Abstract
Ricinoleic acid (12-hydroxyoctadec-cis-9-enoic acid) has many specialized uses in bioproduct industries, while castor bean is currently the only commercial source for the fatty acid. This report describes metabolic engineering of a microbial system (Pichia pastoris) to produce ricinoleic acid using a “push” (synthesis) and “pull” (assembly) strategy. CpFAH, a fatty acid hydroxylase from Claviceps purpurea, was used for synthesis of ricinoleic acid, and CpDGAT1, a diacylglycerol acyl transferase for the triacylglycerol synthesis from the same species, was used for assembly of the fatty acid. Coexpression of CpFAH and CpDGAT1 produced higher lipid contents and ricinoleic acid levels than expression of CpFAH alone. Coexpression in a mutant haploid strain defective in the Δ12 desaturase activity resulted in a higher level of ricinoleic acid than that in the diploid strain. Intriguingly, the ricinoleic acid produced was mainly distributed in the neutral lipid fractions, particularly the free fatty acid form, but with little in the polar lipids. This work demonstrates the effectiveness of the metabolic engineering strategy and excellent capacity of the microbial system for production of ricinoleic acid as an alternative to plant sources for industrial uses.
Keywords: Claviceps purpurea , fatty acid hydroxylase, diacylglycerol acyltransferase
Hydroxy fatty acids are important raw materials with a wide range of industrial uses. In particular, ricinoleic acid (12-hydroxyoctadec-cis-9-enoic acid), a long-chain hydroxy fatty acid produced by castor bean (Ricinus communis), has many specialized uses in the manufacturing of a variety of industrial products such as nylons, lubricants, ink, and paints, as well as pharmaceuticals and cosmetics (1, 2). However, due to the presence of a highly potent toxin (ricin), this native oilseed plant is not considered an ideal source for hydroxy fatty acid production. Therefore, tremendous effort has recently been made in identifying genes involved in the biosynthesis of this fatty acid and using the genes to engineer oilseed crops for producing ricinoleic acid (3–7). However, to date these attempts have met with only partial success. When a hydroxylase gene from a native plant species was introduced into oilseed crops, the hydroxy fatty acid content in transgenic seeds rarely exceeded 20% of the total fatty acids (8–10). In 2008, a new hydroxylase for the biosynthesis of ricinoleic acid was identified from a nonplant origin, and expression of this gene in Arabidopsis resulted in accumulation of a slightly higher level of hydroxy fatty acids in transgenic seeds (11). Very recently, coexpression of genes encoding factors involved in the networks of the hydroxylation or acyl trafficking process along with a hydroxylase has also been attempted (4–7). Although the improved production of hydroxy fatty acids is observed, the amount in transgenic seeds still rarely exceeds 25% of the total fatty acids. It appears that selection of genes from diverse sources and addition of cofactors can make a difference for transgenic production of hydroxyl fatty acids, but production of a single hydroxy fatty acid at the commercially viable level in plants still remains as a challenging task.
Microorganisms have recently emerged as promising systems for producing biofuel for transportation and platform chemicals for biopolymers because some microbes offer high output of biomass with good oil content in a short period of time and are easily used for genetic manipulation to implement a metabolic engineering strategy. Exploitation of metabolic engineering of lipid pathways in microorganisms based on fatty acid biosynthesis and assembly frameworks provides a promising solution to bioproducts from microorganisms to replace petrochemicals, as fatty acids and derivatives share many similar chemical properties with petroleum fuels and petrochemicals such as reduced state of hydrocarbons and high density of energy (12–16). Therefore, metabolic engineering of microbial systems has potential in providing an alternative to plant sources for hydroxy fatty acid for industrial uses (17, 18).
This study describes metabolic engineering of ricinoleic acid in Pichia pastoris, a yeast with a good yield of biomass and oil. Two genes encoding CpFAH, a fatty acid hydroxylase we cloned previously from the fungus Claviceps purpurea (11), and CpDGAT1, a diacylglycerol acyltransferase for the triacylglycerol (TAG) biosynthesis newly cloned from the same species, were coexpressed both in a diploid wild-type strain and in a haploid mutant strain, which resulted in the production of a high amount of ricinoleic acid mainly accumulated in the free fatty acid fraction, followed by the TAG fraction, with very little in the polar lipids. This work highlights the effectiveness of the metabolic engineering strategy and excellent capacity of the yeast for hydroxy fatty acid production. Moreover, it may also suggest a new strategy to produce a higher level of unusual fatty acids in transgenic plants through free fatty acids as intermediate pathways.
MATERIALS AND METHODS
Organisms and culture conditions
C. purpurea was grown and maintained on the potato dextrose agar medium at room temperature. Saccharomyces cerevisiae quadruple mutant (H1246; Matα dga1Δ, lroΔ, are1Δ, are2Δ) (19) used as a heterologous host to study the function of CpDGAT1 was grown on yeast extract-peptone-dextrose medium. Pichia X33 and GS115 (Invitrogen) were used to study the heterologous expression of CpFAH and CpDGAT1. The haploid P. pastoris GS115 (Invitrogen) was used as a host strain for disruption of Δ12 desaturase gene (des12). The des12-Δ::HIS4 disruption cassette was made by cloning 917 bp of 5′ untranslated region and 706 bp of coding region of Δ12 desaturase from P. pastoris GS115 into the upstream and downstream of HIS4 marker, respectively, in pPIC3.5 vector. The Δ12 deletion strain (D12-KO/GS115) was made by introducing the des12-Δ::HIS4 disruption cassette by homologous recombination method. The mutant strain was confirmed by PCR and the inability to produce linoleic acid.
Cloning of full-length cDNA CpDGAT1 from C. purpurea
Total RNA from C. purpurea was isolated using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Genomic DNA contamination was eliminated by on-column DNase I digestion with RNase-free DNase (Qiagen). Poly(A)+ RNA was extracted from the total RNA by Dynabeads mRNA Purification Kit (Invitrogen). Approximately 0.5 μg of Poly(A)+ RNA was used as a template to synthesize first strand for both 5′- and 3′-cDNA end amplification using the Smarter RACE cDNA amplification kit (Clontech). Primers DM268 and DM269 were used to amplify the 5′-end region, and primer DM267 was used for the 3′-end region. The full-length cDNA of CpDGAT1 was amplified by Q5 DNA Polymerase (New England BioLabs) using primers DM303 and DM304. Both primers containing start and stop codons, respectively, were designed, and EcoRI restriction sites added to facilitate subsequent manipulation. Primers used in this study are listed in supplementary Table 1. All the amplified products were directly digested and subcloned into EcoRI sites of the yeast expression vector pYES2.0 under control of Gal1 promoter to generate plasmid, CpDGAT1/pYES2.0. The corrected orientation was confirmed by digestion and sequencing.
