Abstract
Post-translational modification of the histone proteins in chromatin plays a central role in epigenetic control of DNA-templated processes in eukaryotic cells. Developing methods that enable the structure of histones to be manipulated is therefore essential to understand the biochemical mechanisms underlying genomic regulation. Here we present a synthetic biology method to engineer histones bearing site-specific modifications on cellular chromatin using protein trans-splicing. We genetically fused the N-terminal fragment of ultrafast split-intein to the C-terminus of histone H2B, which upon reaction with a complementary synthetic C-intein, generated labeled histone. Using this approach, we incorporated various non-native chemical modifications to chromatin in vivo with temporal control. Furthermore, the time and concentration dependence of protein trans-splicing performed in nucleo enabled us to examine differences in the accessibility of the euchromatin and heterochromatin regions of the epigenome. Finally, we used protein trans-splicing to semi-synthesize a native histone modification, H2BK120 ubiquitination, in isolated nuclei, and show that this can trigger downstream epigenetic cross-talk of H3K79 methylation.
Graphical Abstract

Chromatin serves as the physiologically relevant form of eukaryotic genomes. Modifications to both the DNA and the histone packaging proteins allow chromatin to act as a dynamic signaling platform to regulate genomic DNA access and ultimately establish and maintain cellular phenotypes, so-called epigenetic regulation1. Aberrant chromatin signalling, as a consequence of abnormal inputs and outputs, is associated with many diseases, especially cancer2. A full understanding of individual chromatin signalling processes, and their interconnectivity, is a pre-requisite to designing next generation therapeutic agents that act to ameliorate epigenetic dysregulation3,4.
Approaches to studying chromatin regulation fall into two broad categories; (i) those that approach the problem at a cellular level often employing top-down “omics” methods to derive systems-wide information5 and, (ii) those that take a more reductionist approach and often employ reconstituted biochemical systems to tackle specific mechanistic questions6–8. Both these strategies have attendant strengths and weaknesses. The top-down approach leads to the generation of large amounts of correlative information that points to, in particular, the existence of syntactically (and likely functionally) distinct chromatin states9, but does not allow precise mechanisms underlying the establishment and propagation of these states to be directly deduced. By contrast, biochemical approaches do allow direct cause-and-effect to be studied, often in a highly quantitative fashion, but they are limited in the information they provide on how these individual processes might be tuned at a systems level.
Exploring specific epigenetic mechanisms in the complex milieu of the cell nucleus is extremely challenging and is typically approached by employing genetic or pharmacological manipulation of relevant chromatin modifying factors7,10 or, in suitable cases, by incorporating PTM mimics through mutation of the histones (i.e. glutamic acid for serine phosphorylation and glutamine for lysine acetylation)11. However, these approaches suffer from lack of specificity (histone modifying enzymes are often rather promiscuous and/or have non-histone targets) or lack of chemical control (most histone PTMs cannot be mimicked with canonical amino acids). Methods that bring the precision and flexibility of synthetic chemistry to a native chromatin context provide one possible solution to the problem of how to study chromatin biochemistry in vivo.
Here we introduce a synthetic biology strategy for tailoring the chromatin template. Our approach is based on the use of ultrafast split inteins, naturally fractured proteins that tightly associate and then rapidly catalyze protein trans-splicing12. We demonstrate that suitably engineered split inteins can be used to effect traceless ligation of a wide range of artificial probes to designated chromatin bound histones in cells. The utility of the approach is demonstrated through the in situ generation of site-specifically ubiquitinated chromatin, leading to upregulation of a downstream epigenetic signaling pathway. Moreover, by exploiting the split intein association step preceding the protein splicing reaction, we show that it is possible to interrogate chromatin structure yielding information on the accessibility of genes located in euchromatin and heterochromatin regions of the epigenome. This method bridges together the biochemical and systemic methods to interrogate chromatin by introducing chemically defined histones into native chromatin.
Results
Chemical modification of chromatin in live cells
Our approach exploits split inteins for the in situ generation of semisynthetic chromatin (Fig. 1a). The method is based on expression of the N-terminal fragment of a split intein (IntN) fused to the desired histone and the subsequent cellular delivery of the C-terminal intein fragment (IntC) bearing the synthetic probe of interest. Association of IntN and IntC on the chromatin template leads to the generation of a semi-synthetic histone with expulsion of the intein fragments. A crucial feature of this technology is efficient incorporation of the requisite histone-IntN fusion into chromatin. Preliminary experiments were performed to identify an IntN fragment that is efficiently localized to chromatin when expressed in mammalian cells fused to histone H2B. Of note, this screen included split inteins that have never been explored previously for protein trans-splicing in mammalian cells13,14. Of the various H2B-IntN constructs examined, the fusion containing the IntN fragment of a split intein from the cyanobacterium, Anabaena variabilis (Ava), was consistently found to have the highest expression levels (7–10% of the total H2B) (Supplemental Fig. 1). Fractionation followed by western blotting analysis indicated the majority of the H2B-AvaN fusion is chromatin bound and not freely diffusing in the nucleoplasm (Supplemental Fig. 2). MTT and trypan blue stain assays were used to show that expression of H2B-AvaN did not affect cell viability (Supplemental Fig. 3–5). Moreover, global RNA-seq analysis revealed no major perturbation in the transcriptome upon overexpression of H2B-AvaN (Supplementary Fig. 6).
