Abstract
Lipid droplets (LDs) are the main storage organelles for triglycerides. Elucidation of lipid accumulation mechanisms and metabolism are essential to understand obesity and associated diseases. Adipogenesis has been well studied in murine 3T3-L1 and human Simpson-Golabi-Behmel syndrome (SGBS) preadipocyte cell lines. However, most techniques for measuring LD accumulation are either not quantitative or can be destructive to samples. Here, we describe a novel, label-free LD quantification technique (LipiD-QuanT) to monitor lipid dynamics based on automated image analysis of phase contrast microscopy images acquired during in vitro human adipogenesis. We have applied LipiD-QuanT to measure LD accumulation during differentiation of SGBS cells. We demonstrate that LipiD-QuanT is a robust, nondestructive, time- and cost-effective method compared with other triglyceride accumulation assays based on enzymatic digest or lipophilic staining. Further, we applied LipiD-QuanT to measure the effect of four potential pro- or antiobesogenic substances: DHA, rosiglitazone, elevated levels of D-glucose, and zinc oxide nanoparticles. Our results revealed that 2 µmol/l rosiglitazone treatment during adipogenesis reduced lipid production and caused a negative shift in LD diameter size distribution, but the other treatments showed no effect under the conditions used here.
Keywords: adipocytes, obesity, lipid droplet, triglycerides, omega-3-fatty acids, glucose, SGBS, label-free image analysis, zinc oxide nanoparticles, rosiglitazone
Lipid droplets (LDs) are the main storage organelles for triglycerides in eukaryotic cells (1–3). They are found in several cell types, where they support membrane biosynthesis and aid lipid homeostasis. They also participate in the storage of toxic lipid species in hepatocytes, macrophages, cardiac myocytes, renal glomerular cells muscle, and buccal cells (4, 5). Adipocytes are the main cell type that store the body’s energy reserves as triglycerides, and they do so in larger LDs that serve as highly specialized lipid reservoirs. However, excessive storage of triglycerides leads to obesity and metabolic syndrome, posing serious risks of diet-related noncommunicable diseases such as diabetes mellitus, hypertension, cardiovascular disease, and stroke (6).
The use of animal model systems and comparison with human studies has contributed extensively to our understanding of processes of development of adipose tissue and obesity (reviewed in Ref. 7). Supplementing in vivo systems, in vitro models are invaluable for dissecting molecular mechanisms, including the formation of LDs (8). Adipogenesis processes have been well studied since the establishment of both murine (9) and human (10) preadipocyte cell lines. These models enable controlled investigation of the regulators of adipogenesis and provide mechanistic insight into obesity via the measurement of physiological and molecular responses of adipocytes to specific therapeutic compounds. Since the establishment of human preadipocyte Simpson-Golabi-Behmel syndrome (SGBS) cell line in 2001 (10), it has been used in more than 100 research articles and has proven to be a reproducible model for studying biological mechanisms in preadipocytes and mature adipocytes in the context of human adiposity (11). SGBS cells are neither transformed nor immortalized, have high proliferation and differentiation capacity, and provide unique advantages to study human adipogenesis (12). SGBS cells are easier to maintain and less costly than human primary cell lines.
During adipogenesis, small LDs destabilize to increase storage capacity in two ways: coalescence by fusion of LDs or by Ostwald ripening, in which molecules from one LD diffuse to another (13). Quantification of LD accumulation is the most common measure of differentiation during in vitro adipogenesis. The three most widely used techniques are described below.
Fixed cells are stained with lipophilic dyes, the most commonly used lipophilic dyes being Oil Red O, Sudan III, Nile Red, BODIPY 493/503, and 3,3′-dioctadecyloxacarbocyanine perchlorate (14–16). The absorbed dye can be resolubilized, and the intensity of the stain, measured via spectrophotometry, can be used as a proxy for cellular lipid content.
After cell lysis, lipolytic enzymes are used to digest triglycerides to glycerol and free fatty acids. Additional enzymatic reactions turn glycerol into a colored product, the concentration of which may be measured using spectrophotometry.
Monoclonal antibodies are available against LD-associated proteins such as perilipin, adipophilin, TIP47, and caveolin-1 (17). Fluorescence microscopy after antibody staining makes LDs visible, enabling measurement of LD size and ultimately lipid content.
These techniques are lengthy, require dedicated culture plates to measure the lipid content, and do not permit reuse of cells for other biological or morphological measurements. Moreover, the staining-based methods comprise multiple steps, including fixation, washing, and dehydration, which occasionally result in detachment of cells and lysis of LDs. There is also growing evidence that staining conditions such as dye concentration and fixation time affect the fluorescence intensity of lipophilic dyes, resulting in incorrect correlation between fluorescence intensity and actual total lipid content (18). Additionally, lipophilic dyes have high affinity toward hydrophobic surfaces, which results in background staining when plastic culture dishes are used (19, 20). Finally, when using biochemical assays, a variable fraction of the lysed cells may be retained in the cell culture dishes, which may result in minor errors in the cellular lipid content calculation. Thus, existing assays tend to be more qualitative than quantitative.
More recently, Coherent anti-Stokes Raman Scattering microscopy showed great promise as a technique to study LD dynamics (21). It has been used to identify LDs in Caenorhabditis elegans (22), Drosophila melanogaster (23), murine fibroblasts (24), and human adipose-derived stem cells (25, 26). However, it is currently only available in a handful of research institutes with the appropriate equipment. Therefore, a need still exists for a method of lipid accumulation quantification that is effective and reliable, suitable for use with live cells, and uses common laboratory equipment.
In this contribution, we describe LipiD-QuanT, an automated image analysis tool, to quantify LD accumulation in live cells under phase contrast microscopy. The LipiD-QuanT algorithm depends on the ability of phase contrast microscopy to reveal even small refractive index differences between LDs and the surrounding cytosol by exploiting destructive interference effects. LDs have a well-defined spherical shape; hence, the measurements of LD volume and surface area may be easily obtained on a per-cell basis. Therefore, we used LipiD-QuanT on the human SGBS preadipocyte differentiation model, which provides the ease to study the effect of any treatment of interest on human adipogenic differentiation in short time frame.
We first evaluated the performance of LipiD-QuanT against benchmark Oil Red O staining and biochemical lipid accumulation techniques to monitor LD dynamics during in vitro differentiation of human SGBS preadipocytes over a 21 day time frame. We monitored the gene expression changes in selected adipogenesis genes to assure the reproducibility of SGBS preadipocyte differentiation. We then used LipiD-QuanT to sensitively detect changes in LD growth in response to four potential pro- or antiobesogenic treatments: DHA, high glucose, zinc oxide (ZnO) nanoparticles, and rosiglitazone.
MATERIALS AND METHODS
Cell culture conditions
We used the SGBS cell line, a human-derived preadipocyte cell line isolated from the stromal cell fraction of subcutaneous adipose tissue from an infant with Simpson Behmel Gobali syndrome (10). Proliferation and differentiation media were prepared as previously described (10) with minor changes. In brief, cells were proliferated to 90% confluence in T75 or T150 flasks in DMEM/F-12, GlutaMAX™ (cat#10565; Life Technologies) supplemented with 10% FBS (cat#10099-141; Invitrogen), 10 U/ml penicillin-streptomycin (cat#15070; Invitrogen), 8 mg/l d-biotin (cat#4639; Sigma), and 4 mg/l d-panthothenic acid (cat#P5155; Sigma). Cells were then differentiated in 6-well plates at a concentration of 0.2 million cells/well. The experiments were undertaken with a minimum of three biological replicates at passage number 23. The cells were kept in serum-free quick differentiation media supplemented with 100 nmol/l cortisol (cat#H0888; Sigma), 0.01 mg/ml transferrin (cat#T0665; Sigma), 0.2 nmol/l triiodothyronine (cat#T6397; Sigma), 20 nmol/l insulin (cat#I2643; Sigma), 2 µmol/l rosiglitazone (cat#2408; Sigma), 25 nmol/l dexamethasone (cat#D4902; Sigma), and 0.5 mmol/l 1-methyl-3-isobutylxanthine (cat#I5879; Sigma) for the first 4 days. After 4 days, media was removed and replaced with differentiation media, further excluding rosiglitazone, dexamethasone, and 1-methyl-3-isobutylxanthine for 10 or more days. Media was changed every second day.
During adipogenesis, cells were exposed to the following pro- and antiobesogenic treatments.
DHA.
A single ml of DHA oil emulsion contained 125 mg of DHA (C22:6), 8.5 mg of eicosapentaenoic acid (C20:5, EPA), 9 mg of vitamin C, and 0.19 mg of vitamin E (HiDHA™ oil emulsion: Nu-Mega Ingredients Pty. Ltd.). Over the 14 day course of SGBS differentiation, 10 µmol/l DHA oil emulsion was added, at each change of media, a total of seven times. LDs were monitored at the end of the differentiation period on day 14.