Construction of Pichia expression plasmids of CpFAH and/or CpDGAT1
Both open reading frames (ORFs) of CpFAH and CpDGAT1 were digested from pDM16 (11) and CpDGAT1/pYES2.0 by EcoRI, respectively, and ligated into pPICZ-B (Invitrogen) digested with the same restriction enzyme to generate plasmids CpFAH/pPICZ-B and CpDGAT1/pPICZ-B respectively. To coexpress both CpFAH and CpDGAT1, a whole cassette of CpFAH including AOX1 promoter and terminator was digested with BglII and cloned by blunt end ligation into CpDGAT1/pPICZ-B digested with BamHI.
Yeast expression studies in S. cerevisiae and P. pastoris
The CpDGAT1/pYES2.0 was transformed into S. cerevisiae H1246 using the S.C. EasyComp transformation kit (Invitrogen). For expression studies, yeast transformed with either CpDGAT1 or the empty vector pYES2.0 were grown at 28°C for 1 day in synthetic dropout medium containing 0.17% (w/v) yeast nitrogen base, 0.5% ammonium sulfate, 2% (w/v) dextrose, and 0.06% (w/v) dropout supplement lacking uracil. The overnight cultures were diluted to an optical density (OD)600 of 0.4 in a 10 ml induction medium containing 2% galactose and continued to grow at the same temperature overnight. The cells were then collected for lipid analysis.
For Pichia transformation, 10 μg of linearized plasmid containing one gene construct or 50 μg of uncut plasmid containing two genes constructs was transformed into Pichia by electroporation using an electroporator 2510 (Eppendorf). The transformed cells were selected on selective medium containing either antibiotic (zeocin or hygromycin) or dropout medium. For expression study, the transformed cells were grown at 30°C with 240 rpm in buffered minimal glycerol medium containing 100 mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, and 1% glycerol. After 24 h, the cells were centrifuged and resuspended to an OD600 of 1.0 in induction medium containing 100 mM potassium phosphate, pH 6.0, 1.34% yeast nitrogen base without amino acids, 4 × 10−5% biotin, and 0.5% methanol. The cells were continued to grow at 20°C with 240 rpm. Every 24 h, cells were collected for lipid and total fatty acids analysis, and then 0.5% methanol was added to the culture to maintain induction.
Lipid analysis
Total lipids were extracted from yeast with chloroform-methanol (2:1, v/v) (20). Total lipids were resolved in TLC on silica gel plates (silica gel 60, 20 × 20 cm; EMD Chemicals, Germany) with a solvent system containing hexane-anhydrous ethyl ether-formic acid (70:30:1, v/v). Lipid classes were visualized by spraying with 0.005% primuline in 80% acetone and observing under UV.
Fatty acid analysis
Three different methods were used to synthesize fatty acid methyl esters (FAMEs). In the first method, to analyze total fatty acids including free fatty acid and O-acyl lipids in the cells, acid-catalyzed esterification and transesterification were used as previously described using 1% H2SO4/methanol (11). The samples were heated at 80°C for 2 h. In the second method, esterified fatty acids including glycerolipids and fatty acyl-CoA were transmethylated by base-catalyzed transesterification using 5% (w/v) sodium methoxide (CH3ONa) (Sigma-Aldrich) at room temperature for 30 min (21). For the third method, diazomethane was used as to react with unesterified fatty acids (free fatty acids) in the presence of a little methanol, which catalyzes the reaction to form FAMEs (22). The FAMEs were trimethylsilylated by adding 50 μl of N,O-bis(TMS)-acetamide/pyridine (1:1) and heating at 80°C for 30 min (11). Two microliter samples of total FAME-TMS derivatives were analyzed on an Agilent 6890N gas chromatograph equipped with a DB-23 column (30 m × 0.25 mm) with 0.25 µm film thickness (J and W Scientific). The column temperature was maintained at 160°C for 1 min and then raised to 240°C at a rate of 4°C/min. For mass spectrometry analysis, the mass selective detector was run under standard electron impact conditions (70 eV), scanning an effective m/z range of 40–700 at 2.26 scans/s.
Oil content measurement
Yeast oil content was determined by GC analysis using tripentadecanoin (15:0-TAG) as internal standard (25 μg), which was added to the sample before FAME transmethylation. The 15:0 free fatty acid (5 μg) was used as internal standard for quantifying free fatty acids.
RESULTS
Identification and characterization of CpDGAT1 gene from C. purpurea
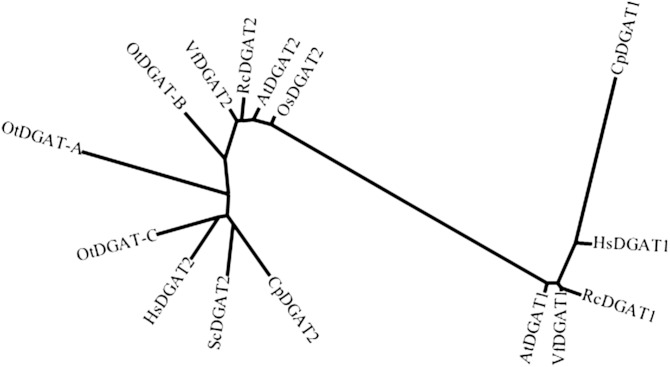
Diacylglycerol acyltransferase (DGAT) catalyzing the committed step of the TAG biosynthesis has been proved to play a critical role in accumulation of unusual fatty acids (4). To clone this gene from C. purpurea, an expressed sequence tag database from the sclerotia was searched using Arabidopsis DGAT1 as a query, which resulted in identification of a partial cDNA sequence of CpDGAT1. The missing 5′ and 3′ cDNA ends were then retrieved using a rapid amplification of cDNA ends method with a combination of primary and nested primers. The full-length ORF of CpDGAT1 showed 1,521 bp encoding a polypeptide of 506 amino acids with a predicted molecular mass of 57.7 kDa. CpDGAT1 belonged to the type I DGAT family, a member of the membrane-bound O-acyl transferase superfamily, which is widely distributed in eukaryotes such as plants and animals as well as fungi (23). The amino acid identity of CpDGAT1 to CpDGAT2, a type II DGAT previously identified from the same species (24), was very low at ∼8% amino acid identity. Phylogenetic analysis of CpDGAT1 with related DGATs from R. communis (RcDGAT1, RcDGAT2), Vernicia fordii (VfDGAT1, VfDGAT2), Arabidopsis thaliana (AtDGAT1), C. purpurea (CpDGAT2), Homo sapiens (HsDGAT1, HsDGAT2), S. cerevisiae (ScDGAT2), Ostreococcus tauri (OtDGAT2A, OtDGAT2B, OtDGAT2C), and Oryza sativa (OsDGAT2) showed that CpDGAT1 from C. purpurea was clustered with a group of other type I DGATs from plants, animals, and fungi, but not with any type II DGATs, including CpDGAT2 from the same species (Fig. 1), indicating that the two DGATs from C. purpurea evolved independently and might have distinct catalytic properties.
