SUMMARY
Combination antiretroviral therapy (ART) is able to suppress HIV-1 replication to undetectable levels. However, the persistence of latent viral reservoirs allows for a rebound of viral load upon cessation of therapy. Thus, therapeutic strategies to eradicate the viral latent reservoir are critically needed. Employing a targeted RNAi screen, we identified the ubiquitin ligase BIRC2 (cIAP1), a repressor of the noncanonical NF-κB pathway, as a potent negative regulator of LTR-dependent HIV-1 transcription. Depletion of BIRC2 through treatment with small molecule antagonists known as Smac mimetics enhanced HIV-1 transcription, leading to a reversal of latency in a JLat latency model system. Critically, treatment of resting CD4+ T cells isolated from ART-suppressed patients with the histone deacetylase inhibitor (HDACi) panobinostat together with Smac mimetics resulted in synergistic activation of the latent reservoir. These data implicate Smac mimetics as useful agents for shock-and-kill strategies to eliminate the latent HIV reservoir.
Graphical Abstract
INTRODUCTION
HIV-1 latency is a state of nonproductive infection in which transcription of viral genes is repressed, likely through the concerted activities of multiple host pathways. While HIV-1 replication can be reduced to undetectable levels using combination antiretroviral therapy (ART), latently infected viral reservoirs can persist for decades (reviewed in Margolis, 2010). In well-suppressed patients, cessation of therapy typically leads to increased viremia within 3–4 weeks, and thus HIV-1-infected individuals must remain on ART throughout their lifetimes. Given the expense and toxicities associated with long-term therapies, pharmacological strategies designed to eradicate the viral latent reservoir represent a critical unmet need. Current “shock and kill” approaches seek to purge this reservoir by treating patients with therapeutics that activate latently infected cells, which are thought to be subsequently eliminated due to viral cytopathic effects or the immune response of the host (Xing and Siliciano, 2013). However, the optimal means for reactivating latent HIV-1 is at present unclear.
The establishment and maintenance of HIV-1 latency is controlled by a multitude of cis- and trans-acting mechanisms that include factors affecting the local chromatin environment or the levels of specific transcription factors, respectively (Donahue and Wainberg, 2013). Histone deacetylase inhibitors (HDACis) are known as general activators of transcription and have also been shown to reverse latency in multiple model systems (Margolis, 2011). Recent studies have indicated that most HDACi are unable to reactivate latent HIV-1 ex vivo with robust and consistent efficacies (Bullen et al., 2014; Wei et al., 2014). Therefore, it is presently not clear if treatment with an HDACi as single agent is sufficient to effectively reactivate latent HIV-1.
In addition to the local chromatin environment, specific transcription factors are also critical regulators of viral latency. A significant body of evidence indicates that NF-κB signaling plays an important role in the reactivation of latent HIV-1, implicating its regulation as an important therapeutic strategy for latency reversal (Nabel and Baltimore, 1987; Williams et al., 2006). In fact, among the most efficient proposed latency reversing agents (LRAs) are protein kinase C (PKC) activators, including bryostatin, ingenol, and phorbol esters such as PMA and pros-tratin. These compounds activate the canonical NF-κB pathway and have been found to reverse latency in cellular models and CD4+ T cells from HIV-1-infected patients (Bullen et al., 2014; Spivak et al., 2015; Xing and Siliciano, 2013). However, clinical uptake of these compounds has been complicated by concerns about tumorigenesis and other toxicities, particularly due to uncontrolled cytokine release (Xing and Siliciano, 2013).
Toxicity associated with agonists of canonical NF-κB signaling, which is governed by TAK1/IKK signalosome activation, is in large part due to the acute, short-lived transcriptional activation that is initiated after pathway activation, resulting in a broad inflammatory response. Although abundant evidence implicates canonical NF-κB signaling in the control of HIV-1 transcription and latency, the influence of the noncanonical NF-κB pathway in regulating HIV-1 transcription and latency has not been established. In contrast to canonical NF-κB signaling, the noncanonical NF-κB pathway is characterized by a slower onset, long-lasting transcriptional response, and higher functional selectivity, restricting its impact to a limited number of cellular processes and cell types (reviewed in Sun, 2012). The activation of the non-canonical NF-κB pathway occurs only through a specific subset of tumor necrosis factor receptors (TNFRs), including lymphotoxin beta receptor (LTβR) and CD40. In the absence of stimulation, a complex of BIRC2 (cIAP1), BIRC3 (cIAP2), TRAF2, and TRAF3 constitutively degrades NF-κB-inducing kinase (NIK). Upon receptor activation, BIRC2 and BIRC3 promote the ubiquitination and subsequent degradation of TRAF3, thereby permitting an accumulation of NIK. In turn, NIK activation results in the phosphorylation IKKα, leading to the subsequent proteolytic processing of p100 to p52. p52 forms a heterodimer with the RELB transcription factor and translocates to the nucleus, inducing the expression of target genes. Crosstalk between various canonical and noncanonical NF-κB signaling components, resulting in both positive and negative pathway crossregulation, is considered to be a critical feature in shaping biological responses to a variety of extracellular stimuli (Basak et al., 2007; Shih et al., 2011; Zarnegar et al., 2008).
Here, using targeted RNAi screening, we have identified BIRC2, a repressor of the noncanonical NF-κB pathway, as a potent regulator of HIV-1 transcription and as a therapeutic target for the reversal of latency. Small molecule antagonists of BIRC2, in combination with HDAC inhibitors, activated latent proviruses in both cell line-based models of latency and in primary CD4+ T cells isolated from ART-suppressed HIV-1-infected patients. The results of this study indicate that the pharmacological activation of the noncanonical NF-κB pathway can be used as a component of a combinatorial regimen to reactivate latent HIV-1, while potentially limiting toxicity risks associated with systemic activation of NF-κB signaling by PKC agonists.