D-glucose.
SGBS cells were differentiated in 10 mM D-glucose (cat#G7021; Sigma), 10 mM D-glucose with 7.5 mM Sorbitol (cat#S1876; Sigma) for osmolarity control, and 17.5 mM D-glucose. The low glucose concentrations were maintained by changing the medium to DMEM, low glucose, GlutaMAX™, and pyruvate (cat#11885; Life Technologies) containing 5.5 mM D-glucose. The LDs were assessed at days 7, 10, and 14.
ZnO nanoparticles.
The differentiated SGBS cells were treated with 1 µg/ml or 10 µg/ml ZnO nanoparticles coated with a dimethoxydiphenylsilane/triethoxycaprylylsilane crosspolymer (batch# FCHE1301; Z-COTE MAX from BASF) on day 6 for 48 h. We have described the extensive physicochemical characterization of Z-COTE MAX elsewhere (27, 28). Briefly, primary particle sizes were 36 ± 2 nm wide and 95 ± 5 nm length but formed larger aggregates in water and cell culture medium. Noncytotoxic concentrations to SGBS cells were selected on the basis of previous work (27, 28). We monitored the change in LD size distribution immediately after the treatment period at day 8 as well on days 10 and 14.
Rosiglitazone treatment.
SGBS cells received an additional of 2 µmol/l rosiglitazone from day 4. The LDs were measured on days 10 and 14.
Biochemical measurement of lipid content with triglyceride accumulation assay
SGBS cells were lysed using 200 µl triglyceride accumulation lysis buffer per well in 6-well plates (cat#TG-1-NC; Zen-Bio Inc.). Biological triplicates were included for each assay point, and the lysates were stored at −80°C until the completion of the differentiation process. Triglyceride esters were converted to glycerol, and glycerol concentration was quantitated enzymatically and measured as per the manufacturer’s protocol, using 15 µl of lysate in technical duplicates.
Staining based measurement of lipid content using Oil Red O
Cells were fixed with 1 ml of 4% paraformaldehyde for 20 min in 6-well plates, washed with 1× PBS twice, and stored at 4°C in 1× PBS supplemented with 0.02% (w/v) sodium azide until processing. The fixed cells were washed with 100% propylene glycol (Astral, cat#CSPL010) before 1 ml of Oil Red O staining for 15 min at room temperature (cat#O1516; Sigma). Cells were washed three times with 1 ml of PBS to remove excess dye, and images were acquired using an inverted microscope fitted with a high-definition, cooled color digital camera DXM1200C (ECLIPSE 90i; Nikon, Japan). The Oil Red O dye was extracted from the stained cells using 500 µl of 100% isopropanol for each well, and two aliquots of 200 µl were transferred to black 96-well plates. The O.D. reading of Oil Red O was measured at 520 nm using a plate reader (POLARstar Omega microplate reader; BMG Labtech, UK).
Immunolabeling for LDs
Cells were differentiated on acid-treated coverslips for immunolabeling of the LD coating protein perilipin. The cells were fixed on coverslips with 1 ml of 4% paraformaldehyde for 20 min in 6-well plates and washed with 1× PBS twice before and after permeabilization in Triton X-100 solution (0.2% Triton X-100 in PBS with 10% goat serum, cat#5425; Cell Signaling Technology) for 10 min. Samples were blocked for 20 min in 10% goat serum, 5% FCS, and 0.5% BSA in PBS and stained with anti-Perilipin (D1D8) XP® Rabbit mAb (cat#9349S; Cell Signaling Technology) overnight at 4°C as per the manufacturer’s recommendations. Samples were washed with 1× PBS twice and reblocked. The secondary antibody staining was completed using Alexa Fluor® 488 goat anti-rabbit IgG (H+L) as per the manufacturer’s instructions (cat#A-11034; Invitrogen). A solution of 25 µg/ml DAPI (cat#D9542; Sigma) was added for nuclear staining. The coverslips were washed twice with 1× PBS and mounted on microscope slides using Fluoroshield mounting media (cat#F6182; Sigma).
RNA extraction
SGBS cells were lysed in 500 µl TRI-reagent (cat#T9424; Sigma) on days 0, 4, 7, 14, and 21, and the lysates were stored at −80°C until completion of sample collection. Total RNA was isolated as described by the manufacturer’s manual. RNA concentration was determined by NanoDrop ND-1000 (USA) spectrophotometer readings.
cDNA synthesis and quantitative real-time PCR
Each sample of first-strand cDNA was synthesized from 600 ng of total RNA using QuantiTect Reverse Transcription Kit (cat#205313; Qiagen). Gene-specific primers (supplementary Table 1) were used to amplify target genes using 10 ng of first-strand cDNA as template in a 15 µl SYBR green-based quantitative RT-PCR reaction performed under the following conditions: 95°C for 2 min, 45 cycles at 95°C for 10 s, 60°C for 30 s, 72°C for 10 s with a melting curve from 65°C to 95°C. The gene expression levels were normalized to GAPDH housekeeping gene expression in each sample.
Image acquisition
Images were acquired on an Olympus IX81 microscope equipped with a 20×/0.40 LCPLANFl Ph1 phase contrast objective (USA) and a Roper Scientific CoolSNAP FX monochrome camera (USA) aligned for positive mode phase contrast microscopy. The image intensity was optimized to span the full camera dynamic range, and the focus was adjusted to maximize the LD morphology displaying sharp edges. Care should be taken in the microscope set-up to ensure that any “halo” effect is homogeneous because inhomogeneity may interfere with the image analysis. Also, without care in set-up, the smallest LDs may have dark boundaries that do not close entirely. A total of six images were taken per well. All the images were saved in uncompressed TIFF format. Pixel size in the object space was 0.3 µm. A comprehensive set of label-free images of human adipocytes were captured during adipogenesis (supplementary File 1).
On average there were 25 cells in a single image, with a variation of two to five cells between image fields. Larger variation will lead to incorrect assumptions about total lipid quantity. Therefore, lipid quantities should preferably be normalized by the cell number. The cells were stained with the nuclear fluorescent dye DAPI and imaged with a DAPI fluorescence filter set immediately before acquiring the phase contrast image (example images are provided in Fig. 1A, B, and C). Nuclei were counted using the Otsu threshold method available in MatlabTM (29). LD amounts per cell were calculated by matching nuclei numbers with the LD numbers produced by LipiD-QuanT (Fig. 1). Although LipiD-QuanT is designed for LD quantification in living cells to allow their further use for other biological measurements, we used images of fixed cells to check the effect of specific stimulants, which were used to compare our method with Oil Red O staining method on the same wells. Other physiological nuclear stains may also be used, such as Hoechst 33342 (available to stain live cells in media as NucBlue® by Life Technologies).
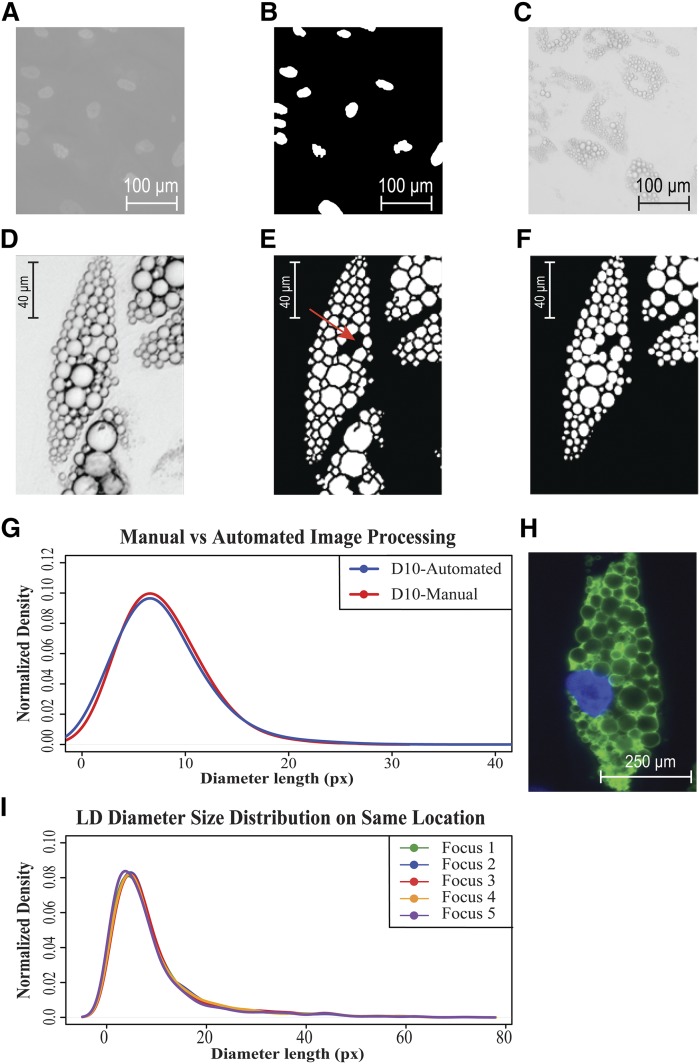
Fig. 1.