Fig. 1.
Phylogenetic analysis of CpDGAT1 and related DGATs from H. sapiens (HsDGAT1, accession #NP_036211), R. communis (RcDGAT1, accession #AY366496), V. fordii (VfDGAT1, accession #ABC94471), A. thaliana (AtDGAT1, accession #NP_179535), C. purpurea (CpDGAT2, accession #ADF29677), S. cerevisiae (ScDGAT2, accession #NP_014888), H. sapiens (HsDGAT2, accession #NP_940914), O. tauri (OtDGAT2A, accession #CAL54993; OtDGAT2B, accession #CAL58088; OtDGAT2C, accession #CAL56438), V. fordii (VfDGAT2, accession #ABC94473), R. communis (RcDGAT2, accession #AAY16324), A. thaliana (AtDGAT1, accession #Q9ASU1), O. sativa (OsDGAT2, accession #NP_001057530).
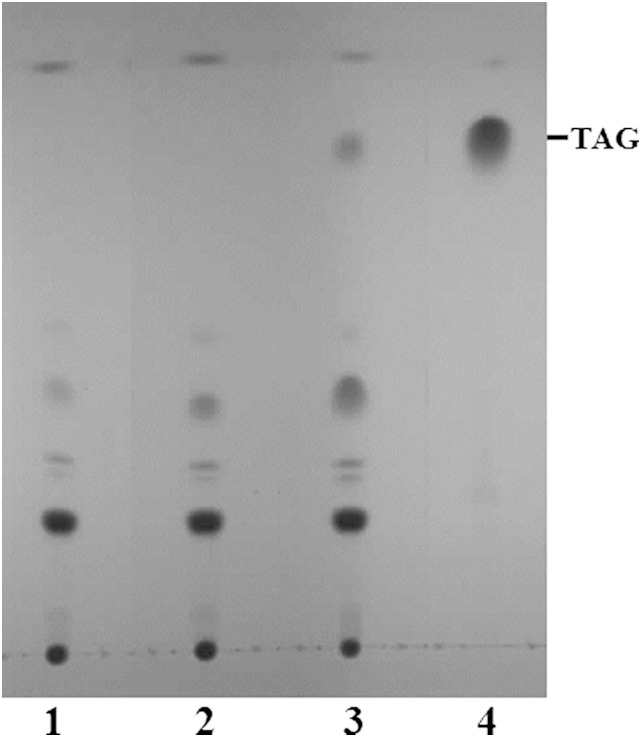
To assess DGAT activity of CpDGAT1, the full-length cDNA was cloned and heterologously expressed in a S. cerevisiae quadruple mutant defective in the synthesis of both TAGs and steryl esters (19), as a similar mutant of P. pastoris was not available for the function analysis. The mutant yeast transformed with the empty vector and the vector with CpDGAT2 was used as a negative and a positive control, respectively. The neutral lipids from transformants expressing the gene were extracted and analyzed by TLC. As shown in Fig. 2, transformant cells expressing CpDGAT1 produced TAGs, while the negative control did not produce any. Comparison of the two DGATs from C. purpurea showed that CpDGAT1 could produce an amount of TAGs substantially higher than CpDGAT2. These results indicate that the CpDGAT1 gene encodes a functional DGAT involved in the biosynthesis of TAGs and that the activity was much higher than that of CpDGAT2 in the yeast.
Fig. 2.
Expression of CpDGAT1 in the yeast S. cerevisae quadruple mutant strain H1246. TLC analysis of neutral lipids from yeast H1246 transformed with empty vector, pYES2.0 (1), CpDGAT2 (2), or CpDGAT1 (3). Solvent system: hexane-diethyl ether-acetic acid (70:30:1). Lipids were visualized by iodine staining. TAG standard (4).
