SUMMARY
Fluoride is a ubiquitous anion that inhibits a wide variety of metabolic processes. Here we report the identification of a series of compounds that enhance fluoride toxicity in Escherichia coli and Streptococcus mutans. These molecules were isolated by using a high-throughput screen (HTS) for compounds that increase intracellular fluoride levels as determined via a fluoride riboswitch-reporter fusion construct. A series of derivatives were synthesized to examine structure-activity relationships, leading to the identification of compounds with improved activity. Thus, we demonstrate that small molecule fluoride toxicity agonists can be identified by HTS from existing chemical libraries by exploiting a natural fluoride riboswitch. In addition, our findings suggest that some molecules might be further optimized to function as binary antibacterial agents when combined with fluoride.
INTRODUCTION
Although fluoride is commonly added to oral hygiene products to increase the strength of tooth enamel, it also has substantial antibacterial effects (Barbier et al., 2010; Li, 2003). For example, when in complex with a divalent metal ion and ADP, fluoride forms a non-functional mimic of ATP that can inhibit enolase (Curran et al., 1994; Qin et al., 2006), and similar complexes can inhibit a large diversity of other metabolic enzymes including phosphatases (Barbier et al., 2010; Nakai et al, 1974). Due to these general mechanisms for enzyme inhibition, fluoride is toxic to organisms from all three domains of life.
Until recently, very little was known about how cells sense and respond to fluoride toxicity. New insights into the genes and mechanisms used by bacterial cells to overcome fluoride toxicity were revealed by the discovery of fluoride-responsive riboswitches that control the expression of a number of genes conferring resistance to fluoride (Baker et al., 2012). Riboswitches are structured RNAs that are typically found in the 5′-untranslated regions of bacterial mRNAs where they regulate the expression of genes in response to binding a small molecule or ion (Breaker, 2011; Peselis and Serganov, 2014; Serganov and Nudler, 2013). The most common gene associated with fluoride riboswitches, termed crcB (also called fex, or fluc), codes for a fluoride-specific channel protein (Baker et al., 2012; Li et al., 2013; Stockbridge et al., 2013). Another commonly controlled gene, eriCF, encodes a fluoride-selective antiporter (Baker et al., 2012; Stockbridge et al., 2012). Numerous other genes presumably involved in mitigating fluoride toxicity are also associated with fluoride riboswitches (Baker et al., 2012).
An E. coli strain in which the fluc gene was deleted (Δfluc) is ~200-fold more sensitive to fluoride (Baker et al., 2012). Similarly, some eukaryotic species use homologous channel proteins to improve resistance to fluoride toxicity (Li et al., 2013), which underscores the importance of fluc to fluoride detoxification. Moreover, the deleterious phenotype resulting from fluc deletion in E. coli can be compensated by expressing an eriCF gene derived from another bacterium (Baker et al., 2012), indicating that the proteins expressed from eriCF appear to be biologically equivalent to those encoded by fluc. These findings demonstrate that a major strategy for overcoming toxicity is for cells to eject fluoride into the environment.
We speculated that small molecules that specifically inhibit the protein products Fluc or EriCF, increase the membrane permeability of fluoride, or increase fluoride retention through some other mechanism could enhance fluoride toxicity. If such molecules could be found, then perhaps they could be optimized and made to function as novel antibacterial chemotherapeutics, particularly when used in combination with high fluoride concentrations. This speculation is supported by the fact that some previously characterized pore-forming antibacterial and antifungal compounds have been demonstrated to enhance the cytotoxic activity of fluoride (Li and Breaker, 2012; Nelson et al., 2014; Zasloff and Steinberg, 1993). However, these existing molecules only modestly increase fluoride toxicity, and no non-peptidic molecules have been identified that increase fluoride toxicity towards bacteria.
To identify novel fluoride toxicity agonists for bacteria, we developed an HTS strategy based on an E. coli strain in which a β-galactosidase reporter gene controlled by a fluoride riboswitch provides a read-out of intracellular fluoride concentration (Baker et al., 2012; Nelson et al., 2014). A number of studies have been conducted to identify compounds that directly target riboswitches and function as antibiotics (Blount and Breaker, 2006; Deigan and Ferré-D’ Amaré, 2011; Kim et al., 2009; Lee et al., 2009; Lünse et al., 2014a; Lünse et al., 2014b; Mulhbacher et al., 2010; Ster et al., 2013). By contrast, our study is designed to exploit a riboswitch as a tool to identify compounds that target other aspects of bacterial physiology. Hit compounds from the HTS were validated via a variety of biochemical assays and were shown to sensitize bacteria to fluoride. A brief survey of analogs of the original hits identified more potent compounds with enhanced fluoride sensitization activity. Overall, this study illustrates the potential for optimization of fluoride toxicity agonists as antibacterial compounds.
RESULTS AND DISCUSSION
Design and Validation of a High-Throughput Screen for Fluoride Toxicity Agonists
To efficiently screen large compound collections for molecules that enhance fluoride uptake and/or retention, a method for the rapid, facile, and inexpensive detection of intracellular fluoride was needed. A number of methods exist for the detection of fluoride, including small-molecule sensors (Cametti and Rissanen, 2009, 2013), direct monitoring of 18F accumulation (Drescher and Suttie, 1972; Li et al., 2013; Quissell and Suttie, 1972), and YFP variants that fluoresce in the presence of fluoride (Jayaraman et al., 2000). However, none of these methods are sufficiently selective or sensitive to rapidly and easily detect intracellular fluoride concentrations in bacteria. Therefore, we designed a screen that utilized the most selective biosensor for fluoride currently known – a recently discovered fluoride sensing riboswitch (Baker et al., 2012).