Figure 1. Modification of native chromatin using protein trans-splicing.
a) Schematic of the approach. A peptide containing the IntC sequence and the desired cargo (purple star) is conjugated to a cell penetrating peptide (CPP) via a disulfide bond and added to cultured live cells. (i) Cellular uptake of the CPP-conjugate via endocytosis. (ii) Endosomal lysis releases the peptide into the cytoplasm. (iii) Reduction of the disulfide bond in the cytosol yields a protein trans-splicing competent version of IntC. (iv) Passive diffusion of the IntC-cargo into the nucleus where it reacts with a histone fused to the complementary IntN fragment, which is embedded in native chromatin. b) Details of the protein constructs used in this study. IntN and IntC refer to the N- and C-terminal fragments of the Ava and Npu DnaE split inteins, respectively. The product as well as the experimental setting of each reaction is noted.
As a first step towards performing protein trans splicing on cellular chromatin, we extracted mononucleosomes from 293T cells expressing H2B-(Ava)IntN (1) (containing a C-terminal Flag epitope for detection purposes) and exposed these to construct 2 comprising the IntC fragment of the closely related Npu split intein (from Nostoc Punctiforme) fused to an HA epitope tag (Fig. 1b, Supplemental Fig. 7). We used the Npu-IntC fragment since it reacts with AvaN as efficiently and rapidly as its native AvaC partner and its synthesis is highly accessible15. Western blotting analysis of the reaction revealed the time-dependent accumulation of the expected splicing product, H2B-HA, as well as the excised IntN fragment (Fig. 2a). We verified that the reaction produces no undesired side-products, in particular the N-terminal cleavage reaction that would result in free histone (Supplemental Fig. 8). Thus, the Ava-IntN and Npu-IntC pair was used for all subsequent studies.
Figure 2. Modification of histone H2B in native chromatin.
a) Western blot analysis of MNase-treated chromatin extracted from 293T cells, with or without H2B-IntN (1) overexpression, and subsequently incubated with 0.5 μM of construct 2, at 37°C, for the indicated times. b) Western blot analysis of cultured 293T cells expressing H2B-IntN (1) that were treated with different concentrations of construct 3 for the indicated times. c) Characterization of the in-cell splicing product by MS. The reaction was performed as in panel b, using 2.5 μM of construct 3 for two hours. Following cell lysis and DNA shearing, chromatin containing the splicing product, H2B-HA was enriched by ChIP against the HA tag. The isolated histones were then trypsinized and analyzed by high-resolution nano-UPLC/MS. Presented here is the CID MS/MS spectrum for one of the identified peptide products from the ligation reaction. Bottom, MS/MS of the [M+2H]2+ peptide FAEYcFNK (Cys is carbamidomethylated) shown with b- and y-ions indicated in red and blue, respectively. Top left, high resolution MS spectrum of the parent ion at m/z 539.73742. Top right, the peptide sequence is annotated to indicate the detected b- and y- ions. d) Fluorescence-labeling of native chromatin detected by confocal microscopy. Top: reaction scheme depicting the formation of TMR (pink star) labeled H2B from the PTS reaction between H2B-IntN (1) and IntC peptide (4) containing TMR, QSY quencher (purple crescent), and a cell penetrating peptide (CPP). Upon splicing, the TMR fluorophore is no longer quenched by QSY. Bottom: 293T cells expressing H2B-IntN (1) were incubated with 2.5 μM of quenched construct 4 for two hours and visualized by confocal microscopy. Representative images from non-transfected (left) and H2B-IntN transfected cells (right). Each panel is composed of a fluorescence image (left), bright field image (middle) and an overlay of both (right). Scale bar corresponds to a size of 10 μm. See supplementary figure 15 for more images.
Encouraged by the in vitro splicing experiment, we next asked if chromatin could be labeled in intact cells. For this, we prepared construct 3 in which a cell penetrating peptide (CPP), corresponding to the TAT sequence fused to an endosome escape motif16, was linked to a key cysteine thiol group in peptide 2 (Fig. 1b, Supplemental Fig. 7). This design ensures that the delivered fragment becomes reactive only upon removal of the CPP through reduction of the disulfide in the cytoplasm (Fig. 1a)17. 293T cells transfected with construct 1 were treated with 4 or 8 μM of construct 3 for 2 and 5 hours. The cells were harvested and lysed in the presence of iodoacetamide, an additive that prevents any post-lysis PTS by alkylating the cysteine residues in the intein required for activity (see Supplemental Fig. 9 for relevant control reactions). Western blotting analysis revealed the formation of a new protein whose size was consistent with the expected splice product, H2B-HA (Fig. 2b). Isolation of this material followed by mass spectrometry confirmed it to be the desired semi-synthetic histone (Fig. 2c). Cell fractionation studies indicated this product is present in the chromatin fraction (Supplemental Fig. 10). Based on additional western blotting analyses we estimate the yield of the in vivo PTS reaction to be in the 60–70% range (Supplemental Fig. 11 & 12). Similar results were obtained upon delivery of 3 into HeLa and U2OS each transfected with 1 (Supplemental Fig. 13), illustrating the generality of the approach.
We next explored whether the PTS approach could be used to incorporate small molecule fluorescent probes into chromatin, thereby complementing existing methods for this purpose that are based on labeling of appended fusion proteins such as DHFR and AGT, which importantly do not self-excise18,19. For this we designed peptide 4 containing a fluorescent reporter, tetramethylrhodamine (TMR), and a HA tag fused to the IntC fragment (Fig. 1b, Supplemental Fig. 7). This construct contained an additional feature, namely a fluorescence quencher moiety (QSY) strategically placed within the IntC sequence such that fluorescence of the TMR is quenched and restored only upon completion of the splicing reaction (Fig. 2d)20,21. Preliminary in vitro studies revealed a QSY-TMR arrangement that resulted in robust activation of fluorescence following protein trans-splicing (Supplemental Fig. 14). Consistent with our design, addition of construct 4 to 293T cells expressing 1 led to a time-dependent increase in nuclear fluorescence that, importantly, was not observed when non-transfected cells were similarly treated (Fig. 2d, Supplemental Fig. 15, 16 & Supplemental Movie 1). The near absence of cytoplasmic fluorescence illustrates the power of coupling fluorescence output to successful protein splicing21, in this case allowing selective imaging of semisynthetic chromatin in living cells.