Production of ricinoleic acid in a diploid yeast by coexpresssing CpFAH and CpDGAT1
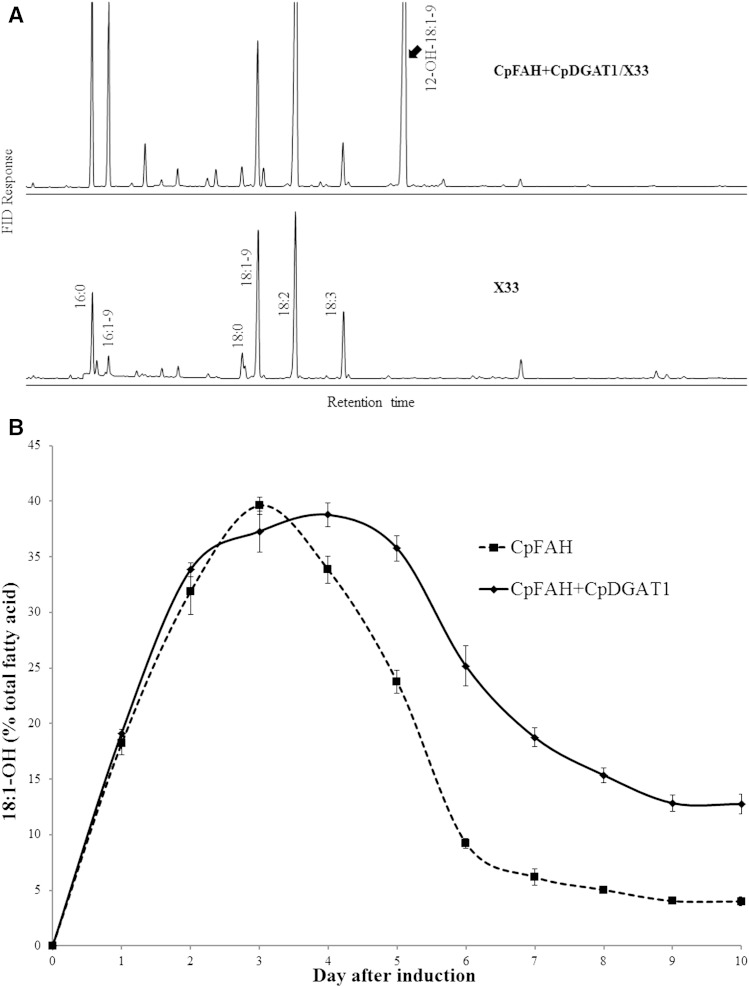
Our initial strategy to produce ricinoleic acid heterologously in Pichia was to coexpress two genes encoding CpFAH and CpDGAT1 involved in both biosynthesis and assembly of the fatty acid. This strategy has proved effective to produce specialty fatty acids heterologously (4, 6, 25). The constructs with one or two gene cassettes of CpFAH and CpDGAT1 were built under the control of an inducible promoter of the yeast expression vector and then introduced into a wild-type diploid yeast P. pastoris strain X33. Through homologous recombination, the gene cassettes were integrated into the yeast chromosome. After confirmation of the stable integration, the strains were induced in a time course for production of ricinoleic acid. Under the standard induction condition, the diploid strains hosting the empty vector, CpFAH, and CpFAH+CpDGAT1 showed similar growth rates (supplementary Fig. 1). As expected, upon induction, both CpFAH/X33 (CpFAH alone) and CpFAH+CpDGAT1/X33 (coexpression) produced a new fatty acid compared with the control, which was identified as ricinoleic acid (Fig. 3A). During a 10-day time course, the amount of ricinoleic acid produced in the transformants increased dramatically from zero after the initial induction of expression, reached the maximum at about the early middle time point, and then decreased substantially. However, compared with CpFAH alone, the coexpression (CpFAH and CpDGAT1) produced a higher amount of ricinoleic acid at almost all time points in the time course except at day 3 when CpFAH alone produced the maximal level of ricinoleic acid (∼40%), which was slightly higher than the coexpression. After that, the amount of ricinoleic acid in CpFAH alone decreased dramatically, and at day 7, it was at ∼6%. At the end of the time course (day 10), the amount of ricinoleic acid remained only at ∼4% of the total fatty acids. In contrast, the amount of ricinoleic acid produced in the coexpression reached the highest level at day 4 at ∼39% of the total fatty acids. After that, the amount of ricinoleic acid decreased, but less dramatically, and at day 7 of the induction, it was at ∼19%. At day 10, the amount of ricinoleic acid remained at ∼13% of the total fatty acids, which was about three times higher than that of CpFAH alone at the same day (Fig. 3B).
Fig. 3.
A: GC analysis of TMS derivatives of the total fatty acids prepared from the diploid P. pastoris strain X33 expressing CpFAH and CpDGAT1 (CpFAH+CpDGAT1/X33) and the wild-type control (X33). B: Production of ricinoleic acid in the diploid strain during a time course of the induction. The solid line is for CpFAH+CpDGAT1/X33, and the dashed line is for CpFAH /X33. Results were calculated from the peak area of FAMEs. Values are means of three replicates with SD. 18:1-OH, ricinoleic acid.
Production of ricinoleic acid in a haploid yeast strain defective in Δ12 desaturase activity
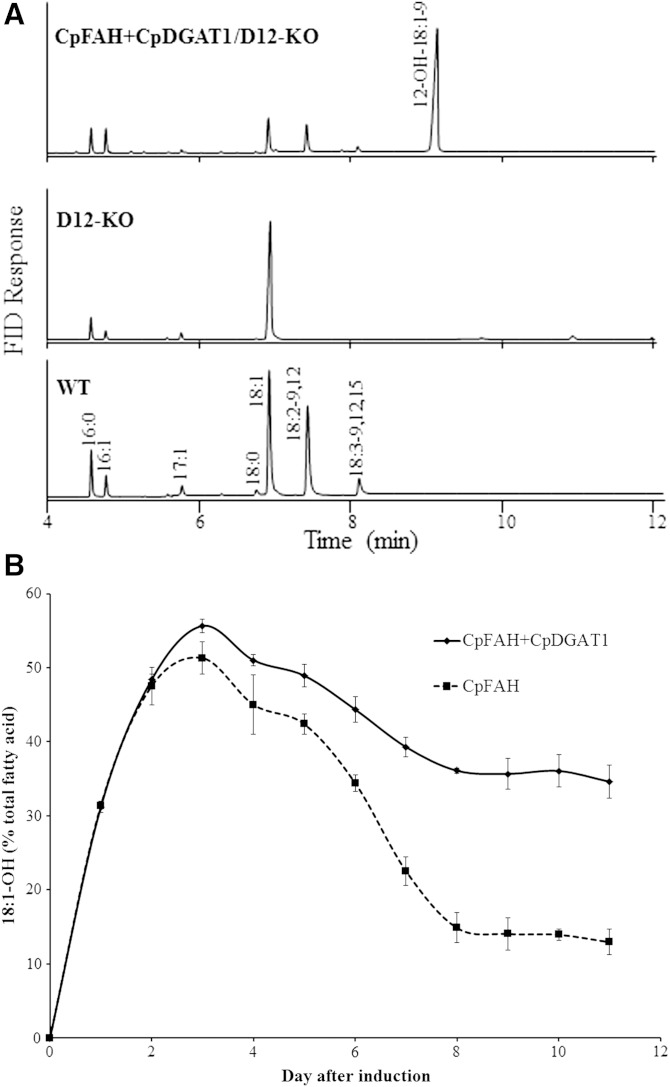
As seen in Fig. 3, although the diploid transformant expressing CpFAH and CpDGAT1 produced a substantial amount of ricinoleic acid, linoleic acid was still one of the major fatty acids in the cell due to a highly active endogenous Δ12 desaturase in P. pastoris. Pichia Δ12 desaturase is a membrane-associated enzyme catalyzing the synthesis of linoleic acid using oleic acid as substrate. As oleic acid is also the substrate for CpFAH to produce ricinoleic acid, competition for the same substrate by the two enzymes would diminish efficiency of the hydroxy fatty acid production in the transformant. To ease the competition and boost the substrate level for the hydroxylation, the endogenous Pichia Δ12 desaturase gene was disrupted by homologous recombination in a haploid P. pastoris strain GS115. The disruption (D12-KO) was confirmed by PCR amplification using sets of specific primers, as well as fatty acid composition in the disrupted strain where both linoleic acid and linolenic acid were absent (Table 1). This mutant strain was then used as a host to express CpFAH or CpFAH and CpDGAT1. Under the standard induction condition, all the haploid strains showed similar growth rates (supplementary Fig. 2). As seen in gas chromatograms and compositions of the fatty acids in the transformant cells (Fig. 4A, Table 1), the haploid wild-type strain (GS115) produced two dominant fatty acids, oleic acid