestinal Lxra activation (Table 1), and the increase in Acsl activity we observed in whole intestine homogenate is a conservative underestimate of increased Acsl3a expression and activity.
Fig. 8.
Overexpression of acls3a in WT enterocytes is sufficient to delay transport of ingested lipids. A: Total ACSL activity was measured in intestine homogenates by monitoring conversion of free [3H]oleic acid to [3H] oleolyl-CoA (n = 5). Asterisk (*) indicates significant differences with the WT group (P < 0.05). B: WT and Tg(fabp2:EGFP-acsl3a) larvae were fed a high-fat meal for 6 h and then fixed and stained with ORO. C: After the 6 h meal, animals were placed in clean embryo water and fixed and stained 24, 48, and 72 h later. The staining patterns were quantified (n = 19–20 per genotype and time point). D: Model for Lxr-driven intestinal storage of absorbed lipids. Cholesterol (C) and plant sterols (PS) are suspended in micelles in the lumen of the intestine. They are absorbed at the apical surface of enterocytes by NPCL1L1. Plant sterols are immediately excreted back into the lumen by the action of ABCG5/ABCG8 heterodimers. FAs are transported across the apical surface by carriers that are not shown. Absorbed C and FAs, which are liberated by luminal lipases, are reassembled in the endoplasmic reticulum (ER) into cholesteryl esters (CE) and triacylglycerol (TAG) by a series of enzymes (not shown). ER-synthesized neutral lipids are packaged with ApoB into chylomicrons by MTP. Nascent chylomicrons are transferred to the Golgi apparatus and then secreted into basolateral space, entering the lymphatic drainage. In parallel, ApoA1 is released along the basolateral surface, where it combines with ABCA1-transported free cholesterol to form nascent HDL particles. Enterocyte Lxra activation induces expression of the lipid droplet-anchored FA-activating enzyme Acls3, which drives FAs into this organelle and away from the secretory machinery.
To demonstrate that the acsl3a induction was a dominant driver of lipid accumulation in Lxra-overexpressing enterocytes, we prepared an intestine-limited acsl3a-overexpressing transgenic line. Similar to Tg(fabp2:EGFP-nr1h3) larvae fed a high-fat meal, 6 dpf Tg(fabp2a:EGFP-acsl3) larvae showed less vascular lipid staining at the conclusion of a 6 h, high-fat meal (Fig. 8B). The appearance (Fig. 8B) and subsequent clearance (Fig. 8C) of absorbed lipids was also delayed in Tg(fabp2a:EGFP-acsl3) larvae. Thus, overexpression of the Lxra target gene acsl3a in enterocytes was sufficient to recapitulate, in part, the phenotype of Lxra overexpression (Fig. 8D).
DISCUSSION
We found that Lxra overexpression in enterocytes delays the appearance of ingested lipids in the vasculature. This delay is observed in zebrafish larvae following a single meal and in adults fed defined diets chronically. The net effect of Lxra activation in enterocytes included protection from hypercholesterolemia and hepatic steatosis when fed HFDs. After excluding the possibilities of impaired Mtp activity and exaggerated FA uptake, we found that enterocytes overexpressing Lxra show increased acsl3a transcript abundance, and that the encoded protein’s forced expression is sufficient to recapitulate, in part, the phenotype of Tg(fabp2:EGFP-nr1h3) transgenic animals. Of course, other Lxr effectors acting in the intestine may contribute to the global control of lipid trafficking; however, our work demonstrates that, among the myriad regulatory events governing lipid droplet biogenesis, site-specific activation of fatty acyl-CoA represents a mechanism for funneling these metabolites to storage (28, 29, 55, 57).
While nr1h3z101/z102 animals showed anticipated gene expression changes and were cholesterol intolerant, they did not show many differences when compared with WT when studied for their intestinal lipid-handling properties other than increased intestinal FA oxidation. This finding is consistent with the ability of young Lxra−/−;Lxrb−/− mice to resist diet-induced obesity, through induction of a thyroid hormone-dependent thermogenesis program (52). Nevertheless, as Lxra−/−;Lxrb−/− mice age, they lose adipose mass on normal diets. This weight loss might be a function of the age-dependent brain lipid accumulation and neurodegeneration that occurs in these animals (27). Introducing the ob (leptin) mutation homozygously into Lxra−/−;Lxrb−/− mice causes modest weight gain, indicating that the “adipostat” signaling system remains intact (59).
