Abstract
Amyloidosis is a disease characterized by the formation of extracellular amyloid deposits. Immunoglobulin light-chain amyloidosis can appear as a local disorder presenting with mild symptoms or as a life threatening systemic disease. The systemic form of immunoglobulin light-chain amyloidosis is the most common type of amyloidosis in western countries although it is a rare disease. Identification of the proteins forming amyloid fibrils is essential for the diagnosis of the disease and knowledge about the overall protein composition of the deposits may lead to a larger understanding of the deposition events thereby facilitating a more detailed picture of the molecular pathology.
In this pilot study, we investigated the protein composition of amyloid deposits isolated from human specimens of the eyelid, conjunctiva, and orbit. Deposits and internal control tissue (patient tissue without apparent deposits) were procured by laser capture microdissection. Proteins in the captured amyloid and control samples were quantified by liquid chromatography tandem mass spectrometry using the label-free exponential modified Protein Abundance Index (emPAI) method.
Immunoglobulin light chain kappa or lambda was found to be the most predominant protein in the amyloid deposits from the eyelid, conjunctiva, and orbit. Five proteins, apolipoprotein A-I, carboxypeptidase B2 (TAFI), complement component C9, fibulin-1 and plasminogen were found solely across all amyloid but not in the control tissue. In addition, the protein profiles identified apolipoprotein E and serum amyloid P component to be associated with the immunoglobulin light chain deposits across all three tissues analyzed. The method used in this study provided high sensitivity and specificity for the type of amyloid and may provide additional information on the pathology of the amyloid deposits in the ocular tissues studied.
Keywords: Amyloidosis, conjunctiva, eyelid, orbit, laser capture microdissection, mass spectrometry
Introduction
Amyloidosis is a disorder associated with the aggregation of soluble amyloid prone protein into insoluble fibrils within the extracellular tissue. More than 25 proteins are known to form amyloid fibrils in humans [1–3]. Amyloidosis results in massive protein accumulation leading to organ and tissue failure or to amyloid induced cell death as believed to occur in Alzheimer’s disease (AD) [4]. The amyloid fibrils exhibit a characteristic β-pleated sheet configuration, which histologically can be visualized using the amyloidophilic dyes Congo red or Thioflavine T [1, 5–7]. Several factors favor the formation of fibrils, such as high local protein concentration, low pH, proteolytic processing and chemical modifications by physiologically formed metabolites [8, 9]. Furthermore, the presence of amyloid associated components like glycosaminoglycans and serum amyloid P component (SAP) are thought to be important for fibrillogenesis and the fibril stability [10–12].
Immunoglobulin light chain (AL) amyloidosis has been identified in most tissues and is caused by the accumulation of immunoglobulin light chains within forming amyloid deposits [13–16]. AL amyloidosis can manifest as a local disorder displayed with mild symptoms or as a life threatening systemic disease [1]. The systemic form of AL amyloidosis represents the most common form of systemic amyloidosis in western countries, but it is a rare disease [17, 18]. An accumulation of immunoglobulin light chain into amyloid deposits and possibly cytotoxic amyloid intermediates are both considered being important in the pathogenesis of this form of amyloidosis [10, 19]. Localized deposition of light chain immunoglobulin is related to focal infiltration of plasma cells secreting amyloid-forming light chain in the immediate vicinity, whereas systemic amyloidosis usually is a rare complication of monoclonal gammopathies [1, 2]. All classes of immunoglobulin light chain can cause AL amyloidosis; however, monoclonal lambda light chains are most frequently involved [20–22].
Amyloidosis can affect the tissue of the orbit, eyelid and conjunctiva although uncommonly [23]. Amyloid deposits in the conjunctiva and orbit are usually localized and not part of a systemic disease, whereas eyelid involvement may be associated with both local and systemic amyloidosis [24, 25]. The presentation of amyloidosis in the ocular tissues is highly variable making the diagnosis and treatment challenging. The most common treatment of amyloid deposits in the conjunctiva, eyelid or orbit, is surgical debulking when observation is no longer feasible [23, 24]. AL amyloidosis has been reported in conjunctival, eyelid and orbital tissue using immunohistochemistry or biochemical methods [23, 25–27]. In one study, the nature of amyloid in two cases of ocular amyloidosis was identified using a recently described diagnostic technique laser capture microdissection (LCM) followed by liquid chromatography tandem mass spectrometry (LC-MS/MS). They identified a polyclonal IgG type amyloid in one case and AL kappa type amyloid in the other [28]. To our knowledge nothing is known of other proteins associated to the immunoglobulin light chain deposits in these eye-related tissues.