and linoleic acid, while linolenic, palmitic, and palmitoleic acids were relatively less abundant forms. On the other hand, the Δ12 desaturase-knockout strain (D12-KO) produced only a single dominant fatty acid, oleic acid (>70%), and linoleic acid and linolenic acid were completely lost compared with the wild-type strain. In the knockout strain expressing CpFAH (CpFAH/D12-KO) or CpFAH and CpDGAT1 (CpFAH+CpDGAT1/D12-KO), ricinoleic acid was produced as a dominant fatty acid with oleic acid, palmitic acid, and palmitoleic acid being relatively less abundant at a high production point in the time course. It was noted that linoleic acid and linolenic acid were also produced in the transformants. Production of linoleic acid was due to the minor Δ12 desaturase activity of CpFAH, while a small amount of linolenic acid resulted from a combination of the Δ12 desaturase activity of CpFAH and an endogenous Δ15 desaturase activity in Pichia. Similar to the production trend of ricinoleic acid in the diploid cells, the hydroxyl fatty acid production in the haploid cells increased dramatically after the initial induction of expression, reached a peak, and then decreased substantially in the time course. The amount of ricinoleic acid produced in CpFAH alone was lower at all the time points of the time course compared with the coexpression. The maximum of ricinoleic acid in CpFAH alone was reached at day 3 at ∼51%; after that, it decreased dramatically and remained at a steady level of ∼13% at day 8. However, in the coexpression of CpFAH and CpDGAT1, the hydroxy fatty acid level was the highest at day 3 at ∼56% of the total fatty acids. After that, it decreased only gradually and remained at the steady level of ∼36% at day 8 (Fig. 4B), which was more than two times that in CpFAH alone at the same day. At the highest production time point (day 3), the total lipid content of the coexpression was measured at ∼495 μg/ml of ricinoleic acid, while the lipid content of CpFAH alone was ∼391 μg/ml and the control was 282 μg/ml (Table 2). The lipid content in the coexpression strain increased by ∼27% relative to CpFAH alone and nearly 76% compared with the control strain. At day 8, when the hydroxy fatty acid remained at the steady level in the transformants, the lipid content in the coexpression was increased by ∼147% and 20%, respectively, compared with CpFAH alone and the control.
TABLE 1.
Fatty acid compositions of Pichia GS115 (wild type), D12-KO (defective in the Δ12 desaturase), CpFAH/D12-KO (CpFAH alone), and CpFAH+CpDGAT1/D12-KO (coexpression) at day 3 of induction
| Fatty Acid | GS115 | D12-KO | CpFAH/D12-KO | CpFAH+CpDGAT1/D12-KO |
| 16:0 | 14.38 ± 0.35 | 10.52 ± 0.10 | 7.7 ± 0.11 | 7.5 ± 0.50 |
| 16:1 | 4.33 ± 0.19 | 4.75 ± 0.36 | 7.6 ± 0.49 | 6.9 ± 0.11 |
| 17:0 | 0 ± 0.00 | 0.56 ± 0.06 | 0.5 ± 0.06 | 0.4 ± 0.06 |
| 17:1 | 0 ± 0.00 | 3.52 ± 0.04 | 1.3 ± 0.13 | 1.5 ± 0.56 |
| 18:0 | 1.32 ± 0.11 | 1.57 ± 0.26 | 0.6 ± 0.11 | 0.8 ± 0.15 |
| 18:1–9 | 50.71 ± 1.56 | 77.85 ± 0.04 | 17.2 ± 1.55 | 14.1 ± 2.06 |
| 18:1–11 | 0 ± 0.00 | 1.23 ± 0.06 | 1.1 ± 0.12 | 0.9 ± 0.25 |
| 18:2 | 22.81 ± 1.07 | 0 ± 0.00 | 10.5 ± 0.06 | 10.2 ± 2.14 |
| 18:3 | 6.46 ± 0.30 | 0 ± 0.00 | 2.1 ± 0.31 | 2.0 ± 0.05 |
| 18:1-OH | 0 ± 0.00 | 0 ± 0.00 | 51.3 ± 2.18 | 55.7 ± 0.94 |
18:1-OH, ricinoleic acid. Results were calculated from the peak area of FAMEs. Values are means of three replicates with SD.
Fig. 4.
A: GC analysis TMS derivatives of the total fatty acids prepared from the haploid mutant P. pastoris strain expressing CpFAH and CpDGAT1. WT, wild-type GS115; D12-KO, GS115 defective in Δ12 desaturase activity; CpFAH+CpDGAT1/D12-KO, D12-KO expressing CpFAH and CpDGAT1. B: Production of ricinoleic acid in the haploid mutant strains during a time course of the induction. The solid line is for CpFAH+CpDGAT1/D12-KO, and the dashed line is for CpFAH /D12-KO. Results were calculated from the peak area of FAMEs. Values are means of three replicates with SD. 18:1-OH, ricinoleic acid.
TABLE 2.
Lipid contents in the haploid mutant strain expressing CpFAH alone (CpFAH/D12-KO) or CpFAH and CpDGAT1 (CpFAH+CpDGAT1/D12-KO) at days 3 and 8 after induction
| Lipid Content (μg/ml ± SD) | ||
| Constructs | Day 3 | Day 8 |
| WT | 282.78 ± 33.95 | 318.42 ± 1.86 |
| CpFAH | 391.73 ± 18.50 | 154.59 ± 13.08 |
| CpFAH+CpDGAT1 | 495.82 ± 26.22 | 381.71 ± 16.95 |
Results were calculated from the peak area of FAMEs. Values are means of four replicates with SD.