Tagging chromatin states in nucleo
Local variation in the structure and dynamics of chromatin are thought to affect access of the underlying DNA to trans-acting proteins such as transcription factors and polymerases22,23. Consequently, we wondered whether this rugged epigenetic landscape might similarly affect the local rate of protein trans-splicing on chromatin, particularly since the IntN and IntC fragments must associate prior to splicing, an event that might be influenced by chromatin state. The difference in yield between PTS performed in live cells and isolated mononucleosomes (Supplemental Fig. 11 & 12) raises the possibility that differential accessibility may be reflected in native chromatin.
To explore this idea, we set about identifying conditions that would be most sensitive to the effects of chromatin structure on the splicing reaction. We reasoned that being able to carefully control the concentration of reactants and the duration of the reaction would be key to this endeavor. This led us to consider the use of isolated nuclei, which are permeable to the IntC –cargo construct, thereby improving our ability to control the aforementioned parameters. To test whether trans-splicing could be conducted in this context, nuclei from 293T cells expressing construct 1 were isolated by hypotonic lysis and then treated with 0.5 μM construct 2 at 37 °C for various times before quenching the reaction by the addition of iodoacetamide (Fig. 3a, Supplemental Fig. 17). Western blotting indicated the time-dependent generation of a new protein consistent with the expected product, H2B-HA (Supplemental Fig. 18). The identity of the splicing product was again confirmed by mass spectrometry (Supplemental Fig. 19). As expected, this product was not observed upon adding construct 2 to nuclei isolated from control cells that either did not express 1 or expressed an inactive version (1*) containing a cysteine to alanine mutation in the intact fragment (Supplemental Fig. 18).
Figure 3. Protein trans-splicing as a function of chromatin state.
a) Schematic of in nucleo procedure for assessing chromatin accessibility using PTS. (b–d) ChIP-qPCR analysis of the in nucleo protein trans-splicing reaction between 1 and 2 under the following set of conditions; 1 hour reaction with 0.5 μM construct 2 (b), 5 minute reaction with 0.5 μM construct 2 (c) and, 5 minute reaction with 0.05 μM construct 2 (d). All reactions were conducted at 37 °C as described under material and methods. The input signal for each gene was normalized to Rhob. e) Same as panel d only using nuclei isolated from 293T cells synchronized with nocodazole. (error bars, s.e.m. *P value ≤ 0.05, n.s. = not significant). Three independent experiments were performed for each condition. Note that labeling of the heterochromatin and euchromatin genomic loci occurs to a similar degree when high concentration (0.5 μM) of the IntC peptide was used (b, c). When lower concentration of peptide was used (0.05 μM), a difference in labeling of these regions was observed (d). Synchronizing the cells into M-phase, where chromatin is in a more compact state, prevents the differential labeling of these regions (e).
Having established that chromatin labeling is possible in isolated nuclei, we turned to the question of how to extract information on chromatin states using this system. For this we imagined a workflow that couples a kinetically-controlled trans-splicing reaction to a subsequent chromatin immunoprecipitation (ChIP) step, thereby allowing the relative rates of product formation at specific genomic loci to be interrogated by, for example, quantitative PCR (qPCR). Several genes were selected, representative of different chromatin states9: three actively transcribed genes (euchromatin; Rhob, ADH5 and GAPDH), two centromeric regions (constitutive heterochromatin; from chromosome 1 and chromosome 4), one telomeric region (constitutive heterochromatin; Tel1/2) and two developmentally repressed genes (Facultative heterochromatin; Oct4 and Nanog). ChIP-qPCR employing an antibody against a known chromatin activating PTM (H3K4me3) was used to verify that the epigenetic landscape is not altered in our isolated nuclei compared to intact cells (Supplemental Fig. 20). ChIP-qPCR was also used to show that H2B-IntN (1) is incorporated into each of the genomic loci in our set, although interestingly we did observe ~50% reduction in the telomeric region compared to the others (Supplemental Fig. 21). Nonetheless, there was sufficient H2B-IntN signal at each locus for us to explore the question of relative reactivity towards 2. An important consideration here was how to discriminate between the splicing product, H2B-HA, and any non-specific complex between the IntC-HA fragment and chromatin since both would, in principle, yield a ChIP-qPCR signal using an anti-HA antibody. Our solution to this problem was to employ control construct, 5, in which the HA detection tag was placed at the N-terminus of the IntC (Fig. 1b, Supplemental Fig. 7). Reversing the order of tag relative to the intein fragment means that ChIP-qPCR signal against HA can only originate from non-specific complexes since the splicing reaction leads to unmodified H2B (Supplemental Fig. 22).