In this pilot study, we investigated the protein composition of expected AL amyloid deposits isolated from human specimens of the eyelid, conjunctiva and orbit. Deposits and internal control tissue (patient tissue without apparent deposits) were procured using LCM and subsequently analyzed by LC-MS/MS. Using the label-free mass spectrometry (MS) quantification method exponential modified Protein Abundance Index (emPAI), either immunoglobulin light chain kappa or lambda was revealed to be the most predominant protein in the amyloid deposits. In addition, with all three ocular tissues analyzed the protein profiles disclosed apolipoprotein E (apoE) and SAP in association with the immunoglobulin light chain deposits. Furthermore, the five proteins, apolipoprotein A-I (apoA-I), carboxypeptidase B2 (thrombin-activable fibrinolysis inhibitor (TAFI)), complement component C9, fibulin-1 and plasminogen were found to be unique to all amyloid tissue analyzed. The LCM/LC-MS/MS method provided high sensitivity and specificity of amyloid types compared to immunohistochemistry, the routinely used method [15, 29].
Materials and methods
Specimens
Examples of eyelid amyloidosis were obtained from three patients (two females and one male). One specimen of orbital amyloidosis and one case of conjunctival amyloidosis were also available for analysis. Both of the later specimens were from patients of the female gender. The affected tissue of all cases was characterized by a slowly enlarging swelling that was excised surgically. Tissue was acquired with the approval of the Institutional Review Board of Duke University Medical Center, according to the Declaration of Helsinki.
Laser capture microdissection
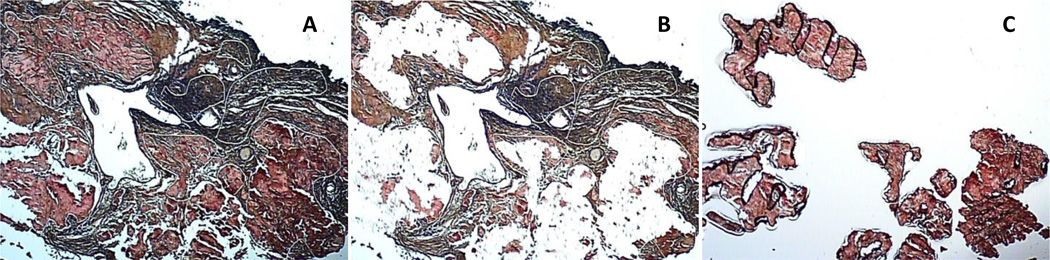
Procuring of amyloid deposits was performed as previously described [30]. Shortly, 8 µm thick tissue sections of formaldehyde fixed and paraffin embedded tissue were cut and placed centrally on non-charged glass slides, followed by deparaffinization, staining with Congo red, and dehydration using standard histological procedures. The Arcturus Autopix Laser Capture Microscope (Applied Biosystem) was used to isolate amyloid deposits appearing red due to the Congo red staining (Figure 1). In addition, tissue with no apparent amyloid deposits was likewise procured and used as control tissue. Tissue was collected on CapSure Macro LCM Caps (Applied Biosystem), subsequently placed in sterile 0.5 ml Eppendorf Biopur Microcentrifuge Tubes (VWR) and stored at −80°C until further processed.
Figure 1.
Illustration of amyloid isolation from the eyelid (specimen 2) by laser capture microdissection. The amyloid was removed from fixed tissue sections by laser capture microdissection and analyzed by LC-MS/MS. A: tissue before laser capture microdissection. B: tissue after laser capture microdissection. C: captured sections of deposits. Amyloid was removed from the conjunctiva and orbit in a comparable way.
Sample preparation
Proteins from the laser capture microdissected samples of amyloid from the eyelid, conjunctiva and orbit were extracted using a modified protocol of the Liquid Tissue MS Protein Prep Kit (Expression Pathology Inc., Gaithersburg, MD). LCM films containing captured protein deposits and control tissue samples were stripped from the caps and incubated with 40 µl extraction buffer (10 mM Tris, pH 7.8, 0.1 % RapiGest (Waters)) at 95 °C for 90 min. Samples were centrifuged at 10.000 × g for 1 min and subsequently cooled on ice. Extracted proteins were incubated with 5 µg porcine trypsin (Sigma) at 37 °C for 16 hours, vortexing every 20 min for the first hour. Samples were then centrifuged at 10.000 × g for 5 min, supernatant isolated and incubated with 10 mM DTT at 95 °C for 5 min followed by incubation with 30 mM iodoacetamide for 15 min in the dark. Peptides generated from each digest were acidified with 5 µl of 5% formic acid and recovered using Stage-tips (Proxeon, Thermo Scientific) [31]. The peptides were eluted with 10 µl 70 % acetonitrile, and subsequently lyophilized before being dissolved in 12 µl 0.1 % formic acid.