RESULTS
Identification of Host Cell Factors that Impede Early Stages of HIV-1 Replication
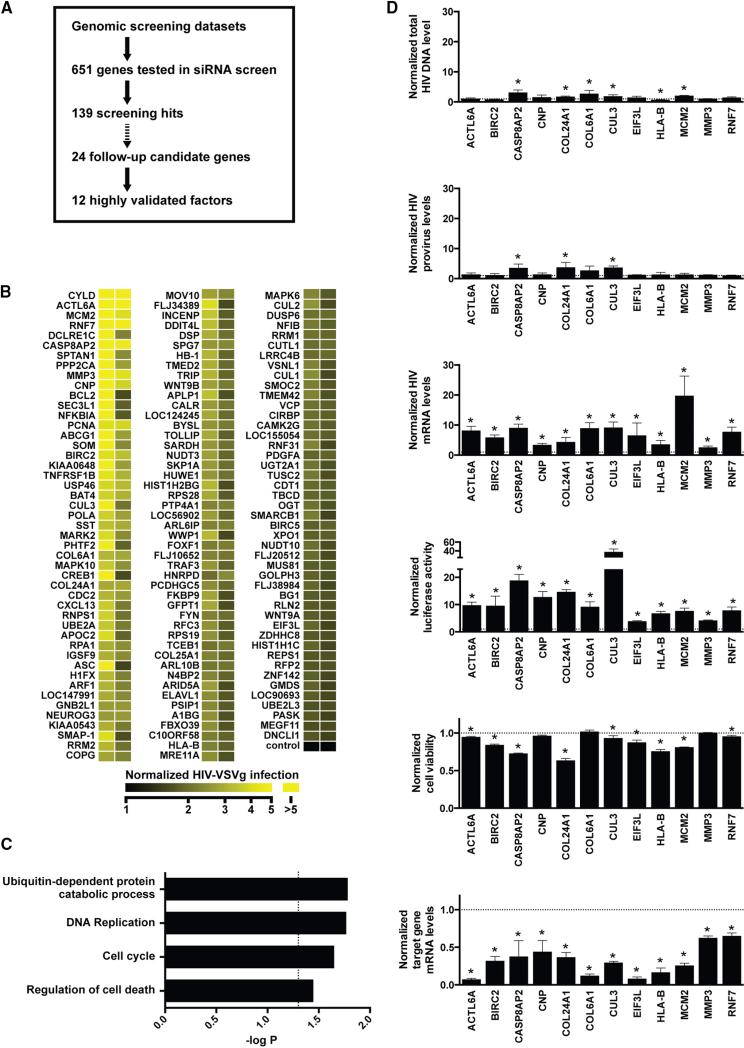
Using a genome-wide siRNA-based loss-of-function screen, we previously identified 295 cellular genes encoding proteins that support viral replication (König et al., 2008). To elucidate cellular factors that interfere with HIV-1 replication, we have reanalyzed this data set along with additional published and unpublished data sets from genome-wide gain- or loss-of-function analyses (Agarwal et al., 2006; Nguyen et al., 2007), identifying 651 genes that were predicted to have a likelihood of impeding HIV-1 replication. Using a triaging strategy based on results from an arrayed siRNA loss-of-function screen (Figure 1A), we evaluated the role of these factors in the early stages of HIV-1 replication, including LTR-mediated transcription. Four distinct siRNAs targeting each candidate gene were evaluated in HEK293T cells infected with a single-cycle, VSV-G-pseudotyped, HIV-1 reporter virus (HIV-1 [VSVg]) in the absence of interferon. Results from this analysis led to the identification of 139 host factors that, when depleted, enhanced viral infection by 50% or more compared to the negative control with at least two independent siRNAs (Figure 1B; see Table S1 available online). Proteins involved in cell cycle regulation, ubiquitination, apoptosis, and DNA replication were found to be enriched in the set of 139 genes (Figure 1C), including CYLD, a gene we recently reported to regulate LTR-dependent transcription (Manganaro et al., 2014).
Figure 1. Identification of Cellular Factors Impeding HIV-1 Replication through Targeted siRNA Screening.
(A) Candidate genes derived from genome-wide screening data sets were evaluated in a siRNA screen in HEK293T cells infected with HIV-1(VSVg). Twenty-four screening hits were selected for further validation, with 12 of these candidate genes being successfully validated using a set of more rigorous criteria (see text).
(B) Heatmap showing the activity of siRNAs targeting 139 genes identified in a siRNA screen described in (A). See also Table S1.
(C) Enrichment analysis of biological processes among screening hits. Shown are selected gene ontology terms enriched with p < 0.05 following Benjamini-Hochberg correction.
(D) Mapping of 12 validated genes to the HIV-1 life cycle. HEK293T cells were treated with siRNAs targeting the indicated genes. Following infection with HIV-1(VSVg), the levels of HIV-1 total DNA, integrated provirus, and HIV-1 mRNA were determined by qPCR. Also shown are luciferase expression levels, target gene knockdown levels, and cell viability upon target gene knockdown. All values are normalized to nontargeting control siRNAs and represent mean ± SD of at least three biological replicates. * indicates statistical significance as determined by Holm-Sidak t test (alpha = 0.05).
Cellular Factors that Limit Viral Transcription
We next performed a more rigorous validation study on 24 of the 139 factors that, when depleted, promoted HIV-1 infection. A gene was considered validated if at least two sequence-independent siRNAs enhanced expression of the luciferase reporter gene by 2.5-fold or more. We excluded genes where RNAi depletion altered cell viability >30% or the mRNA expression level of the targeted gene was not reduced by >35%. We found that 12 of the 24 selected genes met these more rigorous criteria (Table S2), including factors involved in apoptotic signaling (BIRC2, CASP8AP2), ubiquitin-mediated proteolysis (CUL3, RNF7), and antigen presentation (HLA-B). Only one factor induced G2/M cell cycle arrest upon depletion (Figure S1A) (Groschel and Bushman, 2005; Gummuluru and Emerman, 1999). We next used previously established assays to identify the specific steps of the early viral replication cycle that were influenced by these 12 host factors (Figures 1D and S1B). Knockdown of a subset of genes, including CASP8AP2, CUL3, and COL6A1, led to a modest increase in the levels of viral DNA and integrated provirus. Depletion of eight genes enhanced levels of HIV-1 transcription without significantly affecting levels of integrated provirus, among these the ubiquitin ligase BIRC2, a critical regulator of noncanonical NF-κB signaling.