Our finding that nr1h3z101/z102 larvae appear to transport absorbed lipids at the same pace as WT larvae raises the possibility that loss of Lxra function in the intestine might require longer experimental manipulations to reveal additional differences. Alternatively, loss of Lxr globally might mask changes in intestinal lipid transport, with offsetting transcriptional programs driven by such nuclear receptors as peroxisome proliferator-activated receptors (60). We will address these questions in the future with more tissue-specific overexpression models placed on the nr1h3z101/z102 background. Likewise, long-term studies will be required to address whether intestinal Lxr overexpression has effects on longevity, fertility, atherogenesis, and obesity. The last area is of interest because cold adaptation in poikilothermic zebrafish requires evolutionarily conserved induction of thyroid hormone signaling to generate heat (61–63). We anticipate that exploring Lxr’s regulation of heat dissipation could lead to generalizable findings regarding regulation of body mass.
In conclusion, we have shown that Lxra induces accumulation of lipids in the enterocytes by diverting absorbed lipids into cytoplasmic lipid droplets. This diversion from chylomicron assembly and secretion hinges, in part, on induction of the direct Lxra target Acsl3 and has salutary effects on systemic lipid handling. Conditional, cell-type-limited deletion of acsl3a will allow us to further explore the role of the encoded gene in intestinal lipid transport.
Supplementary Material
Acknowledgments
The authors thank Sarah Hugo for technical assistance and Diana Lim for improving Figure 8D.
Footnotes
Abbreviations:
- Acsl3
- acyl-CoA synthetase long-chain family member 3
- BODIPY FL C12
- 4,4-difluoro-5,7-dimethyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoic acid
- Btub or Tubb
- β-tubulin
- dpf
- days post-fertilization
- EGFP
- enhanced green fluorescent protein
- HFD
- high-fat diet
- mpf
- months post-fertilization
- Mtp
- microsomal triglyceride transfer protein
- ORO
- Oil Red O
- PFA
- paraformaldehyde
- TALEN
- transcription activator-like effector nuclease
- TopFluorPC
- 1-palmitoyl-2-(dipyrrometheneboron difluoride)undecanoyl-sn-glycero-3-phosphocholine
This work was supported, in part, by a Pilot & Feasibility Award from the Washington University Diabetes Research Center (P30-DK020579).
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two tables and four figures.
REFERENCES
- 1.Lozano R., Naghavi M., Foreman K., Lim S., Shibuya K., Aboyans V., Abraham J., Adair T., Aggarwal R., Ahn S. Y., et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 380: 2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kannel W. B., Dawber T. R., Friedman G. D., Glennon W. E., McNamara P. M. 1964. Risk factors in coronary heart disease: an evaluation of several serum lipids as predictors of coronary heart disease: the Framingham Study. Ann. Intern. Med. 61: 888–899. [DOI] [PubMed] [Google Scholar]
- 3.Baigent C., Blackwell L., Emberson J., Holland L. E., Reith C., Bhala N., Peto R., Barnes E. H., Keech A., Simes J., et al. 2010. Efficacy and safety of more intensive lowering of LDL cholesterol: a meta-analysis of data from 170,000 participants in 26 randomised trials. Lancet. 376: 1670–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jørgensen A. B., Frikke-Schmidt R., Nordestgaard B. G., Tybjærg-Hansen A. 2014. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N. Engl. J. Med. 371: 32–41. [DOI] [PubMed] [Google Scholar]
- 5.Varbo A., Benn M., Nordestgaard B. G. 2014. Remnant cholesterol as a cause of ischemic heart disease: evidence, definition, measurement, atherogenicity, high risk patients, and present and future treatment. Pharmacol. Ther. 141: 358–367. [DOI] [PubMed] [Google Scholar]