Liquid chromatography tandem mass spectrometry analyses
For each sample two technical replicates were run. The samples were analyzed by LC-MS/MS using nanoflow liquid chromatography on an Easy-nLC system (Thermo Scientific) coupled directly to a TripleTOF 5600+ mass spectrometer (AB Sciex). Samples were loaded on a 0.1×21 mm C18 trap column and a 0.075×150 mm C18 analytical column (NanoSeparations). Peptides were eluted and directly electrosprayed into the mass spectrometer using a 60 min gradient from 5–40% acetonitrile in 0.1% formic acid at a flow rate of 250 nl/min. Data was acquired using an ion spray voltage of 2.5 kV, curtain gas of 30, and an interface heater temperature of 150 °C. For IDA, survey scans were acquired in 250 ms, and as many as 50 product ion scans were collected if exceeding a threshold of 125 counts per second (counts/s) and with a +2 to +5 charge-state. A sweeping collision energy setting of 40 ± 15 eV was applied to all precursor ions for collision-induced dissociation. Dynamic exclusion was set for ½ of peak width (~6 s), and then the precursor was refreshed off of the exclusion list.
Identification and rough relative quantification of proteins
The mass spectrometry data was converted to Mascot generic format (MGF) using the AB SCIEX MS Data Converter beta 1.3 (AB SCIEX) and the "proteinpilot MGF" parameters and used to interrogate the Swiss-Prot (version 2014_02) Homo sapiens (20,264 sequences) database using an in-house Mascot search engine (Matrix Science, London, England version 2.3.02) [32] with a peptide tolerance of +/− 10 ppm, MS/MS tolerance of 0.2 Da, carbamidomethylation of cysteine residues as a fixed modification, methionine oxidation and proline hydroxylation as variable modifications, semiTrypsin as enzyme, 1 missed cleavage was allowed and ESI-QUAD-TOF mass spectrometer was selected as instrument. The Mascot search results were imported to the MS Data Miner (MDM) software v. 1.2.2 [33] with an ions score cut-off value of 45. MS/MS spectra validation of protein identifications based on less than 2 unique peptides with ion scores less than 100 were validated automatically in MDM. Only MS/MS spectra containing a consecutive b or y-ion series of at least 3 fragments were accepted in MDM. The remaining MS/MS spectra of protein identifications were manually validated as only spectra containing a clear sequence tag of at least three consecutive residues with high intensity fragment ions compared to the background were accepted. The data was then exported to excel and contaminating exogenous proteins were removed. Among these human tryptic peptides identical to porcine tryptic peptides were considered as contaminants as porcine trypsin was used for sample processing. The resulting endogenous proteins were listed according to their emPAI values, which reflect their relative abundance. The molar fractions of the endogenous proteins were calculated by dividing the emPAI value for each identified protein with the total emPAI value [34]. The average molar fraction of the two technical replicates was calculated for each sample, and only proteins identified in both runs were accepted.
Results
Amyloid deposits and non-amyloid control tissue were procured from tissue specimens of the eyelid, orbit and conjunctiva using LCM (Figure 1). Peptides obtained by trypsin digestion were recovered from the dissected samples and analyzed by LC-MS/MS. For each sample two technical replicates were run.
Protein profile of eyelid amyloid deposits
The proteomic analysis of amyloid deposits from the three cases of eyelid amyloidosis resulted in 35, 92 and 74 protein hits of which 31, 83 and 64 were endogenous proteins from specimen 1, 2 and 3, respectively (supplementary). Only proteins identified in both technical replicates were accepted. From the eyelid non-amyloid control tissue we obtained 32, 57 and 49 protein hits for specimen 1, 2 and 3, respectively, of which 23, 49 and 38 were endogenous proteins. In the eyelid amyloid deposits several proteins were enriched compared to eyelid control tissue indicating that they accumulated in the deposits. The most abundant component of the eyelid amyloid deposits from specimen 1 was Ig kappa chain C region, whereas Ig lambda-2 chain C regions was accumulated in the amyloid deposits from specimen 2 and 3 (Table 1). We also identified Ig kappa chain C region as the most abundant component of the eyelid control tissue from eyelid specimen 1, suggesting that even though no apparent amyloid deposits are observed by bright field microscopy immunoglobulin light chains may also be present in elevated levels in the surrounding of the amyloid deposits (Table 1).
Table. 1.