Specifically, we employed a previously described wild-type (WT) E. coli strain in which a fluoride-sensing riboswitch from Pseudomonas syringae controls the expression of a lacZ reporter gene in a fluoride concentration-dependent manner (Baker et al., 2012; Nelson et al., 2014). Whereas most riboswitches control gene expression by regulating the formation of an intrinsic transcription terminator stem or by regulating access to the ribosome binding site of the mRNA, the precise mechanism by which this specific riboswitch representative controls gene expression has not been established. The relative amount of fluoride present in cells can be established by quantitatively measuring reporter gene expression. This is achieved by the addition of 4-methyl-umbelliferyl-β-D-galactopyranoside (4-MUG), which is a proto-fluorescent β-galactosidase substrate. A Δfluc E. coli strain containing the riboswitch reporter was utilized as a positive control for cells that uptake or retain an unusually high amount of fluoride. Reporter gene expression in the Δfluc E. coli strain was 15- to 20-fold higher compared to the WT strain.
HTS assays were performed using 1 mM fluoride, which is considerably higher than the concentration of fluoride present in most natural sources of water. However, this fluoride concentration is far below that tolerated by WT E. coli cultures, which can still grow at concentrations as high as 100 mM (Baker et al., 2012). Although fluoride concentrations in the mM range might seem high, amounts are known to vary widely in the environment, and local concentrations will increase greatly as relatively modest amounts of fluoride are concentrated in soils or surface water pools upon evaporation. Moreover, fluoride in the form of HF will dominate under acidic conditions (HF pKa is 3.14), and this acidic form is far more bioavailable. Therefore, bacteria that have genetic and biochemical responses to even rare exposures to high fluoride concentrations or to low concentrations of fluoride under slightly acidic conditions have a large selective advantage. Our chemical screening approach is targeting this considerable capacity of bacteria to overcome the toxic effects of high concentrations of fluoride.
After optimizing screening conditions, approximately 75,000 compounds from Chembridge, Maybridge, ChemDiv, and proprietary Yale small molecule libraries were tested. In all cases, Z' values (Zhang et al., 1999) did not vary beyond 0.5 and 1, and typically ranged between 0.6 to 0.8. The ratio of the fluorescence signal of the positive control well (fluoride-treated Δfluc cells) to background (average fluorescence of all wells on a plate) was higher than 15:1. The action of each compound, when added to WT cells with fluoride, was established by determining a score between 1 and 100, where 1 represents fluorescence of WT cells treated with fluoride alone and 100 represents fluorescence of fluoride-treated Δfluc cells. Hit compounds that yielded a signal greater than 3-fold above background (scores typically above 4) were chosen for further analysis. Hit compounds that are inherently fluorescent were discarded, after which the hit rate was approximately 1%.
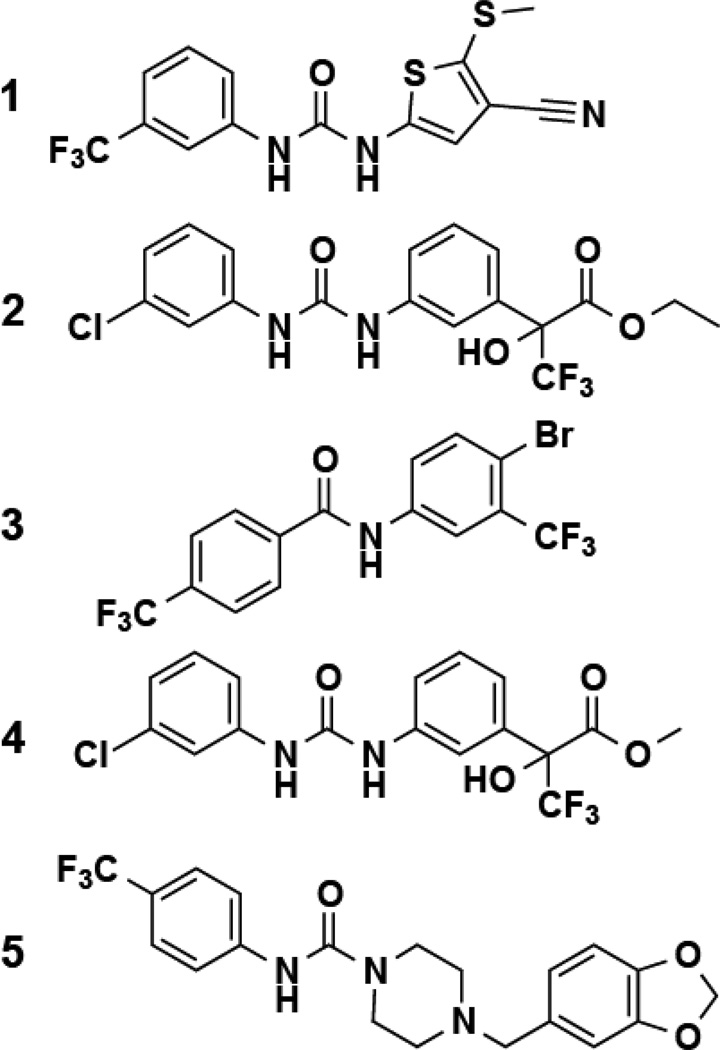
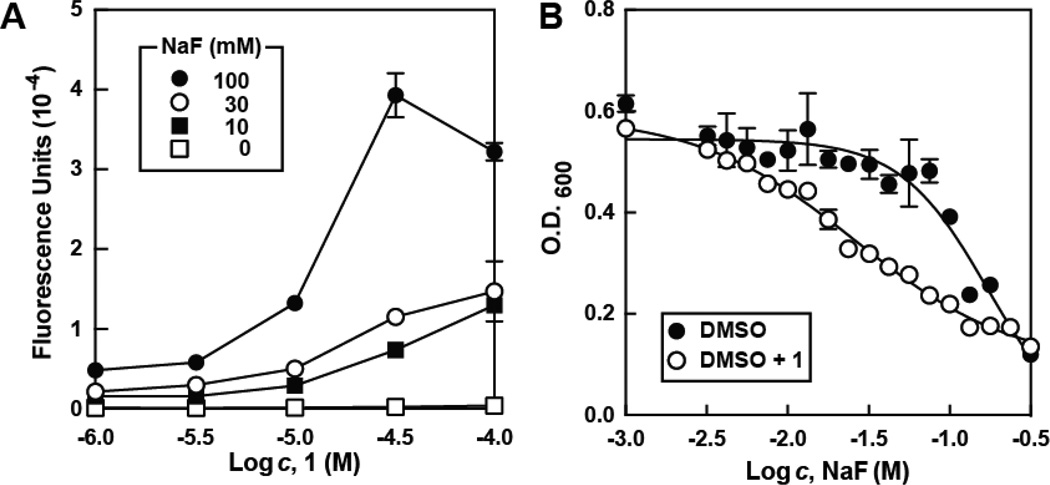
Hit validation consisted of repeating the screening assay and confirming that the signal increase was only observed in the presence of both cells and fluoride, resulting in five molecules that were chosen for future analysis (Figure 1 and Table S1). Of these five hits, compound 1 was the most active, yielding an approximately 8-fold increase in reporter signal at 32 µM when 10 mM sodium fluoride was present relative to the addition of fluoride alone (Figure 2A). These compounds do not increase the activity of the β-galactosidase enzyme in vitro, and there was no increase in fluorescence upon treatment of cells containing a reporter construct carrying a fluoride riboswitch with a disruptive mutation (Figure S1). These results are consistent with our hypothesis that 1 increases intracellular fluoride concentrations.