Distribution of ricinoleic acid in lipid classes of the coexpression strain
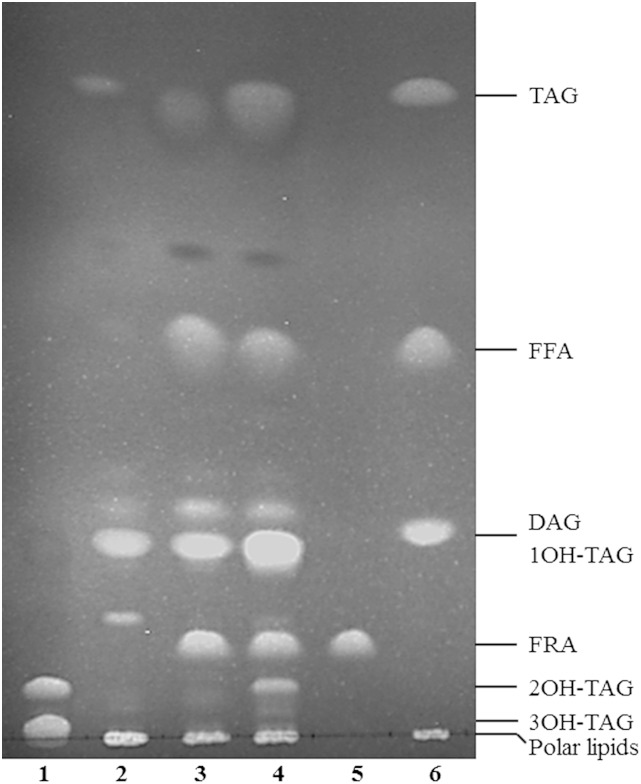
To look into distribution of ricinoleic acid in different lipid classes of the haploid mutant strain coexpressing CpFAH and CpDGAT1, the total lipids of the coexpression strain at day 3 after induction were analyzed by TLC, and fatty acid compositions of different lipid classes were determined. As shown in Fig. 5, compared with the mutant control (D12-KO), the mutant transformants expressing CpFAH alone (CpFAH/D12-KO) or CpFAH and CpDGAT1 (CpFAH+CpDGAT1/D12-KO) could produce two or three new types of neutral lipids with ricinoleic acid, one hydroxy fatty acid TAG (1OH-TAG), FRA, and two hydroxy fatty acid TAGs (2OH-TAG). Among them, 1OH-TAG comigrated with normal DAG in the TLC plate; thus, the proportion of ricinoleic acid in this lipid class could not be determined. 2OH-TAG comigrating with 2-OH-TAG of castor oil contained ∼66% of ricinoleic acid, which was consistent with the nature of TAGs with two ricinoleic acids. The free hydroxyl fatty acid band was large, which possessed ∼99% of total fatty acids as ricinoleic acid, consistent with a single fatty acid in the band (Fig. 5). It was noteworthy that the polar lipids localized in the origin of the TLC plate contained <1% of ricinoleic acid. In addition, no 2OH-TAG was produced in the transformant expressing CpFAH alone, but a substantial amount of 2OH-TAG could be observed in the transformant coexpressing CpFAH and CpDGAT1, which indicates the effectiveness of CpDGAT1 to pull ricinoleic acid to the storage TAGs.
Fig. 5.
TLC analysis of the total lipids extracted from the haploid mutant P. pastoris strain expressing CpFAH and CpDGAT1. 1, Castor oil; 2, D12-KO; 3, CpFAH/D12-KO; 4, CpFAH+CpDGAT1/D12-KO; 5, ricinoleic acid standard (FRA); 6, TLC standard. DAG, diacylglycerol; FRA, free ricinoleic acid.
To further confirm that a substantial amount of FRA remained in the haploid expression strains and the effect of CpDGAT1 in facilitating the flux of ricinoleic acid into storage glycerolipids, the total lipids from the transformant cells of both CpFAH alone and CpFAH+CpDGAT1 at days 3 and 8 in a separate time course were extracted and analyzed by three different trans-esterification methods. The acid-catalyzed esterification analyzed the total lipids, and the base-catalyzed esterification analyzed the esterified lipids, while the diazomethane derivatization analyzed the unesterified free fatty acids (22). As shown in Table 3, at both time points, the coexpression (CpFAH+CpDGAT1) had a higher proportion of ricinoleic acid in the esterified form than CpFAH alone. At day 3, a large majority of ricinoleic acid (>90%) was found in the unesterified form in CpFAH alone, and FRA in the coexpression accounted for ∼80% of the total lipids. At day 8, the proportion of FRA in the CpFAH alone remained at ∼60% of the total lipids, while the amount in the coexpression was ∼50% of the total lipids. The ratio of the amount of esterified ricinoleic acid in the coexpression versus in CpFAH alone at day 3 was about three, while the ratio increased as the time course progressed, and at day 8, the ratio of the amount of esterified ricinoleic acid in the two strains was increased to approximately five. This result indicates that the effect of CpDGAT1 for flux of ricinoleic acid to the storage lipids becomes stronger as the time course progresses.
TABLE 3.
Ricinoleic acid contents in different lipid classes of the haploid mutant strain expressing CpFAH alone (CpFAH/D12-KO) or CpDGAT1 (CpFAH+CpDGAT1/D12-KO) at days 3 and 8 after induction
| Ricinoleic Acid (μg/ml ± SD) | ||||
| Day 3 | Day 8 | |||
| Lipid Classes | CpFAH | CpFAH+CpDGAT1 | CpFAH | CpFAH+CpDGAT1 |
| Total lipids | 125.39 ± 7.28 | 171.44 ± 6.81 | 15.86 ± 1.88 | 69.46 ± 2.69 |
| FRA form | 116.37 ± 8.08 | 142.75 ± 2.24 | 10.55 ± 0.32 | 37.29 ± 3.28 |
| Esterified lipid form | 9.23 ± 1.20 | 31.66 ± 1.83 | 6.88 ± 1.06 | 34.97 ± 4.77 |
Results were calculated from the peak area of FAMEs. Values are means of three replicates with SD.