BIRC2 Antagonist Treatment Enhances HIV-1 Infection
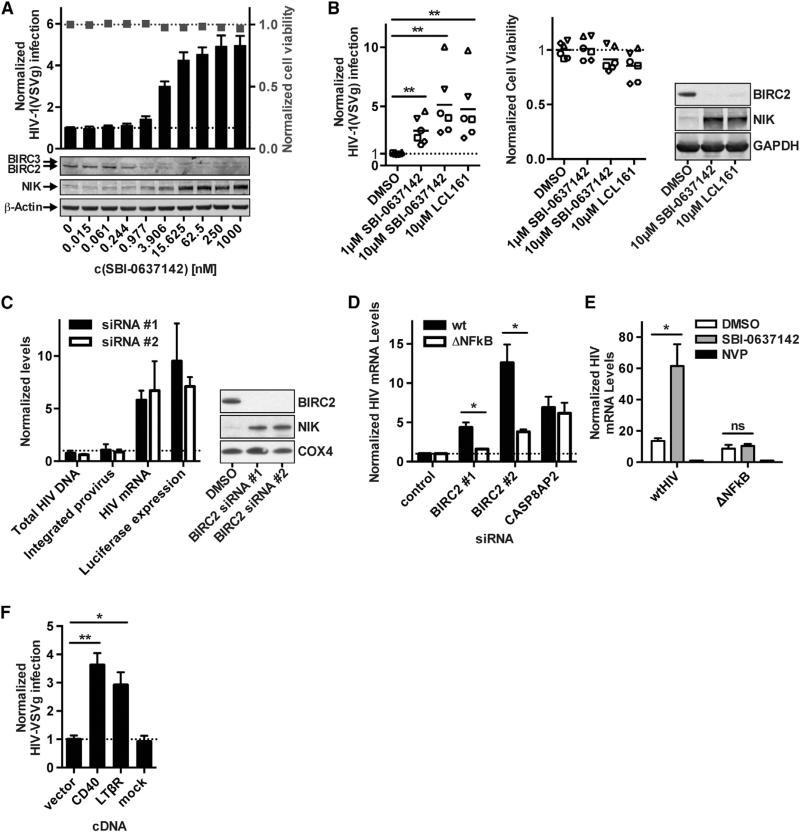
Smac mimetics are synthetic molecules that mimic a critical tetrapeptide sequence from the second mitochondria-derived activator of caspase (Smac/Diablo). Smac binds to the baculoviral IAP repeat (BIR) domain that is common to the eight members of the inhibitor of apoptosis (IAP) family of proteins, which includes XIAP, BIRC2, and BIRC3 (Fulda and Vucic, 2012). The IAP proteins differ in function, and only BIRC2 and BIRC3 are known activators of noncanonical NF-κB signaling. Most Smac mimetics directly compete with caspases for XIAP binding, but also can allosterically activate the E3 ubiquitin ligase activity of BIRC2 and BIRC3, leading to autoubiquitination and subsequent degradation of these proteins. Primarily through their ability to bind XIAP, Smac mimetics can elicit proapoptotic activities, and thus have been developed to treat both solid and hemato-logical cancers (Bai et al., 2014). We have previously described the small molecule SBI-0637142 as a potent Smac mimetic that preferentially targets BIRC2 (Finlay et al., 2014; Vamos et al., 2013). Here, we find that treating 293T cells with SBI-0637142 resulted in enhanced HIV-1 replication, similar to the effects of siRNA-mediated BIRC2 knockdown. This activity was concordant with the depletion of BIRC2 protein, while no change in BIRC3 protein levels was observed (Figure 2A). Furthermore, the loss of BIRC2 led to the accumulation of NIK, indicating that treatment with the Smac mimetic resulted in the activation of the noncanonical NF-κB pathway.
Figure 2. Effects of BIRC2 Depletion on HIV-1 Transcription in HEK293T Cells and Primary Cells.
(A) HEK293T cells were treated with the BIRC2 antagonist SBI-0637142 at the indicated concentrations and infected with HIV-1(VSVg) for 24 hr. Levels of infection were evaluated by measuring luciferase reporter activity. Lysate of SBI-0637142-treated cells was evaluated for BIRC2 and NIK protein levels by western blotting.
(B) Primary activated CD4+ T cells isolated from six healthy donors were treated with SBI-0637142 or LCL161 at the indicated concentrations for 24 hr. Cells were subsequently infected with HIV-1(VSVg) for 48 hr before analysis of luciferase reporter activity. Cell viability was evaluated by measuring cellular ATP levels. Each data point indicates mean of biological triplicates from a single donor. Lines indicate mean of six donors. BIRC2 depletion and NIK accumulation were analyzed by western blotting.
(C) HIV-1(VSVg)-infected HEK293T cells treated with siRNAs targeting BIRC2 were analyzed for levels of total HIV-1 DNA, integrated provirus, and HIV-1 mRNA by qPCR. HIV-1 luciferase levels were evaluated in parallel. All values are normalized to nontargeting control siRNAs. BIRC2 and NIK protein expression levels upon siRNA treatment were analyzed by western blotting.
(D) siRNA-treated HEK293T cells were infected with VSVg-pseudotyped HIV-1 (WT) or a virus mutant lacking functional NF-κB binding sites (ΔNFκB). Viral mRNA was measured by qRT-PCR 24 hr after infection, and values were normalized to nontargeting control siRNA. CASP8AP2-targeting siRNAs are shown as control.
(E) HEK293T cells were treated with 1 μM SBI-0637142 and infected for 24 hr with VSVg-pseudotyped HIV-1 containing either a functional or mutated NF-κB binding site in the viral LTR. HIV-1 mRNA levels were quantified by qPCR and normalized to samples from cells treated with 5 μM nevirapine (NVP).
(F) HEK293T cells transfected with vectors expressing CD40 or LTβR were subsequently infected with HIV-1(VSVg) for 24 hr. Viral infection was quantified by measuring expression of the viral encoded luciferase reporter and normalized to cells transfected with and empty vector as negative control.
All data are represented as mean ± SD of three biological replicates (A, B, and D) or as mean ± SEM of at least three independent experiments (E–G). p values were calculated using an unpaired t test with *p < 0.05 and **p < 0.01.