LipiD-QuanT validation. A: Image of 4% paraformaldehyde-fixed SGBS human preadipocytes on day 7, stained with DAPI to localize nuclei. B: Nuclei in Figure 1A were segmented automatically via LipiD-QuanT in order to derive per-cell statistics for lipid accumulation. C: The same image field as in Figure 1A was acquired under phase contrast microscopy. D: A representative image of differentiated SGBS cells acquired under phase contrast microscopy showing well-contrasted LD boundaries. E: A representative image of differentiated SGBS cells representing LDs segmented by LipiD-QuanT. A region of negative curvature cytoplasmic space between LDs is indicated by a red arrow. F: A representative image of differentiated SGBS cells representing LDs segmentation by manual counting (the lower cell was not analyzed). G: Comparison of smoothed distribution of LD diameter sizes obtained from automated LipiD-QuanT measurements and manual segmentation (n = 1,048 LDs). Pixel size in object space is 0.3 µm. H: The LD identity was confirmed at the molecular level using antiperilipin fluorescence staining of the protein (green channel) decorating their periphery. I: Size distributions are not sensitive to focus choice. Smoothed distribution of LD diameter sizes of mature human adipocytes is reproduced reliably after resetting the focus in independent trials (n = 1,165 LDs).
LipiD-QuanT image processing and analysis
LDs naturally adopt a spherical geometry driven by their surface tension (30). LipiD-QuanT is based on the Laplacian edge detector, which we have previously used to monitor bacteria under phase contrast microscopy (31). The Laplacian edge detector defines level sets (similar to level lines on topography maps) that automatically guarantee the formation of closed contours surrounding the target shapes (32). LipiD-QuanT detected contour lines closely corresponding to individual LDs in human adipocyte images. However, false-positive contours are also formed in the background. These were eliminated because they are associated with low intensity variance along their contour.
Cytoplasmic spaces, surrounded by genuine LDs, tend to have a concave shape, being complementary to a set of limiting convex vesicles. We used the function TRACE_MooreNeighborhood.m contributed by Adam H. Aitkenhead available on “Matlab Central” to trace the LD edges until it generated a closed contour circle (33). We then applied the LineCurvature2D.m function contributed by Dirk-Jan Kroon and available on “Matlab Central” to compute the curvature of the traces generated by the previous function (34).
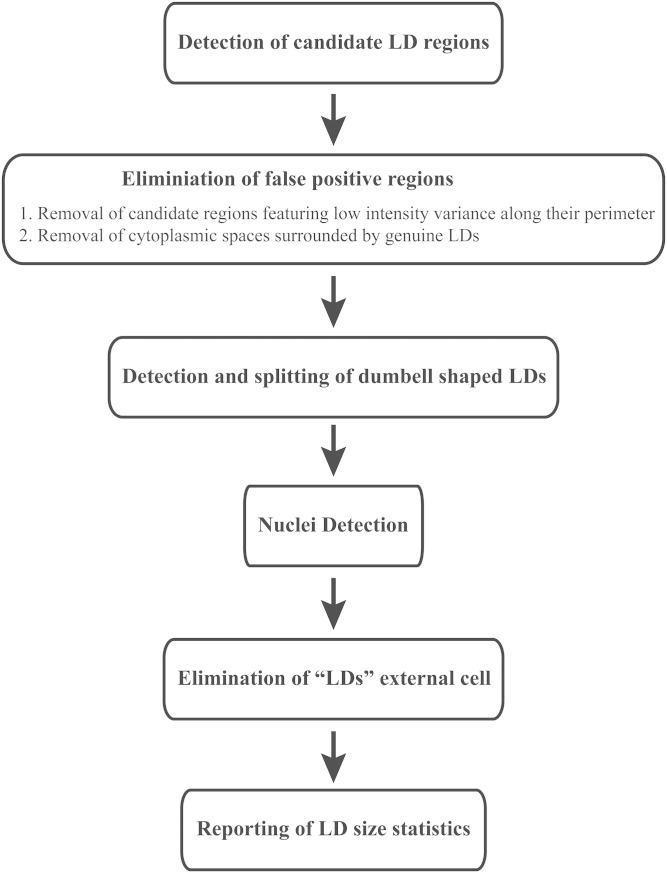
Analysis of the curvature at the outer boundary of the segmented regions and subsequent removal of regions with negative average curvature allowed us to eliminate the false-positive LDs (Fig. 1E, red arrow). LDs often occur as clusters. Occasionally, fused LDs form dumbbell shapes. We have incorporated the watershed transform algorithm to LipiD-QuanT to detect and divide dumbbell-shaped LDs (35) and eliminated LDs external to cells. Additionally, a mask outlining the position of cells was constructed in order to avoid counting LDs in the background (36). The key steps of the LipiD-QuanT algorithm are presented in Fig. 2.
Fig. 2.
Flowchart presenting the key steps of LipiD-QuanT algorithm.
LipiD-QuanT data analysis and output
LipiD-QuanT installer (supplementary File 3) is available on the CSIRO Data Access Portal as well as on GitHub (https://github.com/Varinli/LipiD-QuanT). The details are further explained in the installment instructions included in supplementary File 3. It will automatically process the images saved to the same directory, providing numeric outputs corresponding to LD diameter length in pixels as described in the LipiD-QuanT installer instructions. The script itself is provided in supplementary File 4. LipiD-QuanT is fully automated and can be used even by potential users that are not familiar with MatlabTM.
The images produced by LipiD-QuanT correspond to binary images, where all the pixels of each identified LD are set to 1 and the background pixels are set to 0. We generated the size and shape information pertaining to LDs using the regionprops function in Matlab. LipiD-QuanT is applicable to both fixed and live cells because it uses the “equivalent diameter,” indicating the diameter of the disk that has the same area as the object as the absolute measure of LD diameter length in pixels. Based on the microscope setup, the measurements can be converted to micrometers.
We used the smoothed kernel density distributions to visualize the distribution of LD sizes among treatments and to characterize potential effects. The complete script for producing smooth LD diameter size distributions from LipiD-QuanT output is given in supplementary File 5. The LipiD-QuanT output can also be used to calculate total LD area as an indicative of LD content. We advise to normalize the total LD area for number of cells to remove any technical or biological variance due to cell number among wells.
RESULTS
SGBS preadipocyte differentiation
SGBS preadipocyte cells were cultured and differentiated according to standard procedures (10). Undifferentiated SGBS cells exhibit a fibroblast-cell like structure. However, the cells initiate adipogenesis by pulling their longitudinal structures into more localized single globular structure with the influence of the adipocyte differentiation cocktail. By day 4, tiny LDs, sparsely distributed within the cell, became more apparent. By day 7, a high proportion of the cells appeared terminally differentiated, containing multiple visible LDs. By day 10, the majority of the differentiated cells matured and contained large quantities of lipids. Over the 14-day course of adipogenesis, 95% of the SGBS preadipocytes were differentiated into mature adipocytes. From day 14 to 21, we observed LD enlargement by coalescence and growth.
We monitored changes in the cellular morphology and expression profiles of a set of key genes, such as CEBPB, PPARG, GLUT4, ADIPOQ, PLIN, and FABP4, during adipogenesis. All of these genes except ADIPOQ had the highest relative expression on day 10 (supplementary Fig. 1). Whereas the expression of CEBPB, PPARG, and GLUT4 genes was higher in later stages of adipogenesis, ADIPOQ, PLIN, and FABP4 expression levels were reduced by at least 2-fold (supplementary Fig. 1). The gene expression changes were consistent with previously published data confirming that SGBS preadipocyte differentiation is a reproducible model for the study of human adipogenesis (12, 37–40).
Lipid-QuanT validation
LipiD-QuanT algorithm validation against manually segmented LDs.
In Fig. 1 we show the process of applying LipiD-QuanT. The Fig. 1A shows a phase contrast image of a field of differentiating adipocytes at day 7, and Fig. 1B and C show the DAPI-stained image of the field and nuclei identification used to determine cell numbers per field. We present an example of a phase contrast image of a differentiated adipocyte in Fig. 1D and the processed LipiD-QuanT automated segmentation of LDs in Fig. 1E. The shapes and sizes of LDs measured automatically via LipiD-QuanT closely matched those in original images (compare Fig. 1D and E).