DISCUSSION
Although tremendous efforts have been made in metabolic engineering of ricinoleic acid in oilseed crops as a possible alternative for industrial use, a commercially viable level of this fatty acid has not been achieved in transgenic plants. In this study, we explored producing ricinoleic acid by coexpressing CpFAH and CpDGAT1 in a diploid wild-type strain and a haploid mutant strain of P. pastoris. In particular, the level of ricinoleic acid can reach ∼56% of the total fatty acids at day 3 after expression induction in the haploid mutant strain defective in the Δ12 desaturase activity. This result demonstrates the effectiveness of the metabolic strategy used for metabolic engineering of the hydroxy fatty acid in Pichia. Specifically, utilization of a highly active CpDGAT1 increases not only the total level of ricinoleic acid, but also the lipid content and the amount of esterified ricinoleic acid in the storage lipids. Interestingly, a similar level of hydroxyl fatty acid was also achieved very recently in other yeast species (17, 18). Together, these results highlight the capacity and potential of microbial systems for the production of hydroxyl fatty acid.
Pichia offers several advantages for producing hydrocarbon and fatty acids for bioproducts. It is amenable to genetic engineering, and abundant genetic tools are available for manipulating metabolic pathways. It is grown quickly with a simple medium and has long been used for producing heterologous enzymes and chemicals. In addition, it is well known that the fatty acid biosynthesis exhibits feedback inhibition by long-chain acyl-acyl carrier protein (ACP) and acyl-CoA, two common fatty acid intermediates for the metabolism (12–14). In this study, ricinoleic acid accumulated in Pichia is mainly distributed in free fatty acid form, which offers a benefit without the use of acyl-CoA/ACP thioesterases, a common strategy to relieve the feedback inhibition in the metabolic engineering of biofuels (14). All these advantages make Pichia a promising system for producing hydroxyl fatty acid as an alternative to plant sources for industrial uses.
When two heterologous systems (plant and Pichia) used for metabolic engineering of ricinoleic acid are compared, it appears that the latter is more effective than transgenic plants in producing hydroxyl fatty acid. It is generally believed that ricinoleic acid synthesized by oleate hydroxylase occurs on oleic acid esterified to the sn-2 position of the membrane phospholipids by adding a hydroxyl group to the carbon at the 12th position. The inefficient removal of the unusual fatty acid from phospholipids where it is synthesized (26, 27) and inefficient transfer of the unusual fatty acid from phospholipids to storage lipids are likely the main factors, or “bottlenecks,” limiting the production of this fatty acid in transgenic plants (28–30). As phospholipids are the main components of the membrane structure in eukaryotes, unusual fatty acid remaining in the membrane would severely harm the membrane unity and physiological properties. Heterologous expression of a fatty acid conjugase in Arabidopsis and soybean was found to accumulate a high level of unusual conjugated fatty acids in phosphatidylcholine in developing and mature seeds of transgenic plants (27). On the other hand, transgenic Pichia expressing the two genes accumulates ricinoleic acid mostly in free fatty acid form (>50% of the total), and the level of this fatty acid in phospholipids represents <1% of the total fatty acids. Such an accumulation pattern is surprising, as it is generally believed that free fatty acids are toxic to cells. In addition, the phenomenon has never been reported in transgenic plants producing unusual fatty acids. This result implies that Pichia cells are highly tolerant to free fatty acids and the transformants might have a more effective mechanism than transgenic plants to shuffle freshly synthesized ricinoleic acid from phospholipids to the free fatty acid pool by phospholipase or other enzymes, whereby the membrane integrity would be better maintained or protected from the possible damage caused by excessive accumulation of the hydroxyl fatty acid. If this is case, a future direction for improved production of unusual fatty acids in transgenic plants could focus on the utilization of phospholipase or other enzymes to remove the heterologous fatty acids from phospholipids to the free fatty acid pool, from which these fatty acids are then assembled to TAGs for storage through acyl-CoA intermediates. However, it was noted that a phospholipase A2 from castor bean was recently coexpressed with a hydroxylase in Arabidopsis, which resulted in production of an even lower level of ricinoleic acid compared with transgenic plants with the hydroxylase alone (31). It is possible that the phospholipase tested is not specific to phospholipids with ricinoleic acid in transgenic plants, although it was shown to prefer ricinoleate-phospholipids in yeast by the in vitro assay. In addition, plants might be highly susceptible to the FRA accumulation, for which the factors that can facilitate the entry of the free fatty acid toward the TAG biosynthesis should be provided for transgenic plant production of unusual fatty acids in the future.
Acknowledgments
The authors thank Drs. P. Covello and M. Smith (National Research Council, Canada) for critical reading of the manuscript and the assistance of the National Research Council DNA facility for DNA sequencing and primer synthesis.