Next, CD4+ T cells isolated from six healthy donors were treated with SBI-0637142 or LCL161, a second Smac mimetic that has been evaluated in phase I/II clinical trials for patients with advanced solid tumors (Infante et al., 2014). Treatment with SBI-0637142 and LCL161 both enhanced expression of the viral luciferase reporter gene 2- to 10-fold relative to the DMSO control upon HIV-1(VSVg) infection, without inducing significant cytotoxicity (Figure 2B). As expected, both compounds decreased BIRC2 protein levels and resulted in the stabilization of NIK.
BIRC2 Affects Viral Transcription via NF-κB-Dependent Signaling
The HIV LTR contains two copies of an NF-κB enhancer element known to bind the RELA:p50 heterodimer in response to the activation of canonical NF-κB signaling (Nabel and Baltimore, 1987). Observations using in vitro biochemical systems indicate that the noncanonical RELB:p52 heterodimers also can bind these sequences (Britanova et al., 2008; Fusco et al., 2009). Since knockdown of BIRC2, a negative regulator of noncanonical NF-κB signaling, increased expression of HIV-1 mRNA (Figure 2C), we hypothesized that the effects of BIRC2 depletion were mediated through NIK-dependent activation of NF-κB signaling and subsequent interaction of transcription factors with the NF-κB binding sites in the HIV-1 LTR. To test this hypothesis, siRNA-treated HEK293T cells were infected with VSV-G-pseudotyped HIV-1 that had either mutant or native NF-κB binding sites in the LTR (Figure 2D). We found that knocking down BIRC2 by siRNA treatment had little effect on HIV-1 expression when the NF-κB binding sites were inactivated by mutation. Consistent with these findings, mutating the NF-κB binding sites in the LTR abrogated the effects of SBI-0637142 upon HIV-1 transcription (Figure 2E). Moreover, overexpression of LTβR or CD40, both members of the TNF receptor superfamily that stimulate the noncanonical NF-κB pathway, increased the expression of the viral luciferase re porter gene upon HIV-1(VSVg) infection (Figure 2F). Taken together, these results indicate that BIRC2 affects HIV-1 LTR-dependent transcription through regulation of NF-κB signaling.
BIRC2 Antagonists Act as Latency-Reversing Agents
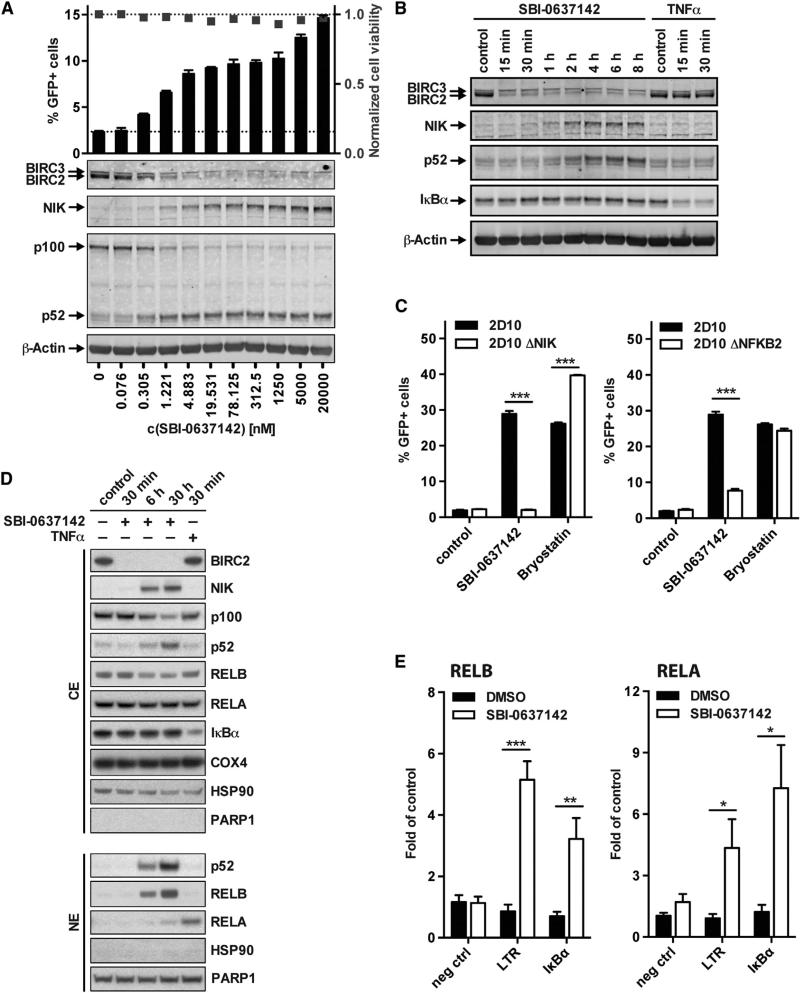
Since transcriptional regulation has been implicated in the maintenance of HIV-1 latency, we investigated whether antagonism of BIRC2 can lead to reactivation of latent infection. Treating the latently infected Jurkat cell line JLat 10.6 with SBI-0637142 led to a dose-dependent reactivation of the provirus with negligible effects on cell viability (Figure 3A). The extent of viral latency reversal was found to be proportional to the depletion of BIRC2 and the activation of the noncanonical NF-κB pathway, as indicated by the accumulation of NIK and the processing of p100 to p52. Importantly, we found that three additional Smac mimetics, which have previously been evaluated in clinical trials (Bai et al., 2014), also showed LRA activity in a Jurkat latency model (Figure S2A). This indicates that latency reversal is not limited to individual compounds, but that Smac mimetics more generally represent a novel class of LRA.
Figure 3. Smac Mimetic-Mediated BIRC2 Depletion Leads to the Reactivation of Latent HIV-1 in JLat Cells.
(A) JLat 10.6 cells were treated with increasing amounts of SBI-0637142 for 36 hr. Reversal of latency was determined by FACS analysis of GPF expression. Depletion of BIRC2, accumulation of NIK, and processing of p100 to p52 were analyzed by western blotting. FACS data are represented as mean ± SEM of three experiments.
(B) Kinetics of NF-κB activation upon treatment of JLat 10.6 cells with 1 μM SBI-0637142 were assessed by western blot analysis. Treatment with 10 ng/ml TNFα served as positive control for canonical NF-κB pathway activation.