Abundant proteins in eyelid amyloid deposits and control tissue
| Specimen 1 | Specimen 2 | Specimen 3 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Amyloid deposits | Control tissue | Amyloid deposits | Control tissue | Amyloid deposits | Control tissue | ||||||
| Protein | mol % | Protein | mol % | Protein | mol % | Protein | mol % | Protein | mol % | Protein | mol % |
| Ig kappa chain C region |
58.3 | Ig kappa chain C region |
61.7 | Ig lambda-2 chain C regions |
55.3 | Hemoglobin subunit beta |
15.8 | Ig lambda-2 chain C regions |
52.8 | Hemoglobin subunit beta |
57.7 |
| Apolipo- protein A-IV |
5.5 | Hemoglobin subunit beta |
6.0 | Ig lambda chain V-III region LOI |
6.1 | Hemoglobin subunit alpha |
14.5 | Hemoglobin subunit beta |
11.1 | Hemoglobin subunit alpha |
35.9 |
| Serum amyloid P- component |
4.4 | Collagen alpha-1(I) chain |
3.7 | Hemoglobin subunit beta |
3.1 | Collagen alpha-1(I) chain |
13.3 | Hemoglobin subunit alpha |
5.6 | Hemoglobin subunit delta |
1.9 |
| Collagen alpha-1(I) chain |
3.6 | Dermcidin | 3.4 | Immunoglobulin lambda-like polypeptide 5 |
3.0 | Collagen alpha-2(I) chain |
7.0 | Collagen alpha-1(I) chain |
3.5 | Collagen alpha-1(I) chain |
1.4 |
| Apolipo- protein E |
3.1 | Hemoglobin subunit alpha |
3.0 | Serum amyloid P-component |
2.4 | Histone H4 | 6.1 | Collagen alpha-2(I) chain |
2.3 | Collagen alpha-2(I) chain |
0.7 |
| Vitronectin | 2.7 | Neutrophil defensin 1 |
2.8 | Collagen alpha- 1(I) chain |
2.2 | Vimentin | 5.1 | Apolipo- protein A-IV |
2.2 | Ig lambda-2 chain C regions |
0.4 |
| Apolipo- protein A-I |
2.7 | Apolipo- protein A-IV |
2.7 | Hemoglobin subunit alpha |
2.0 | Collagen alpha-1(III) chain |
4.7 | Apolipo- protein A-I |
2.2 | Hemoglobin subunit gamma-2 |
0.3 |
| Collagen alpha-2(I) chain |
2.1 | Collagen alpha-2(I) chain |
2.6 | Apolipo- protein A-I |
1.8 | Ig lambda chain V-III region LOI |
4.6 | Hemoglobin subunit delta |
1.9 | Hemoglobin subunit gamma-1 |
0.3 |
| Protein S100-A6 |
2.0 | Apolipo- protein E |
1.7 | Ig kappa chain C region |
1.8 | Hemoglobin subunit delta |
3.8 | Collagen alpha-1(III) chain |
1.6 | Collagen alpha-1(III) chain |
0.3 |
| Neutrophil defensin 1 |
2.0 | Collagen alpha-1(III) chain |
1.5 | Collagen alpha- 2(I) chain |
1.6 | Serum albumin |
3.4 | Vimentin | 1.6 | Carbonic anhydrase 1 |
0.1 |
The table depicts the ten most abundant proteins within the amyloid deposits of the eyelid and control eyelid tissue. Results are from specimen 1, 2 and 3 listed according to their molar % values. The molar % values are averages of two technical replicates [34].
Protein profile of conjunctival amyloid deposits
The one case of conjunctival amyloidosis resulted in 79 protein hits in the amyloid deposits of which 71 were endogenous proteins, whereas 87 proteins were identified in the conjunctival control tissue of which 77 were endogenous proteins (supplementary). The most abundant component of the conjunctival amyloid deposits was Ig kappa chain C region (Table 2).
Table. 2.
Abundant proteins in conjunctival amyloid deposits and control tissue
| Amyloid deposits | Control tissue | ||
|---|---|---|---|
| Protein | mol % | Protein | mol % |
| Ig kappa chain C region | 75.9 | Collagen alpha-1(I) chain | 22.6 |
| Hemoglobin subunit beta | 2.9 | Ig kappa chain C region | 17.1 |
| Collagen alpha-1(I) chain | 1.8 | Collagen alpha-2(I) chain | 10.7 |
| Collagen alpha-2(I) chain | 1.6 | Hemoglobin subunit beta | 3.5 |
| Hemoglobin subunit alpha | 1.4 | Histone H2B type 1-B | 4.6 |
| Vimentin | 1.0 | Vimentin | 5.5 |
| Apolipoprotein A-IV | 1.0 | Collagen alpha-1(III) chain | 5.0 |
| Serum amyloid P-component | 0.9 | Histone H4 | 3.5 |
| Serum albumin | 0.9 | Actin, cytoplasmic 1 | 3.6 |
| Collagen alpha-1(III) chain | 0.8 | Hemoglobin subunit alpha | 2.9 |
The table depicts the ten most abundant proteins in the conjunctival amyloid deposits and control conjunctival tissue listed according to their molar % values.
The molar % values are averages of two technical replicates.
Protein profile of orbital amyloid deposits
For the case of orbital amyloidosis, deposits and non-amyloid control tissue 92 and 70 protein hits were obtained, respectively, of which 85 and 63 were endogenous proteins (supplementary). The most abundant component of the orbital amyloid deposits was Ig kappa chain C region (Table 3).
Table. 3.