With the various controls in place, we next followed the in nucleo splicing reaction between 1 and 2 by ChIP-qPCR using an anti-HA antibody. A series of reaction conditions were explored, varying both the amount of 2 added and the time of reaction, and in each case the corrected ChIP-qPCR data were normalized to the Rhob signal. These experiments confirmed the importance of being able to control the reaction conditions, which is afforded by the in nucleo system; while relatively homogeneous tagging of the loci was observed under more aggressive reaction conditions (Fig. 3b,c), restricting the reaction time and available reactant revealed clear differences in the signals across the set (Fig. 3d). Remarkably, the loci fell into two distinct groups, with the euchromatic regions being significantly (p value <0.05) more reactive than the heterochromatic regions. This separation correlates perfectly with the localization of the H3K4me3 PTM (Supplemental Fig. 20) and is fully consistent with the idea that the rate of trans-splicing is sensitive to chromatin state under the in nucleo experimental conditions. To further validate this idea, we performed the chromatin tagging experiment on nuclei isolated from synchronized cells arrested in G2/M-phase by nocodazole treatment (Supplemental Fig. 23). Since mitotic chromatin condensation would still be expected under these conditions24, we hypothesized that the reactivity of the genomic loci in our set might be altered compared to that in interphase. Indeed, the differences in ChIP-qPCR signals between euchromatin and heterochromatin regions, observed in nuclei from asynchronous cells, was greatly reduced in the G2/M-phase nuclei (Fig. 3e). Thus, the chromatin tagging strategy can be used to assess changes in chromatin structure as a function of cell cycle.
Customizing chromatin states in nucleo
Next, we turned to the question of whether the split-intein system could be used to introduce an endogenous PTM (and by extension analogs thereof) into native chromatin. In principle, this would provide a powerful way to directly explore biochemical mechanisms, for example crosstalk between histone PTMs25 in a physiologically relevant background. To illustrate this point, we chose as a test case the ubiquitination of lysine 120 in H2B (H2B-K120Ub), a modification important for the regulation of transcription26. H2B-K120Ub cannot be mimicked by any simple mutation of H2B and so the tools available for manipulating the levels and structure of this complex PTM on native chromatin are limited. Moreover, the large size and complexity of the H2B-K120Ub PTM was expected to test the limits, from a synthetic chemistry perspective, of chromatin tailoring by split-inteins.
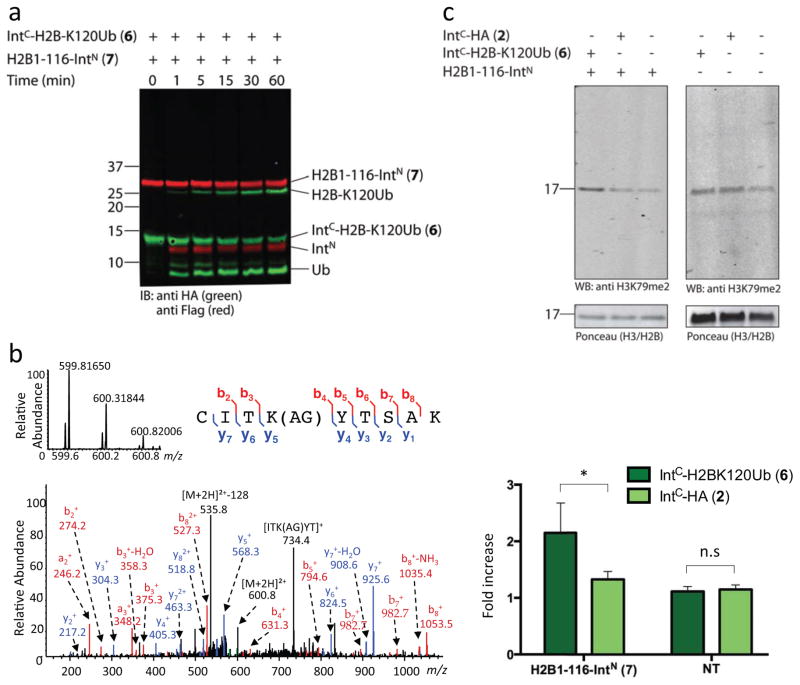
To generate semisynthetic H2B-K120Ub on chromatin, we again turned to the in nucleo system due to the ease by which the requisite reactant can be delivered. We began by preparing reactive construct 6 in which the IntC peptide is fused to a pre-ubiquitinated (bearing an HA tag) fragment of H2B corresponding to C-terminal residues 117–125 (Fig. 1b). Briefly, this involved the expressed protein ligation between a synthetic peptide corresponding to the IntC moiety followed by residues 117–125 of H2B and a HA-tagged ubiquitin α-thioester generated by thyolysis of an intein fusion (Fig. 4a). Following ligation and desulfurization the product, IntC-H2B-K120Ub (6), was purified by RP-HPLC and characterized by mass spectrometry (Fig 4b). In order to generate a native-like product, we expressed a truncated version of H2B (residues 1–116) fused to IntN (7). Reaction between 6 and 7 generates ubiquitinated H2B containing two conservative mutations, namely (i) A117C within H2B which is an unavoidable consequence of the splicing reaction occurring on chromatin, and (ii) G76A within ubiquitin, which is the result of the synthetic route used to generate 6. The G76A mutation has an advantage in our experimental setting as it was shown to reduce sensitivity to cellular ubiquitin hydrolases27. Neither of these mutations was expected to affect the trans-activation functions of ubiquitin on chromatin28,29. We verified that construct 7 expresses and localizes to chromatin similarly to construct 1 (Supplemental Fig. 24). Nuclei from cells expressing 7 were then treated with ubiquitinated construct 6 and analyzed by western blotting using an anti-HA antibody. This revealed the rapid generation of a new protein consistent with the expected splice product, H2B-K120Ub (Fig. 5a, Supplemental Fig. 25). Isolation of the PTS product followed by mass spectrometry confirmed this assignment (Fig. 5b).