(C) 2D10 cells and clones with a knockout of NIK or NFKB2 were incubated with 1 μM SBI-0637142 or 30nM bryostatin for 36 hr. GFP expression was analyzed by FACS. Data are represented as mean ± SD of three biological replicates.
(D) CD4+ T cells from healthy donors were treated with 1 μM SBI-0637142 or 10 ng/ml TNFα. Cytoplasmic (CE) and nuclear (NE) extracts were analyzed by western blot. HSP90 and PARP1 served as control for cytoplasmic and nuclear proteins, respectively.
(E) 2D10 cells were treated with 1 μM SBI-0637142 for 9 hr prior to ChIP analysis using antibodies against RELA, RELB, or IgG as control. RELA- and RELB-specific association with the HIV-1 LTR and the IκBα gene promoter region, or an intergenic region upstream of the PABPC1 gene not known to contain NF-κB binding sites as negative control, was analyzed by qPCR using specific primers and is shown as fold enrichment over IgG control. Data are represented as mean ± SEM of at least three experiments. *p < 0.05, **p < 0.01, ***p < 0.001, determined by unpaired t test.
We also monitored the activation kinetics of NF-κB signaling upon treatment with SBI-0637142. After exposing JLat cells to the Smac mimetic, we observed degradation of BIRC2 within 15 min, followed by the stabilization of NIK after 1 hr, and subsequent increases of the p52 protein levels (Figure 3B), indicating the activation of the noncanonical NF-κB pathway. However, we did not observe a reduction in the levels of IκBα by western blot, which is a hallmark of canonical NF-κB signaling activation (Figure 3B). The kinetics of viral RNA (vRNA) expression following the treatment of latently infected Jurkat cells with SBI-0637142 correlated with the observed induction of NIK-dependent NF-κB signaling (Figure S2B).
To determine whether the observed activation of HIV transcription by Smac mimetics is solely mediated through the NIK signaling axis, we generated a NIK knockout in the latently infected 2D10 cell line using a CRIPSR-based approach (Figure S2C). The loss of NIK abrogated the reactivation of latent HIV-1 through SBI-0637142 treatment, while not impinging upon the activity of PKC agonist bryostatin, an activator of canonical NF-κB signaling (Figure 3C). To further investigate the mechanism by which induction of the noncanonical pathway results in the LTR-dependent transcriptional activation of HIV-1, we have analyzed the nuclear translocation of NF-κB transcription factors upon treatment of CD4+ T cells with SBI-0637142 (Figure 3D). We found that both p52 and RELB translocate to the nucleus following BIRC2 depletion and NIK accumulation. In addition, low levels of RELA translocation were detected as well. Importantly, the translocation of RELA paralleled the delayed kinetics of NF-κB activation via the noncanonical NIK-p100 signaling axis, in contrast to the rapid nuclear accumulation resulting from TNFα stimulation. This suggests that activation of RELA was likely due to pathway crosstalk initiated after the induction of noncanonical signaling.
To further investigate this result, we used chromatin immuno-precipitation (ChIP) to assess the physical occupancy of the HIV-1 LTR by NF-κB transcription factors after induction of noncanonical signaling (Figure 3E). We find that Smac mimetic treatment leads to an association of RELB with the viral LTR, suggesting that noncanonical NF-κB transcription factors can directly influence HIV transcription. Moreover, we also detected binding of RELA to the HIV LTR, implicating a concerted action of both canonical and noncanonical transcription factors in HIV-1 activation upon Smac mimetic treatment. TNFα treatment, by contrast, resulted in RELA binding to the LTR but did not lead to a significant interaction with RELB (Figure S2D). To discern the functional contribution of the noncanonical RELB:p52 heterodimer to LRA activity after Smac mimetic treatment, we created a cell line with a knockout of the NFKB2 gene, resulting in a loss of p100 expression (Figure S2E). In the absence of p100, and thereby p52, the reversal of HIV latency by Smac mimetics was reduced by ~75% (Figure 3C). Taken together, these results indicate that the LRA activity of Smac mimetics is solely initiated through the NIK-dependent noncanonical NF-κB signaling pathway. Induction of this pathway activates both the RELB:p52 heterodimer and, to a lesser extent, the RELA:p50 heterodimer, and results in the activation of LTR-dependent transcription that is primarily, but not exclusively, regulated by the noncanonical NF-κB transcription factor heterodimer.
BIRC2 Antagonist Treatment Acts Synergistically with HDAC Inhibitors to Reverse HIV-1 Latency
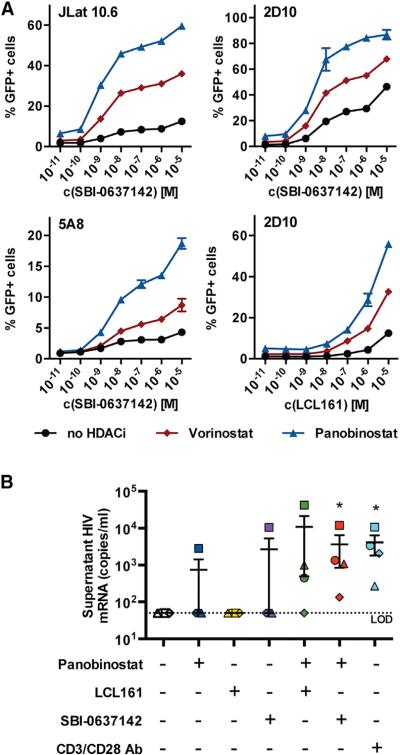
LRAs, such as PKC activators and HDAC inhibitors, have been shown to activate latent provirus synergistically (Burnett et al., 2010; Laird et al., 2015). We thus investigated the effects of Smac mimetics together with HDACi in three Jurkat-derived latency models—JLat 10.6, 2D10, and 5A8 cells (Figures 4A and S3A) (Pearson et al., 2008; Sakane et al., 2011). In each of the three cell lines tested, the combination of the Smac mimetic SBI-0637142, with either of the HDAC inhibitors panobinostat or vorinostat, reactivated latent provirus synergistically (Figure S3B). Although less potent, the Smac mimetic LCL161 showed similar synergy.
Figure 4. Combined Treatment with Smac Mimetics and HDAC In-hibitors Reverses HIV-1 Latency in Jurkat-Based Latency Model and Patient-Derived Resting CD4+ T Cells.