Abundant proteins in orbital amyloid deposits and control tissue
| Amyloid deposits | Control tissue | ||
|---|---|---|---|
| Protein | mol % | Protein | mol % |
| Ig kappa chain C region | 94.4 | Hemoglobin subunit beta | 78.0 |
| Ig kappa chain V-I region DEE | 0.5 | Hemoglobin subunit alpha | 12.6 |
| Hemoglobin subunit beta | 0.5 | Hemoglobin subunit delta | 4.1 |
| Hemoglobin subunit alpha | 0.5 | Ig kappa chain C region | 0.8 |
| Collagen alpha-1(I) chain | 0.4 | Histone H4 | 0.6 |
| Apolipoprotein E | 0.3 | Histone H2B type 1-L | 0.6 |
| Serum amyloid P-component | 0.3 | Vimentin | 0.4 |
| Serum albumin | 0.3 | Collagen alpha-1(I) chain | 0.3 |
| Apolipoprotein A-I | 0.2 | Actin, cytoplasmic 1 | 0.3 |
| Collagen alpha-2(I) chain | 0.2 | Collagen alpha-2(I) chain | 0.2 |
The table depicts the ten most abundant proteins in the orbital amyloid deposits and control orbital tissue listed according to their molar % values.
The molar % values are averages of two technical replicates.
Proteins associated with the amyloid deposits
The most abundant protein in the amyloid deposits of the eyelid, conjunctiva and orbit was a single light chain immunoglobulin. The Ig lambda light chain accumulated in specimen 2 and 3 of the eyelid amyloid deposits while Ig kappa light chain was the most abundant protein in eyelid specimen 1, conjunctival and orbital amyloid deposits as disclosed by the emPAI-based molar fraction of the enriched protein in the amyloid deposits when compared to control tissue. Besides Ig light chains, apoE and SAP were abundant components of the amyloid deposits compared to control tissue (supplementary), correlating well with other findings where apoE and SAP has been shown to interact with several different types of amyloid [2, 12]. In addition, protein comparison across all samples and tissues revealed 22 proteins to be found in all amyloid deposits analyzed across the three tissues (supplementary). Of these proteins, apoA-I, TAFI, complement component C9, fibulin-1 and plasminogen were unique to all amyloid samples, and thus not observed in any of the control samples.
Cytokeratin’s
The proteomic profiling of the amyloid deposits and non-amyloid control tissues resulted in the identification of a number of proteins, of which 13 were considered to be common contaminating exogenous proteins during sample preparation. These were regarded to be keratin, type I cytoskeletal 10 (P13645), keratin, type I cytoskeletal 14 (P02533), keratin, type I cytoskeletal 16 (P08779), keratin, type I cytoskeletal 9 (P35527), keratin, type II cytoskeletal 1 (P04264), keratin, type II cytoskeletal 2 epidermal (P35908), keratin, type II cytoskeletal 4 (P19013), keratin, type II cytoskeletal 5 (P13647), keratin, type II cytoskeletal 6A (P02538), keratin, type II cytoskeletal 6C (P48668), keratin, type II cytoskeletal 7 (P08729), keratin, type I cytoskeletal 13 (P13646) and keratin, type I cytoskeletal 19 (P08727). Swiss-Prot accession numbers are indicated in brackets for each exogenous protein. The resulting endogenous proteins were listed according to emPAI values for each tissue (supplementary).
Discussion
LCM, a technique originally developed to isolate single cells, has proven highly suitable for the isolation of protein deposits as illustrated in this study and by others [15, 16]. Combined with sensitive MS instruments minute amounts of tissue can be captured and analyzed. This makes LCM/LC-MS/MS a useful method for the identification of proteins within pathologic deposits. In addition to identifying the particular proteins forming the amyloid fibrils, the method also adds insight to the overall composition of the “bystander” proteins present in the deposits. This provides forensic evidence facilitating the analysis of the events leading to the deposits and thus helps establish a more detailed picture of the molecular pathology.
In this study, typing and protein profiles of amyloid deposits situated in different eye tissue derived from 5 patients were performed. A striking feature of all of the amyloid deposits studied within the ocular tissues was the detection of immunoglobulin light chains, which were invariably of either the lambda or kappa light chain type, but not a combination of both light chains. A specific Ig light chain was found to be highly enriched in amyloid deposits within the conjunctiva, eyelid and orbit. In the eyelid of two patients Ig lambda light chain was enriched, while Ig kappa light chain accumulated in another example of amyloid deposition in the eyelid as well as in each of the cases of amyloid in the conjunctiva and orbit. Control tissue consisted of apparently unaffected tissue (Congo red-negative tissue) from the patients and in all but one of the cases; the protein profile of the Congo red-negative control tissue was clearly different from the amyloid deposits. Furthermore, protein comparison across all of the samples and the three tissues identified 22 proteins in the amyloid deposits of which the 5 proteins apoA-I, TAFI, complement component C9, fibulin-1 and plasminogen were exclusively found in the amyloid deposits and not in any of the control tissues. ApoA-I is among the most abundant proteins identified in the amyloid deposits (table 1, 3 and supplementary) and is known to form amyloid in systemic amyloidosis [35] and may co-aggregate with immunoglobulin light chain. The finding of the two proteases carboxypeptidase B2 (TAFI) and plasminogen in amyloid tissue may be part of a clearing response to remove aggregated protein. In the context of biomarkers, further analyses might reveal if the proteins, unique to the amyloid tissue are associated with other Ig amyloid deposits and/or other kinds of amyloid deposit. These proteins could be useful as AL amyloidosis biomarkers.