Figure 1. Structures of Hits from a High-throughput Screen for Compounds that Enhance Fluoride Uptake or Retention.
Figure 2. Small Molecules Enhance Fluoride Uptake and Toxicity in E. coli.
(A) Reporter activity in E. coli treated with 1 and NaF. Data points are the average of three replicates, and error bars represent standard deviations. Decreases in reporter expression at high sodium fluoride and 1 are due to cell death. See Figure S1 for additional control experiments.
(B) Growth of bacteria cultured in the absence or presence of 1 and increasing concentrations of sodium fluoride following 16 hours of incubation at 37°C. DMSO (~1% [v/v] in the growth medium) was used to solublize and deliver test compounds. Data points are the average of three replicates, and error bars represent standard deviations.
We next assessed whether 1 enhances the antibacterial activity of fluoride. Although 1 did not significantly change the minimal inhibitory concentration (MIC) of fluoride toward E. coli, it did reproducibly enhance the ability of 1 to slow E. coli growth (Figure 2B). Addition of 1 alone at 3.2 µM causes no decrease in the viability of E. coli but enhances the natural toxicity of fluoride. These results demonstrate that high-throughput screening of a small molecule library with cells carrying a fluoride-dependent reporter gene can reveal compounds that exhibit antibacterial activity in the presence of fluoride. However, given the modest enhancement in fluoride toxicity generated by 1, we sought to identify analogs with improved activity.
Structure-Activity Relationships
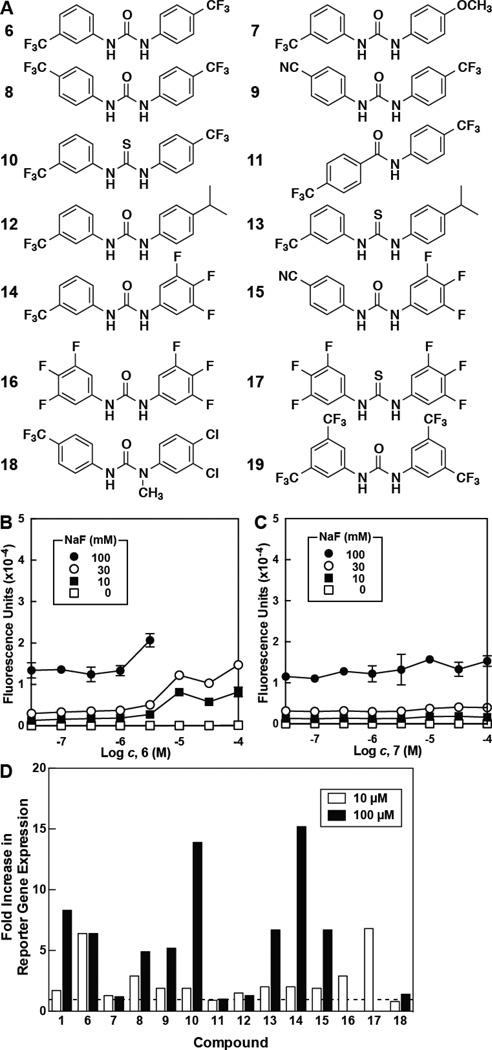
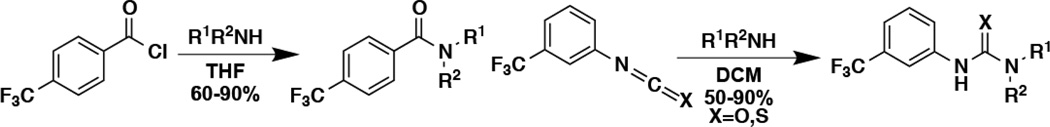
All five selected hits isolated from the HTS contained an electron-deficient aryl group linked by either an amide or urea to another ring system. To better understand the possible shared pharmacophore, and to identify more potent fluoride toxicity agonists, a small library of related compounds was synthesized with high yield and purity. This was achieved via one-step coupling of acyl chlorides, isocyanates, or isothiocyanates to a variety of commercially available aniline derivatives (Scheme 1). The fluoride-sensing riboswitch reporter strain was again used to assess the extent to which each compound increased intracellular fluoride concentration.
Scheme 1. Synthesis of analogs for structure-activity relationship analyses.