We generated ground truth data by manually creating LD outlines in randomly selected two cells in each of a total of six test images taken on day 10 of human adipogenesis, using the region of interest selection tool in MatlabTM (imellipse), which allows precisely overlaying an ellipse shape onto each individual LD by adjusting its center and main axes. An example image where these ellipses were fitted manually is shown in Fig. 1F. The same LDs were also detected by executing LipiD-QuanT on the same six images. LipiD-QuanT detected over 95% of LDs as determined by manual segmentation (Fig. 1E and F). In total, 1,048 LDs were analyzed by the two methods (Fig. 1G). Although there was a minor shift of larger sizes in LD diameter distribution using manual segmentation, the LD size distributions obtained using the two methods were not statistically different according to chi2 and Kolmogorov-Smirnov tests (P > 0.05) (Fig. 1G). The LipiD-QuanT algorithm requires an average of 1 min per image and detects LDs with a minimum diameter of 0.34 µm.
Antiperilipin-stained LDs structures corresponded accurately with stain-free LipiD-QuanT detection.
To confirm correspondence of LDs detected by LipiD-QuanT with fluorescence-stained images, we stained mature adipocytes using an antibody against perilipin, a protein known to decorate the outer surface of LDs (Fig. 1H). The appearance and size of the fluorescently stained LDs corresponded to those detected under phase contrast. We also acquired 3D widefield stacks of antiperilipin-stained human SGBS adipocytes. LDs tended to be approximately spherical and predominantly arranged in a single horizontal plane (supplementary File 2).
LipiD-QuanT robustness against small variations in focal plane selection.
LipiD-QuanT, developed for in vitro studies, relies on the acquisition of a single image for lipid quantification rather than a full 3D stack of images spanning the entire 3D cell volume. We tested LipiD-QuanT’s robustness against variations in focal plane selection by acquiring the same image field five times, each time independently refocusing to maximize the contrast of LDs. The resulting LD size distributions corresponding to individual images were almost indistinguishable (Fig. 1I). Thus, as long as image contrast is maximized by the operator, LipiD-QuanT is resilient against focus variations. The choice of focal plane does not interfere with the results because adipocytes are predominantly arranged flat in a horizontal plane, as shown in supplementary File 5.
LipiD-QuanT consistency across image fields in the same well.
Although a certain biological variability in lipid content is expected for individual cells in the same well, cell-specific variability is expected to be averaged out because images often contained multiple cells. We acquired six images from various locations of the same well in a total of two treatment groups. The 12 images were analyzed by LipiD-QuanT separately. The separate LD diameter size distribution was produced for each image taken from the same well. The results were highly consistent and robust in each treatment group (supplementary Fig. 2A and B).
Comparison of LipiD-QuanT with standard methods
We monitored lipid accumulation in differentiating SGBS cells on days 0, 4, 7, 10, 14, and 21 via biochemical and lipid staining methods (Fig. 3). We used LipiD-QuanT on images acquired from day 7 onward because the LDs were too small to identify with a 20× objective before day 7. A total of 5,670 LDs were characterized to assess the performance of LipiD-QuanT against staining and biochemical-based triglyceride accumulation assays.
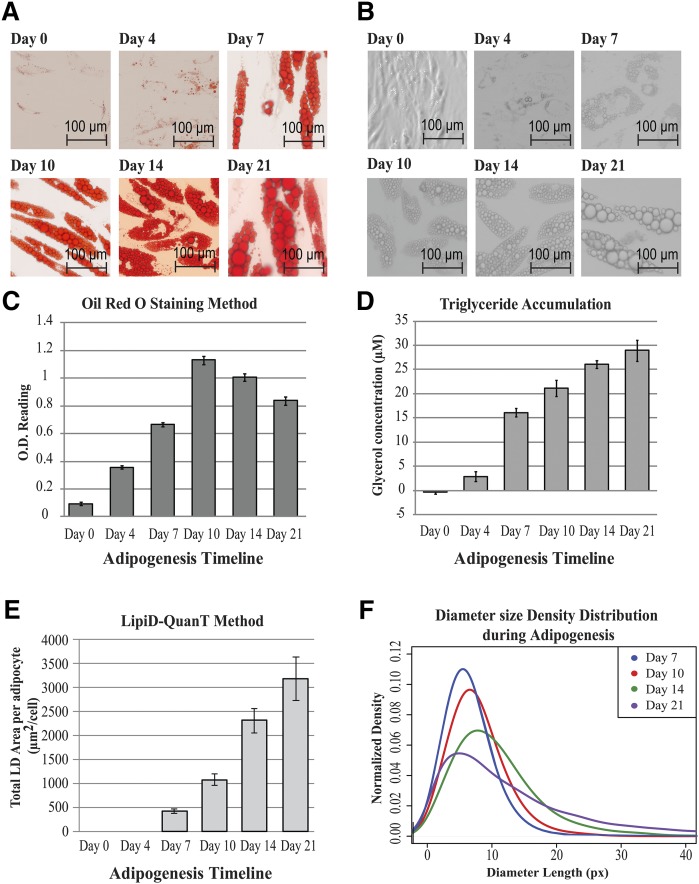
Fig. 3.
Performance of LipiD-QuanT in monitoring LD accumulation during adipogenesis, compared with Oil Red O staining and triglyceride accumulation protocols. A: Representative images of Oil Red O stained SGBS cells over the adipogenesis time course of 21 days. As expected, Oil Red O is seen to partition predominantly in LDs. B: Representative images of label-free differentiating SGBS cells as observed by phase contrast microscopy over a period of 21 days. The average LD diameter is seen to increase spectacularly over this period. C: The Oil Red O dye extractions were measured spectrophotometrically at 520 nm (OD520nm) over a period of 21 days. Data are shown as mean ± SD (three biological replicates with two technical replicates per sample). D: Quantification of total triglyceride accumulation using enzymatic digest method during adipogenesis. The total glycerol concentration is shown in µM. Data are shown as mean ± SD of experiment performed in triplicate wells (two technical replicates per sample). E: Total LD area calculated on days 7, 10, 14, and 21 of human adipogenesis using LipiD-QuanT divided by the total number of differentiated cells used per DAPI-stained nuclei images coupled with phase contrast images. On days 7, 10, 14, and 21, there were a total of 99, 104, 96, and 132 cells, respectively (six images were used per differentiation time point). F: Smoothed distribution of LD diameter sizes obtained using the LipiD-QuanT software during adipogenesis (n = 5,670 LDs). A systematic shift toward larger LD sizes is measured.
Over the process of well-orchestrated adipogenesis, some preadipocytes trigger differentiation more quickly, rapidly reaching their maximum LD storage capacity, whereas others contain a plethora of differentially sized LDs. Similarly, both live and Oil Red O-stained adipocyte images confirmed the structural changes in LDs as adipogenesis progressed (Fig. 3A). New LDs form while larger LDs merge and augment in size to increase lipid storage capacity until day 21 (i.e., there are fewer but larger LDs as the cells mature) (Fig. 3B). Fig. 3A and B provide representative sections of images. More examples of label-free human adipocytes images during differentiation are provided in supplementary File 1.
The overall trend for LD accumulation obtained via the Oil Red O staining method differed from both LipiD-QuanT and triglyceride accumulation results (Fig. 3C, D, and E). The biochemical assay demonstrated a linear increase in lipid content (Fig. 3D) consistent with the previously published results (41). LipiD-QuanT measurements demonstrated a sustained increase in lipid content across the entire differentiation time course (Fig. 3E), whereas Oil Red O staining surprisingly indicated that total lipid content decreased after day 10 (Fig. 3C), possibly due to lysis or detachment during washing and staining procedures. Because LipiD-QuanT measurements are based on individual cells, it is possible to discern features that are obscured in measurements of total lipid content.
The LD size distribution obtained by LipiD-QuanT reflected LD coalescence and ripening as the mean of LD diameter size distributions shifted toward larger LDs as the adipogenesis progressed (Fig. 3F). The total LD area increased 10 times, whereas the average LD diameter size increased 118% from day 7 to day 21 (Fig. 3E and F). Median LD diameter size of SGBS adipocytes increased from 1.72 µm to 2.95 µm from day 7 to day 21. The size range of LDs is similar to that of SGBS cells (42, 43). LipiD-QuanT coupled with nuclear staining also allowed us to calculate the ratio of differentiated cells during adipogenesis. The proportion of differentiated cells increased from 75% to 96% from day 7 to day 21.
LD size distribution in the presence of pro- and antiobesogenic substances
Once we confirmed that LipiD-QuanT was accurately detecting LDs compared with the standard methods, we used LipiD-QuanT to measure the response to four potential pro- or antiobesogenic interventions during adipogenesis.