(A) Latently infected Jurkat cell lines were treated with increasing amounts of SBI-0637142 alone or in combination with panobinostat or vorinostat. JLat 10.6 and 5A8 cells were treated with 10 nM panobinostat and 500 nM vorinostat, where indicated. 2D10 cells were treated with 5 nM panobinostat and 250 nM vorinostat. GFP expression was evaluated after 36 hr by FACS. Data are represented as mean ± SD of three biological replicates.
(B) Resting CD4+ T cells from HIV-1-infected patients under ART were treated with 100 nM panobinostat, 10 μM LCL161, 10 μM SBI-0637142, or a combination thereof for 48 hr. Viral production was subsequently evaluated by detection of viral mRNA in cell supernatants using qPCR. Data are represented as mean ± SEM of four donors. Significance of treatments was evaluated using a ratio paired t test. *p < 0.05.
We next evaluated Smac mimetics for their ability to reverse latency in resting CD4+ T cells collected from HIV-1-infected patients undergoing ART (n = 4). Cells were treated with SBI-0637142 or LCL161 alone, or in combination with the HDACi panobinostat, and release of viral genomic RNA to the supernatant was measured by the recently described REVEAL (rapid ex vivo evaluation of antilatency) assay (Spivak et al., 2015). While none of the small molecules were found to activate latent provirus after 48 hr when used individually, both LCL161 and SBI-0637142 in combination with panobinostat were found to reactivate latent HIV-1 at levels comparable to those achieved upon treatment with antibodies against CD3 and CD28 (Figure 4B). Importantly, these results reach statistical significance (p < 0.05) for SBI-0637142 and CD3/CD28 antibody treatment, and no significant activation of resting CD4+ T cells was observed upon treatment with SBI-0637142 (Figure S3C).
DISCUSSION
Using a targeted siRNA screen, we identified BIRC2, a negative regulator of noncanonical NF-κB signaling, as a regulator of viral transcription. Critically, we observed that induction of non-canonical signaling results in both RELA:p50 and RELB:p52 heterodimeric transcription factors binding to the HIV-1 LTR, with genetic loss-of-function studies indicating that the transcriptional activation by RELB:p52 is the predominant regulator of this activity. Taken together, these results support an unappreciated role for the noncanonical NF-κB signaling machinery, and specifically RELB:p52, in the regulation of HIV-1 LTR-dependent transcription.
There have been considerable efforts focused on understanding the molecular basis of HIV-1 latency and devising pharmacological strategies to activate the latent provirus. Ideally, an LRA should only target cell types that are latently infected so as to avoid toxicity or widespread immune activation. In contrast to strategies targeting canonical NF-κB signaling, it may be preferable to target components of the noncanonical NF-κB pathway, as it is active in a more restricted set of cell lineages. Smac mimetics are considered a promising new class of cancer therapeutics that are well tolerated in vivo, and primarily through their antagonism of XIAP, they promote apoptosis in tumor cells, while normal tissue remains unaffected (Fulda and Vucic, 2012). Many Smac mimetics can also trigger the noncanonical NF-κB signaling pathway through the depletion of BIRC2 or BIRC3. Although constitutive activation of the noncanonical NF-κB pathway through somatic mutations in mice has been associated with hematogenous malignancies including B cell lymphomas (Keats et al., 2007), proposed latency-reversing “shock and kill” approaches entail acute treatment regimens, largely mitigating these risks associated with chronic activation. In fact, six Smac mimetics, including LCL161, have been evaluated in clinical trials and have favorable safety and pharmacokinetic/pharmacodynamic (PK/PD) profiles (Bai et al., 2014). Unlike broadly acting Smac mimetics that target multiple proteins of the IAP family including the caspase inhibitor XIAP, certain compounds, such as SBI-0637142, preferentially target BIRC2 or BIRC3, the only known regulators of noncanonical NF-κB signaling among the IAP proteins (Finlay et al., 2014). Future studies will be required to investigate whether the observed increased potency of SBI-0637142 as an LRA may be related to its selectivity profile.
Many current strategies for reactivating latent HIV-1 focus on the use of PKC agonists to stimulate canonical NF-κB signaling. Due to toxicity risks, safety concerns have dampened enthusiasm for the use of these canonical NF-κB activators as LRAs (Morgan et al., 2012). HDACi have also shown promise as LRAs, with multiple compounds being evaluated in clinical trials (Rasmussen et al., 2013). Although HDACis increase intra-cellular levels of HIV-1 mRNA both in vitro and in vivo, the level of viral outgrowth induced by these compounds is likely insufficient to purge the viral reservoir (Bullen et al., 2014; Wei et al., 2014). Therefore, it is expected that a safe and effective drug regimen to reverse HIV-1 latency will require the combination of multiple agents (Xing and Siliciano, 2013), much like ART. Some HDACi have been shown to synergize with different classes of LRAs including PKC agonists (Laird et al., 2015), indicating that combinatorial use could increase efficacy while reducing required dosage. Consistently, we see similar levels of synergy between Smac mimetics and HDACi as levels reported for combinatorial treatment with HDACi and PKC agonists (Wong et al., 2014).
Given the scarcity of clinical data to date, the optimal strategy for reversing HIV-1 latency in patients is far from certain. Our results demonstrating that Smac mimetics, in conjunction with the HDACi panobinostat, can reverse latency in patient-derived resting CD4+ T cells suggest a promising clinical approach toward the development of a “cure” for patients with HIV-1. Importantly, the established clinical safety and pharmacodynamic profiles of Smac mimetics should enable this class of small molecule antagonists to be readily evaluated as a therapeutic strategy. Taken together, these data indicate that rapid preclinical development and clinical repositioning of Smac mimetics may help provide a safe and effective combinatorial therapeutic regimen to eradicate HIV-1.
EXPERIMENTAL PROCEDURES
siRNA Transfections and Infection with HIV-1
siRNA transfections of HEK293T cells and infections with a single-cycle envelope deleted, VSV-G-pseudotyped HIV-1 reporter virus (HIV-1[VSVg]), were performed as previously described (König et al., 2008). Cells were infected 48 hr after siRNA transfection, and luciferase expression levels were determined 24 hr after infection. Mapping to viral life cycle stages was done by isolating mRNA and DNA from infected cells and quantifying proviral DNA content, total HIV DNA, and HIV mRNA levels by qPCR. Cells infected with VSV-G-pseudotyped HIV-1 containing either wild-type or mutant NF-κB binding sites in the LTR (Bosque and Planelles, 2009) were analyzed by measuring HIV mRNA levels by qPCR. Cell viability was analyzed using the ATPlite cell viability assay (Perkin Elmer).