Many of the identified proteins in the amyloid deposits are well-known amyloidogenic proteins or are known to associate with amyloid in other types of amyloidosis; however these proteins are not exclusively identified in the amyloid deposits, in this study. One of the proteins found in all analyzed samples of amyloid was SAP (supplementary). Even though the function is unknown SAP is thought to interact with all types of amyloid and its presence is often used as criteria for the diagnosis of amyloidosis at the proteomic level using LCM/LC-MS/MS [16]. In addition, apoE that previously has been associated with amyloid deposits [2] was likewise found to be abundant in the amyloid deposits in this study (supplementary). ApoE is suspected of playing an important role in the deposition of fibrillar amyloid-β in AD [36] and perhaps in all types of amyloidosis. Accumulation of Ig light chain and presence of apoE and SAP in the amyloid deposits suggests that the patients were suffering from AL amyloidosis. Besides from apoE and SAP, vitronectin that is present within amyloid deposits in AD and certain systemic amyloidosis [37] is identified in the amyloid deposits (supplementary). Vitronectin has also been shown to form amyloid fibrils in vitro, in one study [37].
Aside from immunoglobulin light chains the amyloidogenic proteins identified with amyloid of the analyzed ocular tissues included apolipoprotein A-IV (apoA-IV) identified in all the amyloid deposits and transthyretin found in all but eyelid specimen 1 amyloid deposits (supplementary). ApoA-IV forming amyloid in systemic amyloidosis has been shown to accumulate within the corneal stroma in amyloid deposits from patients suffering from lattice corneal dystrophy type I [38, 39]. Transthyretin is known to form amyloid in systemic amyloidosis [38]. In addition, protein S100-A6 reported to form amyloid was almost exclusively identified in the amyloid deposits (supplementary) [40]. The finding of several amyloid-forming proteins could suggest co-fibrillation, which may be important parameters in terms of morphological appearance, time of onset and severity.
Altogether, the obtained protein profiles found in this study were comparable to the protein composition of amyloid deposits from patients suffering from other types of amyloidosis, such as lattice corneal dystrophy type I and AD [36, 39]. The presence of both apoE and SAP alongside immunoglobulin light chain suggests the patients were suffering from AL amyloidosis. Furthermore, findings of five proteins unique to the amyloid deposits regardless of the tissue may be used as protein markers for AL amyloidosis. The use of LCM/LC-MS/MS for diagnosis of AL amyloidosis has been used on a wide variety of tissues from among other’s kidney, breast, lymph node, bone to brain [15, 16, 41, 42]. Diagnosis by LCM/LC-MS/MS is highly advantageous compared to traditional immunohistochemical methods as it combines specific sampling by LCM and high sensitivity of LC-MS/MS and can accurately handle complex cases of amyloidosis [15, 16]. In addition, the obtainment of protein profiles by LCM/LC-MS/MS may lead to a better understanding of the pathogenesis of disease.
Supplementary Material
Acknowledgements
This study was supported by the Danish Agency for Science, Technology and Innovation, the Danish National Research Foundation, Fight for Sight - Denmark and the National Eye Institute (R01 EY012712). Finally, we would like to thank the PRIDE team for assistance in data management in the ProteomeXchange Consortium.
Abbreviations
- AD
Alzheimer’s disease
- SAP
serum amyloid P component
- AL amyloidosis
immunoglobulin light chain amyloidosis
- LCM
laser capture microdissection
- LC-MS/MS
liquid chromatography tandem mass spectrometry
- MS
mass spectrometry
- emPAI
exponential modified Protein Abundance Index
- apoE
apolipoprotein E
- apoA-I
apolipoprotein A-I
- TAFI
thrombin-activable fibrinolysis inhibitor (carboxypeptidase B2)
- MGF
Mascot generic format
- MDM
MS data miner
- apoA-IV
apolipoprotein A-IV
Footnotes
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository [43] with the dataset Identifier PXD000743.