- 6.Robertson M. D., Parkes M., Warren B. F., Ferguson D. J. P., Jackson K. G., Jewell D. P., Frayn K. N. 2003. Mobilisation of enterocyte fat stores by oral glucose in humans. Gut. 52: 834–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J., Lee B., Buhman K. K., Cheng J-X. 2009. A dynamic, cytoplasmic triacylglycerol pool in enterocytes revealed by ex vivo and in vivo coherent anti-Stokes Raman scattering imaging. J. Lipid Res. 50: 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carten J. D., Bradford M. K., Farber S. A. 2011. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev. Biol. 360: 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gabbi C., Warner M., Gustafsson J-Å. 2014. Action mechanisms of liver X receptors. Biochem. Biophys. Res. Commun. 446: 647–650. [DOI] [PubMed] [Google Scholar]
- 10.Calkin A. C., Tontonoz P. 2012. Transcriptional integration of metabolism by the nuclear sterol-activated receptors LXR and FXR. Nat. Rev. Mol. Cell Biol. 13: 213–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14: 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grefhorst A., Elzinga B. M., Voshol P. J., Plosch T., Kok T., Bloks V. W., van der Sluijs F. H., Havekes L. M., Romijn J. A., Verkade H. J., et al. 2002. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J. Biol. Chem. 277: 34182–34190. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y., Breevoort S. R., Angdisen J., Fu M., Schmidt D. R., Holmstrom S. R., Kliewer S. A., Mangelsdorf D. J., Schulman I. G. 2012. Liver LXRalpha expression is crucial for whole body cholesterol homeostasis and reverse cholesterol transport in mice. J. Clin. Invest. 122: 1688–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed M. H., Barakat S., Almobarak A. O. 2012. Nonalcoholic fatty liver disease and cardiovascular disease: has the time come for cardiologists to be hepatologists? J. Obes. 2012: 483135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seth A., Stemple D. L., Barroso I. 2013. The emerging use of zebrafish to model metabolic disease. Dis. Model. Mech. 6: 1080–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang L., Green S. R., Baek J. S., Lee S-H., Ellett F., Deer E., Lieschke G. J., Witztum J. L., Tsimikas S., Miller Y. I. 2011. In vivo visualization and attenuation of oxidized lipid accumulation in hypercholesterolemic zebrafish. J. Clin. Invest. 121: 4861–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang L., Harkewicz R., Hartvigsen K., Wiesner P., Choi S-H., Almazan F., Pattison J., Deer E., Sayaphupha T., Dennis E. A., et al. 2010. Oxidized cholesteryl esters and phospholipids in zebrafish larvae fed a high cholesterol diet. J. Biol. Chem. 285: 32343–32351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoletov K., Fang L., Choi S-H., Hartvigsen K., Hansen L. F., Hall C., Pattison J., Juliano J., Miller E. R., Almazan F., et al. 2009. Vascular lipid accumulation, lipoprotein oxidation, and macrophage lipid uptake in hypercholesterolemic zebrafish. Circ. Res. 104: 952–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin W., Carballo-Jane E., McLaren D. G., Mendoza V. H., Gagen K., Geoghagen N. S., McNamara L. A., Gorski J. N., Eiermann G. J., Petrov A., et al. 2012. Plasma lipid profiling across species for the identification of optimal animal models of human dyslipidemia. J. Lipid Res. 53: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlegel A., Stainier D. Y. 2006. Microsomal triglyceride transfer protein is required for yolk lipid utilization and absorption of dietary lipids in zebrafish larvae. Biochemistry. 45: 15179–15187. [DOI] [PubMed] [Google Scholar]
- 22.Hugo S. E., Cruz-Garcia L., Karanth S., Anderson R. M., Stainier D. Y., Schlegel A. 2012. A monocarboxylate transporter required for hepatocyte secretion of ketone bodies during fasting. Genes Dev. 26: 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karanth S., Tran V. M., Balagurunathan K., Schlegel A. 2013. Polyunsaturated fatty acyl-coenzyme As are inhibitors of cholesterol biosynthesis. Dis. Model. Mech. 6: 1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Archer A., Lauter G., Hauptmann G., Mode A., Gustafsson J-Å. 2008. Transcriptional activity and developmental expression of liver X receptor (lxr) in zebrafish. Dev. Dyn. 237: 1090–1098. [DOI] [PubMed] [Google Scholar]
- 25.Cruz-Garcia L., Minghetti M., Navarro I., Tocher D. R. 2009. Molecular cloning, tissue expression and regulation of liver X receptor (LXR) transcription factors of Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss). Comp. Biochem. Physiol. B. 153: 81–88. [DOI] [PubMed] [Google Scholar]