HIV-1 Infection of Human CD4+ T Cells
Following isolation, CD4+ T cells from healthy donors were activated with 4 μg/ml phytohemagglutinin-P (PHA, Sigma) for 48 hr. Activated CD4+ T cells were treated with compounds for 24 hr prior to infection with HIV-1(VSVg). Luciferase expression levels were normalized to mock-treated cells; mean of DMSO-treated cells was defined as 1.
Jurkat HIV Latency Model
Latently infected Jurkat cells were treated with compounds and subsequently analyzed for GFP expression by flow cytometry. Analytical cytometry was performed in the Sanford Burnham Prebys Flow Cytometry Core. Cell viability was determined using the ATPlite cell viability assay.
Chromatin Immunoprecipitation
2D10 cells were stimulated with 1 μM SBI-0637142 or DMSO for 9 hr prior to ChIP using antibodies targeting RELA and RELB.
Treatment of Resting CD4+ T Cells from Aviremic HIV Patients
Resting CD4+ T cells isolated from aviremic HIV-1-infected patients on ART were cultured in the presence or absence of reactivating compounds for 48 hr. Culture supernatants were analyzed by two-step qPCR using a primer and probe set for conserved regions of the 3′ LTR of HIV-1 mRNA as previously described (Spivak et al., 2015).
Please refer to the Supplemental Experimental Procedures for additional information.
Supplementary Material
Highlights.
Targeted RNAi screen identifies host proteins that impede early-stage HIV-1 replication
BIRC2/cIAP1 is a negative regulator of LTR-dependent HIV-1 transcription
BIRC2 depletion by Smac mimetic activates NF-κB signaling and reverses HIV-1 latency
Smac mimetic and HDAC inhibitor synergize to reverse HIV-1 latency in vitro and ex vivo
ACKNOWLEDGMENTS
We are grateful for advice on flow cytometry from D. Chambers and C. O'Connor (Salk Institute) and for assistance with viral stock preparation from The Salk Institute Gene Transfer, Targeting, and Therapeutics Core. We thank Carl Ware (Sanford Burnham Prebys Medical Discovery Institute) for providing cDNA constructs of NF-κB regulators. We would like to thank Warner C. Greene (Gladstone Institutes) for making the cell line 5A8 available to us. We thank Jonathan Karn (Case Western Reserve University) for providing the cell line 2D10. This study was supported by NIH/NIAID grants P01 AI09093 to A.F.-S., V.S., F.D.B., J.A.T.Y., and S.K.C.; by grant R01 DA033773 to S.K.C.; and in part by grant R01 AI087508 to V.P. We acknowledge funding from NIH/NIAID grants 5R01 AI073450 and HHSN272201400008C and from DOD/DARPA grant HR0011-11-C-0094 to A.F.-S. This work was also supported by a generous grant from the James B. Pendleton Charitable Trust to support HIV/AIDS research. J.P.M. is currently an employee of Gilead Sciences. J.A.T.Y. is currently employed by F. Hoffmann-La Roche Ltd. All contributions by J.P.M. and J.A.T.Y. to this study occurred prior to their employment at Gilead Sciences and F. Hoffmann-La Roche Ltd., respectively.
Footnotes
AUTHOR CONTRIBUTIONS
L.P., J.A.T.Y., and S.K.C. conceived the study. L.P., J.M.M., Y.H., A.M.M., L.M., and R.K. conducted screening and validation experiments. L.P., M.S.D., and J.P.M. conducted mechanistic studies. A.M.S., L.J.M., and A.B. conducted patient cell assays. M.V., P.T., and N.D.P.C. developed and synthesized compounds. V.S., A.F.-S., N.D.P.C., F.D.B., J.A.T.Y., V.P., and S.K.C. supervised the studies. L.P. and S.K.C. wrote the manuscript.
SUPPLEMENTAL INFORMATION
Supplemental Information includes three figures, two tables, and Supplemental Experimental Procedures and can be found with this article at http://dx.doi.org/10.1016/j.chom.2015.08.009.
REFERENCES
- Agarwal S, Harada J, Schreifels J, Lech P, Nikolai B, Yamaguchi T, Chanda SK, Somia NV. Isolation, characterization, and genetic complementation of a cellular mutant resistant to retroviral infection. Proc. Natl. Acad. Sci. USA. 2006;103:15933–15938. doi: 10.1073/pnas.0602674103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Smith DC, Wang S. Small-molecule SMAC mimetics as new cancer therapeutics. Pharmacol. Ther. 2014;144:82–95. doi: 10.1016/j.pharmthera.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basak S, Kim H, Kearns JD, Tergaonkar V, O'Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. A fourth IkappaB protein within the NF-kappaB signaling module. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosque A, Planelles V. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood. 2009;113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova LV, Makeev VJ, Kuprash DV. In vitro selection of optimal RelB/p52 DNA-binding motifs. Biochem. Biophys. Res. Commun. 2008;365:583–588. doi: 10.1016/j.bbrc.2007.10.200. [DOI] [PubMed] [Google Scholar]
- Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat. Med. 2014;20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett JC, Lim KI, Calafi A, Rossi JJ, Schaffer DV, Arkin AP. Combinatorial latency reactivation for HIV-1 subtypes and variants. J. Virol. 2010;84:5958–5974. doi: 10.1128/JVI.00161-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue DA, Wainberg MA. Cellular and molecular mechanisms involved in the establishment of HIV-1 latency. Retrovirology. 2013;10:11. doi: 10.1186/1742-4690-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay D, Vamos M, González-López M, Ardecky RJ, Ganji SR, Yuan H, Su Y, Cooley TR, Hauser CT, Welsh K, et al. Small-molecule IAP antagonists sensitize cancer cells to TRAIL-induced apoptosis: roles of XIAP and cIAPs. Mol. Cancer Ther. 2014;13:5–15. doi: 10.1158/1535-7163.MCT-13-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 2012;11:109–124. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- Fusco AJ, Huang DB, Miller D, Wang VY, Vu D, Ghosh G. NF-kappaB p52:RelB heterodimer recognizes two classes of kappaB sites with two distinct modes. EMBO Rep. 2009;10:152–159. doi: 10.1038/embor.2008.