References
- 1.Desport E, Bridoux F, Sirac C, Delbes S, Bender S, et al. Al amyloidosis. Orphanet J Rare Dis. 2012;7:54. doi: 10.1186/1750-1172-7-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinney JH, Hawkins PN. Amyloidosis. Ann Clin Biochem. 2012;49:229–241. doi: 10.1258/acb.2011.011225. [DOI] [PubMed] [Google Scholar]
- 3.Sipe JD, Benson MD, Buxbaum JN, Ikeda S, Merlini G, et al. Amyloid fibril protein nomenclature: 2012 recommendations from the nomenclature committee of the international society of amyloidosis. Amyloid. 2012;19:167–170. doi: 10.3109/13506129.2012.734345. [DOI] [PubMed] [Google Scholar]
- 4.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 5.Bennhold H. Über die ausscheidung intravenös einverleibten kongorotes bei den verschiedensten erkrangungen insbesondere bei amyloidosis (on the elimination of intravenously absorbed congo red in various diseases, in particular in amyloidosis) Deutsches Arch Klin Med. 1923;142:32–46. [Google Scholar]
- 6.Vassar PS, Culling CF. Fluorescent stains, with special reference to amyloid and connective tissues. Arch Pathol. 1959;68:487–498. [PubMed] [Google Scholar]
- 7.Westermark P, Benson MD, Buxbaum JN, Cohen AS, Frangione B, et al. Amyloid fibril protein nomenclature -- 2002. Amyloid. 2002;9:197–200. doi: 10.3109/13506120209114823. [DOI] [PubMed] [Google Scholar]
- 8.Nilsson MR, Driscoll M, Raleigh DP. Low levels of asparagine deamidation can have a dramatic effect on aggregation of amyloidogenic peptides: Implications for the study of amyloid formation. Protein Sci. 2002;11:342–349. doi: 10.1110/ps.48702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q, Powers ET, Nieva J, Huff ME, Dendle MA, et al. Metabolite-initiated protein misfolding may trigger alzheimer's disease. Proc Natl Acad Sci U S A. 2004;101:4752–4757. doi: 10.1073/pnas.0400924101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merlini G, Bellotti V. Molecular mechanisms of amyloidosis. N Engl J Med. 2003;349:583–596. doi: 10.1056/NEJMra023144. [DOI] [PubMed] [Google Scholar]
- 11.Alexandrescu AT. Amyloid accomplices and enforcers. Protein Sci. 2005;14:1–12. doi: 10.1110/ps.04887005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirschfield GM, Hawkins PN. Amyloidosis: New strategies for treatment. Int J Biochem Cell Biol. 2003;35:1608–1613. doi: 10.1016/s1357-2725(03)00169-9. [DOI] [PubMed] [Google Scholar]
- 13.Paccalin M, Hachulla E, Cazalet C, Tricot L, Carreiro M, et al. Localized amyloidosis: A survey of 35 french cases. Amyloid. 2005;12:239–245. doi: 10.1080/13506120500351174. [DOI] [PubMed] [Google Scholar]
- 14.Biewend ML, Menke DM, Calamia KT. The spectrum of localized amyloidosis: A case series of 20 patients and review of the literature. Amyloid. 2006;13:135–142. doi: 10.1080/13506120600876773. [DOI] [PubMed] [Google Scholar]
- 15.Vrana JA, Gamez JD, Madden BJ, Theis JD, Bergen HR, 3rd, et al. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114:4957–4959. doi: 10.1182/blood-2009-07-230722. [DOI] [PubMed] [Google Scholar]
- 16.Sethi S, Vrana JA, Theis JD, Leung N, Sethi A, et al. Laser microdissection and mass spectrometry-based proteomics aids the diagnosis and typing of renal amyloidosis. Kidney international. 2012;82:226–234. doi: 10.1038/ki.2012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinney JH, Smith CJ, Taube JB, Lachmann HJ, Venner CP, et al. Systemic amyloidosis in england: An epidemiological study. British journal of haematology. 2013;161:525–532. doi: 10.1111/bjh.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyle RA, Linos A, Beard CM, Linke RP, Gertz MA, et al. Incidence and natural history of primary systemic amyloidosis in olmsted county, minnesota, 1950 through 1989. Blood. 1992;79:1817–1822. [PubMed] [Google Scholar]
- 19.Bellotti V, Mangione P, Merlini G. Review: Immunoglobulin light chain amyloidosis--the archetype of structural and pathogenic variability. J Struct Biol. 2000;130:280–289. doi: 10.1006/jsbi.2000.4248. [DOI] [PubMed] [Google Scholar]
- 20.Perfetti V, Palladini G, Casarini S, Navazza V, Rognoni P, et al. The repertoire of lambda light chains causing predominant amyloid heart involvement and identification of a preferentially involved germline gene, iglv1-44. Blood. 2012;119:144–150. doi: 10.1182/blood-2011-05-355784. [DOI] [PubMed] [Google Scholar]
- 21.Yao Y, Wang SX, Zhang YK, Qu Z, Liu G, et al. A clinicopathological analysis in a large cohort of chinese patients with renal amyloid light-chain amyloidosis. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2013;28:689–697. doi: 10.1093/ndt/gfs501. [DOI] [PubMed] [Google Scholar]
- 22.Gertz MA. Immunoglobulin light chain amyloidosis: 2013 update on diagnosis, prognosis, and treatment. American journal of hematology. 2013;88:416–425. doi: 10.1002/ajh.23400. [DOI] [PubMed] [Google Scholar]
- 23.Leibovitch I, Selva D, Goldberg RA, Sullivan TJ, Saeed P, et al. Periocular and orbital amyloidosis: Clinical characteristics, management, and outcome. Ophthalmology. 2006;113:1657–1664. doi: 10.1016/j.ophtha.2006.03.052. [DOI] [PubMed] [Google Scholar]