Figure 4. Synthesis of IntC-H2B-K120Ub (construct 6).
a) Semi-synthesis of 6 using expressed protein ligation. A synthetic peptide corresponding to the NpuC intein sequence (green) and the histone H2B C-terminal tail residues 117–125 (blue) was assembled using Fmoc-SPPS. This peptide underwent (i) alloc deprotection with Pd(PPh3)4 followed by (ii) coupling of Fmoc-Cys(Trt)-OH to the newly exposed Lys side chain and (iii) peptide cleavage with TFA. (iv) Chemical ligation of an N-terminally HA-tagged ubiquitin α-thioester. (v) Radical-based desulfurization followed by Acm deprotection with mercury (II) acetate. b) Characterization of purified 6 by RP-HPLC (left) and ESI-MS (right).
Figure 5. In nucleo semi-synthesis of H2B-K120Ub and its effect on H3K79 methylation.
a) Nuclei isolated from 293T cells expressing H2B1-116-IntN (construct 7) were incubated with 1 μM of construct 6, at 37 °C, for the indicated times and then analyzed by western blot with the indicated antibodies. Note, that the ubiquitin within construct 6 contains an HA epitope tag at its N-terminus. In addition to the product of the splicing product, H2B-K120Ub, we also see the generation free HA-tagged ubiquitin presumably generated by de-ubiquitination of the reactant 6 or the PTS product (see Supplemental Fig. 25). b) Characterization of the splicing product, H2B-K120Ub, by MS. Reaction was performed as in a, using 1 μM of construct 6 for 1 hour at 37°C. Chromatin fraction containing the slicing product was enriched and analyzed as in Fig. 2c. Shown is the MS/MS spectrum from the tryptic peptide ion corresponding to the branched peptide from the splice junction. Bottom, MS/MS of the [M+2H]2+ peptide CITK(AG)YTSAK shown with b- and y-ions indicated in red and blue, respectively. Top left, high resolution MS spectrum of the parent ion at m/z 599.81650. Top right, the peptide sequence is annotated to indicate the detected b- and y- ions. The major peak showing neutral loss of Δ128 Da corresponds to the gas-phase loss of Ala-Gly from the lysine side chain. c) Reactions performed on isolated nuclei from non-transfected cells or cells overexpressing H2B1-116-IntN (7) as in a in the presence of SAM. Reactions were quenched after 30 minutes and analyzed by western blot using anti-H3K79me2 (top panel). Membranes were stained with ponceau to account for sample loading. Bottom panel: H3K79me2 signal was quantified by densitometry as fold change relative to the negative control (no peptide, lane 3 of each blot) after normalizing for loading (error bars, s.e.m.;. *P value ≤ 0.05, n.s. = not significant). Three independent experiments were performed for each bar (see Supplemental Fig. 26).
Finally, we asked if semisynthetic H2B-K120Ub, generated on native chromatin, could induce a canonical epigenetic signaling event, thereby validating its functionality. For this we chose the crosstalk with H3K79 methylation; the cognate histone methyltransferase for this PTM, Dot1L, is directly stimulated by H2B-K120Ub in vitro28. Accordingly, we performed the in nucleo reaction between 1 μM of 6 and 7 for 30 minutes at 37°C and, following quenching, the reaction was analyzed by western blot employing an antibody against the downstream crosstalk product, H3K79me2 (Fig. 5c, Supplemental Fig. 26). Gratifyingly, a ~2-fold increase in H3K79me2 levels were observed upon generating exogenous H2B-K120Ub by splicing – note, the 293T cells employed have endogenous H2B-K120Ub and hence an intrinsic level of H3K79me2. No such stimulation was found in control reactions in which 6 was added to nuclei isolated from non-transfected cells, suggesting no trans stimulation of the methyltransferase by the ubiquitinated peptide 6 is occurring (Fig. 5c). Moreover, stimulation of H3K79me2 levels was abrogated when the reaction between 6 and 7 was performed in the presence of the hDot1L inhibitor, EPZ004777 (Supplemental Fig. 27). Collectively, these data establish that semisynthetic H2B-K120Ub is functional and able to induce chromatin signaling in an isolated nuclear environment.
Discussion
The technology described herein both extends and complements existing methods for the biochemical analysis of chromatin. Recent years have seen tremendous progress in our ability to manufacture chemically defined chromatin for use in functional studies7. Methods based on protein semi-synthesis and unnatural amino acid mutagenesis (via amber suppression) allow the preparation of site-specifically modified histones that can then be reconstituted into chromatin in vitro. Extending these highly flexible protein chemistry strategies to a cellular setting would open up many new opportunities for the targeted analysis of epigenetic processes. Indeed, the power of chemically tailoring native chromatin is underlined by the recent use of the amber suppression method to incorporate a photocrosslinker into yeast chromatin for the purposes of studying structural changes30. The split intein approach described in this study complements this method by expanding the range of modifications and probes that can be introduced with temporal control14. It is also worth noting that PTS is its traceless process, i.e. the intein is excised from the final labeled product. This feature may provide advantages over other fusion-protein approaches such as SNAP- and DHFR-tags that have been used to label corresponding histone fusions with synthetic dyes18,19.