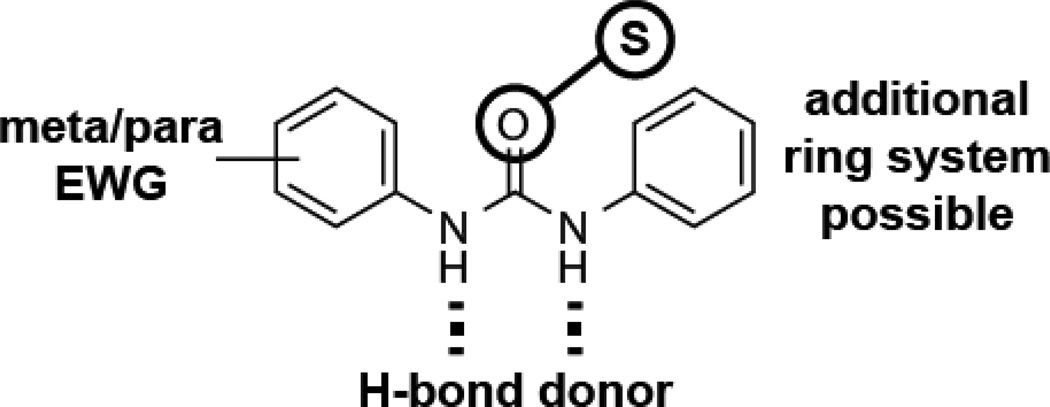
The relative activities of various analogs (Figure 3 and Table S2) confirmed that an electron-deficient aromatic ring, such as those in compounds 6 and 7 (Figure 3B and C), were indeed necessary for enhancing intracellular fluoride concentrations. The locations and identities of the electron-withdrawing groups, however, do not appear to be of critical importance (e.g., see 6, 8, and 9). Notably, carboxamide linked compounds were generally less active than the corresponding urea- or thiourea-linked compounds (e.g., see 9 through 11). The most potent compounds (14 through 17) all include a trifluoro-substituted aryl ring, suggesting that this moiety is particularly beneficial for activity. A graphical summary of the SAR relationships (Figure 4) denotes the importance of these electron-withdrawing moieties, and the presence of hydrogen-bond donor groups in the urea or thiourea linker.
Figure 3. Increase in Reporter Expression for Selected Hit Analogs.
(A) Structures of selected compounds synthesized or purchased for determination of SAR. Analytical data is presented in Supplemental Information.
(B) Reporter gene expression in E. coli treated with 6 and various concentrations of fluoride. When 6 was tested at or above 10 µM, bacterial growth was inhibited in 100 mM fluoride. Data points are the average of three replicates. Error bars are the standard deviation of the measurement.
(C) Reporter gene expression in E. coli treated with 7 and various concentrations of fluoride. Details are as described for B.
(D) Fold increase in reporter gene expression in E. coli treated with compound and 10 mM NaF. Values are normalized to that measured with cells treated with fluoride in the absence of any compound (value set to one, dashed line). White bars indicate the addition of 10 µM compound, while black bars indicate the addition of 100 µM compound, with the exceptions of 1, 14, and 15 (32 µM) and 16 and 17 (3.2 µM). The absence of a bar indicates no substantial bacterial growth was observed for that condition. See Tables S2 for data on additional compounds tested.
Figure 4. Summary of Structure Activity Relationships.
Incorporation of electron-withdrawing groups (EWG) at the meta or para position of either aryl ring results in enhanced activity. Utilization of a thiourea typically has little effect on activity compared to a urea, and likewise the asymmetric incorporation of an additional ring system has little affect. Both amide protons are necessary for activity and might be important for hydrogen bonding (dashed lines).
Interestingly, the SAR data derived from substitutions within the urea-linked series was not the same as with thiourea-linked (e.g., see 12 and 13). This suggests that the determinants of activity for these two series might be different. It is possible that the compounds from the two series might enhance fluoride toxicity via different mechanisms. Alternatively, the preferred conformations of the urea-linked molecules may be distinct from the thiourea-linked examples (Lepore et al., 1973; Bryantsev et al., 2006), which potentially could generate the differences in the activities we observe.
Hit Derivatives with Enhanced Fluoride-dependent Toxicity
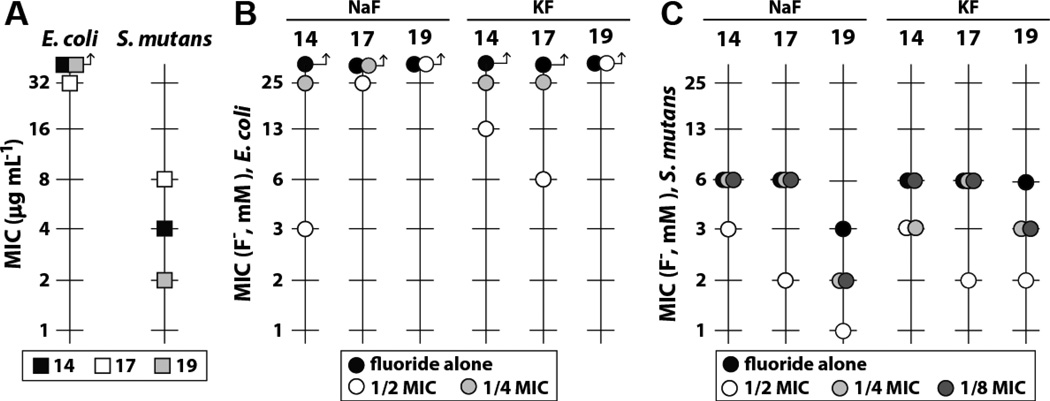
We expected that compounds with improved activity in the fluoride riboswitch reporter assay might also enhance the antibacterial activity of fluoride. Specifically, we sought to identify analogs that stop bacterial growth rather than simply slow replication as is observed for compound 1, our most active initial hit (Figure 2). Accordingly, we chose to examine the ability of our best urea- or thiourea-linked compounds (based on the fluorescence assay) to enhance the toxicity of fluoride to certain bacteria. Treatment with a subinhibitory concentration (0.5× MIC) of 14 resulted in a greater than 8-fold improvement in the MIC of fluoride against E. coli and a ~2-fold improvement against Streptococcus mutans, a causative agent of gingivitis (Figure 5). A subinhibitory concentration of 17 was less effective at increasing fluoride toxicity compared to 14. However, 17 achieves its more modest effects at a lower concentration. Additionally, we observed significant bacterial toxicity for certain compounds in the absence of any added fluoride (Figure 3 and Figure 5). No enhancement of fluoride toxicity was observed upon treatment with ciprofloxacin (Figure S2).
Figure 5. Structural derivatives of hit compounds enhance fluoride toxicity.
(A) MIC of hit compounds for E. coli and S. mutans in the absence of fluoride. Arrows indicate that the values for the analyses denoted by the symbols were not measurable at the highest concentrations tested.