DHA.
An important omega-3 long-chain polyunsaturated fatty acid is reported to have antiobesogenic effects in animal models (44); reduced plasma levels of omega-3 fatty acids have also been linked to obesity (45–47). Higher omega-3 fatty acid levels have been shown to reduce fat cell size in overweight and obese individuals (48).
D-glucose.
Glucose has been shown to act through SREBP-1c to induce de novo lipogenesis in vitro in rat muscle satellite cells (49). Exposure of 3T3-L1 cells to high glucose conditions leads to insulin resistance (50, 51) and to accumulation of significantly increased amounts of lipid compared with low glucose (52).
ZnO nanoparticles.
Zinc homeostasis is involved in dysfunction of insulin metabolism (53). Low levels of erythrocytary zinc are linked to type-2 diabetes and metabolic syndrome outcomes (54–56). Because the therapeutic effects of ZnO nanoparticles in treating lipid-associated diseases show promise (57, 58) but are underexplored, we tested their possible effect on LD dynamics in differentiating human preadipocytes.
Rosiglitazone treatment.
The peroxisome proliferator-activated receptor γ agonist is an insulin sensitizer and triggers new LD formation during adipogenesis (59). However, prolonged treatment is known to be antiobesogenic, to promote lipolysis, and to block LD fusion, resulting adipocytes containing small LDs (60, 61).
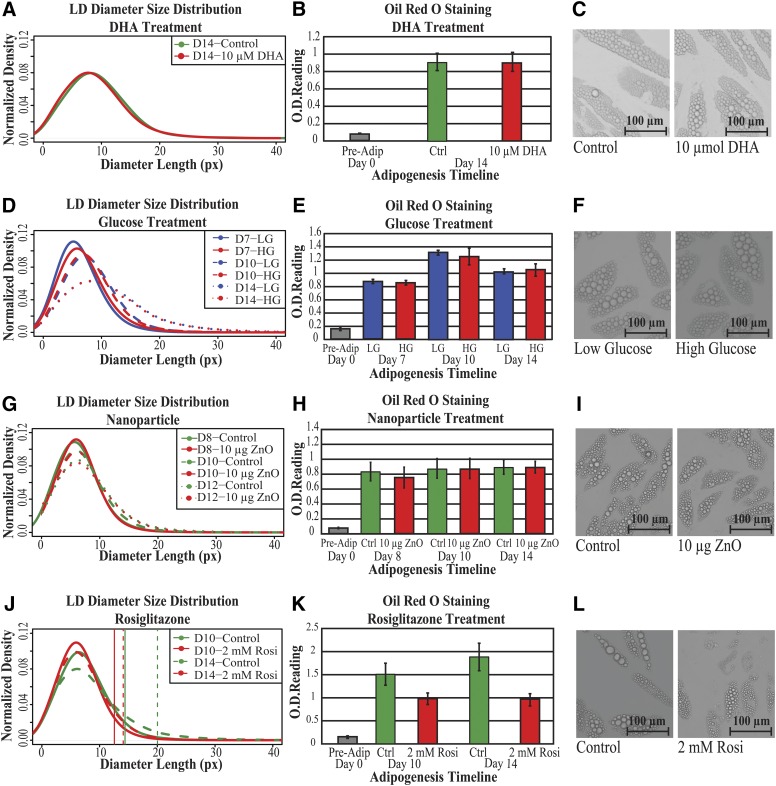
In Fig. 4 we show the smoothed distributions of LD sizes and total lipid quantification obtained using LipD-QuanT and Oil Red O staining methods, respectively, with representative images of adipocytes corresponding to each treatment and control group. The distributions of LD diameter results obtained from independent experiments indicated that measurement of LD accumulation during SGBS adipogenesis was very robust (Fig. 4A, D, G, and J). The chi2 and Kolmogorov-Smirnov tests indicated that DHA and ZnO nanoparticle treatments had neither positive nor negative effects on the overall distribution of LD diameter size during adipogenesis (p > 0.05) (Fig. 4A and G). Whereas elevated glucose concentration also did not result in a statistically significant change in LD size distribution or total lipid content, a consistent slight shift to higher LD was seen (Fig. 4D). The total lipid quantification using the Oil Red O staining method was also unaffected in these treatments (t-test; p >0.05) (Fig. 4B, E, and H).
Fig. 4.
The effect of pro- and antiobesogenic treatments on LD formation and accumulation during adipogenesis. A: Smoothed distribution of LD diameter sizes in terminally differentiated SGBS cells in the presence and absence of 10 µmol/l DHA treatment over a 14 day period (n = 7,444 LDs in eight images). B: The Oil Red O dye extractions were measured spectrophotometrically at 520 nm (OD520nm) in terminally differentiated SGBS cells in the presence and absence of 10 µmol/l DHA treatment over a 14 day period. Data are shown as mean ± SD (three biological replicates with two technical replicates per sample). C: Representative microscopy images of control and 10 µmol/l DHA-treated SGBS cells on day 14. For these and panels F, I, and L, images are only representative of a small portion within a single image. D: The change of smoothed distribution of LD diameter sizes in differentiating SGBS cells in low and high glucose concentrations for a total of 14 days (n = 5,955 LDs). E: The Oil Red O dye extractions were measured spectrophotometrically at 520 nm (OD520nm) in differentiating SGBS cells in low and high glucose concentrations for a total of 14 days. Data are shown as mean ± SD (four biological replicates with two technical replicates per sample). F: Representative microscopy images of SGBS cells fed in low- and high-glucose-concentrated media up to day 14. G: The change of smoothed distribution of LD diameter sizes in differentiating SGBS cells on days 8 and 12 after a 48 h treatment with or without the 10 µg coated ZnO nanoparticles on day 6 (n = 33,626 LDs). H: The Oil Red O dye extractions were measured spectrophotometrically at 520 nm (OD520nm) in differentiating SGBS cells on days 8 and 12 after a 48 h treatment with or without the 10 µg coated ZnO nanoparticles on day 6. Data are shown as mean ± SD (two biological replicates with two technical replicates per sample). I: Representative microscopy images of SGBS cells at day 14 after a 48 h treatment with or without a 48 h treatment the 10 µg coated ZnO nanoparticles on day 6. J: Smoothed distribution of LD diameter sizes in SGBS cells differentiated in the presence and absence of 2 µmol/l roziglitazone until day 14 (n = 33,749 LDs). Vertical lines: 95th percentile; full lines: day 10; dashed lines: day 14. K: The Oil Red O dye extractions were measured spectrophotometrically at 520 nm (OD520nm) in SGBS cells differentiated in the presence or absence of 2 µmol/l roziglitazone until day 14. Data are shown as mean ± SD (three biological replicates with two technical replicates per sample). L: Representative microscopy images of SGBS cells in the presence and absence of 2 µmol/l roziglitazone for a total of 14 days.
For example, under the effect of ZnO nanoparticles exposure (Fig. 4G and H), the tail of LD diameter size distributions fattens, indicating the increase in the proportion of larger LDs in adipocytes as the adipogenesis progress (Figs. 3F, D, G, and J). However, these structural features are lost if measures of total lipid quantification are used (Fig. 4E, H, and K).
Rosiglitazone treatment is essential for the early stages of adipogenesis to promote LD formation and fusion. However, prolonged stimulation is known to influence adipocyte morphology, leading to adipocytes containing small LDs scattered in the cytoplasm. We observed a statistically significant effect of rosiglitazone in LD diameter size distribution at both day 10 (chi2, p < 2.2e-16; Kolmogorov-Smirnov test, p < 2.2e-16) and day 14 adipocytes (chi2, p < 8.62e-15; Kolmogorov-Smirnov test, p < 2.2e-16). There was a negative shift in LD size distributions revealing an abundance of small LDs on both day 10 and day 14 (Fig. 4J). The total lipid quantification using Oil Red O staining method also revealed that there was a significant reduction in the lipid content both on day 10 and day 14 in response to prolonged treatment with rosiglizatone (t-test; p = 1.6e-5 and 1.9e-5, respectively) (Fig. 4K). Rosiglitazone treatment almost stopped LD fusion and enlargement, whereas the increase in LD size was aberrant in the control group: there were fewer LDs under 3 µm diameter in the day 10 control group compared with day 14 (Fig. 4J). We observed the biggest effect of rosiglitazone on larger LDs. The 95th percentile LD diameter size was 0.6 and 1.8 µm smaller in rosiglitazone treatment compared with control group on day 10 and day 14, respectively (vertical lines in Fig. 4J). This indicated that prolonged treatment with rosiglitazone prevented LD enlargement during adipogenesis. The results are discussed further below.