In the course of the current study we were able to customize H2B in several ways, including the installation of the ~8 kDa ubiquitin modification on the relevant lysine side-chain. The large size of this PTM precludes it from being mimicked by a simple amino acid substitution, a strategy that is often employed when studying smaller modifications (e.g. Glu as a mimic of pSer). In suitable cases, it may be possible to use a linear ubiquitin fusion as a surrogate of the normal lysine-modified ubiquitinated species31. Indeed, a recent study in yeast described a clever approach to mimic ubiquitinated H2B in which ubiquitin was fused to the N-terminus of a truncated version of histone H2A32, a strategy that positions ubiquitin close to its normal attachment site on the nucleosome surface. However, this fusion strategy required removal of the functionally important N-terminal tail of H2A, which could affect epigenetic processes, and of course does not present ubiquitin via an isopeptide linkage. Our semisynthesis strategy circumvents these issues. The semi-synthetic H2B-K120Ub generated was not only functional, in that it induced H3K79me crosstalk, but also contained a Gly76Ala mutation within ubiquitin. This highlights a key advantage of using targeted protein chemistry to tailor native chromatin, namely the ability to modify a PTM in ways impossible using the endogenous cellular ubiquitination apparatus. Indeed, the success with ubiquitin sets the stage for future structure-activity studies employing other ubiquitin mutants or analogs designed to explore aspects of its function on chromatin. By extension, we imagine that this technology can be used to install other PTMs (e.g. acetyl-, methyl-, or phospho-groups, as well as analogs thereof) into native chromatin. In principle, any modification that can be introduced into a synthetic peptide can be introduced into a histone via protein trans-splicing. From a practical perspective, the scope of the method is likely dependent on the efficiency of the protein transduction step needed to get the cargo-bearing intein fragment into cells (this is not an issue for in nucleo experiments). While the HA-TAT fusion strategy employed herein clearly worked for the cargoes described in the current study, the moderate efficiency of this transduction method may limit certain applications33. However, cellular protein delivery is a very active area of research34. Indeed, new technologies have recently been disclosed35,36 that appear to greatly improve the efficiency of protein delivery into cells and that could be easily integrated into our overall protocol.
Use of protein trans-splicing restricts semi-synthetic manipulations to the histone termini, in the current example we focused on the C-terminus of H2B. Fortunately, the vast majority of histone PTMs are found within the N- and C-terminal tail regions37,38. Access to the N-terminal regions of histones is also possible using our system; in this case the IntC peptide would be fused the N-terminus of the recombinant histone and the probe-bearing IntN fragment delivered to the cells (or nuclei). Indeed, in proof-of-principle studies we have demonstrated that labeling of the N-terminus of histone H3 is possible using such a scheme (Supplemental Fig. 28). Thus, we expect it will be possible to introduce a range of PTMs or biophysical probes into the N-terminal tails of histones.
Beyond protein labeling, we envisioned the bimolecular nature of the protein trans-splicing reaction could be used to probe chromatin structure. This application rests on the assumption that compact, heterochromatin presents a more hindered steric environment for split intein association than the more open euchromatic regions. Indeed, by exploiting an in nucleo splicing system we were able to identify conditions that allowed differential labeling of euchromatin versus heterochromatin regions. While only a handful of genomic loci were examined in the current study, there is no reason, given the apparent broad incorporation of the H2B-IntC fusion, why the PTS labeling strategy could not be adapted for a more global analysis of chromatin accessibility by using a ChIP-seq format rather ChIP-qPCR. The ‘splice-seq’ protocol we envision would complement existing methods for probing chromatin accessibility39. The most commonly used of these, and arguably the ‘gold standard’40, involves treating isolated cell nuclei with the non-specific endonuclease, DNase I, which preferentially cuts within unprotected genomic DNA regions41. When used in conjunction with a massively parallel sequencing step (so-called Dnase-seq) it is possible to directly map open chromatin (and by inference condensed chromatin) on a genome-wide scale42. The PTS-based method would complement Dnase-seq by probing chromatin accessibility at the histone level. An attractive feature of this approach would be its flexibility; (i) different surfaces of chromatin could be examined simply by moving the location of the split intein fusion on the core histones, and (ii) different steric thresholds could be interrogated by changing the bulk of the cargo attached to the incoming intein fragment (the current example used a very small cargo). In this way, it may be possible to retrieve another layer of information on local chromatin accessibility.
In conclusion, a method has been developed to modify histones on native chromatin using protein trans-splicing. The method allows installation of user-defined chemical functionalities, artificial or native, onto chromatin in a temporally controlled manner, with great specificity and no background. We illustrate this by installing a fluorophore as well as a large native PTM, ubiquitin. The resulting modified histone products can be used to directly image chromatin, interrogate biochemical signaling pathways, and to probe chromatin accessibility to trans-acting factors. We imagine this technology, which for the first time extends established protein semi-synthesis methods for chromatin tailoring to an in vivo setting, will have a range of uses in the epigenetic area.
Methods
Solid Phase Peptide Synthesis (SPPS)
Standard Fmoc-based SPPS was used for the synthesis of all peptides in this study on ChemMatrix resins with Rink Amide or Wang linkers. Peptides were either synthesized using manual addition of the reagents or on a Liberty Peptide Synthesizer equipped with a Discovery microwave module (CEM, Matthews, NC). Peptides were cleaved and isolated by RP-HPLC and verified by ESI-MS. For the specific procedures used for the synthesis of the different peptides as well as the amino acid sequence, see supplemental information.
Constructs
The H2B-IntN fusions were generated using an H2B-EGFP43 construct (in pEGFP-N1 vector, Clontech Laboratories Inc., CA, USA) as a cloning template. Standard sub-cloning strategies were used to replace the EGFP sequence with the IntN sequence from the intein of interest. We also inserted a short linker, GGKFAEY, between the C-terminus of H2B and the N-terminus of the IntN fragments, which contains the 4 native extein residues for the DnaE inteins (FAEY) and a di-Gly linker (for full amino acid sequence of each of the fusions see Supplementary Table 5).