(B) Decrease in the MIC of sodium or potassium fluoride for E. coli in the presence of varying concentrations of hit compounds. For compounds whose MIC in the absence of fluoride was higher than 32 µg ml−1, half and quarter MIC values were calculated as if the MIC were 64 µg ml−1. Values are the average of three replicates.
(C) Decrease in the MIC of sodium or potassium fluoride for S. mutans in the presence of varying concentrations of hit compounds. Values are the average of three replicates.
Our findings are consistent with the hypothesis that these molecules might enhance fluoride toxicity by targeting a cellular process that is critical for cell growth even in the absence of fluoride. However, their antibacterial effects are enhanced by the presence of fluoride. Importantly, the concentrations of fluoride used in our study are substantially higher than bacteria typically experience in a natural environment, except for those exposed to fluoride-containing oral hygiene products. Thus, any use of such compounds as potential antibacterial agents would likely require the concomitant application of additional fluoride.
There are several possible mechanisms by which these molecules may be acting. The simplest might be that the compounds disrupt bacterial membranes in a manner that allows fluoride to more rapidly enter cells. There is precedence for compounds that disrupt membrane integrity to enhance fluoride toxicity (Li and Breaker, 2012; Nelson et al., 2014; Zasloff and Steinberg, 1993). Compounds that create non-specific ion pores in cell membranes should permit the rapid equilibrium between fluoride outside and inside cells.
Curiously, evidence presented to date convincingly demonstrates that Fluc-type ion channels permit the selective and passive diffusion of fluoride through membranes (Stockbridge et al., 2013; Ji et al., 2014), and so members of this protein class theoretically allow fluoride transport both into and out of cells. However, the large negative membrane potential of bacterial cells should greatly favor fluoride efflux through the channel (Stockbridge et al., 2013). Also, under slightly acidic conditions, fluoride is expected to accumulate in cells relative to the environment (Stockbridge et al., 2013; Li et al., 2013), and therefore fluoride outflow should again be favored.
By contrast, amphotericin B increases fluoride toxicity to fungal cells presumably by allowing fluoride to pass either way across cell membranes, resulting in enhanced fluoride toxicity (Zasloff and Steinberg, 1993; Li and Breaker, 2012). One key difference between Fluc proteins and compounds such as amphotericin B is that the protein channel is highly specific for fluoride. Formation of non-selective ion pores, or general membrane disruption will also disrupt the membrane potential of cell, which is critical for the activity of the productive use of Fluc channel proteins to expel fluoride from inside the cell. In this regard, it is interesting to note that various sodium-proton antiporters are believed to be controlled by fluoride riboswitches, and this might be critical for maintaining a negative membrane potential to facilitate fluoride movement out of cells. The disruption of membranes in a non-specific fashion via various externally added compounds will also disrupt membrane potential. Thus, these compounds might either directly facilitate the transport of fluoride into cells, or, by disrupting membrane potential, inhibit its transport out (Nelson et al., 2013).
Some similarities exist between our molecules and known inhibitors of bacterial cell wall biosynthesis (Li et al., 2003; Francisco et al., 2004) and lipid biosynthesis (Russell, 2004). It is possible that the disruption of these processes could potentially enhanced fluoride transport into cells. Furthermore, the specific nature of the hits obtained via our high-throughput screen would suggest these compounds might function by interacting with a specific target, rather than by non-specifically disrupting membranes. General membrane disruptors should be far more tolerant of changes to chemical substructures than our SAR data indicates.
Alternatively, the compounds could inhibit Fluc or some other transporter that is necessary to eject fluoride as it slowly enters cells, resulting in an increase in fluoride retention. However, given that so little is known about fluoride toxicity mitigation mechanisms in bacteria, it is not yet possible to rule out a mechanism involving the inhibition of some other process that is critical for fluoride exclusion or expulsion from cells.
Intriguingly, some of our hits are also structurally similar to known small molecule fluoride receptors (Boiocchi et al., 2004). Receptors for fluoride and other anions often utilize diaryl-ureas or thioureas as hydrogen-bond donors to bind anions (Davis et al., 2010), and the removal of one such potential hydrogen-bond donor results in a dramatic decrease in compound activity (Figure 3, 18). Furthermore, incorporation of fluorine and trifluoromethyl substituents onto aryl rings of small molecule chloride binders is known to improve their ability to transport chloride across phospholipid membranes (Busschaert et al., 2011; Busschaert et al., 2012).
Since we observe that the incorporation of similar substituents onto the aryl rings of our molecules enhances fluoride uptake and/or retention E. coli (Figure 3 and Table S2), it is tempting to speculate that the compounds identified in the current study function as membrane-permeant fluoride binders that directly facilitate the transport of fluoride into bacteria. To evaluate this hypothesis we examined 19 (Figure 3), which is a compound known to promote the transport of chloride out of lipid vesicles (Busschaert et al., 2012). This compound exhibits the greatest ability to increase fluoride toxicity when tested with S. mutans (Figure 5). This finding suggests that the compound series identified in our HTS might indeed function by directly facilitating the transport of fluoride into bacteria. However, additional studies are needed to convincingly establish the true mechanism of action.
Concluding Remarks
Our findings indicate that small, non-peptidic molecules can be used to increase fluoride toxicity against bacteria and that such molecules might be frequently identified as hit compounds via high-throughput screening. Compounds that could find utility as fluoride toxicity agonists should have a number of characteristics. First, for the development of broad-spectrum agents, a good hit compound should have a provable mechanism that disrupts a key component of the fluoride defense system used by many bacteria or fungi. Second, a hit compound should be a member of a molecular series that has the potential to be improved through the synthesis and testing of analogs. Third, the optimized agent to be used in a useful formulation should induce strong fluoride toxicity against the pathogen, but remains non-toxic to the host when used alone or in combination with fluoride.