DISCUSSION
There is growing public interest in pro- and antiobesogenic substances to potentially combat the global obesity epidemic, yet little is known about the effect of many substances on LD dynamics. Identification of specific interactions between fat storing cells and environmental signals will potentially delineate intrinsic mechanisms leading to fat depot expansion either through increase cell size or numbers (62). LipiD-QuanT provides a readily applicable method to monitor LD formation and growth during adipogeneisis and to study the impact of exogenous factors. We have demonstrated here that LipiD-QuanT is a suitable approach for measuring LD content and for determining whether a substance of interest can modulate LD accumulation, that it delivers a detailed picture of LD dynamics at the single LD/cell resolution, and that it can measure LDs down to a size of a single pixel. LipiD-QuanT generates highly reproducible results and is sensitive enough to determine small differences in LD sizes and size distributions during in vitro human adipogenesis. It compares favorably with biochemical or staining-based techniques (Figs. 3 and 4), whereas the results of biochemical and staining-based techniques can be dependent on fixation or lysis conditions and lack information about structural features of LDs (supplementary Fig. 2C, D, and E).
We have used LipiD-QuanT to monitor the impact of four treatments across adipogenesis. Antidiabetic effects of ZnO nanoparticles had been observed in a diabetic rat model in which nanoparticles were absorbed by the fat tissue when consumed orally and reduced serum levels of free fatty acid and triglycerides over 40% (58). This prompted us to examine whether ZnO nanoparticles had a direct effect on LD formation and lipid accumulation. The lack of a significant effect, even at the relatively high dose used, suggests that the observed lowering of serum-free fatty acids and triglycerides is not due to a direct effect of ZnO nanoparticles on adipocytes.
In a number of systems, including stem cells from bone marrow, muscle, and adipose tissue, high glucose exposure has been found to stimulate adipogenenic differentiation of the stem cells (63). In vitro studies in 3T3-L1 cells and rat muscle satellite cells have also shown that hyperglycemic conditions lead to increased lipogenesis and lipid accumulation (52). Across differentiation of SGBS cells in the presence of high (17.5 mM) compared with low (5 mM) glucose, we did not see a significant change in lipid accumulation measured using LipiD-QuanT or Oil Red O staining, although a consistent slight shift to higher LD size was seen. It is possible that we may have seen greater effects with a broader range of glycemic conditions, such as 4–25 mM glucose used in 3T3-L1 cells (52), but the lower concentration was chosen because SGBS cells do not reliably proliferate or differentiate below 5 mM glucose, and 17.5 mM was chosen as being at the high end of the potential physiological range.
We studied the effect of DHA on differentiating SGBS cells at a 10 µM concentration, which is relatively low compared with physiological plasma levels (64), but higher levels were toxic to SGBS cells. Neither LipiD-QuanT nor Oil Red O measurements showed any significant effect of DHA supplementation. Although our results contrast with a DHA-induced dose-dependent decrease in LD numbers and area in 3T3-L1 cells at higher (25–200 µM) levels of DHA (65), a more recent study also found no change in lipid accumulation of 3T3-L1 cells due to 2 day treatments of either 100 µM EPA or 50 µM DHA (66).
It has been shown in animal models that rosiglitazone lowers circulating triglyceride and free fatty acids and hence improves insulin sensitivity (67, 68) and reduces adipocyte size and fat accumulation in muscle (59, 60). In cultured human subcutaneous adipocytes and in combination with insulin, rosiglitazone stimulated lipolysis in vitro (69). Moreover, on mature 3T3-L1 cells kept in high glucose, 2 day rosiglitazone treatment reduced the measured lipid content by 10% (70). Consistent with this, we have demonstrated that prolonged exposure to rosiglitazone has an antiadipogenic effect during human adipogenesis, and LipiD-QuanT measurements demonstrate that it prevents enlargement of LDs, leading to smaller LD size distribution (Fig. 4J).
Although the emphasis of this manuscript has been LD dynamics with the specific focus on human obesity, the application of Lipid-QuanT is not limited to a specific cell or organism. LDs contain well-conserved features among all organisms (71). Recently, commercial software has been used to measure LD dynamics in murine 3T3-L1 cells after fixation and Oil Red O staining (72). The size distribution of LDs in 3T3-L1 cells is very similar to that of SGBS cells measured using LipiD-QuanT. Thus, Lipid-QuanT should readily be able to be applied to the murine 3T3-L1 adipocyte cell line. Additionally, HepG2 cells are reported to contain LDs ranging from 0.2 µm to 1.4 µm (73). Visualization of the smaller lipid droplets (<0.3 µm) is feasible but would require capturing of images at higher magnification (2×) than the microscopy set-up that is described. Thus, LipiD-QuanT adds further knowledge to previous research that investigated LD accumulation and lipid metabolism in the context of health and disease or due to the influence of various treatments during adipogenesis (74–76).
LipiD-QuanT complements staining and biochemical methods for the measurement of lipid content and LD dynamics. It has the advantages that it can provide measures of LD content and size distribution on a per cell basis, it is nondestructive to cells because the cells do not need to be fixed, and it is user friendly.
In addition, we believe LipiD-QuanT can be adapted to quantify lipid emulsions or lipid research in other systems where the contrast of LD contours is maximized and potentially be used for studies of LD biophysical characters such as stability, shape, and dynamics.
Supplementary Material
Acknowledgments
The authors thank Stephen Bradford and David James’ laboratory at the Garvan Institute of Medical Research for providing the SGBS cell line; Musarat Ishaq for providing PCR reagents; Susan van Dijk and Jing Zhou for providing HiDHA™ tuna oil; Penny Bean, Meg Evans, and Vijay Vaithilingam for assistance with their microscopes; and Michael Gillings, Kim Fung, Wayne Leifert, and Lance Macaulay for critical reading of the manuscript.
Footnotes
The study is part of the EpiSCOPE project supported by Science and Industry Endowment Fund (Australia) grant RP03-064 and is supported by iMQRES and CSIRO OCE Doctoral scholarships (H.V).
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Brown D. A. 2001. Lipid droplets: proteins floating on a pool of fat. Curr. Biol. 11: R446–R449. [DOI] [PubMed] [Google Scholar]
- 2.Haemmerle G., Zimmermann R., Zechner R. 2003. Letting lipids go: hormone-sensitive lipase. Curr. Opin. Lipidol. 14: 289–297. [DOI] [PubMed] [Google Scholar]
- 3.Londos C., Sztalryd C., Tansey J. T., Kimmel A. R. 2005. Role of PAT proteins in lipid metabolism. Biochimie. 87: 45–49. [DOI] [PubMed] [Google Scholar]
- 4.Francois M., Leifert W., Hecker J., Faunt J., Martins R., Thomas P., Fenech M. 2014. Altered cytological parameters in buccal cells from individuals with mild cognitive impairment and Alzheimer’s disease. Cytometry A. 85: 698–708. [DOI] [PubMed] [Google Scholar]
- 5.Fujimoto T., Parton R. G. 2011. Not just fat: the structure and function of the lipid droplet. Cold Spring Harb. Perspect. Biol. 3: pii: a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller M., Stone N. J., Ballantyne C., Bittner V., Criqui M. H., Ginsberg H. N., Goldberg A. C., Howard W. J., Jacobson M. S., Kris-Etherton P. M., et al. 2011. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 123: 2292–2333. [DOI] [PubMed] [Google Scholar]
- 7.Lutz T. A., Woods S. C. 2012. Overview of animal models of obesity. Curr. Protoc. Pharmacol.5.61. [Google Scholar]
- 8.Poulos S. P., Dodson M. V., Hausman G. J. 2010. Cell line models for differentiation: preadipocytes and adipocytes. Exp. Biol. Med. (Maywood). 235: 1185–1193. [DOI] [PubMed] [Google Scholar]