- 26.Bertrand S., Thisse B., Tavares R., Sachs L., Chaumot A., Bardet P-L., Escrivà H., Duffraisse M., Marchand O., Safi R., et al. 2007. Unexpected novel relational links uncovered by extensive developmental profiling of nuclear receptor expression. PLoS Genet. 3: e188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., Schuster G. U., Hultenby K., Zhang Q., Andersson S., Gustafsson J. A. 2002. Liver X receptors in the central nervous system: from lipid homeostasis to neuronal degeneration. Proc. Natl. Acad. Sci. USA. 99: 13878–13883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poppelreuther M., Rudolph B., Du C., Großmann R., Becker M., Thiele C., Ehehalt R., Füllekrug J. 2012. The N-terminal region of acyl-CoA synthetase 3 is essential for both the localization on lipid droplets and the function in fatty acid uptake. J. Lipid Res. 53: 888–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kassan A., Herms A., Fernandez-Vidal A., Bosch M., Schieber N. L., Reddy B. J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203: 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., et al. 2007. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 48: 1280–1292. [DOI] [PubMed] [Google Scholar]
- 31.Dahlem T. J., Hoshijima K., Jurynec M. J., Gunther D., Starker C. G., Locke A. S., Weis A. M., Voytas D. F., Grunwald D. J. 2012. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8: e1002861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parant J. M., George S. A., Pryor R., Wittwer C. T., Yost H. J. 2009. A rapid and efficient method of genotyping zebrafish mutants. Dev. Dyn. 238: 3168–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwan K. M., Fujimoto E., Grabher C., Mangum B. D., Hardy M. E., Campbell D. S., Parant J. M., Yost H. J., Kanki J. P., Chien C-B. 2007. The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev. Dyn. 236: 3088–3099. [DOI] [PubMed] [Google Scholar]
- 34.Her G. M., Yeh Y-H., Wu J-L. 2004. Functional conserved elements mediate intestinal-type fatty acid binding protein (I-FABP) expression in the gut epithelia of zebrafish larvae. Dev. Dyn. 230: 734–742. [DOI] [PubMed] [Google Scholar]
- 35.Siccardi A. J., III, Garris H. W., Jones W. T., Moseley D. B., D’Abramo L. R., Watts S. A. 2009. Growth and survival of zebrafish (Danio rerio) fed different commercial and laboratory diets. Zebrafish. 6: 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Collymore C., Rasmussen S., Tolwani R. J. 2013. Gavaging adult zebrafish. J. Vis. Exp. 78: e50691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Folch J., Lees M., Sloane Stanley G. H. 1957. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 38.Luchoomun J., Hussain M. M. 1999. Assembly and secretion of chylomicrons by differentiated Caco-2 cells: nascent triglycerides and preformed phospholipids are preferentially used for lipoprotein assembly. J. Biol. Chem. 274: 19565–19572. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl M. W. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandoval A., Fraisl P., Arias-Barrau E., DiRusso C. C., Singer D., Sealls W., Black P. N. 2008. Fatty acid transport and activation and the expression patterns of genes involved in fatty acid trafficking. Arch. Biochem. Biophys. 477: 363–371. [DOI] [PubMed] [Google Scholar]
- 41.Dong B., Kan C. F. K., Singh A. B., Liu J. 2013. High-fructose diet downregulates long-chain acyl-CoA synthetase 3 expression in liver of hamsters via impairing LXR/RXR signaling pathway. J. Lipid Res. 54: 1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirschey M. D., Shimazu T., Goetzman E., Jing E., Schwer B., Lombard D. B., Grueter C. A., Harris C., Biddinger S., Ilkayeva O. R., et al. 2010. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 464: 121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Field H. A., Kelley K. A., Martell L., Goldstein A. M., Serluca F. C. 2009. Analysis of gastrointestinal physiology using a novel intestinal transit assay in zebrafish. Neurogastroenterol. Motil. 21: 304–312. [DOI] [PubMed] [Google Scholar]