The fluoride riboswitch reporter system used in this study is sufficiently robust that it can be used to reveal hit compounds by HTS that only weakly increase the intracellular concentration of fluoride in bacteria. Our chemical library was modest in compound number and diversity, and therefore it is not surprising that nearly all identified hits were based on the same chemical scaffold. Although this collection of hits likely operate by a similar mechanism, it is likely that many other compounds could be identified by our HTS process that increase fluoride toxicity by different mechanisms. Although additional chemically distinct molecules were rarely observed to increase fluoride reporter expression, fluoride toxicity agonists with different chemical scaffolds and mechanisms likely remain to be discovered. Novel compounds with diverse mechanisms might be more easily identified in larger and more diverse libraries.
Regardless, we have succeeded in identifying novel small molecules able to render both Gram-positive and Gram-negative bacteria susceptible to fluoride. Optimization of the ability of these molecules to enhance fluoride toxicity via additional rounds of SAR studies could yield antimicrobial agents that when combined with fluoride offer a novel mechanism of action. The fluoride riboswitch reporter system described in this study could be used throughout this optimization process to determine whether new compounds in the lead series can increase fluoride concentration in cells. There are numerous challenges ahead for those who seek to create compounds that could conceivably be used in topical formulations in conjunction with fluoride in a novel antibacterial approach, but, given the spread of antibiotic-resistant bacteria, such novel approaches are needed.
SIGNIFICANCE
Herein we describe a high-throughput screen that exploits a fluoride-sensing riboswitch to regulate reporter gene expression in E. coli. This screen was successfully used to isolate compounds that increase intracellular fluoride concentrations. Hit compounds identified with this screen reproducibly increase reporter gene expression and enhance the ability of fluoride to slow bacterial growth. Based on SAR data, additional and more potent analogs were identified by the synthesis of analogs. The most potent derivatives substantially increase the antibacterial potency of fluoride toward representative Gram-negative and Gram-positive bacteria. There are numerous potential mechanisms of action for these compounds and for other such agents that might be identified in the future, including the disruption of cell barriers or the direct transport of fluoride across bacterial membranes. Additional molecules that enhance fluoride toxicity could conceivably be optimized for use as antibacterial or antifungal agents when used in conjunction with this anion.
EXPERIMENTAL PROCEDURES
Riboswitch Reporter Expression Analyses
Reporter gene expression analyses were performed as described previously with some modifications (Nelson et al., 2014). E. coli transformed with fluoride reporter constructs in pRS414 (Baker et al., 2012) were grown with aeration overnight in lysogeny broth (LB) supplemented with 75 µg mL−1 carbenicillin at 37°C. 8 µL of either the overnight culture or the culture diluted 1:10 with fresh LB were then added to 96-well plates containing 36 µL of 2× modified LB (20 g L−1 tryptone and 10 g L−1 yeast extract) and 36 µL of various concentrations of aqueous NaF. Cultures not supplemented with NaF were instead supplemented with 1 mM NaCl (final concentration). 0.8 µL of compound dissolved in DMSO were added, the plates sealed with parafilm and grown, with shaking, at 37°C for either four hours (1:10 diluted; data presented in Table S1 and S2) or overnight (1:100; all other data). Cultures grown without compound contained 0.8 µL of DMSO alone.
Following incubation, bacterial growth was determined via measuring the optical density at 600 nm. Reporter gene expression was established by first adding 80 µL of modified Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, pH 7.0 at 25°C), followed by 40 µL 4-methylumbelliferyl-β-D-galactopyranoside (1 mg mL−1 in 50:50 v/v deionized H2O:DMSO). Samples were mildly agitated and allowed to stand at room temperature for 15 min. The reaction was halted by the addition of Na2CO3 and gene expression was measured via fluorescence (ex 360, em 460).
High-throughput screening was performed largely as described above, with some modifications. LB supplemented with 1 mM sodium fluoride was added in 10 µL volumes to each well of 384-well plates. 20 nL of a 10 mM solution of each compound to be tested was added via pronging, followed by the addition of 10 µL of an overnight culture of E. coli containing the fluoride reporter. WT and Δfluc E. coli cells, as well as media alone, were included on each plate as controls. Bacteria were grown by shaking in a humidified incubator at 37°C for 24 hours, at which time reporter gene expression was assayed as described above.
Chemical Synthesis
Diaryl ureas were synthesized by the one-step coupling of various anilines with various isocyanates in pre-weighed one dram (3.7 mL) vials. Each aniline (0.05 mmol) was placed in the vial and dissolved in dichloromethane (1.5 mL), to which the isocyanate (0.05 mmol) was added. The vials were sealed and stirred overnight at room temperature. In most cases the product urea precipitated or crystalized from the reaction. The solid was isolated from the reaction by drawing the solution away from the solid with a fine tipped glass pipette. The solid was washed with dichloromethane and the liquid was again withdrawn. In the few cases where solid did not precipitate from the reaction, the reaction was evaporated under a stream of nitrogen and the resulting solid was washed with 1:1 ether/hexanes to yield the purified product. Solids were air dried in the vial and then placed under high vacuum. Proton and fluorine NMR and LC/MS were obtained on each compound to verify identity and purity, which was greater than 90% for all compounds tested. Isolated yields were 60% to 90%.
Diarylthioureas were synthesized exactly as described above using isothiocyanates in place of isocyanates, except reactions were stirred for three days to ensure completion. Many of the thiourea products were soluble in dichloromethane necessitating the above-described evaporation and trituration procedure. Solids were air dried in the vial and then placed under high vacuum. Proton and fluorine NMR and LC/MS were obtained on each compound to verify identity and purity, which was greater than 90% for all compounds tested. Isolated yields were 50% to 90%.
Arylbenzamides were synthesized by adding the aniline (0.29 mmol) to one-dram vials. Silicycle morpholine resin (1.3 mmol) was added followed by the addition of dry tetrahydrofuran (1.5 mL) and, subsequently, 4-trifluoromethyl benzoyl chloride (49 µL, 0.29 mmol) was added. The reaction was stirred overnight before Silicycle amine resin (0.30 mmol) was added to quench any excess acyl chloride. Additional dry tetrahydrofuran was added as needed to aid stirring. After 24 hours the reactions were filtered through a pipette with a cotton plug with the aid of tetrahydrofuran and the filtrates were further filtered using a syringe and a 0.45 um PTFE syringe tip filter. The solutions were evaporated under a stream of nitrogen and dried under high vacuum. Proton and fluorine NMR and LC/MS were obtained on each compound to verify identity and purity, which was greater than 90% for all compounds tested. Isolated yields were 60% to 90%. Compounds that were not synthesized were either purchased from Chembridge, Maybridge, or Sigma Aldrich (compound 19).