- 9.Green H., Kehinde O. 1975. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 5: 19–27. [DOI] [PubMed] [Google Scholar]
- 10.Wabitsch M., Brenner R. E., Melzner I., Braun M., Moller P., Heinze E., Debatin K. M., Hauner H. 2001. Characterization of a human preadipocyte cell strain with high capacity for adipose differentiation. Int. J. Obes. Relat. Metab. Disord. 25: 8–15. [DOI] [PubMed] [Google Scholar]
- 11.Allott E. H., Oliver E., Lysaght J., Gray S. G., Reynolds J. V., Roche H. M., Pidgeon G. P. 2012. The SGBS cell strain as a model for the in vitro study of obesity and cancer. Clin. Transl. Oncol. 14: 774–782. [DOI] [PubMed] [Google Scholar]
- 12.Fischer-Posovszky P., Newell F. S., Wabitsch M., Tornqvist H. E. 2008. Human SGBS cells: a unique tool for studies of human fat cell biology. Obes. Facts. 1: 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiam A. R., Antonny B., Wang J., Delacotte J., Wilfling F., Walther T. C., Beck R., Rothman J. E., Pincet F. 2013. COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. USA. 110: 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukumoto S., Fujimoto T. 2002. Deformation of lipid droplets in fixed samples. Histochem. Cell Biol. 118: 423–428. [DOI] [PubMed] [Google Scholar]
- 15.Gan W. B., Grutzendler J., Wong W. T., Wong R. O. L., Lichtman J. W. 2000. Multicolor “DiOlistic” labeling of the nervous system using lipophilic dye combinations. Neuron. 27: 219–225. [DOI] [PubMed] [Google Scholar]
- 16.Listenberger L. L., Ostermeyer-Fay A. G., Goldberg E. B., Brown W. J., Brown D. A. 2007. Adipocyte differentiation-related protein reduces the lipid droplet association of adipose triglyceride lipase and slows triacylglycerol turnover. J. Lipid Res. 48: 2751–2761. [DOI] [PubMed] [Google Scholar]
- 17.Robenek H., Robenek M. J., Troyer D. 2005. PAT family proteins pervade lipid droplet cores. J. Lipid Res. 46: 1331–1338. [DOI] [PubMed] [Google Scholar]
- 18.Cirulis J. T., Strasser B. C., Scott J. A., Ross G. M. 2012. Optimization of staining conditions for microalgae with three lipophilic dyes to reduce precipitation and fluorescence variability. Cytometry A. 81: 618–626. [DOI] [PubMed] [Google Scholar]
- 19.Kacmar J., Carlson R., Balogh S. J., Srienc F. 2006. Staining and quantification of poly-3-hydroxybutyrate in Saccharomyces cerevisiae and Cupriavidus necator cell populations using automated flow cytometry. Cytometry A. 69: 27–35. [DOI] [PubMed] [Google Scholar]
- 20.Loudet A., Burgess K. 2007. BODIPY dyes and their derivatives: syntheses and spectroscopic properties. Chem. Rev. 107: 4891–4932. [DOI] [PubMed] [Google Scholar]
- 21.Hellerer T., Axang C., Brackmann C., Hillertz P., Pilon M., Enejder A. 2007. Monitoring of lipid storage in Caenorhabditis elegans using coherent anti-Stokes Raman scattering (CARS) microscopy. Proc. Natl. Acad. Sci. USA. 104: 14658–14663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Le T. T., Duren H. M., Slipchenko M. N., Hu C. D., Cheng J. X. 2010. Label-free quantitative analysis of lipid metabolism in living Caenorhabditis elegans. J. Lipid Res. 51: 672–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dou W., Zhang D. L., Jung Y., Cheng J. X., Umulis D. M. 2012. Label-free imaging of lipid-droplet intracellular motion in early drosophila embryos using femtosecond-stimulated Raman loss microscopy. Biophys. J. 102: 1666–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong C. S. Y., Robinson I., Ochsenkuhn M. A., Arlt J., Hossack W. J., Crain J. 2011. Changes to lipid droplet configuration in mCMV-infected fibroblasts: live cell imaging with simultaneous CARS and two-photon fluorescence microscopy. Biomed. Opt. Express. 2: 2504–2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mouras R., Bagnaninchi P. O., Downes A. R., Elfick A. P. D. 2012. Label-free assessment of adipose-derived stem cell differentiation using coherent anti-Stokes Raman scattering and multiphoton microscopy. J. Biomed. Opt. 17: 116011. [DOI] [PubMed] [Google Scholar]
- 26.Jungst C., Klein M., Zumbusch A. 2013. Long-term live cell microscopy studies of lipid droplet fusion dynamics in adipocytes. J. Lipid Res. 54: 3419–3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osmond-McLeod M. J., Osmond R. I. W., Oytam Y., McCall M. J., Feltis B., Mackay-Sim A., Wood S. A., Cook A. L. 2013. Surface coatings of ZnO nanoparticles mitigate differentially a host of transcriptional, protein and signalling responses in primary human olfactory cells. Part. Fibre Toxicol. 10: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osmond-McLeod MJ, Oytam Y., Osmond R. I. W., Sobhanmanesh F., McCall MJ M. J. 2014. Surface coatings protect against the in vitro toxicity of zinc oxide nanoparticles in human hepatic stellate cells. J Nanomed Nanotechnol 5: 232. [Google Scholar]
- 29.Ostwald W. 1897. Studien uber die bildung und umwandlung fester korper. Z. Phys. Chem. 22: 289. [Google Scholar]
- 30.Ollila O. H. S., Lamberg A., Lehtivaara M., Koivuniemi A., Vattulainen I. 2012. Interfacial tension and surface pressure of high density lipoprotein, low density lipoprotein, and related lipid droplets. Biophys. J. 103: 1236–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallotton P. 2013. Size matters: filamentous bacteria drive interstitial vortex formation and colony expansion in Paenibacillus vortex. Cytometry A. 83: 1105–1112. [DOI] [PubMed] [Google Scholar]
- 32.Kimmel R., Bruckstein A. Regularized laplacian zero crossings as optimal edge integrators. 2001. [Google Scholar]
- 33.Aitkenhead A. H. 2010Boundary tracing using the Moore Neigh-bourhood. Matlab Central File Exchange Accessed April 3, 2012, at http://www.mathworks.com/matlabcentral/fileexchange/27639-boundary-tracing-using-the-moore-neighbourhood..
- 34.Kroon D.-J. 2011. 2D Line Curvature and Normals. Matlab Central File Exchange. Accessed April 3, 2012, at http://www.mathworks.com/matlabcentral/fileexchange/27639-boundary-tracing-using-the-moore-neighbourhood.
- 35.Soille P. Morphological image analysis: principles and applications. Springer-Verlag, New York. [Google Scholar]
- 36.Otsu N. 1979. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 9: 62–66. [Google Scholar]
- 37.Lahnalampi M., Heinäniemi M., Sinkkonen L., Wabitsch M., Carlberg C. 2010. Time-resolved expression profiling of the nuclear receptor superfamily in human adipogenesis. PLoS One. 5: e12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindroos J., Husa J., Mitterer G., Haschemi A., Rauscher S., Haas R., Gröger M., Loewe R., Kohrgruber N., Schrögendorfer K. F., et al. 2013. Human but not mouse adipogenesis is critically dependent on LMO3. Cell Metab. 18: 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murholm M., Isidor M. S., Basse A. L., Winther S., Sorensen C., Skovgaard-Petersen J., Nielsen M. M., Hansen A. S., Quistorff B., Hansen J. B. 2013. Retinoic acid has different effects on UCP1 expression in mouse and human adipocytes. BMC Cell Biol. 14: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weaver R. E., Donnelly D., Wabitsch M., Grant P. J., Balmforth A. J. 2008. Functional expression of glucose-dependent insulinotropic polypeptide receptors is coupled to differentiation in a human adipocyte model. Int. J. Obes. (Lond.) 32: 1705–1711. [DOI] [PubMed] [Google Scholar]
- 41.Calzadilla P., Sapochnik D., Cosentino S., Diz V., Dicelio L., Calvo J. C., Guerra L. N. 2011. N-acetylcysteine reduces markers of differentiation in 3T3–L1 adipocytes. Int. J. Mol. Sci. 12: 6936–6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paar M., Jungst C., Steiner N. A., Magnes C., Sinner F., Kolb D., Lass A., Zimmermann R., Zumbusch A., Kohlwein S. D., et al. 2012. Remodeling of lipid droplets during lipolysis and growth in adipocytes. J. Biol. Chem. 287: 11164–11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suzuki M., Shinohara Y., Ohsaki Y., Fujimoto T. 2011. Lipid droplets: size matters. J. Electron Microsc. (Tokyo). 60: S101–S116. [DOI] [PubMed] [Google Scholar]