Cell-lines and transfections
All cell culture and transfection reagents were obtained from Thermo Fisher Scientific (Grand Island, NY) unless otherwise indicated. 293T, HeLa cells (ATCC) and U2OS (courtesy of Cristea lab, Princeton University) were cultured in DMEM supplemented with 10% FBS (Sigma-Aldrich), 2 mM L-Glutamine, and 500 units/ml penicillin and streptomycin. Transfections were performed using lipofectamine 2000 (Invitrogen) as directed by the manufacturer.
Peptide delivery into cells
IntC–cargo peptides conjugated to CPP were delivered into living cells using a modified protein transduction methodology44. Peptide conjugate stocks were prepared by dissolving lyophilized peptides in DMSO up to 1 mM. Prior to delivery experiments, peptide stocks were diluted to their indicated delivery concentration in Opti-MEM medium (Life technologies). CPP-conjugated IntC peptides were then added to H2B-IntN expressing 293T, HeLa or U2OS cells grown to ~70–90% confluency, previously washed with OptiMEM, and incubated at 37 °C for 1 h. Cells were then supplemented with an equal volume of full cell culture media containing DMEM and FBS and incubated at 37°C for an additional 1 to 4 hours as indicated. Post-lysis PTS was prevented by the addition of 80 mM iodoacetamide, which alkylates cysteines, including the active cysteines involved in splicing (see Supplemental Fig. 9).
Nuclear isolation and in nucleo peptide delivery
293T cells (107) were harvested on ice in cold PBS. The cell pellet was resuspended in hypotonic lysis buffer (10 mM Tris base, 15 mM sodium chloride, 1.5 mM magnesium chloride and protease inhibitors; pH 7.6) followed by homogenization with 10 strokes of a loose pestle Dounce homogenizer. Nuclei were centrifuged and equilibrated in nucleus delivery buffer (20 mM Hepes, 1.5 mM magnesium chloride, 150 mM potassium chloride, 1 mM DTT, 1 mg/ml BSA, 1 mM ATP and protease inhibitors, pH7.6) and then treated with the appropriate IntC-cargo peptides and experiment-specific biochemical factors. Reactions were allowed to proceed for the indicated times at 37 °C before being quenched by the addition of 80mM final concentration of iodoacetamide.
Chromatin ImmunoPrecipitation (ChIP) and qPCR
In vivo ChIP was conducted essentially as described45 using approximately 3x107 cells per condition. For full description, see Supplemental information.
In nucleo ChIP was performed similarly to that above, but with the following modification. Following the protein trans-splicing reaction, nuclei were washed with nuclear cross-linking buffer (10 mM Tris base, 0.25 M sucrose, 3 mM calcium chloride and protease inhibitors, pH 7.5) and cross-linked by addition of 1% final paraformaldehyde at room temperature for 15 minutes. Next, glycine (125 mM of final concentration) was added and incubated for 5 minutes at room temperature. Nuclei were then lysed and the rest of the ChIP procedure was carried out as for in vivo ChIP. Each qPCR reaction contained a 1:100 dilution of the DNA in Power SYBR green PCR Master Mix (Applied Biosystems, Fisher brand) and 0.25 μM final concentration of each of the primers (see Table 3 for primer sequences). qPCR reactions were performed using an Applied Biosystems ABI 7900 instrument.
Supplementary Material
Acknowledgments
The authors thank the current and former members of the Muir laboratory for many valuable discussions. We further thank D. H. Perlman (Princeton Proteomics & Mass Spectrometry Core, Princeton University) for the mass spectrometry data and G. Laevsky (Confocal core facility, Princeton University) for help with microscopy experiments. We also thank Wei Wang, Donna Storton and Jennifer Miller (Microarray facility, Princeton university) for help with qPCR and RNA-seq experiments. Finally we thank Steven Josefowicz, Charles Li and David Allis (Rockefeller University) for help with the ChIP assays. This research was supported by the U.S. National Institutes of Health (grants R37-GM086868 and R01 GM107047).
Footnotes
Author contributions Y.D, M.V.-P. and T.W.M. conceived and designed the research. Y.D, M.V.-P. and S.V. prepared reagents and performed experiments. Y.D. and T.W.M. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Badeaux AI, Shi Y. Emerging roles for chromatin as a signal integration and storage platform. Nature reviews. Molecular cell biology. 2013;14:211–224. doi: 10.1038/nrm3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Sturm D, et al. Paediatric and adult glioblastoma: multiform (epi)genomic culprits emerge. Nature reviews. Cancer. 2014;14:92–107. doi: 10.1038/nrc3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodriguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nature medicine. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 5.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allis CD, Muir TW. Spreading chromatin into chemical biology. Chembiochem : a European journal of chemical biology. 2011;12:264–279. doi: 10.1002/cbic.201000761. [DOI] [PubMed] [Google Scholar]
- 7.Fierz B, Muir TW. Chromatin as an expansive canvas for chemical biology. Nature chemical biology. 2012;8:417–427. doi: 10.1038/nchembio.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang H, et al. Regulation of transcription by the MLL2 complex and MLL complex-associated AKAP95. Nature structural & molecular biology. 2013;20:1156–1163. doi: 10.1038/nsmb.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nature biotechnology. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole PA. Chemical probes for histone-modifying enzymes. Nature chemical biology. 2008;4:590–597. doi: 10.1038/nchembio.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki W, et al. Comprehensive structural analysis of mutant nucleosomes containing lysine to glutamine (KQ) substitutions in the H3 and H4 histone-fold domains. Biochemistry. 2011;50:7822–7832. doi: 10.1021/bi201021h. [DOI] [PubMed] [Google Scholar]