- 44.Athar H., Iqbal J., Jiang X. C., Hussain M. M. 2004. A simple, rapid, and sensitive fluorescence assay for microsomal triglyceride transfer protein. J. Lipid Res. 45: 764–772. [DOI] [PubMed] [Google Scholar]
- 45.Soengas J. L., Moon T. W. 1998. Transport and metabolism of glucose in isolated enterocytes of the black bullhead Ictalurus melas: effects of diet and hormones. J. Exp. Biol. 201: 3263–3273. [DOI] [PubMed] [Google Scholar]
- 46.Htun H., Barsony J., Renyi I., Gould D. L., Hager G. L. 1996. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc. Natl. Acad. Sci. USA. 93: 4845–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans R. M., Mangelsdorf D. J. 2014. Nuclear receptors, RXR, and the Big Bang. Cell. 157: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Iqbal J., Hussain M. M. 2009. Intestinal lipid absorption. Am. J. Physiol. Endocrinol. Metab. 296: E1183–E1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fisher E. A. 2012. The degradation of apolipoprotein B100: multiple opportunities to regulate VLDL triglyceride production by different proteolytic pathways. Biochim. Biophys. Acta. 1821: 778–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abumrad N. A., Davidson N. O. 2012. Role of the gut in lipid homeostasis. Physiol. Rev. 92: 1061–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kampf J. P., Kleinfeld A. M. 2007. Is membrane transport of FFA mediated by lipid, protein, or both? Physiology (Bethesda). 22: 7–14. [DOI] [PubMed] [Google Scholar]
- 52.Kalaany N. Y., Gauthier K. C., Zavacki A. M., Mammen P. P., Kitazume T., Peterson J. A., Horton J. D., Garry D. J., Bianco A. C., Mangelsdorf D. J. 2005. LXRs regulate the balance between fat storage and oxidation. Cell Metab. 1: 231–244. [DOI] [PubMed] [Google Scholar]
- 53.Soupene E., Fyrst H., Kuypers F. A. 2008. Mammalian acyl-CoA:lysophosphatidylcholine acyltransferase enzymes. Proc. Natl. Acad. Sci. USA. 105: 88–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soupene E., Kuypers F. A. 2008. Mammalian long-chain acyl-CoA synthetases. Exp. Biol. Med. (Maywood). 233: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ellis J. M., Frahm J. L., Li L. O., Coleman R. A. 2010. Acyl-coenzyme A synthetases in metabolic control. Curr. Opin. Lipidol. 21: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mashek D. G., Li L. O., Coleman R. A. 2007. Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol. 2: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Füllekrug J., Ehehalt R., Poppelreuther M. 2012. Outlook: membrane junctions enable the metabolic trapping of fatty acids by intracellular acyl-CoA synthetases. Front. Physiol. 3: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mashek D. G., Li L. O., Coleman R. A. 2006. Rat long-chain acyl-CoA synthetase mRNA, protein, and activity vary in tissue distribution and in response to diet. J. Lipid Res. 47: 2004–2010. [DOI] [PubMed] [Google Scholar]
- 59.Beaven S. W., Matveyenko A., Wroblewski K., Chao L., Wilpitz D., Hsu T. W., Lentz J., Drew B., Hevener A. L., Tontonoz P. 2013. Reciprocal regulation of hepatic and adipose lipogenesis by liver X receptors in obesity and insulin resistance. Cell Metab. 18: 106–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Anderson S. P., Dunn C., Laughter A., Yoon L., Swanson C., Stulnig T. M., Steffensen K. R., Chandraratna R. A., Gustafsson J. A., Corton J. C. 2004. Overlapping transcriptional programs regulated by the nuclear receptors peroxisome proliferator-activated receptor alpha, retinoid X receptor, and liver X receptor in mouse liver. Mol. Pharmacol. 66: 1440–1452. [DOI] [PubMed] [Google Scholar]
- 61.Little A. G., Seebacher F. 2014. Thyroid hormone regulates cardiac performance during cold acclimation in zebrafish (Danio rerio). J. Exp. Biol. 217: 718–725. [DOI] [PubMed] [Google Scholar]
- 62.Little A. G., Seebacher F. 2013. Thyroid hormone regulates muscle function during cold acclimation in zebrafish (Danio rerio). J. Exp. Biol. 216: 3514–3521. [DOI] [PubMed] [Google Scholar]
- 63.Little A. G., Kunisue T., Kannan K., Seebacher F. 2013. Thyroid hormone actions are temperature-specific and regulate thermal acclimation in zebrafish (Danio rerio). BMC Biol. 11: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.