- 44.Buckley J. D., Howe P. R. C. 2009. Anti-obesity effects of long-chain omega-3 polyunsaturated fatty acids. Obes. Rev. 10: 648–659. [DOI] [PubMed] [Google Scholar]
- 45.Burrows T., Collins C. E., Garg M. L. 2011. Omega-3 index, obesity and insulin resistance in children. Int J Pediatr Obes 6(2–2): e532–539. [DOI] [PubMed] [Google Scholar]
- 46.Micallef M., Munro I., Phang M., Garg M. 2009. Plasma n-3 polyunsaturated fatty acids are negatively associated with obesity. Br. J. Nutr. 102: 1370–1374. [DOI] [PubMed] [Google Scholar]
- 47.Saito E., Okada T., Abe Y., Kuromori Y., Miyashita M., Iwata F., Hara M., Ayusawa M., Mugishima H., Kitamura Y. 2011. Docosahexaenoic acid content in plasma phospholipids and desaturase indices in obese children. J. Atheroscler. Thromb. 18: 345–350. [DOI] [PubMed] [Google Scholar]
- 48.Garaulet M., Hernandez-Morante J. J., Lujan J., Tebar F. J., Zamora S. 2006. Relationship between fat cell size and number and fatty acid composition in adipose tissue from different fat depots in overweight/obese humans. Int. J. Obes. (Lond.) 30: 899–905. [DOI] [PubMed] [Google Scholar]
- 49.Guillet-Deniau I., Pichard A. L., Kone A., Esnous C., Nieruchalski M., Girard J., Prip-Buus C. 2004. Glucose induces de novo lipogenesis in rat muscle satellite cells through a sterol-regulatory-element-binding-protein-1c-dependent pathway. J. Cell Sci. 117: 1937–1944. [DOI] [PubMed] [Google Scholar]
- 50.Lu B., Ennis D., Lai R., Bogdanovic E., Nikolov R., Salamon L., Fantus C., Le-Tien H., Fantus I. G. 2001. Enhanced sensitivity of insulin-resistant adipocytes to vanadate is associated with oxidative stress and decreased reduction of vanadate (+5) to vanadyl (+4). J. Biol. Chem. 276: 35589–35598. [DOI] [PubMed] [Google Scholar]
- 51.Tang S., Le-Tien H., Goldstein B. J., Shin P., Lai R., Fantus I. G. 2001. Decreased in situ insulin receptor dephosphorylation in hyperglycemia-induced insulin resistance in rat adipocytes. Diabetes. 50: 83–90. [DOI] [PubMed] [Google Scholar]
- 52.Lin Y., Berg A. H., Iyengar P., Lam T. K., Giacca A., Combs T. P., Rajala M. W., Du X., Rollman B., Li W., et al. 2005. The hyperglycemia-induced inflammatory response in adipocytes: the role of reactive oxygen species. J. Biol. Chem. 280: 4617–4626. [DOI] [PubMed] [Google Scholar]
- 53.Saper R. B., Rash R. 2009. Zinc: an essential micronutrient. Am. Fam. Physician. 79: 768–772. [PMC free article] [PubMed] [Google Scholar]
- 54.Mateo M. C., Bustamante J. B., Cantalapiedra M. A. 1978. Serum, zinc, copper and insulin in diabetes mellitus. Biomedicine (Taipei). 29: 56–58. [PubMed] [Google Scholar]
- 55.Melchior T., Simonsen K. W., Johannessen A. C., Binder C. 1989. Plasma zinc concentrations during the 1st 2 years after diagnosis of insulin-dependent diabetes-mellitus: a prospective study. J. Intern. Med. 226: 53–58. [DOI] [PubMed] [Google Scholar]
- 56.Zargar A. H., Shah N. A., Masoodi S. R., Laway B. A., Dar F. A., Khan A. R., Sofi F. A., Wani A. I. 1998. Copper, zinc, and magnesium levels in non-insulin dependent diabetes mellitus. Postgrad. Med. J. 74: 665–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alkaladi A., Afifi M. 2014. Antidiabetic activity of zinc oxide and silver nanoparticles on streptozotocin-induced diabetic rats. Int. J. Mol. Sci. 15: 2015–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Umrani R. D., Paknikar K. M. 2014. Zinc oxide nanoparticles show antidiabetic activity in streptozotocin-induced Type 1 and 2 diabetic rats. Nanomedicine (Lond.). 9: 89–104. [DOI] [PubMed] [Google Scholar]
- 59.Albrektsen T., Frederiksen K. S., Holmes W. E., Boel E., Taylor K., Fleckner J. 2002. Novel genes regulated by the insulin sensitizer rosiglitazone during adipocyte differentiation. Diabetes. 51: 1042–1051. [DOI] [PubMed] [Google Scholar]
- 60.Johnson J. A., Trasino S. E., Ferrante A. W., Vasselli J. R. 2007. Prolonged decrease of adipocyte size after rosiglitazone treatment in high- and low-fat-fed rats. Obesity (Silver Spring). 15: 2653–2663. [DOI] [PubMed] [Google Scholar]
- 61.Molero J. C., Lee S., Leizerman I., Chajut A., Cooper A., Walder K. 2010. Effects of rosiglitazone on intramyocellular lipid accumulation in Psammomys obesus. Biochim. Biophys. Acta. 1802: 235–239. [DOI] [PubMed] [Google Scholar]
- 62.Fève B. 2005. Adipogenesis: cellular and molecular aspects. Best Pract. Res. Clin. Endocrinol. Metab. 19: 483–499. [DOI] [PubMed] [Google Scholar]
- 63.Aguiari P., Leo S., Zavan B., Vindigni V., Rimessi A., Bianchi K., Franzin C., Cortivo R., Rossato M., Vettor R., et al. 2008. High glucose induces adipogenic differentiation of muscle-derived stem cells. Proc Natl Acad Sci U S A 105: 1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maki K. C., Reeves M. S., Farmer M., Griinari M., Berge K., Vik H., Hubacher R., Rains T. M. 2009. Krill oil supplementation increases plasma concentrations of eicosapentaenoic and docosahexaenoic acids in overweight and obese men and women. Nutr. Res. 29: 609–615. [DOI] [PubMed] [Google Scholar]
- 65.Kim H. K., Della-Fera M., Lin J., Baile C. A. 2006. Docosahexaenoic acid inhibits adipocyte differentiation and induces apoptosis in 3T3–L1 preadipocytes. J. Nutr. 136: 2965–2969. [DOI] [PubMed] [Google Scholar]
- 66.Prostek A., Gajewska M., Kamola D., Balasinska B. 2014. The influence of EPA and DHA on markers of inflammation in 3T3–L1 cells at different stages of cellular maturation. Lipids Health Dis. 13: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oakes N. D., Kennedy C. J., Jenkins A. B., Laybutt D. R., Chisholm D. J., Kraegen E. W. 1994. A new antidiabetic agent, BRL 49653, reduces lipid availability and improves insulin action and glucoregulation in the rat. Diabetes. 43: 1203–1210. [DOI] [PubMed] [Google Scholar]
- 68.Yki-Järvinen H. 2004. Thiazolidinediones. N. Engl. J. Med. 351: 1106–1118. [DOI] [PubMed] [Google Scholar]
- 69.McTernan P. G., Harte A. L., Anderson L. A., Green A., Smith S. A., Holder J. C., Barnett A. H., Eggo M. C., Kumar S. 2002. Insulin and rosiglitazone regulation of lipolysis and lipogenesis in human adipose tissue in vitro. Diabetes. 51: 1493–1498. [DOI] [PubMed] [Google Scholar]
- 70.Wang P., Renes J., Bouwman F., Bunschoten A., Mariman E., Keijer J. 2007. Absence of an adipogenic effect of rosiglitazone on mature 3T3–L1 adipocytes: increase of lipid catabolism and reduction of adipokine expression. Diabetologia. 50: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thiel K., Heier C., Haberl V., Thul P. J., Oberer M., Lass A., Jackle H., Beller M. 2013. The evolutionarily conserved protein CG9186 is associated with lipid droplets, required for their positioning and for fat storage. J. Cell Sci. 126: 2198–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rizzatti V., Boschi F., Pedrotti M., Zoico E., Sbarbati A., Zamboni M. 2013. Lipid droplets characterization in adipocyte differentiated 3T3–L1 cells: size and optical density distribution. Eur. J. Histochem. 57: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hur W., Kim S. W., Lee Y. K., Choi J. E., Hong S. W., Song M. J., Bae S. H., Park T., Um S. J., Yoon S. K. 2012. Oleuropein reduces free fatty acid-induced lipogenesis via lowered extracellular signal-regulated kinase activation in hepatocytes. Nutr. Res. 32: 778–786. [DOI] [PubMed] [Google Scholar]
- 74.Koves T. R., Sparks L. M., Kovalik J. P., Mosedale M., Arumugam R., DeBalsi K. L., Everingham K., Thorne L., Phielix E., Meex R. C., et al. 2013. PPARgamma coactivator-1alpha contributes to exercise-induced regulation of intramuscular lipid droplet programming in mice and humans. J. Lipid Res. 54: 522–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Robciuc A., Hyotylainen T., Jauhiainen M., Holopainen J. M. 2012. Hyperosmolarity-induced lipid droplet formation depends on ceramide production by neutral sphingomyelinase 2. J. Lipid Res. 53: 2286–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Russell T. D., Palmer C. A., Orlicky D. J., Fischer A., Rudolph M. C., Neville M. C., McManaman J. L. 2007. Cytoplasmic lipid droplet accumulation in developing mammary epithelial cells: roles of adipophilin and lipid metabolism. J. Lipid Res. 48: 1463–1475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.