- 24.Al-Nuaimi D, Bhatt PR, Steeples L, Irion L, Bonshek R, et al. Amyloidosis of the orbit and adnexae. Orbit. 2012;31:287–298. doi: 10.3109/01676830.2012.707740. [DOI] [PubMed] [Google Scholar]
- 25.Demirci H, Shields CL, Eagle RC, Jr, Shields JA. Conjunctival amyloidosis: Report of six cases and review of the literature. Surv Ophthalmol. 2006;51:419–433. doi: 10.1016/j.survophthal.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 26.Hamidi Asl K, Liepnieks JJ, Nunery WR, Yazaki M, Benson MD. Kappa iii immunoglobulin light chain origin of localized orbital amyloidosis. Amyloid. 2004;11:179–183. doi: 10.1080/1350-6120400000707. [DOI] [PubMed] [Google Scholar]
- 27.Kaplan B, Martin BM, Cohen HI, Manaster J, Kassif Y, et al. Primary local orbital amyloidosis: Biochemical identification of the immunoglobulin light chain kappaiii subtype in a small formalin fixed, paraffin wax embedded tissue sample. Journal of clinical pathology. 2005;58:539–542. doi: 10.1136/jcp.2004.022517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Al Hussain H, Edward DP. Anterior orbit and adnexal amyloidosis. Middle East African journal of ophthalmology. 2013;20:193–197. doi: 10.4103/0974-9233.114789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenzweig M, Landau H. Light chain (al) amyloidosis: Update on diagnosis and management. J Hematol Oncol. 2011;4:47. doi: 10.1186/1756-8722-4-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulsen ET, Runager K, Risor MW, Dyrlund TF, Scavenius C, et al. Comparison of two phenotypically distinct lattice corneal dystrophies caused by mutations in the transforming growth factor beta induced (tgfbi) gene. Proteomics Clinical applications. 2013 doi: 10.1002/prca.201300058. 10.1002/prca.201300058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and lc/ms sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 32.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 33.Dyrlund TF, Poulsen ET, Scavenius C, Sanggaard KW, Enghild JJ. Ms data miner: A web-based software tool to analyze, compare, and share mass spectrometry protein identifications. Proteomics. 2012;12:2792–2796. doi: 10.1002/pmic.201200109. [DOI] [PubMed] [Google Scholar]
- 34.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, et al. Exponentially modified protein abundance index (empai) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Ramella NA, Schinella GR, Ferreira ST, Prieto ED, Vela ME, et al. Human apolipoprotein a-i natural variants: Molecular mechanisms underlying amyloidogenic propensity. PloS one. 2012;7:e43755. doi: 10.1371/journal.pone.0043755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Donahue JE, Johanson CE. Apolipoprotein e, amyloid-beta, and blood-brain barrier permeability in alzheimer disease. Journal of neuropathology and experimental neurology. 2008;67:261–270. doi: 10.1097/NEN.0b013e31816a0dc8. [DOI] [PubMed] [Google Scholar]
- 37.Shin TM, Isas JM, Hsieh CL, Kayed R, Glabe CG, et al. Formation of soluble amyloid oligomers and amyloid fibrils by the multifunctional protein vitronectin. Molecular neurodegeneration. 2008;3:16. doi: 10.1186/1750-1326-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergstrom J, Murphy CL, Weiss DT, Solomon A, Sletten K, et al. Two different types of amyloid deposits--apolipoprotein a-iv and transthyretin--in a patient with systemic amyloidosis. Laboratory investigation; a journal of technical methods and pathology. 2004;84:981–988. doi: 10.1038/labinvest.3700124. [DOI] [PubMed] [Google Scholar]
- 39.Karring H, Poulsen ET, Runager K, Thogersen IB, Klintworth GK, et al. Serine protease htra1 accumulates in corneal transforming growth factor beta induced protein (tgfbip) amyloid deposits. Mol Vis. 2013;19:861–876. [PMC free article] [PubMed] [Google Scholar]
- 40.Botelho HM, Leal SS, Cardoso I, Yanamandra K, Morozova-Roche LA, et al. S100a6 amyloid fibril formation is calcium-modulated and enhances superoxide dismutase-1 (sod1) aggregation. The Journal of biological chemistry. 2012;287:42233–42242. doi: 10.1074/jbc.M112.396416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.D'Souza A, Theis J, Quint P, Kyle R, Gertz M, et al. Exploring the amyloid proteome in immunoglobulin-derived lymph node amyloidosis using laser microdissection/tandem mass spectrometry. American journal of hematology. 2013;88:577–580. doi: 10.1002/ajh.23456. [DOI] [PubMed] [Google Scholar]
- 42.Klein CJ, Vrana JA, Theis JD, Dyck PJ, Dyck PJ, et al. Mass spectrometric-based proteomic analysis of amyloid neuropathy type in nerve tissue. Archives of neurology. 2011;68:195–199. doi: 10.1001/archneurol.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, et al. The proteomics identifications (pride) database and associated tools: Status in 2013. Nucleic acids research. 2013;41:D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.