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groschel B, Bushman F. Cell cycle arrest in G2/M promotes early steps of infection by human immunodeficiency virus. J. Virol. 2005;79:5695–5704. doi: 10.1128/JVI.79.9.5695-5704.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gummuluru S, Emerman M. Cell cycle- and Vpr-mediated regulation of human immunodeficiency virus type 1 expression in primary and transformed T-cell lines. J. Virol. 1999;73:5422–5430. doi: 10.1128/jvi.73.7.5422-5430.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Dees EC, Olszanski AJ, Dhuria SV, Sen S, Cameron S, Cohen RB. Phase I dose-escalation study of LCL161, an oral inhibitor of apoptosis proteins inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 2014;32:3103–3110. doi: 10.1200/JCO.2013.52.3993. [DOI] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE, et al. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49–60. doi: 10.1016/j.cell.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird GM, Bullen CK, Rosenbloom DI, Martin AR, Hill AL, Durand CM, Siliciano JD, Siliciano RF. Ex vivo analysis identifies effective HIV-1 latency-reversing drug combinations. J. Clin. Invest. 2015;125:1901–1912. doi: 10.1172/JCI80142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manganaro L, Pache L, Herrmann T, Marlett J, Hwang Y, Murry J, Miorin L, Ting AT, König R, García-Sastre A, et al. Tumor suppressor cylindromatosis (CYLD) controls HIV transcription in an NF-κB-dependent manner. J. Virol. 2014;88:7528–7540. doi: 10.1128/JVI.00239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DM. Mechanisms of HIV latency: an emerging picture of complexity. Curr. HIV/AIDS Rep. 2010;7:37–43. doi: 10.1007/s11904-009-0033-9. [DOI] [PubMed] [Google Scholar]
- Margolis DM. Histone deacetylase inhibitors and HIV latency. Curr. Opin. HIV AIDS. 2011;6:25–29. doi: 10.1097/COH.0b013e328341242d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan RJ, Jr., Leong L, Chow W, Gandara D, Frankel P, Garcia A, Lenz HJ, Doroshow JH. Phase II trial of bryostatin-1 in combination with cisplatin in patients with recurrent or persistent epithelial ovarian cancer: a California cancer consortium study. Invest. New Drugs. 2012;30:723–728. doi: 10.1007/s10637-010-9557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- Nguyen DG, Yin H, Zhou Y, Wolff KC, Kuhen KL, Caldwell JS. Identification of novel therapeutic targets for HIV infection through functional genomic cDNA screening. Virology. 2007;362:16–25. doi: 10.1016/j.virol.2006.11.036. [DOI] [PubMed] [Google Scholar]
- Pearson R, Kim YK, Hokello J, Lassen K, Friedman J, Tyagi M, Karn J. Epigenetic silencing of human immunodeficiency virus (HIV) transcription by formation of restrictive chromatin structures at the viral long terminal repeat drives the progressive entry of HIV into latency. J. Virol. 2008;82:12291–12303. doi: 10.1128/JVI.01383-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TA, Tolstrup M, Winckelmann A, Østergaard L, Søgaard OS. Eliminating the latent HIV reservoir by reactivation strategies: advancing to clinical trials. Hum. Vaccin. Immunother. 2013;9:790–799. doi: 10.4161/hv.23202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakane N, Kwon HS, Pagans S, Kaehlcke K, Mizusawa Y, Kamada M, Lassen KG, Chan J, Greene WC, Schnoelzer M, Ott M. Activation of HIV transcription by the viral Tat protein requires a demethylation step mediated by lysine-specific demethylase 1 (LSD1/KDM1). PLoS Pathog. 2011;7:e1002184. doi: 10.1371/journal.ppat.1002184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih VF, Tsui R, Caldwell A, Hoffmann A. A single NFkB system for both canonical and non-canonical signaling. Cell Res. 2011;21:86–102. doi: 10.1038/cr.2010.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivak AM, Bosque A, Balch AH, Smyth D, Martins L, Planelles V. Ex Vivo Bioactivity and HIV-1 Latency Reversal by Ingenol Dibenzoate and Panobinostat in Resting CD4+ T Cells from Aviremic Patients. Antimicrob. Agents Chemother. 2015 doi: 10.1128/AAC.01077-15. http://dx.doi.org/10.1128/AAC.01077-15, AAC.01077-15. [DOI] [PMC free article] [PubMed]
- Sun SC. The noncanonical NF-κB pathway. Immunol. Rev. 2012;246:125–140. doi: 10.1111/j.1600-065X.2011.01088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vamos M, Welsh K, Finlay D, Lee PS, Mace PD, Snipas SJ, Gonzalez ML, Ganji SR, Ardecky RJ, Riedl SJ, et al. Expedient synthesis of highly potent antagonists of inhibitor of apoptosis proteins (IAPs) with unique selectivity for ML-IAP. ACS Chem. Biol. 2013;8:725–732. doi: 10.1021/cb3005512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei DG, Chiang V, Fyne E, Balakrishnan M, Barnes T, Graupe M, Hesselgesser J, Irrinki A, Murry JP, Stepan G, et al. Histone deacetylase inhibitor romidepsin induces HIV expression in CD4 T cells from patients on suppressive antiretroviral therapy at concentrations achieved by clinical dosing. PLoS Pathog. 2014;10:e1004071. doi: 10.1371/journal.ppat.1004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong VC, Fong LE, Adams NM, Xue Q, Dey SS, Miller-Jensen K. Quantitative evaluation and optimization of co-drugging to improve anti-HIV latency therapy. Cell. Mol. Bioeng. 2014;7:320–333. doi: 10.1007/s12195-014-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing S, Siliciano RF. Targeting HIV latency: pharmacologic strategies toward eradication. Drug Discov. Today. 2013;18:541–551. doi: 10.1016/j.drudis.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh WC, Mak TW, et al. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat. Immunol. 2008;9:1371–1378. doi: 10.1038/ni.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.