- 12.Shah NH, Dann GP, Vila-Perello M, Liu Z, Muir TW. Ultrafast protein splicing is common among cyanobacterial split inteins: implications for protein engineering. Journal of the American Chemical Society. 2012;134:11338–11341. doi: 10.1021/ja303226x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood DW, Camarero JA. Intein Applications: From Protein Purification and Labeling to Metabolic Control Methods. The Journal of biological chemistry. 2014;289:14512–14519. doi: 10.1074/jbc.R114.552653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah NH, Muir TW. Inteins: Nature’s Gift to Protein Chemists. Chem Sci. 2014;5:446–461. doi: 10.1039/C3SC52951G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vila-Perello M, et al. Streamlined expressed protein ligation using split inteins. Journal of the American Chemical Society. 2013;135:286–292. doi: 10.1021/ja309126m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nature medicine. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 17.Giriat I, Muir TW. Protein semi-synthesis in living cells. Journal of the American Chemical Society. 2003;125:7180–7181. doi: 10.1021/ja034736i. [DOI] [PubMed] [Google Scholar]
- 18.Wombacher R, et al. Live-cell super-resolution imaging with trimethoprim conjugates. Nature methods. 2010;7:717–719. doi: 10.1038/nmeth.1489. [DOI] [PubMed] [Google Scholar]
- 19.Klein T, et al. Live-cell dSTORM with SNAP-tag fusion proteins. Nature methods. 2011;8:7–9. doi: 10.1038/nmeth0111-7b. [DOI] [PubMed] [Google Scholar]
- 20.Pellois JP, Hahn ME, Muir TW. Simultaneous triggering of protein activity and fluorescence. Journal of the American Chemical Society. 2004;126:7170–7171. doi: 10.1021/ja0499142. [DOI] [PubMed] [Google Scholar]
- 21.Borra R, Dong D, Elnagar AY, Woldemariam GA, Camarero JA. In-cell fluorescence activation and labeling of proteins mediated by FRET-quenched split inteins. Journal of the American Chemical Society. 2012;134:6344–6353. doi: 10.1021/ja300209u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 23.Woodcock CL, Ghosh RP. Chromatin higher-order structure and dynamics. Cold Spring Harbor perspectives in biology. 2010;2:a000596. doi: 10.1101/cshperspect.a000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salina D, et al. Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell. 2002;108:97–107. doi: 10.1016/s0092-8674(01)00628-6. [DOI] [PubMed] [Google Scholar]
- 25.Voigt P, Reinberg D. Histone tails: ideal motifs for probing epigenetics through chemical biology approaches. Chembiochem : a European journal of chemical biology. 2011;12:236–252. doi: 10.1002/cbic.201000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochimica et biophysica acta. 2014;1839:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 27.Hodgins RR, Ellison KS, Ellison MJ. Expression of a ubiquitin derivative that conjugates to protein irreversibly produces phenotypes consistent with a ubiquitin deficiency. The Journal of biological chemistry. 1992;267:8807–8812. [PubMed] [Google Scholar]
- 28.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McGinty RK, et al. Structure-activity analysis of semisynthetic nucleosomes: mechanistic insights into the stimulation of Dot1L by ubiquitylated histone H2B. ACS chemical biology. 2009;4:958–968. doi: 10.1021/cb9002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkins BJ, et al. A cascade of histone modifications induces chromatin condensation in mitosis. Science. 2014;343:77–80. doi: 10.1126/science.1244508. [DOI] [PubMed] [Google Scholar]
- 31.Matsushita N, et al. A FancD2-monoubiquitin fusion reveals hidden functions of Fanconi anemia core complex in DNA repair. Molecular cell. 2005;19:841–847. doi: 10.1016/j.molcel.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Vlaming H, et al. Flexibility in crosstalk between H2B ubiquitination and H3 methylation in vivo. EMBO reports. 2014;15:1220–1221. doi: 10.15252/embr.201471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwarze SR, Hruska KA, Dowdy SF. Protein transduction: unrestricted delivery into all cells? Trends in cell biology. 2000;10:290–295. doi: 10.1016/s0962-8924(00)01771-2. [DOI] [PubMed] [Google Scholar]
- 34.van den Berg A, Dowdy SF. Protein transduction domain delivery of therapeutic macromolecules. Current opinion in biotechnology. 2011;22:888–893. doi: 10.1016/j.copbio.2011.03.008. [DOI] [PubMed] [Google Scholar]
- 35.Erazo-Oliveras A, et al. Protein delivery into live cells by incubation with an endosomolytic agent. Nature methods. 2014;11:861–867. doi: 10.1038/nmeth.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuris JA, et al. Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nature biotechnology. 2014 doi: 10.1038/nbt.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Perceiving the epigenetic landscape through histone readers. Nature structural & molecular biology. 2012;19:1218–1227. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan M, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsompana M, Buck MJ. Chromatin accessibility: a window into the genome. Epigenetics & chromatin. 2014;7:33. doi: 10.1186/1756-8935-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crawford GE, et al. DNase-chip: a high-resolution method to identify DNase I hypersensitive sites using tiled microarrays. Nature methods. 2006;3:503–509. doi: 10.1038/nmeth888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford GE, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome research. 2006;16:123–131. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kanda T, Sullivan KF, Wahl GM. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Current biology : CB. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 44.Sugita T, et al. Improved cytosolic translocation and tumor-killing activity of Tat-shepherdin conjugates mediated by co-treatment with Tat-fused endosome-disruptive HA2 peptide. Biochemical and biophysical research communications. 2007;363:1027–1032. doi: 10.1016/j.bbrc.2007.09.077. [DOI] [PubMed] [Google Scholar]
- 45.Goldberg AD, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.