Solubility of selected compounds in LB and 0.1 M Tris (pH 7.0 at 25°C) were determined via nephelometry. NMR spectroscopy data for key compounds (those in Figure 3, with the exception of 19, which was purchased from Sigma Aldrich) were obtained using an Agilent 400 mHz spectrometer equipped with an auto tunable probe. LC/MS spectra were obtained using an Agilent 1260 infinity liquid chromatograph attached to an Agilent 6120 quadrupole mass spectrometer. The compounds were eluted using an increasing gradient of acetonitrile (0.1% formic acid) and water. The gradient varied from 10% acetonitrile to 100% acetonitrile over 2 min with a total run time of 5 min. Compounds were separated using an Agilent Eclipse plus C18 5.1 × 50 mm column and monitored at 254 nm.
Bacterial Growth Curves and MIC Determinations
Bacteria were grown as described above, overnight, and diluted 1:10 in 2× LB. 8 µL of the resulting solution was then added to 42 µL of 2× modified LB and 42 µL of aqueous NaF, to which 0.8 µL of either DMSO or 1 dissolved in DMSO was added. Cultures were then incubated in a 100-well Honeycomb plate at 37°C in a Bioscreen C system (Growth Curves USA) as described previously (Nelson et al., 2014).
MIC assays were conducted in a final volume of 100 µL according to methods established by the Clinical Laboratory Standards Institute (CLSI, 2012). Briefly, test or control compound dissolved in DMSO or water as appropriate was serially diluted (1:2) into successive tubes of the same solvent. A 5 µL aliquot of each dilution was transferred to the appropriate wells of 96-well clear round-bottom plates. To each dilution series, 95 µL of a freshly prepared bacterial suspension was added with mixing to provide a final bacterial inoculum of approximately 105–106 CFU per well and a final DMSO concentration of 5%. DMSO was added regardless of whether compound was dissolved in water or DMSO. Cation-adjusted Mueller-Hinton broth (Fluka) was used for all MIC assays, and lysed horse blood (Cleveland Scientific) was included at a final concentration of 5% for assays with S. mutans. Assay cultures were incubated 18–24 hours at 37°C and with an atmosphere enriched to 5% CO2 for S. mutans. The MIC is defined as the lowest concentration of antimicrobial agent that completely inhibits growth of the organism as detected by the unaided eye. Ciprofloxacin was used for comparison to a known antibiotic and as a benchmark for each screening assay.
Supplementary Material
HIGHLIGHTS.
A riboswitch was used to report increased fluoride levels inside bacteria.
Molecules that trigger riboswitch-reporter activity were isolated via HTS.
Improved hit compounds enhance fluoride toxicity in bacteria.
ACKNOWLEDGMENTS
We thank Mariya Kolesnikova, Janie Merkel, and the entire Yale Center for Molecular Discovery for their assistance in performing high-throughput screening, and Edward Bickham, Daniel Moonan, and Meredith Redick for assistance in acquiring the SAR data. We also thank Timothy Newhouse at Yale University for bringing to our attention compound 19. This work was supported by the National Institutes of Health (5R01DE022340) and the Howard Hughes Medical Institute.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information includes one figure, two tables, and characterization of synthesized compounds.
AUTHOR CONTRIBUTIONS
T.D.A. and R.R.B. designed the high-throughput screen. T.D.A. performed it with J.W.N. M.S.P. synthesized and verified all other molecules. J.W.N. conducted reporter assay and growth curve experiments. K.F.B. determined the MIC of selected compounds with and without fluoride. J.W.N., K.F.B., and R.R.B analyzed the data and wrote the paper.
REFERENCES
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier O, Arreola-Mendoza L, Del Razo LM. Molecular mechanisms of fluoride toxicity. Chem. Biol. Interact. 2010;188:319–333. doi: 10.1016/j.cbi.2010.07.011. [DOI] [PubMed] [Google Scholar]
- Boiocchi M, Del Boca L, Gomez DE, Fabbrizzi L, Licchelli M, Monzani E. Nature of urea-fluoride interaction: incipient and definitive proton transfer. J. Am. Chem. Soc. 2004;126:16507–16514. doi: 10.1021/ja045936c. [DOI] [PubMed] [Google Scholar]
- Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SJ, Edwards PR, Gale PA, Light ME. Carboxylate complexation by a family of easy-to-make ortho-phenylenediamine based bis-ureas: studies in solution and the solid state. New J. Chem. 2006;30:65–70. [Google Scholar]
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Bryantsev VS, Hay BP. Conformational preferences and internal rotation in alkyl- and phenyl-substituted thiourea derivatives. J. Phys. Chem. A. 2006;110:4678–4688. doi: 10.1021/jp056906e. [DOI] [PubMed] [Google Scholar]
- Busschaert N, Wenzel M, Light ME, Iglesias-Hernandez P, Perez-Tomas R, Gale PA. Structure-activity relationships in tripodal transmembrane anion transporters: the effect of fluorination. J. Am. Chem. Soc. 2011;133:14136–14148. doi: 10.1021/ja205884y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busschaert N, Kirby IL, Young S, Coles SJ, Horton PN, Light ME, Gale PA. Squaramides as potent transmembrane anion transporters. Angew. Chem. Int. Ed. 2012;51:4426–4430. doi: 10.1002/anie.201200729. [DOI] [PubMed] [Google Scholar]
- Cametti M, Rissanen K. Recognition and sensing of fluoride anion. Chem. Comm. 2009;20:2809–2829. doi: 10.1039/b902069a. [DOI] [PubMed] [Google Scholar]
- Cametti M, Rissanen K. Highlights on contemporary recognition and sensing of fluoride anion in solution and in the solid state. Chem. Soc. Rev. 2013;42:2016–2038. doi: 10.1039/c2cs35439j. [DOI] [PubMed] [Google Scholar]