Footnotes
Abbreviations:
- CpDGAT1
- diacylglycerol acyltransferase from Claviceps purpurea
- CpFAH
- fatty acid hydroxylase from Claviceps purpurea
- DGAT
- diacylglycerol acyltransferase
- FAME
- fatty acid methyl ester
- FRA
- free ricinoleic acid
- TAG
- triacylglycerol
The data discussed in this publication have been deposited in the National Institutes of Health database GenBank (Meesapyodsuk et al., 2015) and are accessible through the accession number for C. purpurea CpDGAT1, KT581979.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.McKeon T. A., Lin J. T., Stafford A. E. 1999. Biosynthesis of ricinoleate in castor oil. Adv. Exp. Med. Biol. 464: 37–47. [DOI] [PubMed] [Google Scholar]
- 2.Jaworski J., Cahoon E. B. 2003. Industrial oils from transgenic plants. Curr. Opin. Plant Biol. 6: 178–184. [DOI] [PubMed] [Google Scholar]
- 3.van de Loo F. J., Broun P., Turner S., Somerville C. 1995. An oleate 12-hydroxylase from Ricinus communis L. is a fatty acyl desaturase homolog. Proc. Natl. Acad. Sci. USA. 92: 6743–6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgal J., Shockey J., Lu C., Dyer J., Larson T., Graham I., Browse J. 2008. Metabolic engineering of hydroxy fatty acid production in plants: RcDGAT2 drives dramatic increases in ricinoleate levels in seed oil. Plant Biotechnol. J. 6: 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu C., Fulda M., Wallis J. G., Browse J. 2006. A high-throughput screen for genes from castor that boost hydroxy fatty acid accumulation in seed oils of transgenic Arabidopsis. Plant J. 45: 847–856. [DOI] [PubMed] [Google Scholar]
- 6.van Erp H., Bates P. D., Burgal J., Shockey J., Browse J. 2011. Castor phospholipid:diacylglycerol acyltransferase facilitates efficient metabolism of hydroxy fatty acids in transgenic Arabidopsis. Plant Physiol. 155: 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snapp A. R., Kang J., Qi X., Lu C. 2014. A fatty acid condensing enzyme from Physaria fendleri increases hydroxy fatty acid accumulation in transgenic oilseeds of Camelina sativa. Planta. 240: 599–610. [DOI] [PubMed] [Google Scholar]
- 8.Broun P., Somerville C. 1997. Accumulation of ricinoleic, lesquerolic, and densipolic acids in seeds of transgenic Arabidopsis plants that express a fatty acyl hydroxylase cDNA from castor bean. Plant Physiol. 113: 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broun P., Boddupalli S., Somerville C. 1998. A bifunctional oleate 12-hydroxylase: desaturase from Lesquerella fendleri. Plant J. 13: 201–210. [DOI] [PubMed] [Google Scholar]
- 10.Smith M. A., Moon H., Chowrira G., Kunst L. 2003. Heterologous expression of a fatty acid hydroxylase gene in developing seeds of Arabidopsis thaliana. Planta. 217: 507–516. [DOI] [PubMed] [Google Scholar]
- 11.Meesapyodsuk D., Qiu X. 2008. An oleate hydroxylase from the fungus Claviceps purpurea: cloning, functional analysis, and expression in Arabidopsis. Plant Physiol. 147: 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lennen R. M., Pfleger B. F. 2013. Microbial production of fatty acid-derived fuels and chemicals. Curr. Opin. Biotechnol. 24: 1044–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., McClure A., Del Cardayre S. B., Keasling J. D. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 463: 559–562. [DOI] [PubMed] [Google Scholar]
- 14.Runguphan W., Keasling J. D. 2014. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab. Eng. 21: 103–113. [DOI] [PubMed] [Google Scholar]
- 15.Xu P., Gu Q., Wang W., Wong L., Bower A. G., Collins C. H., Koffas M. A. 2013. Modular optimization of multi-gene pathways for fatty acids production in E. coli. Nat. Commun. 4: 1409. [DOI] [PubMed] [Google Scholar]
- 16.Xu P., Li L., Zhang F., Stephanopoulos G., Koffas M. 2014. Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. USA. 111: 11299–11304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holic R., Yazawa H., Kumagai H., Uemura H. 2012. Engineered high content of ricinoleic acid in fission yeast Schizosaccharomyces pombe. Appl. Microbiol. Biotechnol. 95: 179–187. [DOI] [PubMed] [Google Scholar]
- 18.Beopoulos A., Verbeke J., Bordes F., Guicherd M., Bressy M., Marty A., Nicaud J. M. 2014. Metabolic engineering for ricinoleic acid production in the oleaginous yeast Yarrowia lipolytica. Appl. Microbiol. Biotechnol. 98: 251–262. [DOI] [PubMed] [Google Scholar]
- 19.Sandager L., Gustavsson M. H., Stahl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277: 6478–6482. [DOI] [PubMed] [Google Scholar]
- 20.Bligh E. G., Dyer W. J. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 21.Pan X., Siloto R. M., Wickramarathna A. D., Mietkiewska E., Weselake R. J. 2013. Identification of a pair of phospholipid:diacylglycerol acyltransferases from developing flax (Linum usitatissimum L.) seed catalyzing the selective production of trilinolenin. J. Biol. Chem. 288: 24173–24188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meesapyodsuk D., Qiu X. 2011. A peroxygenase pathway involved in the biosynthesis of epoxy fatty acids in oat. Plant Physiol. 157: 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang H., Damude H. G., Yadav N. S. 2012. Three diacylglycerol acyltransferases contribute to oil biosynthesis and normal growth in Yarrowia lipolytica. Yeast. 29: 25–38. [DOI] [PubMed] [Google Scholar]
- 24.Mavraganis I., Meesapyodsuk D., Vrinten P., Smith M., Qiu X. 2010. Type II diacylglycerol acyltransferase from Claviceps purpurea with ricinoleic acid, a hydroxyl fatty acid of industrial importance, as preferred substrate. Appl. Environ. Microbiol. 76: 1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai M., Stephanopoulos G. 2013. Engineering the push and pull of lipid biosynthesis in oleaginous yeast Yarrowia lipolytica for biofuel production. Metab. Eng. 15: 1–9. [DOI] [PubMed] [Google Scholar]
- 26.Singh S., Thomaeus S., Lee M., Stymne S., Green A. 2001. Transgenic expression of a delta 12-epoxygenase gene in Arabidopsis seeds inhibits accumulation of linoleic acid. Planta. 212: 872–879. [DOI] [PubMed] [Google Scholar]
- 27.Cahoon E. B., Dietrich C. R., Meyer K., Damude H. G., Dyer J. M., Kinney A. J. 2006. Conjugated fatty acids accumulate to high levels in phospholipids of metabolically engineered soybean and Arabidopsis seeds. Phytochemistry. 67: 1166–1176. [DOI] [PubMed] [Google Scholar]
- 28.Napier J. A. 2007. The production of unusual fatty acids in transgenic plants. Annu. Rev. Plant Biol. 58: 295–319. [DOI] [PubMed] [Google Scholar]
- 29.Cahoon E. B., Shockey J. M., Dietrich C. R., Gidda S. K., Mullen R. T., Dyer J. M. 2007. Engineering oilseeds for sustainable production of industrial and nutritional feedstocks: solving bottlenecks in fatty acid flux. Curr. Opin. Plant Biol. 10: 236–244. [DOI] [PubMed] [Google Scholar]
- 30.Bates P. D., Browse J. 2011. The pathway of triacylglycerol synthesis through phosphatidylcholine in Arabidopsis produces a bottleneck for the accumulation of unusual fatty acids in transgenic seeds. Plant J. 68: 387–399. [DOI] [PubMed] [Google Scholar]
- 31.Bayon S., Chen G., Weselake R. J., Browse J. 2015. A small phospholipase A2-alpha from castor catalyzes the removal of hydroxy fatty acids from phosphatidylcholine in transgenic Arabidopsis seeds. Plant Physiol. 167: 1259–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]