- CLSI. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically; Approved Standard – Ninth Edition. Wayne, PA: Clinical and Laboratory Standards Institute; 2012. [Google Scholar]
- Curran TM, Buckley DH, Marquis RE. Quasi-irreversible inhibition of enolase of Streptococcus mutans by fluoride. FEMS Microbiol. Lett. 1994;119:283–288. doi: 10.1111/j.1574-6968.1994.tb06902.x. [DOI] [PubMed] [Google Scholar]
- Davis JT, Okunola O, Quesada R. Recent advances in the transmembrane transport of anions. Chem. Soc. Rev. 2010;39:3843–3862. doi: 10.1039/b926164h. [DOI] [PubMed] [Google Scholar]
- Deigan KE, Ferré-D’Amaré AR. Riboswitches: discovery of drugs that target bacterial gene-regulatory RNAs. Acc. Chem. Res. 2011;44:1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher M, Suttie JW. Intracellular fluoride in cultured mammalian cells. Exp. Biol. Med. 1972;139:228–230. doi: 10.3181/00379727-139-36115. [DOI] [PubMed] [Google Scholar]
- Francisco GD, et al. Phenyl thiazolyl urea and carbamate derivatives as new inhibitors of bacterial cell-wall biosynthesis. Bioorg. Med. Chem. Lett. 2004;14:235–238. doi: 10.1016/j.bmcl.2003.09.082. [DOI] [PubMed] [Google Scholar]
- Jayaraman S, Haggie P, Wachter RM, Remington SJ, Verkman AS. Mechanism and cellular applications of a green fluorescent protein-based halide sensor. J. Biol. Chem. 2000;275:6047–6050. doi: 10.1074/jbc.275.9.6047. [DOI] [PubMed] [Google Scholar]
- Ji C, Stockbridge RB, Miller C. Bacterial fluoride resistance, Fluc channels, and the weak acid effect. J. Gen. Phys. 2014;144:257–261. doi: 10.1085/jgp.201411243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Blount KF, Puskarz I, Lim J, Link KH, Breaker RR. Design and antimicrobial action of purine analogues that bind guanine riboswitches. ACS Chem. Biol. 2009;4:915–927. doi: 10.1021/cb900146k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol. 2009;6:187–194. doi: 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepore G, Migdal S, Blagdon DE, Goodman M. Conformations of substituted aryl ureas in solution. J. Org. Chem. 1973;38:2590–2594. [Google Scholar]
- Li L. The biochemistry and physiology of metallic fluoride: action, mechanism, and implications. Crit. Rev. Oral Biol. Med. 2003;14:100–114. doi: 10.1177/154411130301400204. [DOI] [PubMed] [Google Scholar]
- Li S, Breaker RR. Fluoride enhances the activity of fungicides that destabilize cell membranes. Bioorg. Med. Chem. Lett. 2012;22:3317–3322. doi: 10.1016/j.bmcl.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Smith KD, Davis JH, Gordon PB, Breaker RR, Strobel SA. Eukaryotic resistance to fluoride toxicity mediated by a widespread family of fluoride export proteins. Proc. Natl. Acad. Sci. USA. 2013;110:19018–19023. doi: 10.1073/pnas.1310439110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z. 2-Phenyl-5,6-dihydro-2H-thieno[3,2-c]pyrazol-3-ol derivatives as new inhibitors of bacterial cell wall biosynthesis. Bioorg. Med. Chem. Lett. 2003;13:2591–2594. doi: 10.1016/s0960-894x(03)00471-2. [DOI] [PubMed] [Google Scholar]
- Lünse CE, Scott FJ, Suckling CJ, Mayer G. Novel TPP-riboswitch activators bypass metabolic enzyme dependency. Front. Chem. 2014;2:53. doi: 10.3389/fchem.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lünse CE, Schüller A, Mayer G. The promise of riboswitches as potential antibacterial drug targets. Int. J. Med. Microbiol. 2014;304:79–92. doi: 10.1016/j.ijmm.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Mulhbacher J, Brouillette E, Allard M, Fortier L, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog. 2010;6:e1000865. doi: 10.1371/journal.ppat.1000865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai C, Thomas JA. Properties of a phosphoprotein phosphatase from bovine heart with activity on glycogen synthase, phosphorylase, and histone. J. Biol. Chem. 1974;249:6459–6467. [PubMed] [Google Scholar]
- Nelson JW, Zhou Z, Breaker RR. Gramicidin D enhances the activity of fluoride. Bioorg. Med. Chem. Lett. 2014;24:2969–2671. doi: 10.1016/j.bmcl.2014.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peselis A, Serganov A. Themes and variations in riboswitch structure and function. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbagrm.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Chai G, Brewer JM, Lovelace LL, Lebioda L. Fluoride inhibition of enolase: crystal structure and thermodynamics. Biochemistry. 2006;45:793–800. doi: 10.1021/bi051558s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quissell DO, Suttie JW. Development of a fluoride-resistant strain of L cells: membrane and metabolic characteristics. Am. J. Physiol. 1972;223:596–603. doi: 10.1152/ajplegacy.1972.223.3.596. [DOI] [PubMed] [Google Scholar]
- Russell AD. Whither triclosan? J. Antimicrob. Chemother. 2004;53:693–695. doi: 10.1093/jac/dkh171. [DOI] [PubMed] [Google Scholar]
- Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge RB, Lim HH, Otten R, Williams C, Shane T, Weinberg Z, Miller C. Fluoride resistance and transport by riboswitch-controlled CLC antiporters. Proc. Natl. Acad. of Sci. USA. 2012;109:15289–15294. doi: 10.1073/pnas.1210896109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockbridge RB, Robertson JL, Kolmakova-Partensky L, Miller C. A family of fluoride-specific ion channels with dual-topology architecture. eLife. 2013;2:e01084. doi: 10.7554/eLife.01084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zasloff M, Steinberg WH. Composition and treatment with biologically active peptides and certain anions. 5,217,956 U.S. Patent. 1993
- Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.